Abstract
Huntington's disease (HD) is a fatal neurodegenerative disease caused by mutant huntingtin (htt) protein, and there are currently no effective treatments. Recently, we and others demonstrated that silencing mutant htt via RNA interference (RNAi) provides therapeutic benefit in HD mice. We have since found that silencing wild-type htt in adult mouse striatum is tolerated for at least 4 months. However, given the role of htt in various cellular processes, it remains unknown whether nonallele-specific silencing of both wild-type and mutant htt is a viable therapeutic strategy for HD. Here, we tested whether cosilencing wild-type and mutant htt provides therapeutic benefit and is tolerable in HD mice. After treatment, HD mice showed significant reductions in wild-type and mutant htt, and demonstrated improved motor coordination and survival. We performed transcriptional profiling to evaluate the effects of reducing wild-type htt in adult mouse striatum. We identified gene expression changes that are concordant with previously described roles for htt in various cellular processes. Also, several abnormally expressed transcripts associated with early-stage HD were differentially expressed in our studies, but intriguingly, those involved in neuronal function changed in opposing directions. Together, these encouraging and surprising findings support further testing of nonallele-specific RNAi therapeutics for HD.
Introduction
Huntington's disease (HD) is one of nine dominant neurodegenerative diseases caused by a CAG-trinucleotide repeat expansion (>35 repeats) in a coding exon. In HD, the resulting polyglutamine expansion confers a toxic gain-of-function to the mutant huntingtin (htt) protein. Mutant htt forms insoluble aggregates and causes transcriptional dysregulation, perturbations in protein homeostasis and cell death.1,2,3,4,5,6 Pathological features of HD include cortical thinning and a striking progressive loss of striatal neurons.7 Disease onset usually begins during the third to fourth decade of life with patients exhibiting choreiform movements, impaired coordination, progressive dementia, and other psychiatric disturbances.8 HD is ultimately fatal with death occurring 10–15 years after symptom onset. To date, no effective therapies exist for HD patients.
Current therapeutic strategies being investigated for HD aim to reverse neuronal dysfunction and promote cell survival, and typically involve chemical compounds which are disease-modifying or neuroprotective. These compounds include histone deacetylase inhibitors which ameliorate transcriptional dysfunction, minocycline which inhibits apoptotic factors, and creatine and coenzyme Q10 which increase cellular energy stores required to cope with mutant htt-induced toxicity. In preclinical studies, these compounds have shown therapeutic efficacy in HD mice.9,10,11,12 Although these results are encouraging, gene-targeting therapeutics aim to inhibit the cause of disease and provide attractive alternative or complementary treatments for HD. The expectation being that suppressing mutant htt expression will prevent its many downstream cytotoxic effects.
Recently, the application of RNA interference (RNAi) technologies to silence mutant htt has shown therapeutic potential in HD transgenic mouse models expressing mutant human htt.13,14,15 In these proof-of-principal studies, the therapeutic effect on disease phenotype was evaluated by knocking down a mutant human htt transgene in the setting of two normal mouse htt alleles. Thus, the effects of nonallele-specific htt silencing (i.e., knockdown of both wild-type and mutant alleles) remain unknown. Allele-specific targeting of mutant htt transcripts may be ideal when considering the essential role of wild-type htt in developmental and cellular processes.3,6,16,17,18,19,20 Recent reports highlight the potential of allele-specific silencing of mutant htt using inhibitory RNAs;21 however as of yet, no polymorphism has been identified that would allow specific targeting of the mutant CAG-expanded transcript in all HD patients, even though some prevalent single-nucleotide polymorphisms (SNPs) have been identified.22 We therefore assessed whether therapeutic benefit is attainable via nonallele-specific silencing of mutant and wild-type htt in a mouse model for HD. We expressed inhibitory RNAs, capable of silencing both human and mouse htt transcripts, within the striata of HD-N171-82Q mice. These mice express an N-terminal fragment of human htt containing 82 glutamine repeats and exhibit rapidly progressive motor deficits, striatal neuropathology, and weight loss, thus serving as a valuable model for evaluating potential treatments for HD.23 Interestingly, RNAi-treated HD mice displayed improved behavior and prolonged survival, despite significantly reduced levels of endogenous wild-type mouse htt within the striatum. To evaluate the consequences of knocking down wild-type htt in adult mouse striatum, we performed transcriptional profiling and observed changes in gene expression that are concordant with the known functions of htt, and have interesting implications for HD.
Results
Artificial microRNAs demonstrate improved safety over small-hairpin RNAs in HD-N171-82Q mice
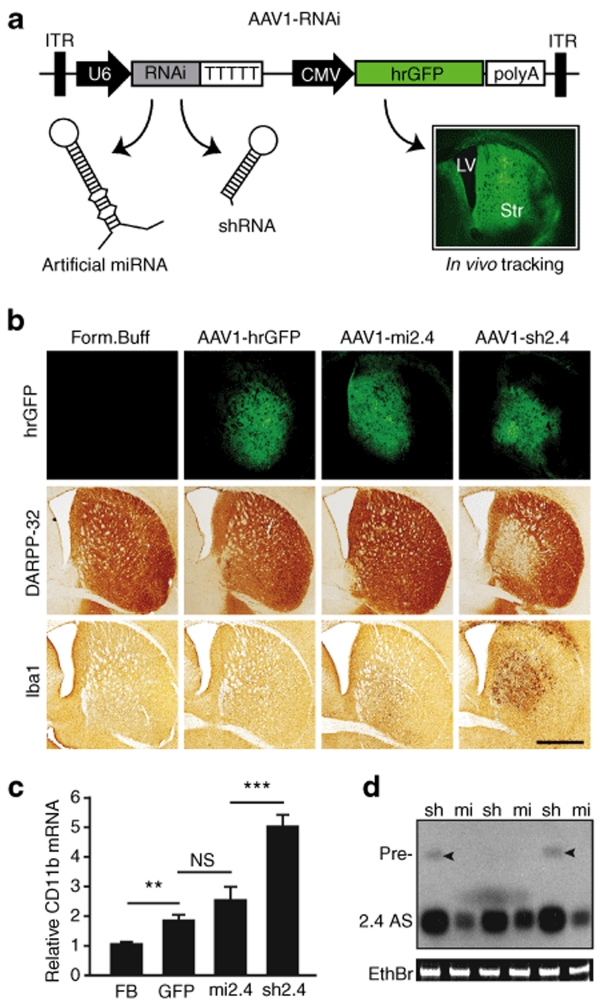
Previously, we compared the efficacy and safety of short-hairpin RNA (shRNA)- and miRNA-based RNAi expression vectors. We found that shRNAs are more potent but induce toxicity in cell cultures and in mice,24,25 whereas artificial miRNAs are expressed at lower levels and display better safety profiles. In dosing studies, we observed a loss of silencing efficiency when lowering shRNA vector doses to minimize toxicity; this probably resulted from reduced cell transduction.26 With relevance to HD, we reported that viral-mediated expression of an artificial miRNA (mi2.4; targets exon 2 of mouse and human htt transcripts) in mouse striata maintained effective gene silencing and was well-tolerated, relative to a corresponding toxic shRNA, sh2.4.26 Sh2.4-induced toxicity was consistent in both wild-type and mutant CAG140 HD knock-in mice on the C57BL/6 background. Here, we evaluated the safety of sh2.4 and mi2.4 in HD-N171-82Q mice, which are maintained on the B6C3F1/J background, to test for strain-to-strain variations in susceptibility to RNAi-induced toxicity. Seven-week-old HD-N171-82Q mice were striatally injected with adeno-associated viral vectors [AAV serotype 2/1 (AAV1); Figure 1a] that express either sh2.4 or mi2.4 along with a humanized renilla green fluorescent protein (hrGFP) reporter to allow for identification of transduced cells within the brain. Of note, we have previously shown that AAV1 vectors primarily transduce medium spiny neurons in mouse striatum.26 At 3 months after injection, we evaluated neurotoxicity by immunohistochemical staining for DARPP-32 (striatal neuron marker) and Iba1 (microglial marker). Similar to our previous findings in C57BL/6 mice, AAV1-sh2.4-treated HD-N171-82Q mice showed a clear loss of DARPP-32 immunoreactivity and intense microglial activation in regions of the striatum which correlated with hrGFP-positivity (i.e., transduction). Conversely, these hallmarks for neurotoxicity were markedly decreased in AAV1-mi2.4-treated HD-N171-82Q mice and absent in HD-N171-82Q mice injected with formulation buffer (FB) (Figure 1b). We also performed quantitative real-time PCR (QPCR) analysis for CD11b mRNA, a readout for microglial activation. Striata treated with AAV1-sh2.4 showed nearly a fivefold increase of CD11b mRNA relative to FB-treated striata (Figure 1c, P < 0.001). Also, AAV1-hrGFP treatment caused a small, but significant CD11b upregulation, as did AAV1-mi2.4 to a slightly greater degree. These findings are dissimilar from our previous report, where CD11b activation was not observed after striatal injection of AAV1-mi2.4 into wild-type C57BL/6 mice.26 Whether B6C3F1/J mice or HD brains are more susceptible to viral- or RNAi-induced toxicity remains to be evaluated.
Figure 1.
Mi2.4 demonstrates improved safety, relative to sh2.4, in HD-N171-82Q mice. (a) Diagram of recombinant AAV2/1 viral vectors which express htt-specific RNAi and hrGFP; the latter allows tracking of in vivo transduction within mouse brain (Str = striatum; LV = lateral ventricle). (b) HD-N171-82Q mice were injected with either formulation buffer (FB) or viral vectors into the striatum, and 3 months later, histological analyses were performed to assess for striatal toxicity. Photomicrographs representing hrGFP autofluorescence and immunohistochemical staining of DARPP-32-positive neurons and IbaI-positive microglia are shown for each treatment group. Scale bar = 500 µm for each photomicrograph. (c) Microglial activation was also evaluated by quantitative real-time PCR analyses measuring the endogenous mouse CD11b mRNA levels in striatal RNA samples. Normalized results are shown as mean ± SEM (n ≥ 5; ***, ** and NS indicate P < 0.001, P < 0.01 and no significance, respectively). (d) Small transcript northern blot analysis was performed to assess the levels of htt-specific antisense (2.4 AS) RNA present in AAV1-RNAi-treated striata (sh = sh2.4, mi = mi2.4; n = 3 treated striata). Sh2.4 treatment yielded a build-up of unprocessed precursor RNAs (Pre-, arrowheads) in 2 of 3 samples. Ethidium bromide (EthBr) staining was performed as a loading control. hrGFP, humanized renilla green fluorescent protein; ITR, inverted terminal repeat; RNAi, RNA interference.
To determine whether the differential toxicity profiles of sh2.4 and mi2.4 may be attributed to HD2.4 inhibitory RNA levels, we performed northern blot analysis. As was observed in C57BL/6 mice,26 AAV1-sh2.4 treatment generated substantially more HD2.4 antisense RNA, relative to AAV1-mi2.4 (Figure 1d). Moreover, we observed slight build-up of shRNA precursors, an indication of saturating the endogenous RNAi processing machinery, in some sh2.4-treated striata. Together, these results corroborate many of our previous findings and support that artificial miRNAs show improved safety over shRNAs, in at least two unique mouse strains. Although this is an exciting step toward developing safer vectors, the therapeutic efficacy of artificial miRNAs in a mouse model of neurodegenerative disease has not yet been reported.
Mi2.4 silences both human and mouse htt mRNAs
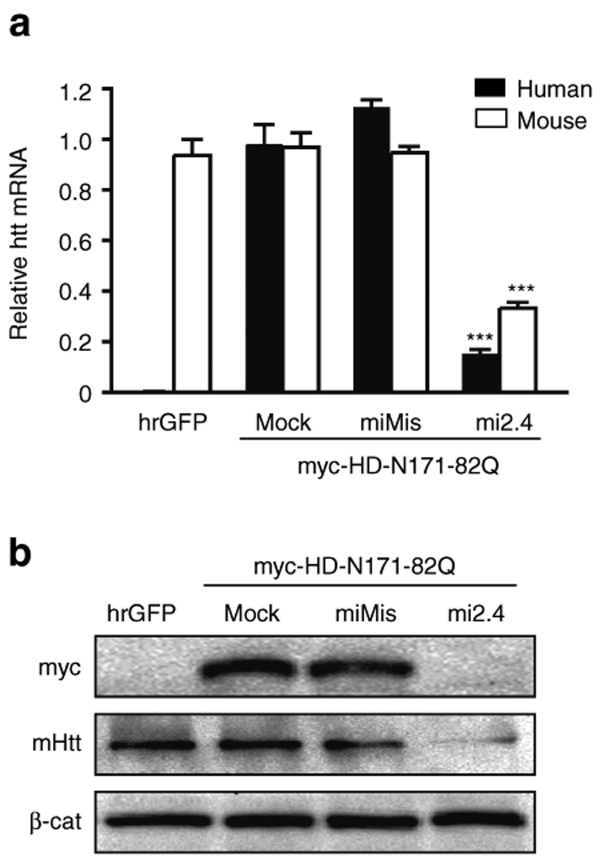
We have previously shown that mi2.4 silences both wild-type human and mouse htt transcripts in separate experimental systems (HEK293 cells and mouse striatum).26 Here, we assessed the capacity of mi2.4 to effectively silence both mutant human and wild-type mouse htt alleles in the same experimental setting. To recapitulate the genetic make-up of our HD model of interest (HD-N171-82Q mice), we overexpressed a myc-tagged human HD-N171-82Q htt fragment in mouse C2C12 cells, which are known to express endogenous mouse htt.26 RNAi expression plasmids were co-transfected into this setting, and gene silencing was measured by QPCR and western blot analyses. Of note, we found the human and mouse QPCR primer/probe sets to be exceptionally specific. Virtually no signal for human htt mRNAs was observed in cells that did not receive the myc-HD-N171-82Q expression plasmid (i.e., hrGFP-treated), and no additional signal for mouse htt mRNAs was observed in cells that did (Figure 2a). In cells co-transfected with myc-HD-N171-82Q and mi2.4 expression plasmids, we observed significant reductions in the mutant human and endogenous wild-type mouse htt transcripts (~85 and 65% silencing, respectively, P < 0.001). Western blot analyses revealed that these levels of mRNA knockdown resulted in a clear decrease of the respective htt proteins (Figure 2b).
Figure 2.
Mi2.4 silences both mutant human and wild-type mouse htt in vitro. RNAi expression plasmids were co-transfected into mouse C2C12 cells along with a plasmid expressing a myc-tagged mutant human HD-N171-82Q transgene, and gene silencing was assessed 24 hours later. (a) QPCR analyses were performed to measure the levels of mutant human and endogenous wild-type mouse htt mRNAs. Results are shown as mean ± SEM (n = 4; *** indicates P < 0.001). (b) Silencing of the human and mouse htt proteins (myc and mHtt, respectively) was assessed by western blot analyses. β-catenin (β-cat) served as the loading control. The mismatch control (miMis) contains multiple base-pair changes that render the miRNA ineffective against htt. Mock-treated cells received promoter-only plasmid (i.e., no miRNA). hrGFP-treated cells served as a control for evaluating the specificity of the human and mouse htt QPCR primer/probes and antibodies. hrGFP, humanized renilla green fluorescent protein; QPCR, quantitative real-time PCR.
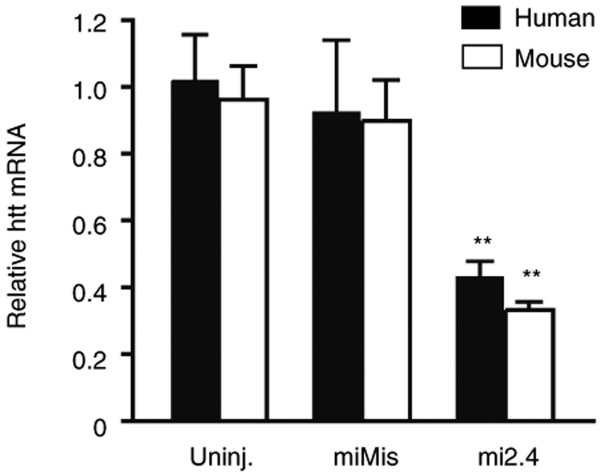
We subsequently tested mi2.4 for simultaneous silencing of both mutant and wild-type htt alleles in HD-N171-82Q mice. Mice were injected with FB, AAV1-mi2.4 or AAV1 control vectors into the striatum, a primary site of neurodegeneration in HD. At 4 weeks after treatment, we observed a significant reduction of mutant human and wild-type mouse htt transcripts (~60%, P < 0.01) following injection with AAV1-mi2.4 compared to control-treated striata (Figure 3). Notably, we have previously demonstrated, in separate experiments, that ~60% silencing of either mutant human or wild-type htt transcripts in mouse striatum leads to marked reductions in the respective proteins.13,26 Together, these results corroborate our in vitro findings regarding the specific activity of mi2.4 and demonstrate that mi2.4 is capable of mediating nonallele-specific silencing of all htt alleles present in HD-N171-82Q mice.
Figure 3.
Mi2.4 silences both mutant human and wild-type mouse htt mRNAs in HD-N171-82Q mice. HD-N171-82Q mice were injected unilaterally into the striatum with AAV1-miMis, or AAV1-mi2.4, and quantitative real-time analyses were performed on striatal RNA samples to measure the levels of mutant human and wild-type mouse htt mRNAs at 4 weeks after treatment. Results are shown as mean ± SEM (n ≥ 3; ** indicates P < 0.01) relative to the uninjected contralateral striata.
Nonallele-specific silencing of htt provides therapeutic benefit in HD-N171-82Q mice
To date, RNAi-based therapies have shown promise in HD transgenic mice;13,14,15 however, these studies tested the therapeutic efficacy of targeting only mutant htt in the presence of endogenous wild-type mouse htt. Thus, whether phenotypic recovery depends on normal wild-type htt levels remains unknown. In this study, we investigated whether therapeutic benefit is attainable via nonallele-specific silencing of mutant and wild-type htt in HD-N171-82Q mice. We measured therapeutic efficacy by monitoring the development of behavioral deficits in HD-N171-82Q mice. Mutant mice were injected bilaterally into the striatum with FB, AAV1-hrGFP or AAV1-mi2.4, and wild-type littermates were injected with FB in the same manner. All mice were treated at 7 weeks of age, weighed and tested on the rotarod apparatus at 10, 14, and 18 weeks and ultimately killed at 20 weeks for QPCR and histological analyses (Figure 4a).
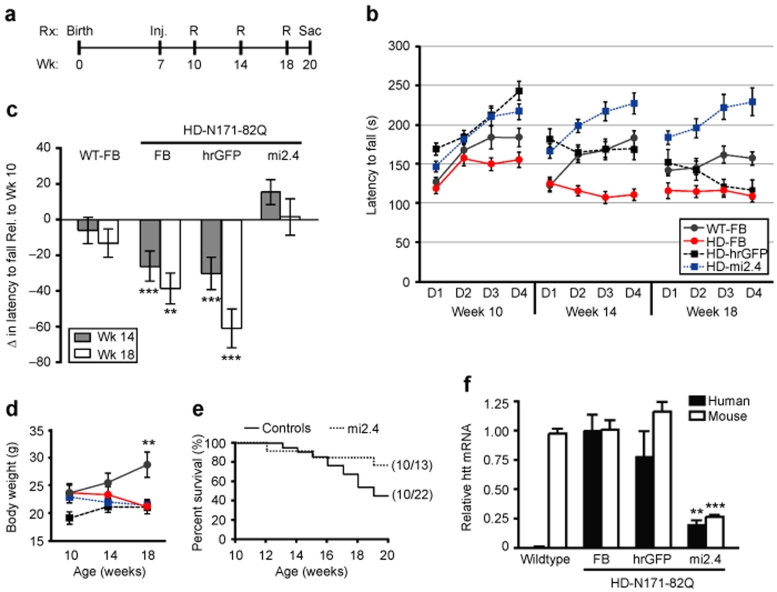
Figure 4.
Mi2.4 provides therapeutic benefit in HD-N171-82Q mice. (a) Timeline for testing the therapeutic efficacy of AAV1-mi2.4 in HD-N171-82Q mice. HD-N171-82Q mice were injected bilaterally into the striatum with formulation buffer (FB), AAV1-hrGFP, or AAV1-mi2.4. Similarly, wild-type littermates were treated with formulation buffer. All mice were injected at 7 weeks of age, weighed and tested on the rotarod (R) apparatus at 10, 14, and 18 weeks and ultimately sacrificed (Sac) at 20 weeks for QPCR and histological analyses. (b) Rotarod data from four consecutive days at 10, 14, and 18 weeks of age is shown as latency to fall (mean ± SEM of trials 1–3 for each group per day). (c) To evaluate the progress of each group over the time-course of the therapeutic trial, the rotarod data is shown as the mean change ± SEM in average weekly performance relative to week 10 (*** and ** indicate P = 0.002 and P = 0.02, respectively). (d) The average body weights for mice in each group are shown for the indicated ages (mean ± SEM; ** represents P < 0.01). Note: legend shown in b applies. (e) Kaplan–Meier survival analysis up to 20 weeks of age (i.e., the time of killing) is shown. In parentheses is the fraction of surviving mice at the end-point per the initial total for each group. The control group consists of HD-N171-82Q mice treated with formulation buffer or AAV1-hrGFP. (f) Mice were killed at week 20, and QPCR analyses were performed on striatal RNA samples to measure mutant human and wild-type mouse htt mRNA levels. Normalized results are shown as mean ± SEM (n ≥ 5; *** and ** indicate P < 0.001 and P < 0.01, respectively). HD, Huntington's disease; hrGFP, humanized renilla green fluorescent protein; QPCR, quantitative real-time PCR; WT, wild type.
Mice were subjected to an accelerating rotarod assay to test for prevention of behavioral deficits in HD-N171-82Q mice following AAV1-mi2.4 treatment. Over the time course of the study, control-treated HD-N171-82Q mice developed a progressive impairment in rotarod performance, relative to their week 10 performance (Figure 4b,c). These results are consistent with previous reports, including the Harper et al. study from our laboratory which tested the therapeutic efficacy of a human-specific htt shRNA, shHD2.1, in HD-N171-82Q mice.13,27,28 Interestingly, HD-N171-82Q mice injected with AAV1-mi2.4 demonstrated striking improvements in rotarod performance, as compared to control-treated HD-N171-82Q mice, at 14 and 18 weeks (P = 0.002 and P = 0.02 respectively, Figure 4c). AAV1-mi2.4-treated HD-N171-82Q mice also performed better than wild-type mice at the 18-week time point. However, this unexpected observation probably resulted from the assay parameters (e.g., allowing mice to hang on for one rotation without running), which provided advantages to smaller mice. Of note, at 18 weeks of age, wild-type mice weighed considerably more than HD-N171-82Q mice, and AAV1-mi2.4 treatment to the striatum failed to normalize the weight-loss observed in the HD-N171-82Q mice (Figure 4d). Indeed, we identified an inverse correlation between weight and rotarod performance in 10-week-old mice, before the onset of HD-related deficits (data not shown; group comprised 20 wild type and 21 mutant mice, P < 0.01).
HD-N171-82Q mice display reduced life-span relative to wild-type littermates; ~50% of HD-N171-82Q mice die by 20 weeks of age.29,30,31 Thus, we halted our experiments at this time point for two reasons: (i) so that sufficient numbers of tissue samples could be harvested for end-point QPCR analyses, and (ii) so that the capacity of AAV1-mi2.4 treatment to improve survivability, within the 50% window, could be evaluated. Similar to previously reported data, we observed that only 45% of control-treated HD-N171-82Q mice remained alive by week 20. By contrast, AAV1-mi2.4-treated HD-N171-82Q mice trended toward improved survival; >75% of them lived to the point-of-sacrifice at week 20 (P = 0.1).
At 20 weeks of age, QPCR analyses were performed to evaluate long-term suppression of mutant human and wild-type mouse htt transcripts. HD-N171-82Q mice treated with AAV1-mi2.4 showed significant reductions (~75%) in human and mouse htt mRNA levels, relative to control-treated littermates (Figure 4f, P < 0.01). These results, together with data from our rotarod and survival analyses, demonstrate that nonallele-specific silencing of mutant and wild-type htt provides therapeutic benefit in HD-N171-82Q mice.
Transcriptional profile changes after wild-type htt silencing in mouse striatum
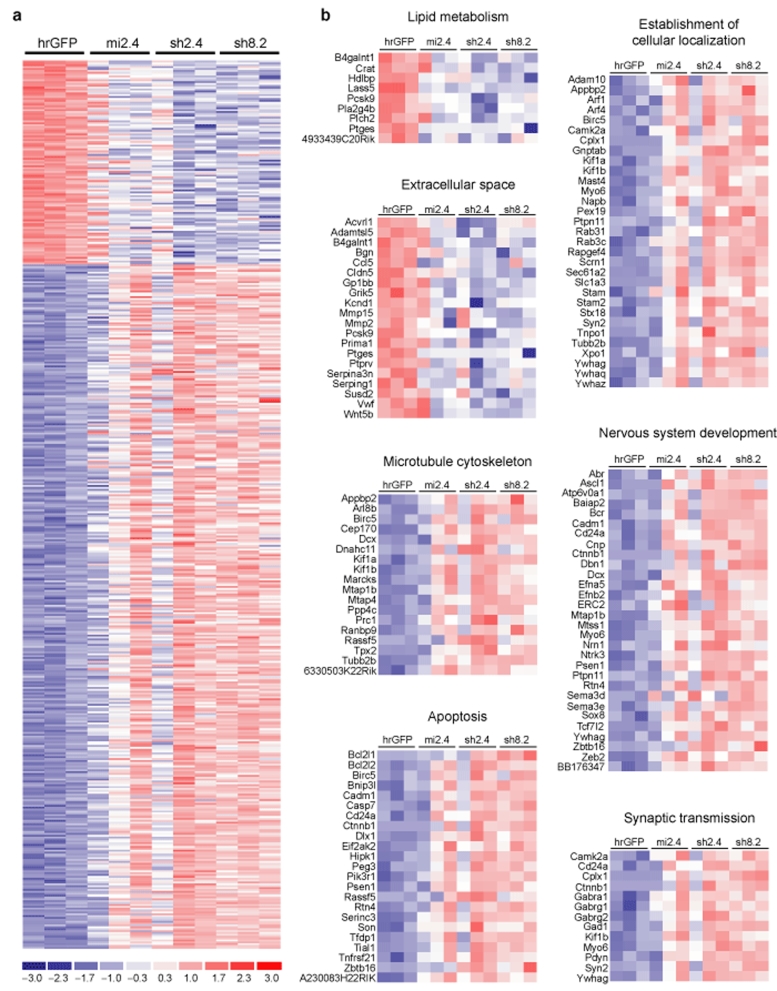
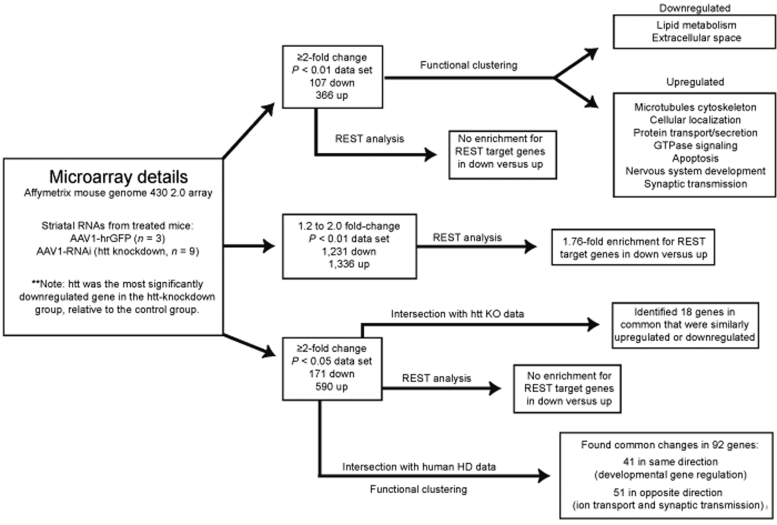
We have observed that a nonallele-specific therapeutic RNAi strategy for HD is efficacious in HD mice. This is interesting given the important role for wild-type htt in several cellular processes.3,6,16,17,18,19,20 Recently, we have demonstrated that silencing wild-type htt in adult mouse striatum was tolerated for at least 4 months, the last time point reported.26 However, it remains unknown whether subtle changes in striatal neuron function occur in adult mouse brain as a result of reduced htt levels. We therefore evaluated the transcriptional profile changes resulting from RNAi-mediated silencing of striatal htt, with the purpose being twofold: (i) to gain additional insight into the functions of wild-type htt, and (ii) to further evaluate the safety of suppressing wild-type htt in adult mouse striatum. Eight-week-old wild-type C57BL/6 mice were injected into the striatum with either AAV1-hrGFP or AAV1 vectors which express htt-specific RNAi [mi2.4, sh2.4, or sh8.2 (another previously reported therapeutic candidate for HD)]. Four weeks later, before the onset of shRNA-induced toxicity, we performed microarray analysis on striatal RNA samples, and looked for transcriptional changes between the AAV1-hrGFP group (n = 3) and AAV1-RNAi group (i.e., htt knockdown group; n = 9, 3 samples for each RNAi vector). We used three unique RNAi vectors for the htt knockdown group to minimize the likelihood of detecting false-positives resulting from RNAi-mediated off-targeting, as opposed to suppression of htt. Silencing of endogenous mouse htt mRNA in each of the AAV1-RNAi-treated striatal RNA samples was confirmed by QPCR analysis (Supplementary Figure S1). These results were consistent with our microarray data, which revealed htt as the most significantly downregulated gene (~3.5-fold, P = 4.678e-07). We found 473 transcripts (with identifiable gene symbols) to be differentially expressed by ≥2.0-fold (P < 0.01, 107 downregulated and 366 upregulated) between the htt-knockdown and control-treated groups (Figures 5a and 6 and Supplementary Table S1). Here, we applied a less stringent P value cutoff to yield a sufficient number of genes for downstream gene set analyses.
Figure 5.
Transcriptional changes resulting from knockdown of wild-type htt in mouse striatum. (a) Heat map depicting the relative fold change in expression levels of the 473 transcripts that were differentially expressed by ≥2.0-fold (P < 0.01, 107 downregulated and 366 upregulated) between the control hrGFP-treated group and the htt-knockdown group (consisting of samples treated with mi2.4, sh2.4, or sh8.2). Data for each experimental sample (n = 3 per indicated treatment) are shown. The scale bar indicates fold change with blue and red tones signifying downregulated and upregulated, respectively. (b) Functional gene annotation clustering performed on the 107 downregulated and 366 upregulated transcripts identified enrichments in the indicated cellular components and processes. Heat maps depicting the fold change in expression of the genes within each enriched category are shown. The scale bar in a applies. hrGFP, humanized renilla green fluorescent protein.
Figure 6.
Microarray analyses workflow and summary of results. hrGFP, humanized renilla green fluorescent protein; RNAi, RNA interference.
To investigate cellular processes which may be affected by htt knockdown, we performed functional gene annotation clustering on the set of 473 genes that showed altered gene expression by ≥2.0-fold (P < 0.01) between the htt-knockdown and control-treated groups. This analysis was done using the David Bioinformatics Resources web-server.32 Among the 107 downregulated transcripts, there were enrichments for genes involved in lipid metabolism (9 genes, P < 0.05) and for protein products destined for the extracellular space (20 genes, P < 0.05) (Figure 5b). These findings are consistent with microarray data from prior studies which evaluated the effects of htt knockout in various cell lines.33,34 By contrast, among the 366 upregulated transcripts, we detected enrichments primarily in intracellular components and processes, which included microtubule cytoskeleton (18 genes, P < 0.001), establishment of cellular localization (31 genes, P < 0.01), intracellular protein transport (19 genes, P < 0.01), secretion (15 genes, P < 0.05), small GTPase-mediated signal transduction (23 genes, P < 0.0001), cell cycle (28 genes, P < 0.01), apoptosis (23 genes, P < 0.05), nervous system development (30 genes, P < 0.001), and synaptic transmission (13 genes, P < 0.01) among others (Figure 5b and Supplementary Figure S2). These results support the identified roles of htt in vesicular trafficking, maintenance of the endoplasmic reticulum, inhibition of apoptosis, and development.3,16,17,18,19,20,35
Effect of htt knockdown on REST-regulated genes
Wild-type htt interacts with the transcriptional corepressor REST in the cytoplasm.3 This interaction prevents REST from entering the nucleus, where it represses genes containing RE1/NRSE (repressor element 1/neuron-restrictive silencer element) binding sites; these genes are typically neuron-specific. Thus, one might hypothesize that suppressing htt levels may result in downregulation of REST target genes. We evaluated our set of differentially expressed genes to determine whether downregulated transcripts were enriched for genes containing RE1 binding sites. For cross comparison to our gene set, we used an RE1 gene set consisting of ~1,000 unique gene symbols which mapped to consensus RE1 binding sites within the mouse genome (retrieved from the RE1 database—http://bmbpcu36.leeds.ac.uk/RE1db_mkII/).36,37 We found no enrichment for RE1-containing genes among the transcripts that were downregulated by ≥2.0-fold (P < 0.01, 107 genes) in the htt-knockdown group versus the control group, as compared to transcripts that were upregulated by ≥2.0-fold (P < 0.01, 366 genes). Similarly, there was no enrichment for RE1-containing genes in downregulated versus upregulated transcripts when using expanded data sets with a less stringent P value cutoff (≥2.0-fold change, P < 0.05, 171 downregulated and 590 upregulated; Supplementary Table S2) for this cross-comparison analysis. However, we reasoned that increased REST activity may impart more subtle transcriptional decreases, considering that gene expression is typically regulated by many factors. Therefore, we looked for enrichment of RE1-containing genes among transcripts that were differentially expressed by 1.2- to 2.0-fold (P < 0.01, 1,231 downregulated and 1,336 upregulated transcripts with identifiable gene symbols; Supplementary Table S3) between the htt-knockdown and control-treated groups. Among these downregulated transcripts, we discovered that 91 mapped to consensus RE1 binding sites (7.4%); this was a 1.76-fold increase relative to the upregulated transcripts (4.2%, P < 0.001 with Fisher's exact test). These results suggest that silencing htt expression may subtly disrupt REST-regulated gene networks; however, the effects were not global, meaning that some known REST target genes were upregulated (e.g., Pdyn, Lrrtm4, and Cntnap2) and others showed no change in expression (e.g., Bdnf, Chrm4, and Penk1) as a result of htt knockdown.
Transcriptional changes in common with htt knockout studies
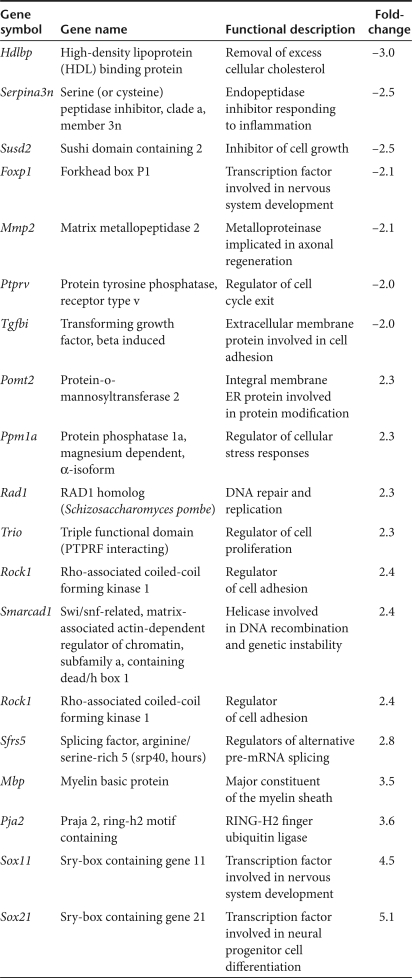
There have been two prior independent reports describing the use of transcriptional profiling to assess the effects of htt knockout in various cell lines.33,34 We compared our data sets to the available data from these two studies to identify common changes in gene expression associated with loss of htt. To increase the probability of finding intersection with these data sets, we used our expanded set of differentially expressed genes with a P value cutoff of P < 0.05. This data set comprised 761 transcripts (with identifiable gene symbols; 171 downregulated and 590 upregulated) that showed altered expression by ≥2.0-fold (P < 0.05) between the htt-knockdown and control-treated groups. Among this data set, we identified 18 genes that were similarly downregulated or upregulated in either of the two htt knockout microarray data sets (Table 1). Some of these genes may have implications in HD; for example, (i) the significant reduction in Hdlbp could have relevance to the perturbations in cholesterol homeostasis observed in HD,38,39 (ii) alterations in transcription factors (Foxp1, Sox11, and Sox21) involved in neuronal development may interfere with maintenance of the postmitotic neuronal phenotype, and (iii) increases in Mbp may relate to the remyelination attempts observed in HD brain that are thought to cause free-radical-induced tissue damage.40 Among these 18 genes there are likely others with relevance to HD research, and further investigation is warranted, given that these genes were identified in two independent microarray analyses assessing the role of htt in very different contexts (in vitro versus in vivo).
Table 1.
Common transcriptional changes in htt knockout (in vitro) and knockdown (in vivo) studies
Relation to transcriptional changes in HD
In comparing our gene sets to other HD-related gene expression data, we noted that several transcripts showing altered expression as a result of htt knockdown were also found to change in early-stage HD in humans and various mouse models for HD.41,42 Therefore, we performed an intersection of our data set consisting of 761 transcripts that were differentially expressed by ≥2.0-fold (P < 0.05) following knockdown of htt in mouse striatum to a data set of 973 unique-identifiable genes (1,616 probe sets) that showed altered expression by ≥1.4-fold (P < 0.001) in early-stage HD (grades 0–2) patient striatum relative to normal human striatum.41 Ninety-two transcripts (with identifiable gene symbols) were present in both data sets; 41 of these genes were similarly upregulated or downregulated in both sets, whereas 51 changed in opposite directions (Supplementary Table S4). We performed functional gene annotation clustering (as described above) on these subsets to look for enrichment in cellular components and processes. Among the 41 transcripts that changed in the same direction, there was enrichment for genes involved in developmental regulation of gene transcription (P < 0.01). Because these HD-related transcriptional changes coincided with our htt-knockdown group, they may be attributed to the loss-of-function component of HD. By contrast, the 51 genes which changed in opposite directions were strongly enriched for proteins involved in ion transport and synaptic transmission (P < 0.0001), many of which are downregulated in human HD. It is interesting to speculate that these transcriptional changes result from the toxic gain-of-function aspect of HD, and that knocking down both mutant and wild type htt may revert these changes.
Discussion
The application of therapeutic RNAi is being investigated for a variety of diseases which affect the central nervous system, where the delivery of inhibitory RNAs by systemic means has proven difficult due to the blood–brain barrier. Thus, many researchers have focused on using viral-based delivery systems to express hairpin RNAs in the brain. Recently, we and others have evaluated hairpin-based RNAi expression strategies for both efficacy and safety in vitro and in vivo.24,25,43,44 Here, we provide additional evidence supporting the improved safety of viral-expressed artificial miRNAs, relative to shRNAs, in mammalian brain.25,26 Furthermore, we present our novel findings that artificial miRNAs show therapeutic potential in treating a mouse model for neurodegenerative disease.
In some diseases, it is possible to use RNAi-based therapeutics to specifically target disease-linked SNPs that are present on the mutant transcripts.45,46 Recently, some prevalent SNPs residing mainly on the mutant htt transcript have been identified in human HD populations;22 however, it remains uncertain whether targeting these SNPs will achieve allele-specific silencing in a therapeutic setting. Unfortunately, no SNP has yet been identified which would allow specific silencing of the mutant htt allele in all HD patients. Therefore, the application of allele-specific SNP targeting for the remaining HD populations may involve developing unique inhibitory RNA sequences on a per patient basis. This is disadvantageous considering that each inhibitory RNA sequence would need to undergo the appropriate safety and efficacy studies. By contrast, nonallele-specific silencing provides an attractive alternative when considering the costs—both time and money—of developing therapeutic RNAi. We therefore tested whether a nonallele-specific RNAi strategy is capable of providing therapeutic benefit for HD. We demonstrate that striatal injection of AAV1-mi2.4 into HD mice effectively silenced both mutant and wild-type htt alleles. Interestingly, this treatment resulted in improved performance in behavioral tests, despite significant reductions of wild-type htt within the striatum.
We have previously reported that silencing wild-type htt in mouse striatum is well-tolerated for at least 4 months after treatment. These findings are surprising given the important roles for htt in various developmental and cellular processes.3,6,16,17,18,19,20,35 Htt knockout mice are embryonic lethal,18,19 and conditional knockout (loss of htt after birth) or hypomorphic htt mice display abnormalities in neurogenesis.20,47,48 But as of yet, the effects of reducing wild-type htt in adult postmitotic neurons in the brain remains to be thoroughly investigated. As a first step toward addressing this, we performed microarray to assess the transcriptional changes associated with silencing wild-type striatal htt in adult mice. In this analysis, we observed that htt-related cellular pathways are disrupted after htt-specific RNAi treatment. However, further investigation is necessary to determine whether these changes affect neuronal function or whether there are compensatory mechanisms which prevent these changes from manifesting into abnormal neurological conditions.
In summary, we demonstrate that nonallele-specific silencing of both mutant and wild-type htt can dramatically improve HD-related behavioral abnormalities, in a mouse model of HD. These findings provide additional support for the feasibility of treating HD using RNAi-based approaches. Moreover, our data are encouraging and suggest that the mammalian brain can tolerate ~75% decrease in wild-type htt mRNA within the striatum for several months, despite changes in the transcriptome. Our results support further and more stringent investigation of the consequences of long-term RNAi-mediated silencing of wild-type htt expression in adult brain.
Materials and Methods
Plasmids and viral vectors. Recombinant AAV serotype 2/1 vectors (AAV1-hrGFP, AAV1-mi2.4, AAV1-sh2.4, and AAV1-sh8.2) have been previously described.26 Each viral vector expresses CMV-driven hrGFP to track cell transduction, and AAV1-RNAi vectors express inhibitory RNAs from the mouse U6 promoter. AAV1 vectors were resuspended in Formulation Buffer 18 (HyClone, Logan, UT), and titers were determined by QPCR.
The plasmids expressing U6-driven mi2.4 or miMis (also known as mi2.4mis) were previously described.26 Myc-tagged HD-N171-82Q and hrGFP were expressed from the pCMV-HD-N171-82Q plasmid23 and pAAV-CMV-hrGFP13 plasmids, respectively.
Animals. All animal protocols were approved by the University of Iowa Animal Care and Use Committee. Wild-type C57BL/6 and HD-N171-82Q mice were obtained from Jackson Laboratories (Bar Harbor, ME)23 and the latter were maintained on a B6C3F1/J background. Mice were genotyped using primers specific for the mutant human htt transgene, and hemizygous and age-matched wild-type littermates were used for the indicated experiments. In the therapeutic trial, the treatment groups comprised approximately equal numbers of male and female mice. Mice were housed in a controlled temperature environment on a 12-hour light/dark cycle. Food and water were provided ad libitum.
AAV injections and brain tissue isolation. Wild-type or HD-N171-82Q mice were injected with AAV1 vectors as previously reported.13,26 For all studies, unless indicated otherwise, mice were injected bilaterally into the striatum (coordinates: 0.86 mm rostral to bregma, ±1.8 mm lateral to midline, 3.5 mm ventral to the skull surface) with 5 µl of FB or AAV1 virus (at 4 × 1012 viral genomes/ml). Mice injected for QPCR analyses at 4 weeks after treatment were injected unilaterally into the right striatum. Mice used in histological analyses were anesthetized with a ketamine/xylazine mix and transcardially perfused with 20 ml of 0.9% cold saline, followed by 20 ml of 4% paraformaldehyde in 0.1 mol/l PO4 buffer. Mice were decapitated, and the brains were removed and postfixed overnight in 4% paraformaldehyde. Brains were stored in a 30% sucrose solution at 4 °C until cut on a sliding knife microtome at 40 µm thickness and stored at −20 °C in a cryoprotectant solution. Mice used for QPCR or microarray analyses were perfused with 20 ml of 0.9% cold saline. Brains were removed and sectioned into 1 mm thick coronal slices using a brain matrix (Roboz, Gaithersburg, MD). Tissue punches were taken from the striatum using a tissue core (1.4 mm in diameter) and titrated in 50 µl of TRIzol (Invitrogen, Carlsbad, CA). RNA was isolated from striatal punches using 1 ml of TRIzol.
Immunohistochemical analyses. Free-floating, coronal brain sections (40 µm thick) were processed for immunohistochemical visualization of striatal neurons (DARPP-32, 1:100; Cell Signaling Technology, Danvers, MA), microglia (Iba1, 1:1,000; WAKO, Richmond, VA). All staining procedures were carried out as previously described,26 using goat antirabbit IgG secondary antibody (1:200) and Vectastain ABC-peroxidase reagent (both from Vector Laboratories, Burlingame, CA). Stained or unstained (the latter for visualization of hrGFP autofluorescence) sections were mounted onto Superfrost Plus slides (Fisher Scientific, Pittsburgh, PA) and coverslipped with Gelmount (Biomeda, Foster City, CA) or Vectashield (Vector Laboratories). Images were captured using an Olympus BX60 light microscope and DP70 digital camera, along with Olympus DP Controller software (Olympus, Melville, NY).
QPCR. Random-primed first-strand cDNA synthesis was performed using 500 ng total RNA (TaqMan reverse transcription reagents; Applied Biosystems, Foster City, CA) per manufacturer's protocol. Assays were performed on a sequence detection system using primers/probe sets specific for human or mouse htt, or mouse CD11b (Itgam) and β-actin (Prism 7900HT and TaqMan 2× Universal Master Mix; Applied Biosystems). Relative gene expression was determined by using the standard curve or ΔΔCT method, normalizing to β-actin mRNA levels.
Northern blot analyses. Striatal RNAs were isolated from tissue punches, and 1–3 µg of total RNA was resolved on a 15% acrylamide gel. Loading was assessed by ethidium bromide stain. RNA was transferred to Hybond-XL membrane (Amersham, Piscataway, NJ) at 400 mA for 2 hours and UV-crosslinked using the auto-crosslink function on a Stratalinker 1800 (Stratagene, La Jolla, CA). Blots were prehybridized using UltraHyb-Oligo (Ambion, Austin, TX), probed with γ-32P-labeled oligonucleotides (Ready-To-Go T4 polynucleotide kinase; Amersham) at 32 °C overnight, washed three times (5 minutes each) in 2× sodium citrate, 0.1% sodium dodecyl sulfate at 32 °C, and exposed to film.
Cell culture and transfection. Mouse C2C12 cells were grown in 24-well plates coated with poly-l-ornithine (0.1 mg/ml; Sigma, St Louis, MO) and transfected in quadruplicate with the indicated plasmids using Lipofectamine 2000 (Invitrogen). Cells were co-transfected with either 75 ng of hrGFP or HD-N171-82Q expression plasmids along with 300 ng of promoter-only or RNAi plasmids. At 24 hours after treatment, cells were lysed in 100 µl of Cell Disruption Buffer (PARIS kit, Ambion), and 50 µl of the lysate was added to 1 ml of TRIzol for RNA isolation. The remaining lysates were kept for western blot analyses.
Western blot analyses. Protein concentrations in C2C12 cell lysates were quantified with the DC protein assay (Bio-Rad, Melville, NY). Then, 30 µg of total protein was separated on a 7.5% sodium dodecyl sulfate–polyacrylamide gel and transferred to 0.45 µm polyvinylidene fluoride membrane (Millipore, Billerica, MA). The membrane was blocked with 2% milk in PBS-Tween 20 (0.05%) and incubated with either antimyc (1:1,000; Abcam, Cambridge, MA), anti-htt (1:5,000, MAB2166; Chemicon, Billerica, MA) or anti- β-catenin (1:10,000, Abcam) antibodies followed by incubation with horseradish peroxidase-conjugated goat antimouse or goat antirabbit secondary antibodies (1:10,000 and 1:50,000, respectively; Jackson ImmunoResearch, West Grove, PA). Blots were developed using ECL-Plus substrate (Amersham) and then exposed to film.
Rotarod performance testing. Mice were tested on an accelerating rotarod apparatus (model 47600; Ugo Basile, Comerio, Italy) at 10, 14, and 18 weeks of age. At week 10, mice were first habituated on the rotarod for 4 minutes. Mice were then tested three trials per day (with at least 30 minutes of rest between trials) for four consecutive days and retested in this fashion twice more at 4-week intervals. For each trial, acceleration was from 4 to 40 rpm over 4 minutes, and then speed maintained at 40 rpm. Latency to fall (or if mice hung on for two consecutive rotations without running) was recorded for each mouse per trial. The trials were stopped at 400 seconds, and mice remaining on the rotarod at that time were scored as 400 seconds. Data from the three trials for each group on each day are presented as means ± SEM (week 10: WT-FB, 8 mice; HD-FB, 13 mice; HD-GFP, 8 mice; HD-mi2.4, 14 mice; week 14: WT-FB, 8 mice; HD-FB, 11 mice; HD-GFP, 8 mice; HD-mi2.4, 13 mice; week 18: WT-FB, 8 mice; HD-FB, 7 mice; HD-GFP, 5 mice; HD-mi2.4, 12 mice).
Survival analyses. The dates of birth were recorded for all mice in the therapeutic trial. Mice were observed once daily during the mid-morning. For mice that died before the point-of-sacrifice at week 20, the date of death was recorded.
Microarray analyses. Microarray analysis was done with assistance from the University of Iowa DNA Facility (Iowa City, IA). Fifty nanograms of total RNA template were used to produce amplified cDNA using the Ovation Biotin RNA Amplification System, v2 (NuGEN Technologies, San Carlos, CA) following the manufacturer's protocol. Amplified cDNA product was purified with DNA Clean and Concentrator-25 (Zymo Research, Orange, CA). Amplified cDNA, 3.75 µg, were processed using the FL-Ovation cDNA Biotin Module v2 (NuGEN Technologies) to produce biotin-labeled antisense cDNA in 50–100 bp fragments. Following denaturation at 99 °C for 2 minutes, fragmented, labeled cDNA were combined with hybridization control oligomer (b2) and control cRNAs (BioB, BioC, BioD, and CreX) in hybridization buffer and hybridized to the Mouse Genome 430 2.0 GeneChip (Affymetrix, Santa Clara, CA) containing 45,000 probe sets capable of detecting >34,000 genes. Following an 18-hour incubation at 45 °C, the arrays were washed, stained with streptavidinphycoerythrin (Molecular Probes, Eugene, OR), and then amplified with an antistreptavidin antibody (Vector Laboratories) using the Fluidics Station 450 (Affymetrix). Arrays were scanned with the Affymetrix Model 3000 scanner and data collected using GeneChip operating software (GCOS) v1.4. Each sample and hybridization underwent a quality control evaluation, including percentage of probe sets reliably detecting between 40 and 60% of the present call and 3′–5′ ratio of the GAPDH gene <3.
Preprocessing, normalization, and gene-level summarization of the microarray data were conducted by GC-RMA49 as implemented in the BioConductor package.50 Differentially expressed genes were identified using two-sample unpaired t-tests of log2 expression values, by requiring a fold change of at least 2.0 between htt-knockdown and hrGFP-treated control groups and a P value of <0.05. A total of 761 genes were identified based on these criteria. A more stringent P value cutoff of 0.01 was used to obtain a smaller list of 473 differentially expressed genes.
For intersection analyses, gene symbols were obtained by submitting Affymetrix probe identification numbers to the Gene ID Conversion Tool at the David Bioinformatics Resources web-server.32
Statistical analyses. For all studies, unless indicated otherwise, statistical values were obtained by using one-way analysis of variance followed by Tukey–Kramer post hoc analyses to assess for significant differences between individual groups. Student's t-test was used for the pair-wise comparisons done for CD11b QPCR analyses. Statistical analysis of microarray data is described in the previous section. In all statistical analyses, P < 0.05 was considered significant.
Supplementary MaterialFigure S1. Htt expression levels in RNA samples used for transcriptional profiling studies.Figure S2. Functional gene annotation clustering performed on the 107 down-regulated and 366 up-regulated transcripts identified enrichments in the indicated cellular components and processes.Table S1. List of the 475 transcripts (including two htt probes, highlighted in yellow) that changed by twofold or greater (P < 0.01) in the htt-knockdown group versus the hrGFP control group.Table S2. List of the 763 transcripts (including two htt probes, highlighted in yellow) that changed by twofold or greater (P < 0.05) in the htt-knockdown group versus the hrGFP control group.Table S3. List of transcripts that changed between 1.2 to 2.0-fold (P < 0.01) in the htt-knockdown group versus the hrGFP control group.Table S4. List of transcripts that intersected with early transcriptional changes in human HD striatum. The direction of change relative to the human HD data is indicated.
Supplementary Material
Htt expression levels in RNA samples used for transcriptional profiling studies.
Functional gene annotation clustering performed on the 107 down-regulated and 366 up-regulated transcripts identified enrichments in the indicated cellular components and processes.
List of the 475 transcripts (including two htt probes, highlighted in yellow) that changed by twofold or greater (P < 0.01) in the htt-knockdown group versus the hrGFP control group.
List of the 763 transcripts (including two htt probes, highlighted in yellow) that changed by twofold or greater (P < 0.05) in the htt-knockdown group versus the hrGFP control group.
List of transcripts that changed between 1.2 to 2.0-fold (P < 0.01) in the htt-knockdown group versus the hrGFP control group.
List of transcripts that intersected with early transcriptional changes in human HD striatum. The direction of change relative to the human HD data is indicated.
Acknowledgments
We thank the B.L.D and McCray laboratories for feedback and discussion, Alex Mas Monteys for assistance with managing the mouse colony and genotyping, and David Borchelt for furnishing the myc-tagged human HD-N171-82Q expression plasmid. This research was supported by funds from the NIH (NS-50210, HD-44093, DK-54759), the Hereditary Disease Foundation, and the Roy J. Carver Trust. R.L.B. is supported by the Lori C. Sasser Fellowship.
REFERENCES
- Schaffar G, Breuer P, Boteva R, Behrends C, Tzvetkov N, Strippel N, et al. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol Cell. 2004;15:95–105. doi: 10.1016/j.molcel.2004.06.029. [DOI] [PubMed] [Google Scholar]
- Saudou F, Finkbeiner S, Devys D., and , Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- Zhai W, Jeong H, Cui L, Krainc D., and , Tjian R. In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell. 2005;123:1241–1253. doi: 10.1016/j.cell.2005.10.030. [DOI] [PubMed] [Google Scholar]
- Qiu Z, Norflus F, Singh B, Swindell MK, Buzescu R, Bejarano M, et al. Sp1 is up-regulated in cellular and transgenic models of Huntington disease, and its reduction is neuroprotective. J Biol Chem. 2006;281:16672–16680. doi: 10.1074/jbc.M511648200. [DOI] [PubMed] [Google Scholar]
- Benn CL, Sun T, Sadri-Vakili G, McFarland KN, DiRocco DP, Yohrling GJ, et al. Huntingtin modulates transcription, occupies gene promoters in vivo, and binds directly to DNA in a polyglutamine-dependent manner. J Neurosci. 2008;28:10720–10733. doi: 10.1523/JNEUROSCI.2126-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas HD, Liu AK, Hersch S, Glessner M, Ferrante RJ, Salat DH, et al. Regional and progressive thinning of the cortical ribbon in Huntington's disease. Neurology. 2002;58:695–701. doi: 10.1212/wnl.58.5.695. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED., and , Richardson EP., Jr Neuropathological classification of Huntington's disease. J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- Ferrante RJ, Kubilus JK, Lee J, Ryu H, Beesen A, Zucker B, et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington's disease mice. J Neurosci. 2003;23:9418–9427. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante RJ, Andreassen OA, Dedeoglu A, Ferrante KL, Jenkins BG, Hersch SM, et al. Therapeutic effects of coenzyme Q10 and remacemide in transgenic mouse models of Huntington's disease. J Neurosci. 2002;22:1592–1599. doi: 10.1523/JNEUROSCI.22-05-01592.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantubungi K, Jacquard C, Greco A, Pintor A, Chtarto A, Tai K, et al. Minocycline in phenotypic models of Huntington's disease. Neurobiol Dis. 2005;18:206–217. doi: 10.1016/j.nbd.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Andreassen OA, Dedeoglu A, Ferrante RJ, Jenkins BG, Ferrante KL, Thomas M, et al. Creatine increase survival and delays motor symptoms in a transgenic animal model of Huntington's disease. Neurobiol Dis. 2001;8:479–491. doi: 10.1006/nbdi.2001.0406. [DOI] [PubMed] [Google Scholar]
- Harper SQ, Staber PD, He X, Eliason SL, Martins I, Mao Q, et al. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc Natl Acad Sci USA. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Lebron E, Denovan-Wright EM, Nash K, Lewin AS., and , Mandel RJ. Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington's disease transgenic mice. Mol Ther. 2005;12:618–633. doi: 10.1016/j.ymthe.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, Liu W, Wada E, Murata M, Wada K., and , Kanazawa I. Clinico-pathological rescue of a model mouse of Huntington's disease by siRNA. Neurosci Res. 2005;53:241–249. doi: 10.1016/j.neures.2005.06.021. [DOI] [PubMed] [Google Scholar]
- Gauthier LR, Charrin BC, Borrell-Pages M, Dompierre JP, Rangone H, Cordelieres FP, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Rigamonti D, Bauer JH, De-Fraja C, Conti L, Sipione S, Sciorati C, et al. Wild-type huntingtin protects from apoptosis upstream of caspase-3. J Neurosci. 2000;20:3705–3713. doi: 10.1523/JNEUROSCI.20-10-03705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir J, Floresco SB, O'Kusky JR, Diewert VM, Richman JM, Zeisler J, et al. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- Duyao MP, Auerbach AB, Ryan A, Persichetti F, Barnes GT, McNeil SM, et al. Inactivation of the mouse Huntington's disease gene homolog Hdh. Science. 1995;269:407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- Zeitlin S, Liu JP, Chapman DL, Papaioannou VE., and , Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nat Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- van Bilsen PH, Jaspers L, Lombardi MS, Odekerken JC, Burright EN., and , Kaemmerer WF. Identification and allele-specific silencing of the mutant huntingtin allele in Huntington's disease patient-derived fibroblasts. Hum Gene Ther. 2008;19:710–719. doi: 10.1089/hum.2007.116. [DOI] [PubMed] [Google Scholar]
- Liu W, Kennington LA, Rosas HD, Hersch S, Cha JH, Zamore PD, et al. Linking SNPs to CAG repeat length in Huntington's disease patients. Nat Methods. 2008;5:951–953. doi: 10.1038/nmeth.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling G, Becher MW, Sharp AH, Jinnah HA, Duan K, Kotzuk JA, et al. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- Boudreau RL, Mas Monteys A., and , Davidson BL. Minimizing variables among hairpin-based RNAi vectors reveals the potency of shRNAs. RNA. 2008;14:1834–1844. doi: 10.1261/rna.1062908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau RL, Martins I., and , Davidson BL. Artificial MicroRNAs as siRNA Shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol Ther. 2008;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci USA. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JL, Ramaswamy S, Gasmi M, Bartus RT, Herzog CD, Brandon EP, et al. Viral delivery of glial cell line-derived neurotrophic factor improves behavior and protects striatal neurons in a mouse model of Huntington's disease. Proc Natl Acad Sci USA. 2006;103:9345–9350. doi: 10.1073/pnas.0508875103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norflus F, Nanje A, Gutekunst CA, Shi G, Cohen J, Bejarano M, et al. Anti-inflammatory treatment with acetylsalicylate or rofecoxib is not neuroprotective in Huntington's disease transgenic mice. Neurobiol Dis. 2004;17:319–325. doi: 10.1016/j.nbd.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Gardian G, Browne SE, Choi DK, Klivenyi P, Gregorio J, Kubilus JK, et al. Neuroprotective effects of phenylbutyrate in the N171-82Q transgenic mouse model of Huntington's disease. J Biol Chem. 2005;280:556–563. doi: 10.1074/jbc.M410210200. [DOI] [PubMed] [Google Scholar]
- Masuda N, Peng Q, Li Q, Jiang M, Liang Y, Wang X, et al. Tiagabine is neuroprotective in the N171-82Q and R6/2 mouse models of Huntington's disease. Neurobiol Dis. 2008;30:293–302. doi: 10.1016/j.nbd.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan W, Peng Q, Masuda N, Ford E, Tryggestad E, Ladenheim B, et al. Sertraline slows disease progression and increases neurogenesis in N171-82Q mouse model of Huntington's disease. Neurobiol Dis. 2008;30:312–322. doi: 10.1016/j.nbd.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, et al. The DAVID Gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehlow AN, Li JZ., and , Myers RM. Wild-type huntingtin participates in protein trafficking between the Golgi and the extracellular space. Hum Mol Genet. 2007;16:391–409. doi: 10.1093/hmg/ddl467. [DOI] [PubMed] [Google Scholar]
- Zhang H, Das S, Li QZ, Dragatsis I, Repa J, Zeitlin S, et al. Elucidating a normal function of huntingtin by functional and microarray analysis of huntingtin-null mouse embryonic fibroblasts. BMC Neurosci. 2008;9:38. doi: 10.1186/1471-2202-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omi K, Hachiya NS, Tokunaga K., and , Kaneko K. siRNA-mediated inhibition of endogenous Huntington disease gene expression induces an aberrant configuration of the ER network in vitro. Biochem Biophys Res Commun. 2005;338:1229–1235. doi: 10.1016/j.bbrc.2005.10.061. [DOI] [PubMed] [Google Scholar]
- Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, et al. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci USA. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Gamblin RJ, Ooi L, Bruce AW, Donaldson IJ, Westhead DR, et al. Identification of the REST regulon reveals extensive transposable element-mediated binding site duplication. Nucleic Acids Res. 2006;34:3862–3877. doi: 10.1093/nar/gkl525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipione S, Rigamonti D, Valenza M, Zuccato C, Conti L, Pritchard J, et al. Early transcriptional profiles in huntingtin-inducible striatal cells by microarray analyses. Hum Mol Genet. 2002;11:1953–1965. doi: 10.1093/hmg/11.17.1953. [DOI] [PubMed] [Google Scholar]
- Trushina E, Singh RD, Dyer RB, Cao S, Shah VH, Parton RG, et al. Mutant huntingtin inhibits clathrin-independent endocytosis and causes accumulation of cholesterol in vitro and in vivo. Hum Mol Genet. 2006;15:3578–3591. doi: 10.1093/hmg/ddl434. [DOI] [PubMed] [Google Scholar]
- Bartzokis G, Lu PH, Tishler TA, Fong SM, Oluwadara B, Finn JP, et al. Myelin breakdown and iron changes in Huntington's disease: pathogenesis and treatment implications. Neurochem Res. 2007;32:1655–1664. doi: 10.1007/s11064-007-9352-7. [DOI] [PubMed] [Google Scholar]
- Hodges A, Strand AD, Aragaki AK, Kuhn A, Sengstag T, Hughes G, et al. Regional and cellular gene expression changes in human Huntington's disease brain. Hum Mol Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- Kuhn A, Goldstein DR, Hodges A, Strand AD, Sengstag T, Kooperberg C, et al. Mutant huntingtin's effects on striatal gene expression in mice recapitulate changes observed in human Huntington's disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum Mol Genet. 2007;16:1845–1861. doi: 10.1093/hmg/ddm133. [DOI] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Castanotto D, Sakurai K, Lingeman R, Li H, Shively L, Aagaard L, et al. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35:5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VM, Xia H, Marrs GL, Gouvion CM, Lee G, Davidson BL, et al. Allele-specific silencing of dominant disease genes. Proc Natl Acad Sci USA. 2003;100:7195–7200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Alegre P, Bode N, Davidson B., and , Paulson HL. Silencing primary dystonia: lentiviral-mediated RNA interference therapy for DYT1 dystonia. J Neurosci. 2005;25:10502–10509. doi: 10.1523/JNEUROSCI.3016-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JK, Auerbach W, Duyao MP, Vonsattel JP, Gusella JF, Joyner AL, et al. Huntingtin is required for neurogenesis and is not impaired by the Huntington's disease CAG expansion. Nat Genet. 1997;17:404–410. doi: 10.1038/ng1297-404. [DOI] [PubMed] [Google Scholar]
- Auerbach W, Hurlbert MS, Hildtich-Maguire P, Wadghiri YZ, Wheeler VC, Cohen SI, et al. The HD mutation causes progressive lethal neurological disease in mice expressing reduced levels of huntingtin. Human Molecular Genetics. 2001;10 22:2515–2523. doi: 10.1093/hmg/10.22.2515. [DOI] [PubMed] [Google Scholar]
- Wu Z., and , Irizarry RA. Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J Comput Biol. 2005;12:882–893. doi: 10.1089/cmb.2005.12.882. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Htt expression levels in RNA samples used for transcriptional profiling studies.
Functional gene annotation clustering performed on the 107 down-regulated and 366 up-regulated transcripts identified enrichments in the indicated cellular components and processes.
List of the 475 transcripts (including two htt probes, highlighted in yellow) that changed by twofold or greater (P < 0.01) in the htt-knockdown group versus the hrGFP control group.
List of the 763 transcripts (including two htt probes, highlighted in yellow) that changed by twofold or greater (P < 0.05) in the htt-knockdown group versus the hrGFP control group.
List of transcripts that changed between 1.2 to 2.0-fold (P < 0.01) in the htt-knockdown group versus the hrGFP control group.
List of transcripts that intersected with early transcriptional changes in human HD striatum. The direction of change relative to the human HD data is indicated.