Abstract
Recent evidence suggests that cancer stem cells (CSCs) play an important role in cancer, as these cells possess enhanced tumor-forming capabilities and are resistant to current anticancer therapies. Hence, novel cancer therapies will need to be tested for both tumor regression and CSC targeting. Herein we show that oncolytic reovirus that induces regression of human breast cancer primary tumor samples xenografted in immunocompromised mice also effectively targets and kills CSCs in these tumors. CSCs were identified based on CD24−CD44+ cell surface expression and overexpression of aldehyde dehydrogenase. Upon reovirus treatment, the CSC population was reduced at the same rate as non-CSCs within the tumor. Immunofluorescence of breast tumor tissue samples from the reovirus- and mock-treated mice confirmed that both CSCs and non-CSCs were infectible by reovirus, and terminal deoxynucleotidyl transferase biotin-dUTP nick end labeling (TUNEL) assay showed that both populations died by apoptosis. Ras, which has been shown to mediate reovirus oncolysis, was found to be present at similar levels in all cell types, and this is consistent with their comparable sensitivity to reovirus. These experiments indicate that oncolytic reovirus has the potential to induce tumor regression in breast cancer patients. More important, the CSC population was equally reduced and was as susceptible to reovirus treatment as the non-CSC population.
Introduction
As early as 1977, it was first observed that certain transformed cell lines had increased susceptibility to the human reovirus.1 However, it was not until two decades later that the cancer-killing implications were fully realized when it was observed that murine cells transformed with the Ras oncogene manifested enhanced susceptibility to reovirus infection and killing.2 Subsequent experiments showed that reovirus was able to replicate efficiently in a number of established human cancer cell lines, including brain-, breast-, lymphoma-, ovarian-, bladder-, spinal-, and colon-derived cells.3,4,5,6,7,8 In vivo data validated the potential use of reovirus as a cancer therapy as a single intratumoral injection of reovirus-induced tumor regression in immunocompromised mice with established tumors from a number of human-derived cancer cell lines.3,4,5,6,8 These studies have led to phase I/II clinical trials presently underway for a variety of human cancers.9
Research in cancer has resulted in increased detection, improved treatments, and enhanced prevention of metastasis. Despite these advances, however, when metastatic cancer occurs, it is generally resistant to therapeutics and the prognosis is poor. Therefore, there is an urgent need for the development of new therapies and novel approaches that, when applied, significantly reduce the chance of metastatic cancer from occurring.
Solid tumors are composed of a heterotypic population of cells. Increasing evidence suggests that only a small percentage of these cells have tumorigenic potential.10,11 In the example of breast cancer, these tumorigenic breast cells were originally isolated based on both expression and nonexpression of distinct cell surface markers (CD24−CD44+ breast cancer cells). These highly tumorigenic cells share with normal stem cells the ability to proliferate and give rise to diversified tumor cell types including those with the capacity for self-renewal.11 These cells are termed cancer stem cells (CSCs), and it takes only a relatively small number of them (~102) to form tumors in immunocompromised mice.
The characterization and isolation of breast CSCs based on cell surface expression of CD44 and CD24 has been controversial as neither of these markers is known for their expression on stem cells. Again, eight of nine patient samples used for the initial isolation of CD44+CD24− cells were from pleural effusions (late stage metastatic breast cancer cells found in the lungs),12 raising some doubt as to how reflective these cells are of the CSCs in the primary tumor. More recently, Ginestier et al. isolated breast cancer cells based on high expression of aldehyde dehydrogenase 1 A1 (ALDH1),13 which is found at high levels in hematopoietic and neural stem cells and is responsible for oxidizing aldehydes to carboxylic acids.14,15 Using the Aldefluor assay to identify ALDH1-expressing cells from human primary breast tumor transplanted and passaged in immunocompromised mice, Ginestier et al. showed that Aldefluor+ cells were highly tumorigenic.13 Further, breast cancer patients with higher percentages of ALDH1+ cells had the worst outcome.13 The Aldefluor assay has since been successfully used to isolate CSCs in other cancers such as hepatocellular cancer16 It needs to be pointed out, however, that both of these CSC detection methods identify cell populations that are enriched for CSCs and that not every CD24−CD44+ cell or Aldefluor+ cell is a CSC.11,13
The revelation that breast cancer tumorigenesis is sustained by a minority subset of cells (i.e., CSCs) has important implications on diagnosis, treatment, and long-term prognosis of cancer patients. First, even if tumor regression is induced, unless the CSCs are eradicated, the long-term therapeutic benefit could be minimal.17 Second, evidence suggests that CSCs are resistant to current chemotherapeutic drugs and radiation therapy.13,17,18,19,20,21,22,23 If that is indeed the case, treating cancer with current therapies could have the long-term detrimental effect of enriching for this minority tumor cell population that has the potential to cause cancer recurrence and metastasis. Therefore, new treatment strategies will need to be tested not only for tumor regressing capabilities but also their efficacy in CSC targeting.
We show that oncolytic reovirus is able to induce regression of tumors generated from a primary human breast cancer core sample xenografted to the mammary fat pad of immunocompromised mice. Furthermore, we show that reovirus is effective at targeting CSCs identified in these xenografted tumors.
Results
Reovirus induces tumor regression of primary breast cancer patient xenografts
Previously reovirus was shown to be highly cytopathic to a number of breast cancer cell lines (MCF7, MDA-MV-468, SK-BR-3, and T-47D) while a normal mammary cell line, Hs-578Bst, remained resistant to reovirus.6 Furthermore, both single intratumoral injection and hind flank tumor/systemic reovirus delivery systems in mouse xenograft tumors established from these human breast cancer cells resulted in tumor regression,6 suggesting the potential use of reovirus as an antibreast cancer therapy. However, it is becoming increasingly clear that in addition to tumor regression, a novel therapy should ideally also be able to target and kill CSCs, thereby preventing inadvertent enrichment for these cells while inducing tumor regression.24
To test the efficacy of oncolytic reovirus to target and kill breast CSCs in vivo, we established palpable tumors in the mammary fat pads of immunodeficient NOD/SCID mice using a core biopsy sample of a primary infiltrating ductal carcinoma obtained from a breast cancer patient at the time of her primary surgery. (NOD/SCID rather than nude mice were used as unlike the breast cancer cell lines previously studied; these primary tumors failed to grow in the less immune-tolerant nude mice.) Previous reports indicate that primary breast cancer patient samples passaged in mice have ~5–30% CD24low/-CD44+ identified CSCs.11 Similarly, 5–10% of these highly tumorigenic cells were identified in breast cancer tumors passaged in miced by using the Aldefluor assay as a marker of breast CSCs.13
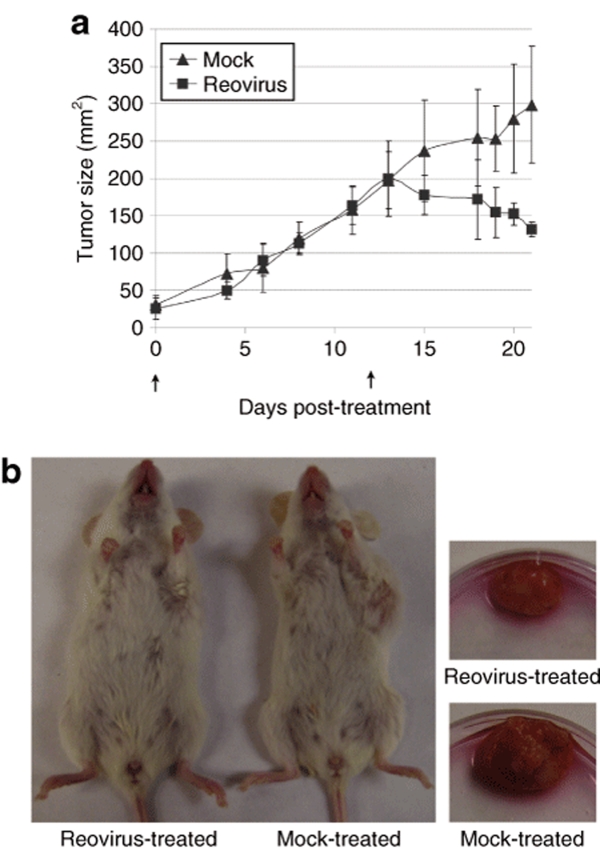
We established palpable tumors in eight mice and subdivided them into two groups for either reovirus or mock treatment, and tumor growth was measured at various times post-treatment. Aggressive growth rate was demonstrated up to day 12 post-treatment, with reovirus noticeably inducing tumor regression 15 days post-treatment (Figure 1). This is the first time reovirus has been shown to successfully treat tumors generated from a solid human cancer specimen (not cell lines) in a mouse model, which better mimics the clinical setting and is arguably a more accurate representation of the effectiveness of reovirus as an oncolytic agent.4,5,6 We noted, however, that regression of these tumors after reovirus treatment was somewhat delayed compared to that previously reported with tumors from cell lines.4,5,6 This was likely due to the aggressive nature of these tumors as their growth rates far exceeded those observed previously with cell line–induced tumors.6 This aggressive tumor growth also prevented us from extending the study beyond 21 days (adherence to tumor endpoint guidelines set by the Canadian Council on Animal Care). We also refrained from using higher doses of reovirus as NOD/SCID mice are far less tolerant of reovirus than nude mice previously used for studies on cell line–derived tumors.11,13,25,26 Nevertheless, at day 21 the tumors treated with reovirus were significantly smaller compared to mock-treated tumors (P = 0.03), and we had achieved the objective of partial tumor reduction that would allow us to assess the in vivo reovirus susceptibility of breast CSC.
Figure 1.
Intratumoral injection of reovirus induces tumor regression of solid tumor xenografts from a breast cancer patient. (a) Passaged primary tumor core samples implanted in the mammary fat pads of immunodeficient mice were injected with reovirus (n = 4, closed squares), or phosphate-buffered saline (mock, n = 4, closed triangles), on days 0 and 12 (upward arrow). Tumor size (mm2) was determined based on calliper measurements taken of the two longest dimensions (width × length) of the tumor. Each data point is the average of all the tumors in each group and error bars represent standard deviation. At day 21 post-treatment the P value = 0.0327 as determined by an unpaired T-test. (b) An example of one of the reovirus and mock-treated mice (left panel) and subsequently excised tumors (right panel) 21 days post-treatment initiation.
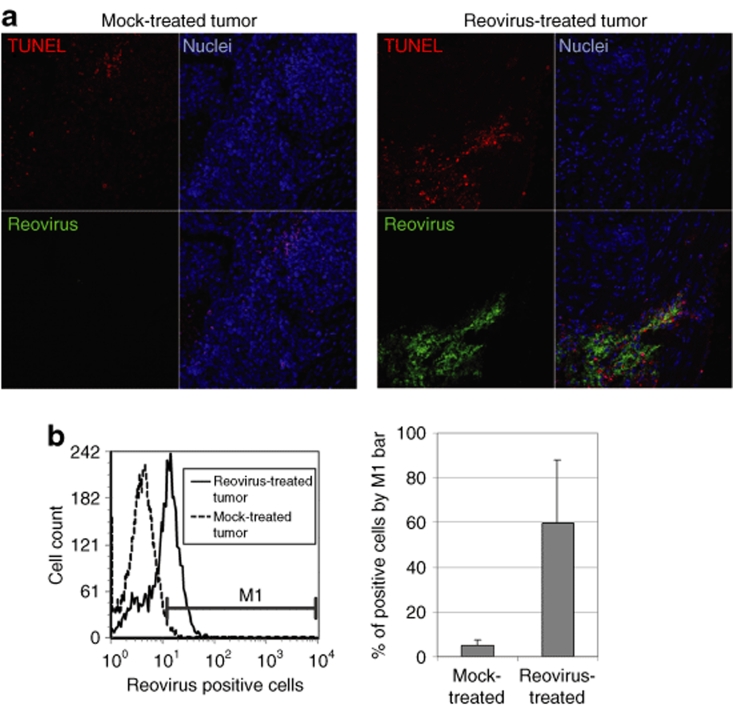
The mice were then euthanized and tumor tissue harvested for postmortem analysis (Figure 1). A portion of each tumor was fixed for paraffin embedding and immunohistochemical analysis. Thin sections were stained with polyclonal antireovirus antibody to detect reovirus-infected cells, and terminal deoxynucleotidyl transferase biotin-dUTP nick end labeling (TUNEL) was used to detect apoptotic cells (Figure 2a). Images captured by confocal microscopy show areas of reovirus-positive cells coinciding with apoptotic cells, confirming that reovirus infection results in cell death (Figure 2a). The remaining tumor sample portions not fixed for immunohistochemistry analysis were processed to generate single-cell suspensions for fluorescence-activated cell-sorting (FACS) analysis. Generated cell suspensions were fixed, permeablized, and labeled with antireovirus antibody to quantify the percentage of reovirus-infected cells in the treated tumors (Figure 2b). Tumors treated with reovirus had on average 60% of breast cancer cells positive for reovirus proteins, which is again indicative of reovirus infection.25
Figure 2.
Reovirus treatment of breast cancer solid tumor xenografts results in virus-infected apoptotic tumor cells. (a) Formalin-fixed thin sections of mock- and reovirus-treated tumors were stained with antireovirus antibody (green) and TUNEL (red) to detect virally infected and apoptotic cells, respectively. Nuclei (blue) were stained with ToPro-3. (b) Single-cell suspensions generated from the tumors (mock and reovirus treated) were fixed and stained with antireovirus antibody to quantify the percentage of reovirus infected cells by fluorescence-activated cell sorting (left panel shows the result of one of the mock- and reovirus-treated tumor samples). The M1-bar was set to determine percentage of reovirus-positive cells and the average of all four tumors in both mock- and reovirus-treated tumors and is shown in the right panel. TUNEL, terminal deoxynucleotidyl transferase biotin-dUTP nick end labeling.
Reovirus infects and kills CD24−CD44+ cells in vivo
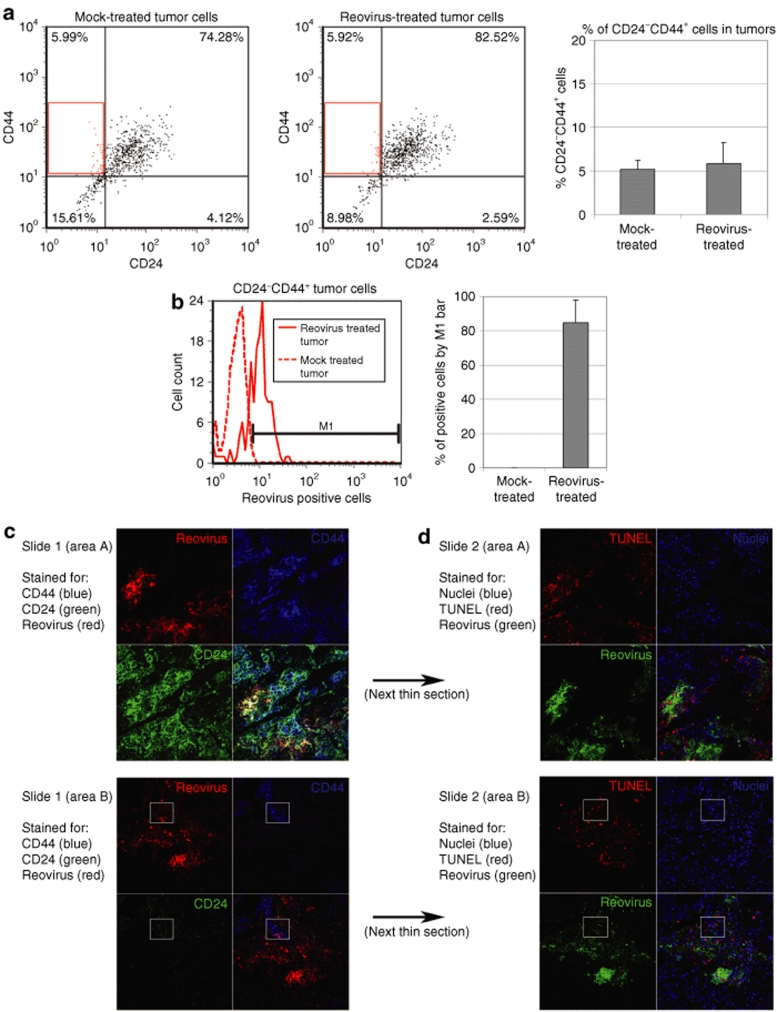
Next, we quantified the percentage of putative breast CSCs (CD24−CD44+ cells) in the mock- and reovirus-treated tumors by FACS. We noted that the percentage of CD24−CD44+ cells was between 5 and 6% (P = 0.63) in both mock- and reovirus-treated tumors (Figure 3a). This finding is important as it demonstrates that treatment with oncolytic reovirus resulted in a proportionate reduction in the total tumor mass and CD24−CD44+ cells and that there was no enrichment for CD24−CD44 cells in the reovirus-treated tumors. We also determined the percentage of infected CD24−CD44+ cells by costaining the fixed cells with antireovirus antibody, and found that they were on average 85% positive for reovirus infection (Figure 3b). This number compares favorably to the 60% total infected cells (Figure 2b).
Figure 3.
CD24−CD44+ cells are susceptible to reovirus infection in vivo. (a and b) Single-cell suspensions of tumors from mock- and reovirus-treated mice were fixed and stained for expression of human CD24, CD44, and reovirus proteins. (a) CD24−CD44+ cells are quantified by fluorescence-activated cell sorting (left panel) and the average percentage of CD24−CD44+ cells in reovirus- and mock-treated tumors calculated (right panel, P = 0.63, unpaired T-test). (b) CD24−CD44+ cells (gated in a, dot plot) were analyzed for expression of reovirus proteins (reovirus-infected cells) with costaining of antireovirus antibody (left panel). The M1 bar was set to determine percentage of reovirus-positive CD24−CD44+ cells and the average of all four tumors in both mock- and reovirus-treated tumors is shown in the right panel. (c and d) Sequential formalin-fixed tumor tissue sections of a reovirus-treated mouse were stained for cell markers, reovirus infection, and apoptotic cells. (c) The first sequential thin section (slide 1) was stained for CD44 (blue), CD24 (green), and reovirus infection (red). Area A (top panel) shows an area of tissue that has predominantly CD24+CD44+ cells. Area B (bottom panel) shows an area with some CD24−CD44+ cells. A white box identifies a reovirus-infected area of CD24−CD44+ cells in the bottom panel. (d) The next sequential thin section (slide 2) was stained for nuclei (blue), TUNEL-positive apoptotic cells (red) and reovirus infection (green). The white box corresponds to CD24−CD44+ cells identified in the bottom panel of c. TUNEL, terminal deoxynucleotidyl transferase biotin-dUTP nick end labeling.
As an alternative approach, we immunohistochemically stained the tumor sections with anti-CD24, anti-CD44, and antireovirus antibodies in an attempt to identify areas comprising primarily CD24− (or low) CD44+ cells and determine whether these cells were susceptible to reovirus infection. Considering the low abundance of CD24−CD44 cells in breast tumors, identification of such cells by immunohistochemical staining was not an easy task. Nonetheless, we were able to identify two different areas of the tumor (Figure 3c, area A and area B) that were representative of predominantly CD24+CD44+ cells and CD24lowCD44+ cells, respectively. We found both cell populations to be susceptible to reovirus infection (box in area B highlights colocalization of CD44 and reovirus proteins with little or no CD24). Figure 3d represents the same areas of the next thin section of the tumor and showed that CD24− (or low) CD44+ breast cancer cells infected by reovirus were also TUNEL positive. This confirms that like all reovirus-infected breast cancer cells (Figure 2a), reovirus-infected CD24− (low) CD44+ cells also died by apoptosis (Figure 3d).
Reovirus infects and kills ALDH1+ breast CSCs
The Aldefluor assay is a viable cell assay used to identify and isolate ALDH1+ cells. Recently, when the assay was used on breast cancer tumor specimens, the isolated Aldefluor+ (ALDH1+) cells were shown to be highly tumorigenic.13 Therefore, in addition to CD24−CD44+, Aldefluor+ is now considered another putative breast CSC marker. Breast cancer cells that overexpress ALDH1 have increased Aldefluor activity.13 The addition of inhibitor DEAB in the assay blocks ALDH1 activity and allows for positive identification of Aldefluor+ cells. Accordingly, we compared the percentage of Aldefluor+ cells in mock- and reovirus-treated tumors (Figure 4a). We noted that although the mock-treated tumors had a greater range of Aldefluor+ cells, on average there was no significant difference between the relative percentage of Aldefluor+ cells in mock-treated and reovirus-treated tumors. This suggests reovirus-induced reduction in total tumor mass (and cells) is associated with a proportionate reduction in the amount of Aldefluor+ cancer cells.
Figure 4.
Aldefluor+ cells are susceptible to reovirus infection in vivo and in vitro. (a) Viable single-cell suspension of a tumor from a reovirus-treated mouse was quantified for ALDH1+ cells by the Aldefluor assay. The left panel shows how Aldefluor+ cells are quantified by FACS analysis in a dot plot (SSC versus FL1, fluorogenic substrate BAAA), of a reovirus-treated tumor samples. The addition of DEAB (blocks ALDH1 activity), right panel, allows the positive identification of ALDH1+ cells. (b) The average percentage of Aldefluor+ cells in reovirus- and mock-treated tumors as quantified by the Aldefluor assay (P = 0.90, unpaired T-test). (c) An in vitro assay comparing the reovirus infection susceptibility of total (left) and Aldefluor+ (right)-viable tumor cells isolated from an untreated tumor. These sorted populations were infected with reovirus in cell culture and quantified for reovirus infection (production of reovirus proteins) by FACS. (d) A formalin-fixed tumor tissue section of a reovirus-treated mouse was stained for ALDH1 expression (green), reovirus infection (blue), and apoptotic cells (TUNEL, red), top. The next sequential thin section (bottom panel) was stained for nuclei (blue). FACS, fluorescence-activated cell sorting; MOI, multiplicity of infection; TUNEL, terminal deoxynucleotidyl transferase biotin-dUTP nick end labeling.
The Aldefluor assay used for the detection of ALDH1+ cells requires the use of viable, unfixed cells, whereas assessment of reovirus infection requires that cells be fixed and permeablized. Therefore, we were unable to directly assess the percentage of Aldefluor+ tumor cells that were infected by reovirus compared to the total cells in vivo, as we did with CD24−CD44+ cells (Figure 3b). To circumvent this problem, we used an indirect approach and compared the infectability of Aldefluor+ cells to total cells in vitro by isolating Aldefluor+ breast cancer cells and total live breast cancer cells from an uninfected tumor and then infecting the respective isolated populations overnight with reovirus (Figure 4b). We noted that Aldefluor+ cells were similarly infectible by reovirus as total breast cancer cells were, confirming that Aldefluor+ identified breast CSCs were permissive to reovirus infection and agreeing with the in vivo data (Figure 4a).
Next, we carried out immunohistochemical staining of tumor samples to assess the susceptibility of ALDH1+ cells in these samples to reovirus infection and reovirus-induced apoptosis (Figure 4c). We noted that areas of tumor staining positive for ALDH1 and reovirus were also TUNEL positive (Figure 4c), confirming that like all reovirus-infected breast cancer cells (Figure 2), reovirus-infected ALDH1+ cells also died by apoptosis (Figure 4c).
Finally, we wanted to determine the Ras status of CD24−CD44+ and ALDH1+ cells compared to the bulk of the tumor cells. Ras mutations are common in cancer and play a critical role in the transformation process.2,26,27 Previous studies in this laboratory have shown that Ras transformation of cells results in increased reovirus susceptibility and is one of proposed mechanisms of reovirus oncolysis.2,25 We isolated Aldefluor+ (ALDH1+) cells and CD24−CD44+ cells from the tumor of an untreated mouse and compared the Ras levels of these putative breast CSCs to the bulk of tumor cells (live, H2Kd− cells) by western blot analysis. When normalized for differences in actin levels, the total Ras levels were similar in all three cell types (Figure 5). This result is consistent with the observation that the identified CSC populations and bulk tumor cells manifest comparable sensitivity to reoviurs (Figures 3 and 4). Although the Ras levels were similar, it is possible that the Ras-activation status could differ between the cell types. Unfortunately, the low number of recoverable Aldefluor+ and CD24−CD44+ cells (~105) precluded an accurate assessment of Ras activation using the Raf-1 pull-down assay (Upstate Biotechnology, Billerica, MA) which typically requires a minimum of 106 cells.28
Figure 5.
Western blot analysis of Ras levels in isolated total (7AAD−, H2Kd-), ALDH1+ (Aldefluor+, 7AAD−, H2Kd−), or CD24−CD44+ (7AAD−, H2Kd−) breast tumor cells. The blots were probed with anti-Ras antibody (top) or anti-β-actin antibody as a loading control (bottom).
Discussion
CSCs are proposed to be initiators of cancer and are probably responsible for cancer recurrence and development of metastases. With breast cancer specifically, comparative studies have shown that putative CSC populations are significantly more resistant to γ irradiation, the commonly used cancer therapy.18 Similarly, in studies with glioblastomas, Bao et al. also found that populations enriched for CSCs were highly resistant to irradiation, and the treatment was relatively ineffective in preventing CSCs from initiating tumor growth in xenograft studies.23 Other groups have also reported that CSCs originating from various cancers are comparatively resistant to commonly used chemotherapeutics like gemcitabine, temozolomide, carboplatin, etoposide, paclitaxel, fluorouracil, paclitaxel, daunorubicin, and mitoxantrone.19,20,21,22,29
There are several suggested possibilities for the apparent resistance of CSCs to current anticancer therapies. First, if it we are to assume that CSCs are similar to non-CSCs that are slow growing and generally in the G0 phase of the cell cycle, then commonly used chemotherapeutics designed to target highly proliferative cycling cells would not kill CSCs.10 Second, at least for glioblastoma CSCs, Bao et al. reported that resistance to irradiation appeared to be a result of increased activation of the DNA damage checkpoint response,23 thereby increasing the survival of these cells. Third, one of the proposed methods for isolating CSCs, exclusion of Hoechst stain,30 is likely based on increased expression in CSCs of adenosine triphosphate-binding cassette proteins or ABC transporters that efflux the stain. The transporters are also known to efflux chemotherapeutic drugs and are a common cause of cancer resistance to chemotherapies.31 It is, therefore, likely that CSC resistance to chemotherapeutics is due to an increased expression of these transporters.32 Finally, with the isolation of CSCs based on increased expression of ALDH1 (Aldefluor+ cells), it is possible that resistance to chemotherapeutic agents is a result of aldehyde dehydrogenase-specific activity that metabolizes chemotherapeutics such as cyclophosphamide.33
Given the emerging evidence illustrating the importance of CSCs in both cancer development and resistance to current cancer therapies, it is becoming increasingly important to test novel therapies for both tumor regression capabilities and CSC targeting efficacy. Oncolytic viruses, engineered or naturally occurring (as in the case of reovirus), were originally identified as having anticancer properties based on their ability to shrink tumors. As with other potential anticancer therapies, reovirus is being evaluated for its ability to cause tumor regression in phase I/II clinical trials.9 However, in view of emerging evidence of CSCs being involved in cancer initiation, tumor regression alone may not be a true measure of a drug's ability to engender long-term cancer remission, and the specific targeting of CSCs should also be included for evaluation.
Recently, using breast cancer cells isolated from the pleural effusions of patients, Eriksson et al.34 purified CD24−CD44+ cells and evaluated the ability of oncolytic adenoviruses Ad5/3-Δ24 and Ad5.pk7-Δ24 to kill these cells in vitro and in vivo. They showed that these oncolytic adenoviral strains were able to block tumor formation induced by CD24−CD44+ cells implanted in immunodeficient mice. In the present study, we also evaluated the ability of an oncolytic virus to kill putative breast CSCs; however, our study differs from theirs in several major aspects. First, to our knowledge, we are the first group to evaluate oncolytic reovirus specifically for its efficacy to kill CSCs. Second, our model utilized tumors established from a fresh core tumor sample obtained directly from a patient diagnosed with infiltrating ductal carcinoma and taken at the time of her surgery. This is significant because the previous study used breast cancer cells isolated from malignant pleural effusions and although pleural effusions are indeed a source of breast cancer cells,12 it is not clear as to how reflective these late stage cancer cells are of the original primary tumor. Third, utilizing the recently described Aldefluor assay to identify highly tumorigenic breast CSCs, we tested the ability of reovirus to target both CSCs identified as CD24−CD44+ cells and ALDH1+ (Aldefluor+) cells. Finally, a major objective of our study design was to establish an experimental system that more closely mimics the clinical setting. We first assessed the ability of reovirus to effect solid tumor regression and then evaluated the percentage reduction of putative CSCs following the treatment. If the CSC population had been resistant to reovirus, we would have detected an enrichment or increase in the percentage of these cells postvirus treatment. Indeed, CSC enrichment has been reported by Dylla et al. who treated mice bearing colorectal xenogenic tumors with chemotherapeutic agents and observed an enrichment of CSCs in the colorectal tumors.24 This highlights the potential dangers of treating patients with a drug that is ineffective at targeting these potent tumor-initiating cells.
With the observation that CSCs are not only highly tumorigenic but also resistant to killing by conventional means, it is necessary to evaluate potential cancer therapeutic candidates for their efficacy to target and kill the CSC population of tumors. This study and the previous adenoviral report34 indicate that oncolytic viruses kill CSCs with equal efficacy to non-CSC cancer cells.
Materials and Methods
Virus propagation and purification. L-929 (ATCC, Manassas, VA) cells were cultivated as adherent monolayers or in suspension in Minimal Essential Medium (MEM) or Joklik's Modified Eagle's Medium (Sigma-Aldrich, Oakville, Ontario, Canada), respectively, supplemented with 5% fetal bovine serum (FBS; Invitrogen, Burlington, Ontario, Canada). Reovirus (Serotype 3 Dearing) was propagated in L-929 suspension culture and purified through a CsCl gradient following established procedures.35 Activity was determined using standard plaque assay on L-929 monolayers and particle number by absorbance at 260 nm.
Establishment of primary human breast cancer solid tumor xenografts in mice. A core tumor sample of infiltrating ductal carcinoma was obtained from a breast cancer patient at the time of her primary surgery (patient consent given and ethics approval obtained from Research Ethics Board). The tumor sample was cut into 2 × 2 mm pieces and individual pieces implanted into the upper left mammary fat region of 3–8-week-old female, nonobese, diabetic-severe, combined immunodeficiency (NOD/SCID) mice purchased from Charles River, Pointe-Claire, Quebec, Canada. A 5-mm incision was made on the upper left-side flank of each mouse just above the mammary fat pad area. The tumor piece was then sutured to the mammary fat with one stitch. Accessing the same incision, a 90-daytime-release 17β-estradiol 3.0 mm 0.72 mg pellet (Innovative Research of America, Sarasota, FL) was implanted on the other side of the mouse using forceps to direct the pellet to the proper position. The incision was closed with 4.0 suture and mice monitored weekly for tumor growth. Three months postimplantation a tumor measuring 196 mm2 had grown in one of the mice attached to the upper left mammary fat pad. This tumor was harvested and above procedure repeated for passaging in multiple mice to achieve a statistically significant number for later described tumor regression studies.
Of 12 mice with passaged tumor xenografts, 8 developed palpable tumors (~20 mm2) by 5 weeks postimplantation. Tumors of each mouse were measured and the mice divided into two groups of four.
Intratumoral reovirus injection into palpable tumors. Groups of four mice were either injected with 50 µl of 1 × 107 plaque forming units (pfu) of reovirus in phosphate-buffered saline (PBS) or PBS alone intratumorally. Tumor size was measured every 2 days postinjection. A second intratumoral injection of reovirus or PBS was administered to the mice 12 days after the first injection. Final tumor measurements were taken, mice were sacrificed, and tumor tissue was harvested for analysis 21 days postinjection.
Tumor tissue analysis by FACS. Tumor tissue was sectioned and a portion fixed and saved for immunofluorescence (described below). The remaining tissue was minced and digested for 3 hours at 37 °C with a 225 U/ml solution of collagenase III (BioShop, Burlington, Ontario, Canada) in Hank's balanced salt solution (Invitrogen). Digested tissue was strained with a 40 µmol/l cell strainer (BD Falcon, Mississauga, Ontario, Canada). Resulting cell suspension was washed with PBS and red blood cells lysed.
The washed suspension was divided and a portion was fixed with 4% paraformaldehyde solution for percentage of reovirus infection analysis in combination with CD24 and CD44 cell surface expression analysis. After fixation, blocking and permeablization in 5% bovine serum albumin, 0.1% Triton X-100 in PBS, cells were probed with antireovirus rabbit antibody25 and goat antirabbit IgG conjugated with Cy5 (Jackson ImmunoResearch, Burlington, Ontario, Canada), antihuman CD24-FITC conjugated, and antihuman CD44-PE conjugated (BD Pharmingin, Mississauga, Ontario, Canada). Washed cells were quantified with a FACSCalibur flow cytometer (Becton Dickinson, Mississauga, Ontario, Canada) and analyzed using WinDMI Version 2.9 (Scripps Research Institute, La Jolla, CA).
The other unfixed portion of the collagenase-generated tumor cell suspensions were saved for ALDH1 expression analysis using the Aldefluor assay (Stemcell Technologies, Vancouver, British Columbia, Canada), following the manufacturer's instructions.
Isolation of Aldefluor+, CD24−CD44+ cell, and live breast cancer cells. As described previously, single-cell suspensions generated from an untreated mouse passaged tumor were either stained for isolation of live total human breast cancer cells, live CD24−CD44+ breast cancer cells, or for live Aldefluor+ human breast cancer cells. Cells were stained with anti-H2Kd (mouse histocompatibility class I) APC conjugated (ebioscience, San Diego, CA) to eliminate contaminating noncancer cells of mouse origin and viability dye 7AAD (BD Pharmingin) to discard dead cells. Side and forward scatter was used to eliminate debris. A portion of the cells were further stained with either Aldefluor assay to identify and isolate Aldefluor+ cells or anti-CD24 and −CD44 antibodies to isolate CD24−CD44+ cells. Desired cell populations were isolated using a FACSAria flow cytometer (Becton Dickinson).
In vitro reovirus infection of isolated Aldefluor+ and live breast cancer cells. A 100,000 isolated cells (Aldefluor+ or live total human breast cancer cells) were mock or infected with 20 multiplicity of infection of reovirus in mammosphere media (DMEM/F12; Invitrogen, supplemented with basic fibroblast growth factor, epidermal growth factor and insulin; Sigma-Aldrich, as previously described in ref. 30). And at 18 hours postinfection, cells were washed, fixed, permeablized, labeled with antireovirus antibody, and analyzed with a FACSCalibur as described earlier.
Western blot analysis of Ras levels. In this process, the 100,000 isolated Aldefluor+, CD24−CD44+, and total (7AAD−, H2Kd−) cells were lysed, nuclear debris removed by high speed centrifugation, and lysates separated by 10% SDS-PAGE, electrophoretically transferred to PolyScreen transfer polyvinylidene fluoride membranes (Perkin Elmer, Woodbridge, Ontario, Canada), and membranes were probed with anti-Ras monoclonal mouse antibody (1:500 dilution, clone Ras10; Upstate Biotechnology) or antiactin (1:1,000 dilution; Sigma Aldrich), followed by HRP-conjugated goat antimouse or antirabbit IgG (1:1,000; Jackson Laboratories, Burlington, Ontario, Canada). The immuno-reactive protein bands were detected using enhanced chemiluminescence (Amersham, Oakville, Ontario, Canada) and visualized with a Kodak Image Station 4,000 mm Pro (Mandel, Guelph, Ontario, Canada).
Tumor tissue analysis by immunofluorescence microscopy. For immunofluorescence, formalin-fixed tissue was paraffin embedded in blocks and 5 µmol/l sequential sections were cut and mounted on microscope slides. After antigen retrieval and blocking, slides were stained with combinations of the following primary antibodies: antihuman CD44 (mouse IgG2a; Labvision, Nepean, Ontario, Canada), antihuman CD24 (mouse IgM; Labvision), antihuman ALDH1 (mouse IgG1; BD Pharmingin), and rabbit polyclonal antireovirus antibody.25 Secondary antibodies specific to the primary antibody Ig subclass for dual labeling were conjugated to either Alexafluor 488 or 546 (Invitrogen) or Cy5 (Jackson ImmunoResearch). Nuclei were stained with ToPro-3 (Invitrogen, Nepean, Ontario, Canada). Washed slides were mounted in 90% glycerol, 100 mmol/l Tris-HCl pH 8, 2.5% w/v DABCO, 1 µg/ml bisbenzimide H 33342 trihydrochloride (Sigma Aldrich). Images were captured with a Zeiss LSM 510 laser scanning confocal microscope.
To detect apoptotic cells in the fixed tumor tissue, slides were stained with an in situ cell death detection kit, TMR red, via terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL; Roche Applied Science, Laval, Quebec, Canada).
Acknowledgments
This work was supported by operating grants from the Canadian Breast Cancer Foundation, Atlantic Chapter and Cancer Care Nova Scotia to P.W.K.L. and C.A.G. P.M. was supported by a postdoctoral fellowship from the Alberta Heritage Foundation for Medical Research. We thank Pat Colp, Alexander Edgar, and Stephen Whitefield of Dalhousie University for technical assistance on the project.
REFERENCES
- Hashiro G, Loh PC., and , Yau JT. The preferential cytotoxicity of reovirus for certain transformed cell lines. Arch Virol. 1977;54:307–315. doi: 10.1007/BF01314776. [DOI] [PubMed] [Google Scholar]
- Strong JE, Coffey MC, Tang D, Sabinin P., and , Lee PW. The molecular basis of viral oncolysis: usurpation of the Ras signaling pathway by reovirus. EMBO J. 1998;17:3351–3362. doi: 10.1093/emboj/17.12.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox ME, Yang W, Senger D, Rewcastle NB, Morris DG, Brasher PM, et al. Reovirus as an oncolytic agent against experimental human malignant gliomas. J Natl Cancer Inst. 2001;93:903–912. doi: 10.1093/jnci/93.12.903. [DOI] [PubMed] [Google Scholar]
- Alain T, Hirasawa K, Pon KJ, Nishikawa SG, Urbanski SJ, Auer Y, et al. Reovirus therapy of lymphoid malignancies. Blood. 2002;100:4146–4153. doi: 10.1182/blood-2002-02-0503. [DOI] [PubMed] [Google Scholar]
- Hirasawa K, Nishikawa SG, Norman KL, Alain T, Kossakowska A., and , Lee PW. Oncolytic reovirus against ovarian and colon cancer. Cancer Res. 2002;62:1696–1701. [PubMed] [Google Scholar]
- Norman KL, Coffey MC, Hirasawa K, Demetrick DJ, Nishikawa SG, DiFrancesco LM, et al. Reovirus oncolysis of human breast cancer. Hum Gene Ther. 2002;13:641–652. doi: 10.1089/10430340252837233. [DOI] [PubMed] [Google Scholar]
- Kilani RT, Tamimi Y, Hanel EG, Wong KK, Karmali S, Lee PW, et al. Selective reovirus killing of bladder cancer in a co-culture spheroid model. Virus Res. 2003;93:1–12. doi: 10.1016/s0168-1702(03)00045-5. [DOI] [PubMed] [Google Scholar]
- Yang WQ, Senger D, Muzik H, Shi ZQ, Johnson D, Brasher PM, et al. Reovirus prolongs survival and reduces the frequency of spinal and leptomeningeal metastases from medulloblastoma. Cancer Res. 2003;63:3162–3172. [PubMed] [Google Scholar]
- Reolysin Clinical Trials. Oncolytics Biotech Inc 2008 . < http://www.oncolyticsbiotech.com/clinical.html >.
- Dalerba P, Cho RW., and , Clarke MF. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267–84. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ., and , Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B. Malignant effusions: from diagnosis to biology. Diagn Cytopathol. 2004;31:246–254. doi: 10.1002/dc.20133. [DOI] [PubMed] [Google Scholar]
- Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong L, Stojkovic M, Dimmick I, Ahmad S, Stojkovic P, Hole N, et al. Phenotypic characterization of murine primitive hematopoietic progenitor cells isolated on basis of aldehyde dehydrogenase activity. Stem Cells. 2004;22:1142–1151. doi: 10.1634/stemcells.2004-0170. [DOI] [PubMed] [Google Scholar]
- Chute JP, Muramoto GG, Whitesides J, Colvin M, Safi R, Chao NJ, et al. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci USA. 2006;103:11707–11712. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Chan KW, Lee TK, Tang KH, Wo JY, Zheng BJ, et al. Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations. Mol Cancer Res. 2008;6:1146–1153. doi: 10.1158/1541-7786.MCR-08-0035. [DOI] [PubMed] [Google Scholar]
- Eyler CE., and , Rich JN. Survival of the fittest: cancer stem cells in therapeutic resistance and angiogenesis. J Clin Oncol. 2008;26:2839–2845. doi: 10.1200/JCO.2007.15.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TM, McBride WH., and , Pajonk F. The response of CD24(-/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98:1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Lee TK, Zheng BJ, Chan KW., and , Guan XY. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene. 2008;27:1749–1758. doi: 10.1038/sj.onc.1210811. [DOI] [PubMed] [Google Scholar]
- Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Dylla SJ, Beviglia L, Park IK, Chartier C, Raval J, Ngan L, et al. Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS ONE. 2008;3:e2428. doi: 10.1371/journal.pone.0002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcato P, Shmulevitz M, Pan D, Stoltz D., and , Lee PW. Ras transformation mediates reovirus oncolysis by enhancing virus uncoating, particle infectivity, and apoptosis-dependent release. Mol Ther. 2007;15:1522–1530. doi: 10.1038/sj.mt.6300179. [DOI] [PubMed] [Google Scholar]
- Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- Malumbres M., and , Pellicer A. RAS pathways to cell cycle control and cell transformation. Front Biosci. 1998;3:d887–d912. doi: 10.2741/a331. [DOI] [PubMed] [Google Scholar]
- Onodera S, Nishihira J, Iwabuchi K, Koyama Y, Yoshida K, Tanaka S, et al. Macrophage migration inhibitory factor up-regulates matrix metalloproteinase-9 and -13 in rat osteoblasts. Relevance to intracellular signaling pathways. J Biol Chem. 2002;277:7865–7874. doi: 10.1074/jbc.M106020200. [DOI] [PubMed] [Google Scholar]
- Wulf GG, Wang RY, Kuehnle I, Weidner D, Marini F, Brenner MK, et al. A leukemic stem cell with intrinsic drug efflSux capacity in acute myeloid leukemia. Blood. 2001;98:1166–1173. doi: 10.1182/blood.v98.4.1166. [DOI] [PubMed] [Google Scholar]
- Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- Gottesman MM, Fojo T., and , Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- Chapuy B, Koch R, Radunski U, Corsham S, Cheong N, Inagaki N, et al. Intracellular ABC transporter A3 confers multidrug resistance in leukemia cells by lysosomal drug sequestration. Leukemia. 2008;22:1576–1586. doi: 10.1038/leu.2008.103. [DOI] [PubMed] [Google Scholar]
- Magni M, Shammah S, Schiró R, Mellado W, Dalla-Favera R., and , Gianni AM. Induction of cyclophosphamide-resistance by aldehyde-dehydrogenase gene transfer. Blood. 1996;87:1097–1103. [PubMed] [Google Scholar]
- Eriksson M, Guse K, Bauerschmitz G, Virkkunen P, Tarkkanen M, Tanner M, et al. Oncolytic adenoviruses kill breast cancer initiating CD44+CD24-/low cells. Mol Ther. 2007;15:2088–2093. doi: 10.1038/sj.mt.6300300. [DOI] [PubMed] [Google Scholar]
- Smith RE, Zweerink HJ., and , Joklik WK. Polypeptide components of virions, top component and cores of reovirus type 3. Virology. 1969;39:791–810. doi: 10.1016/0042-6822(69)90017-8. [DOI] [PubMed] [Google Scholar]