Abstract
Achievement of specific tumor cell targeting remains a challenge for glioma gene therapy. We observed that the human high mobility group box2 (HMGB2) gene had a low level of expression in normal human brain tissues, but was significantly upregulated in glioblastoma tissues. With progressive truncation of a 5′-upstream sequence of the HMGB2 gene, we identified a 0.5-kb fragment displaying a high transcriptional activity in glioblastoma cells, but a low activity in normal brain cells. To test the feasibility of using the HMGB2 promoter sequence in targeted cancer therapy, we constructed a baculoviral vector expressing the herpes simplex virus thymidine kinase (HSVtk) gene driven by the HMGB2 promoter. Transduction with the viral vector induced cell death in glioblastoma cell lines in the presence of ganciclovir (GCV), but did not affect the survival of human astrocytes and neurons. In a mouse xenograft model, intratumor injection of the baculoviral vector suppressed the growth of human glioblastoma cells and prolonged the survival of tumor-bearing mice. Our results suggest that the novel 5′ sequence of HMGB2 gene has a potential to be used as an efficient, tumor-selective promoter in targeted vectors for glioblastoma gene therapy.
Introduction
Glioma is the most common type of primary brain tumor. Being highly invasive and aggressive, high-grade malignant gliomas, glioblastoma multiforme, are one of the most lethal forms of human cancer.1 At present, conventional treatment of glioblastoma multiforme, such as surgery, γ-irradiation, and chemotherapy are ineffective in eradicating the cancer as evidenced by poor prognosis for glioma patients, with a mean survival time of <1 year after diagnosis. Hence, there is an urgent need to improve the efficacy of therapies for this fatal disease.
Selective targeting of cancer cells has been explored to enhance the effectiveness of cancer therapy by eliminating cancer cells specifically, while sparing nontarget normal cells. Among different approaches, transcriptional targeting using a tissue-specific or tumor selective cellular promoter is proving to be a powerful means for cancer gene therapies. This strategy is particularly appealing to cancer suicide gene therapies, which use either toxic genes or genes encoding enzymes that turn prodrugs into toxic compounds. Distinguishing between a tissue-specific promoter and a tumor selective promoter relies on the relative activity of the promoter in normal and tumor tissues and the dividing line between them is rather blurred.2 As a key issue here, a selected promoter should have an expression profile that differs significantly between normal and cancer cells in a given organ.
Although many tumor selective promoters have been reported and reviewed in literature,2,3,4,5 the work on glioma selective promoters has been lagging behind. Currently used promoters for transcriptional targeting to gliomas are those that display tissue specificity. For example, the promoter for the gene encoding glial fibrillary acidic protein (GFAP) has been extensively tested for transgene expression in glioma cells.6,7,8,9,10,11 This promoter, like many other tissue-specific promoters, has an expression profile that does not differ much between normal and tumor cells of the same lineage. When the promoter is used to drive expression of the suicide gene encoding herpes simplex virus thymidine kinase (HSVtk), the gene products promote the ablation of GFAP-positive glial cells in the presence of ganciclovir (GCV) in the small intestine, causing lethal changes in animals.12 In the central nervous system, HSVtk expression from the GFAP promoter followed by GCV treatment has been used to ablate proliferating astrocytes.12,13,14,15
This study aimed at identifying a promoter that can drive transgene expression preferentially in glioblastoma cells and at a high level. In this respect, the gene encoding the human high mobility group box2 (HMGB2) has been shown to have high-level expression in glioblastoma.16 Moreover, HMGB2 is widely expressed in the nervous system during mouse embryogenesis, but becomes undetectable in the adult mouse brain.17 In view of these findings, we selected the 5′ flanking region of the HMGB2 gene for detailed characterization. We report here that a 0.5-kb HMGB2 promoter is effective in driving transgene expression in glioblastoma cells, whereas in human neurons and normal human astrocytes this promoter shows negligible activity.
Results
HMGB2 messenger RNA levels in normal and tumor human brain cells and tissues
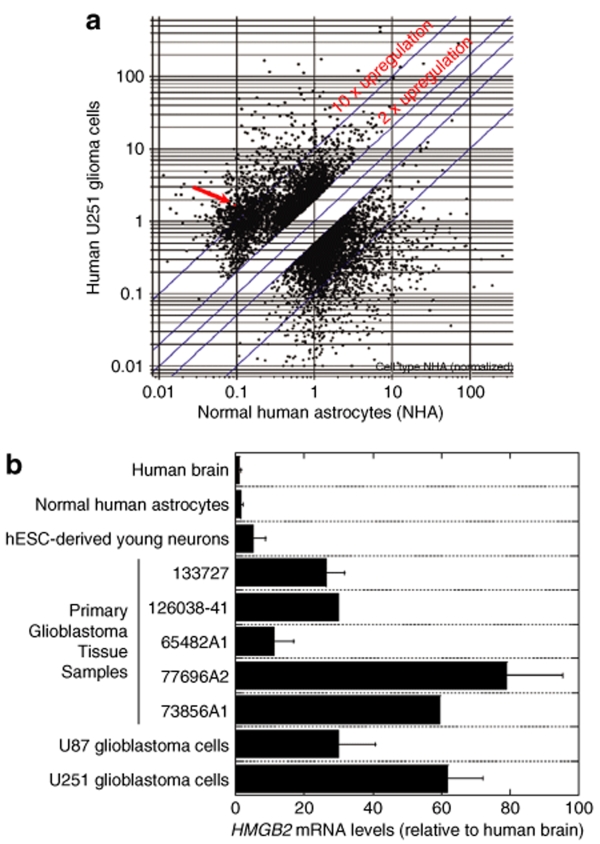
To explore the possibility of using the HMGB2 promoter for transcriptional targeting of glioblastoma, we first performed data mining on the Gene Expression Omnibus at the National Center for Biotechnology Information18 to find out the expression characteristics of the HMGB2 messenger RNA (mRNA) in the normal human brain and primary human glioma tissues. Several data sets within Gene Expression Omnibus (Gene Expression Omnibus data set GDS1962, 1815 and 1816) illustrate a very low level of HMGB2 expression in nontumor human brain samples, a moderate elevation of HMGB2 message in astrocytomas and a significant elevation in glioblastomas, with ~75% of grade 4 glioblastoma samples having abundant HMGB2 expression (GDS1962), indicating a correlation between HMGB2 upregulation and glioma grade. To evaluate HMGB2 expression at a cellular level, rather than in a tissue with mixed cell populations, we performed mRNA expression profiling of U251 human malignant glioblastoma cells and normal human astrocytes using the Affymetrix Human Genome U-133 plus 2.0 GeneChip. Eleven probe sets corresponding to the HMGB2 gene are present in the chip. Figure 1a demonstrates the data from the microarray assay, showing that HMGB2 belonged to a group of genes that had a low level of expression in normal human astrocytes and are upregulated significantly in U251 glioblastoma cells. The complete normalized microarray dataset from the study is available at Gene Expression Omnibus microarray data repository (http://www.ncbi.nlm.nih.gov/geo/), with the accession number GSE12305.
Figure 1.
Expression of HMGB2 mRNA in brain tissues and cells. (a) DNA microarray analysis of HMGB2 mRNA levels in U251 glioblastoma cells and normal human astrocytes. Three replicate Affimetrix HG_U133A_Plus_2 GeneChip arrays were used for each type of cells. Microarray results were analyzed using GeneSpring GX 7.3. The arrow indicates HMGB2 mRNA expression levels, which were low in normal human astrocytes, but more than tenfold upregulated in U251 glioblastoma cells. (b) Real-time PCR quantification of HMGB2 mRNA expression. Normal human brain tissue, primary glioblastoma tissues, normal human astrocytes and neurons, and U87 and U251 glioblastoma cell lines were used. hESC-derived young neurons were collected 1 week after neuronal differentiation of neural stem cells generated from hES1 human embryonic stem cells. The values represent the fold change in HMGB2 expression, normalized to β-actin mRNA levels and relative to HMGB2 expression in the normal brain tissue (means ± SD, n = 2–6). HMGB2, human high mobility group box2; mRNA, messenger RNA; hESC, human embryonic stem cells.
To validate microarray results and to have a more accurate assessment, we performed quantitative real-time reverse-transcriptase PCR analysis, which demonstrates a significant elevation of HMGB2 expression in glioblastoma cells and tissues compared to normal human astrocytes and brain tissues. Relative to the value in the normal human brain, HMGB2 mRNA levels were found to be 11- to 79-fold higher in primary glioblastoma tissue samples, 30-fold higher in U87 glioblastoma cell line and 62-fold higher in U251 glioblastoma cell line (Figure 1b). We also included young human neurons derived from human embryonic stem cells for comparison and detected that HMGB2 mRNA levels in these neurons were fivefold higher relative to the level in normal human brain tissue (Figure 1b). This result is in agreement with the previous observation in mice that showed a relatively high level expression of HMGB2 in developing cells.17 Nevertheless, HMGB2 is upregulated in glioblastoma cells and tissues to a level much higher than that in the young neurons.
The activity and selectivity of the HMGB2 promoter
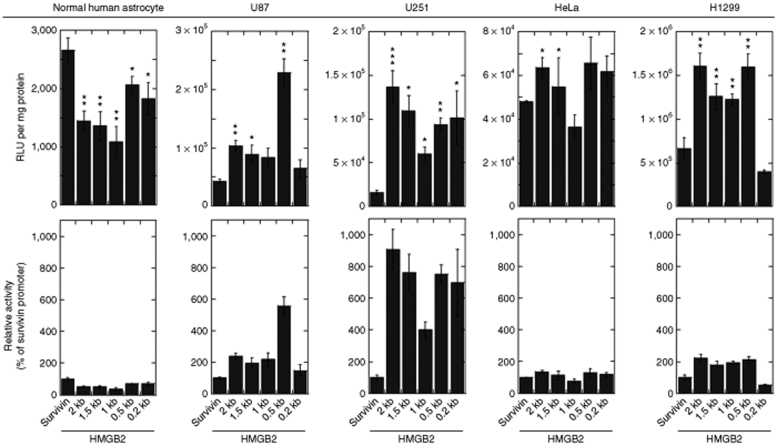
High expression levels of HMGB2 could be attributed to a higher transcription rate or stabilization/delay in degradation of HMGB2 mRNA. To distinguish between these two possibilities, we next examined whether the high levels of HMGB2 expression in glioblastoma cells correlate with a high activity of the HMGB2 promoter in these cells. A 2-kb 5′ sequence upstream of the transcription start site of the HMGB2 gene was cloned from the genomic DNA of U251 glioblastoma cells and normal human astrocytes. We compared the 2-kb fragments from the two types of cells and did not detect any sequence difference, thus excluding the possibility that polymorphism in the HMGB2 promoter region is a mechanism underlying the HMGB2 upregulation in the glioblastoma cells. We then generated the truncated versions (1.5-, 1-, 0.5-, and 0.2-kb) of the 2-kb 5′ sequence to determine the minimal length that is associated with maximum activity. Such minimal promoters can be favorably used in an expression vector without occupying a large region. We placed these truncated promoter constructs into a promoterless luciferase vector and tested their promoter activity in U87 and U251 human glioblastoma cell lines, H1299 human lung cancer cell line and HeLa human cervical cancer cell line. Normal human astrocytes are included as a reference. The survivin promoter is a widely used tumor selective promoter, with a high activity in glioblastomas, but a low activity in terminally differentiated cells.19 Thus, we also included the survivin promoter to compare its activity with that offered by the HMGB2 promoters in parallel experiments.
The upper panels in Figure 2 indicate the relative light units of luciferase per mg protein, whereas the lower panels indicate HMGB2 promoter activity relative to the survivin promoter. In normal human astrocytes, the activities of the 2-kb construct and the truncated versions of the HMGB2 promoter were very low, as demonstrated by the absolute relative light unit readings being between one and three thousands only. These readings were lower than that from the survivin promoter. For example, the activity of the 0.5-kb construct, which has the highest activity among the tested HMGB2 promoter constructs in normal astrocytes, was 64% of that provided by the survivin promoter. This observation is important because a lower activity in normal cells would be related to less off-target effects when the promoter is used for cancer gene therapy.
Figure 2.
HMGB2 promoter activity in normal and tumor cells. Normal human astrocytes, U87 and U251 human glioma cell lines, HeLa human cervical cancer cell line and H1299 human lung cancer cell line were transfected with plasmid expression vectors accommodating a luciferase gene under the control of a HMGB2 promoter construct. A plasmid vector with a survivin promoter was included for comparison. Cells were lysed in a reporter lysis buffer 24 hours post transfection and promoter activity was measured using a luciferase assay system. Data are presented as relative light units per mg protein (upper panels) and as HMGB2 promoter activity relative to survivin promoter activity (lower panels). All data sets are a compilation of at least three independent experiments and expressed as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 vs. the survivin group by Student's t-test. HMGB2, human high mobility group box2.
In the U87 glioblastoma cell line, the 0.5-kb version of the HMGB2 promoter displayed the highest activity among all the tested HMGB2 promoter constructs, being 5.28-fold higher than that from the survivin promoter. Other constructs had activities around twofold higher than the survivin promoter. The activity difference between the HMGB2 and the survivin promoters was even more obvious in U251 glioblastoma cell line, four of the five HMGB2 promoter constructs displaying promoter activity at least sixfold higher as compared to the survivin promoter and the rest one being approximately fourfold higher. To address the question as to whether the high activity of the HMGB2 promoter is an event specific to glioblastoma cell lines, we evaluated the activities of the HMGB2 promoter constructs in other cancer cell lines. In H1299 human lung cancer cell line, we observed that the absolute relative light unit readings from the HMGB2 promoter constructs were almost tenfold higher than those in glioblastoma cell lines. However, in HeLa human cervical cancer cell line, the readings were approximately tenfold lower than those in glioblastoma cell lines. Relative to the survivin promoter, the HMGB2 promoter constructs showed a moderately increased activity in these two nonglioblastoma cell lines, around 1.3- and 2-fold higher in HeLa and H1299 cells, respectively. These findings indicate that a high level of HMGB2 expression in glioblastoma cells is related to the high activity of the gene promoter and this activity is stronger than that of the survivin promoter, particularly in glioblastoma cells.
Transcriptional targeting to glioblastoma cells using the HMGB2 promoter
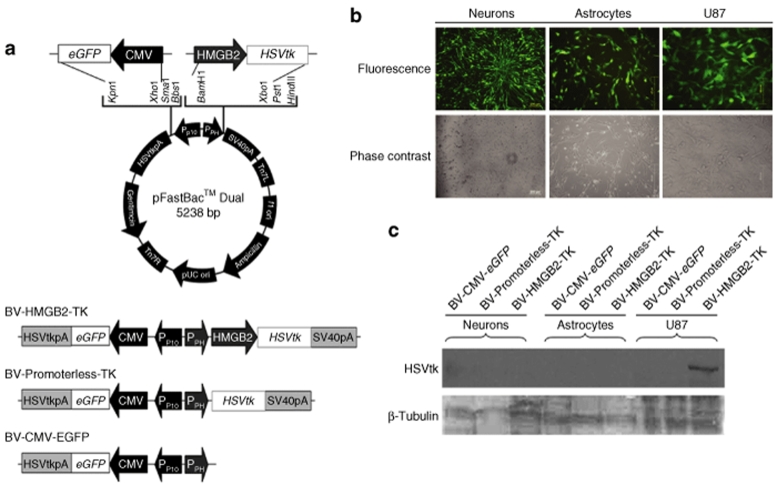
With a high activity in glioblastoma cells and low activity in normal astrocytes, the HMGB2 promoter is a promising regulatory element for transcriptional targeting to gliomas in the brain. To test the hypothesis, we constructed a baculoviral vector accommodating the HSVtk gene under the control of a 0.5-kb HMGB2 promoter (BV-HMGB2-TK). HSVtk expression followed by treatment of the prodrug GCV is widely tested in cancer suicide gene therapy and often used together with a transcriptional targeting promoter to reduce its cell killing effect on nontarget, normal cells.20 pFastBac Dual was used as the vector backbone, which allows the insertion of the emerald green fluorescent protein as a reporter in one of the two multiple cloning sites to monitor transduction efficiency. Two control baculoviral vectors were also constructed, one with the HSVtk gene but without the HMGB2 promoter (BV-Promoterless-TK) and one with neither the HSVtk gene nor the HMGB2 promoter (BV-CMV-eGFP). Figure 3a illustrates the schematic diagrams of the vector constructs used in this study.
Figure 3.
HSVtk suicide gene expression driven by a HMGB2 promoter placed into a baculoviral vector. (a) Schematic representation of baculoviral constructs tested in this study. pFastBac Dual was used as the backbone for baculoviral vector construction. CMV-driven eGFP expression is used to monitor transduction efficiency and BV-Promoterless-TK is used as a negative control for HSVtk. (b) Baculoviral transduction in human neural cells and glioma cells. Human neurons derived from hES1 neural stem cells (Neurons) were transduced with BV-Promoterless-TK at an MOI 100. Normal human astrocytes (Astrocytes) and U87 glioblastoma cells (U87) were transduced BV-Promoterless-TK at an MOI 10. Fluorescence and phase contrast images taken 24 hours after transduction are shown. (c) HSVtk protein expression in vitro after baculoviral transduction. Cell lysates were prepared 24 hours after transduction. Ten µg of total protein from the cell lysates was used for western blotting. CMV, human cytomegalovirus immediate-early gene enhancer/promoter; eGFP, an enhanced green fluorescent protein gene; HMGB2, a 0.5-kb human high mobility group box 2 gene promoter; HSVtk, herpes simplex virus thymidine kinase gene; HSVtkpA, HSVtk polyadenylation signal; PPH, polyhedrin insect cell promoter; PP10, Insect cell p10 promoter; SV40pA, SV40 polyadenylation signal.
As demonstrated earlier in animal studies, baculovirus is capable of transducing several types of brain cells, although the virus is more prone to transducing cells of glial origin, including glioma cells.21,22,23,24 Figure 3b confirmed that the baculoviral vectors constructed in this study were also able to transduce normal and tumor human brain cells efficiently, and bright emerald green fluorescent protein fluorescence was easily detectable in all three types of cells tested, neurons derived from human embryonic stem cells, human primary astrocytes, and U87 glioblastoma cells. When the above three baculoviral vectors were tested for the selective expression of the HSVtk protein from the HMGB2 promoter, we detected the protein only in U87 glioblastoma cells transduced with BV-HMGB2-TK (Figure 3c). No detectable expression of the HSVtk protein in human astrocytes after BV-HMGB2-TK expression is consistent with our finding that the HMGB2 promoter is not active in normal astrocytes (Figure 2). Although the HMGB2 expression level in young neurons derived from human embryonic stem cells was fivefold higher than that in normal human astrocytes (Figure 1b), the activity of the HMGB2 promoter in these neurons was not yet high enough to drive the detectable expression of the HSVtk protein. Overall, these findings further confirm that the HMGB2 promoter has a high glioblastoma cell-selectivity.
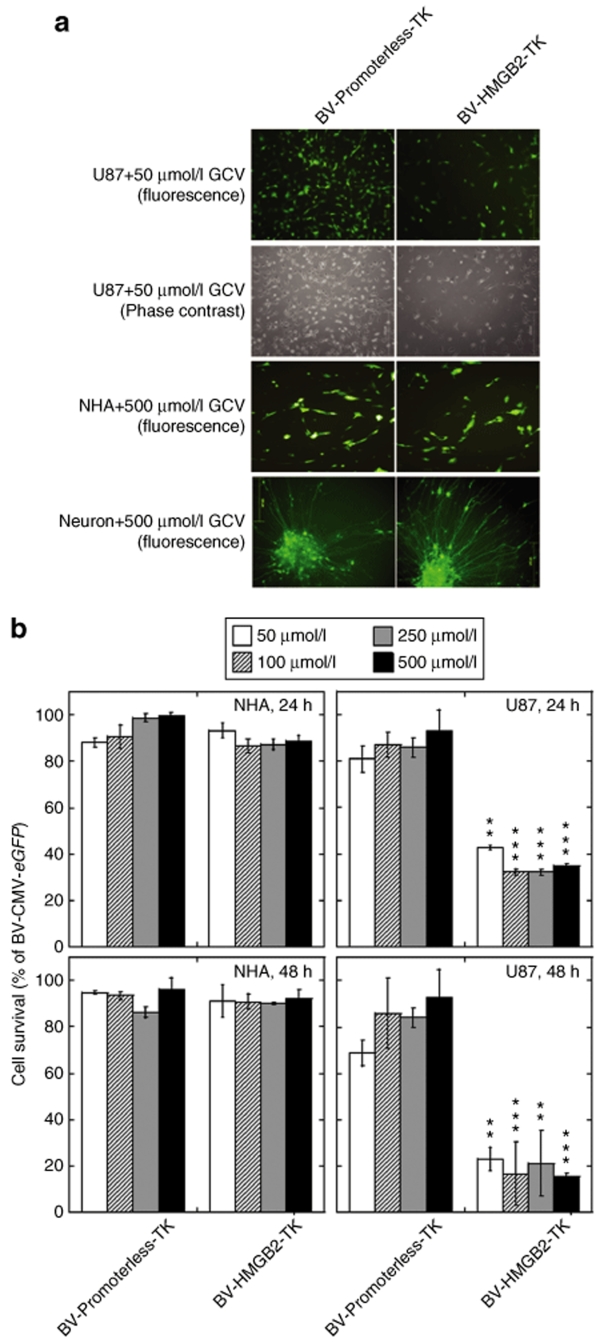
Encouraged by the finding of selective expression of the HSVtk protein in glioblastoma cells, we then examined cellular effects of baculoviral transduction followed by GCV treatment on U87 human glioblastoma cells, normal human astrocytes, and neurons derived human embryonic stem cells. These cells did not show any observable changes in morphology after BV-Promoterless-TK transduction plus GCV treatment (Figure 4a). However, transduction of BV-HMGB2-TK reduced the number of U87 glioblastoma cells at a GCV concentration of 50 µmol/l, whereas transduction of this viral vector produced no visible effects on normal astrocytes and neurons even at a GCV concentration of 500 µmol/l (Figure 4a). To quantify the cell death caused by BV-HMGB2-TK/GCV treatment, MTS assays were performed. A range of GCV concentrations from 50 to 500 µmol/l were tested. As shown in Figure 4b, cell viability dropped to a level of ~30% of control in BV-HMGB2-TK transduced U87 cells after GCV treatment for 24 hours and to <20% of control after GCV treatment for 48 hours. A low dose of GCV at 50 µmol/l was enough to kill BV-HMGB2-TK transduced U87 cells. We did not observe any changes in cell viability in BV-HMGB2-TK transduced normal human astrocytes and BV-Promoterless-TK transduced normal astrocytes or U87 cells.
Figure 4.
Selective cytotoxic effects of HMGB2 promoter-driven suicide gene expression. (a) Cellular effects as visualized under a microscope. Cells were transduced for 24 hours with either BV-HMGB2-TK or BV-Promoterless-TK, at an MOI 10 for astrocytes and U87 cells and at an MOI 100 for neurons, followed by treatment with the prodrug ganciclovir GCV (50 or 500 µmol/l) for another 24 hours. (b) Cell viability analysis of BV-HMGB2-TK transduced U87 glioblastoma cells and normal human astrocytes (NHAs) using quantitative MTS assays. Cells were transduced in the same way as above, followed by treatment with the prodrug ganciclovir GCV (50, 100, 250, or 500 µmol/l) for another 24 or 48 hours. Results are expressed as % BV-CMV-eGFP transduced cells. *P < 0.05, **P < 0.01, ***P < 0.001 vs. BV-Promoterless-TK category treated at the same contraction of GCV by ANOVA. ANOVA, analysis of variance; HMGB2, human high mobility group box2; GCV, ganciclovir; NHA, normal human astrocyte.
Effects of the HMGB2 promoter-driven HSVtk expression on intracranial tumor growth
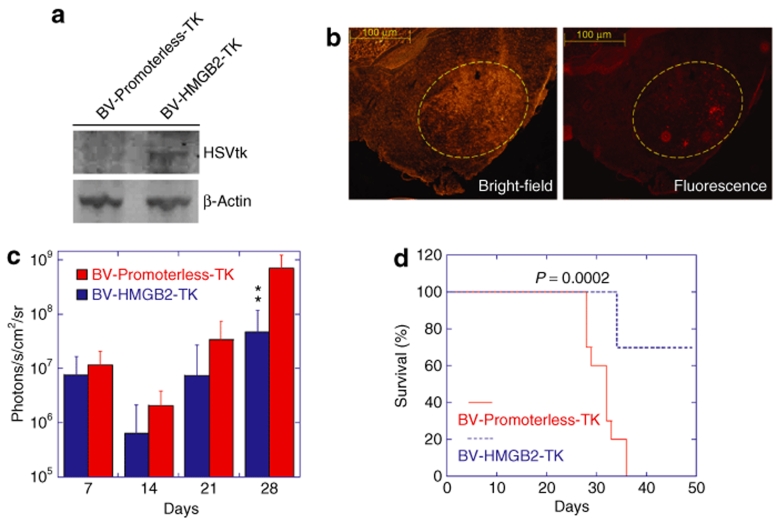
Having tested the efficiency of the HMGB2 promoter in selectively eliminating U87 cells in vitro, we next examined the in vivo efficacy of HSVtk expression under the control of this promoter. We established glioma xenografts in nude mice using U87-luciferase cells, followed by single injection of baculoviral vectors into the xenografts at day 7 post tumor inoculation. We examined HSVtk expression using western blotting and confirmed that the HSVtk protein was detectable in the brain of tumor-bearing mice killed 3 days after injection of BV-HMGB2-TK, but not BV-Promoterless-TK (Figure 5a). Immunohistochemistry staining showed that HSVtk-positive cells distributed throughout the tumor xenograft bed (Figure 5b). In another group of mice, GCV was administered intraperitoneally once a day for 2 weeks after baculoviral injection. The bioluminescence intensities from U87-luciferase cells, indicative of tumor volume, were measured 7, 14, 21, and 28 days after tumor inoculation using an in vivo imaging system. The results are summarized in Figure 5c. At day 14, there was a decrease in tumor burden in both BV-HMGB2-TK- and BV-Promoterless-TK-injected mice, possibly due to damage caused by injection. The tumor burden in control animals injected with BV-Promoterless-TK increased ~60-fold from day 7 to 28. In BV-HMGB2-TK-injected animals, the tumor burden at day 28 was calculated as sixfold higher relative to the value at day 7. Thus, although the HMGB2 promoter-driven suicide gene expression did not completely eliminate tumors in the mice, the promoter shows an activity high enough to inhibit in vivo tumor growth. Furthermore, all ten control animals received BV-Promoterless-TK injection died by day 36, whereas in the BV-HMGB2-TK group, three mice died and seven survived for 50 days, the longest period investigated (Figure 5d). There was a significant difference between survival curves (log rank P = 0.0002).
Figure 5.
In vivo anti-glioma effects of HMGB2 promoter-driven suicide gene expression. Human glioma U87-luciferase cells were inoculated into the brain of nude mice. Seven days later, BV-HMGB2-TK or BV-Promoterless-TK viruses were injected into tumor xenografts. (a) Western blot analysis of HSVtk protein expression in vivo. The brain region with the tumor xenograft was collected 3 days after virus injection to prepare protein lysates. Fifty micrograms of total protein was used for western blotting. (b) Bright-field and fluorescence images of HSVtk immunostaining. The sections of the brain of tumor-bearing mice killed 3 days after virus injection were probed with rabbit polyclonal, anti-HSVtk antibody, followed by anti-rabbit IgG conjugated to TRITC for visualization. The yellow circle indicates the location of the U87 tumor xenograft. (c) Quantitative analysis of tumor burden from IVIS bioluminescence images. The data are presented as the mean of tumor burden ± SD from 10 mice per group imaged 7, 14, 21, and 28 days post tumor cell inoculation. Following viral injection, GCV was administered daily for 2 weeks. **P < 0.01 vs. the BV-Promoterless-TK group at the same time point by ANOVA. (d) Prolonged survival of mice treated with HSVtk/GCV. The mice were observed until 50 days post tumor inoculation. The statistical analysis was performed using the log rank test. ANOVA, analysis of variance; GCV, ganciclovir; HMGB2, human high mobility group box2; HSVtk, herpes simplex virus thymidine kinase; TRITC, tetramethyl rhodamine iso-thiocyanate.
Discussion
This study provided the first evidence that the HMGB2 promoter can be used for transcriptional targeting to glioblastoma cells. HMGB2 is one member in a family of high mobility group nonhistone chromatin proteins that regulate the processes of transcription, replication, recombination, and DNA repair (reviewed in ref. 25). HMGB2 is highly expressed during embryogenesis, but has a limited expression in adult organs, being mainly detected in lymphoid organs and testes.17 Mice lacking HMGB2 continue to survive into adulthood, although spermatogenesis has been impaired.17 In terms of expression profiles of HMGB2 in cancer tissues, a large-scale analysis of 40 cancer microarray data sets has indentified HMGB2 as one of the meta-signatures of undifferentiated cancers, including gliomas.16 HMGB2 is also overexpressed in undifferentiated lung adenocarcinoma, bladder carcinoma, breast ductal carcinoma, and prostate carcinoma relative to well-differentiated cancer.16 Other small-scale studies indicate that HMGB2 expression increased moderately in gastrointestinal tumors26 and was higher in oxiplatin resistant ovarian carcinoma cell lines.27 Our current study confirmed a high level of HMGB2 expression in high-grade glioblastoma samples. We further demonstrated that this high level expression was associated with a high activity of the HMGB2 promoter in the cancer cells, and this high activity could be favorably used for transcriptional targeting to glioblastoma.
The HMGB2 promoter was benchmarked in this study against the survivin promoter, which drives the expression of survivin, a protein belonging to the inhibitor of apoptosis family of proteins and a known mitosis regulator.28 Levels of the survivin protein are upregulated in most cancers and have been associated with resistance to chemotherapy, increased tumor recurrence and shorter patient survival.19 Survivin has been reported as being upregulated in glioblastomas and high cytoplasmic survivin expression is related to progression of low-grade lesions to a secondary glioblastoma multiforme.29 Hence, the survivin promoter has been tested for transcriptional targeting of adenovirus vectors for malignant gliomas.30 Several previous studies have also demonstrated the use of the survivin promoter in conditionally replicative oncolytic viruses as an efficient means to target gliomas.30,31,32 Although much work has been done to devise therapeutic vectors using the survivin promoter, the role of survivin in normal cells, particularly those in the nervous system, is just beginning to emerge. In the normal human brain, survivin is strongly expressed in the choroid plexus and ependymal cells and is also detectable in neurons, astrocytes, and oligodendrocytes.33 More importantly survivin is expressed in the neural precursor cells of the subventricular zone in adult humans34 and conditional deletion of survivin in neuronal precursor cells induces significant apoptosis in the cerebrum, cerebellum, brain stem, and spinal cord, indicating that an antiapoptotic function of survivin in neuronal development and differentiation.35 In light of these reports, transcriptional targeting using the survivin promoter in the brain might have adverse effects on normal central nervous system cells. However, HMGB2 transcriptional profiling, both by microarray analysis as well as the real-time PCR quantification has revealed extremely low levels of transcription of the HMGB2 gene in normal brain tissues.
A recent study compared the activities of three tumor selective promoters, the survivin, midkine, and CXCR4 promoters, in glioma cells and reported the survivin promoter as the one with the highest level of transcriptional activity.36 Although the 0.5-kb HMGB2 promoter provides an activity similar to that from the survivin promoter in H1299 human lung cancer cell line and HeLa human cervical cancer cell line, the promoter was much more active than the survivin promoter in the two tested human glioblastoma cell lines, indicative of the tumor-type selectivity of the HMGB2 promoter. Also interestingly, the activity of the HMGB2 promoter in normal human astrocytes was lower than that provided by the survivin promoter. The high transcriptional activity and the cell specificity of the HMGB2 promoter in glioblastoma cells are unique among tumor selective promoters characterized to date and make it an attractive candidate in driving suicide gene expression to eliminate glioblastoma cells while sparing normal brain cells, thus achieving a high therapeutic index.
The HSVtk gene is one of the most studied suicide genes and has been tested in many clinical gene therapy trials. The HSVtk proteins phosphorylate GCV, converting the prodrug into toxic nucleotide analogs that inhibit DNA replication in the cell nucleus. As such, HSVtk/GCV combination efficiently kills highly proliferating cancer cells. GFAP-positive normal glial cells have a relatively lower rate of cell division than cancer cells, thus are less sensitive to DNA synthesis inhibition by phosphorylated GCV. This difference has promoted the use of the GFAP promoter in transcriptional targeting to gliomas.6,7,8,9,10,11 However, HSVtk/GCV combination also inhibits DNA replication in the mitochondria.37,38 Indeed, it is well known that HSVtk/GCV combination affects normal nondividing but metabolically active hepatocytes, provoking severe liver dysfunction and mortality.39,40,41,42 In adult transgenic mice, GCV ablates HSVtk-expressing, nonproliferating thyrocytes.43 In adult transgenic mice that express HSVtk from a GFAP promoter, although central nervous system astrocytes were not killed in detectable numbers, GCV ablates enteric glial cells that play a fundamental role in bowel function, causing degeneration of myenteric neurons and inflammatory bowel disease.12 Moreover, proliferating astrocytes in the central nervous system of these transgenic mice are susceptible to GCV killing effects and ablation of these cells by GCV is commonly used to study their physiological importance.12,13,14,15 As injury caused by tumor growth would invariably induce proliferation of normal glial cells in regions adjacent to tumor tissues, HSVtk expression from the GFAP promoter followed by GCV treatment will also kill these cells, which play important roles in protecting neurons and repairing the blood–brain barrier.14 The HMGB2 promoter has a low activity in cultured human astrocytes and should be less likely to cause collateral damage to nearby astrocytes in case of leakage of intratumorally injected vectors into normal tissues.
We used baculoviral vectors to test the transcriptional targeting by the HMGB2 promoter. This virus provides high transduction efficiency in glioma cells lines, capable of transducing up to 98% of cells.10 The results we obtained in this study clearly indicate that after being placed into a baculoviral vector, the 0.5-kb HMGB2 promoter was transcriptionally active and able to drive the expression of the HSVtk gene to a level to inhibit glioma cell growth both in vitro and in vivo and to prolong the survival of tumor-inoculated mice. However, the tumor mass was not completely eradicated in our in vivo experiment after one single injection of the baculoviral vector, followed by 2 weeks of treatment with GCV. The quantum of decrease in tumor mass should be proportional to the expression of HSVtk in the tumor cells. One possible explanation for the lack of complete elimination of the tumor in the present experimental setup could be a rapid decrease in expression of the HSVtk gene over a time span of 2 weeks, due to the transient transgene expression mediated by baculoviral vectors. Engineering the HMGB2 promoter to enhance its transcriptional activity and repeated injection of the viral vectors could be the possible ways to improve the therapeutic efficacy. Alternatively, conditionally replication-competent oncolytic viruses under the control of the HMGB2 promoter could be generated and used to maintain viral replication in glioma cells.
In conclusion, the HMGB2 promoter is highly activated in glioblastoma cells, possibly also in several other types of cancers such as lung, bladder, breast, and prostate cancers. Current literature on HMGB2 suggests that in normal adult tissues the gene is expressed only in the testis and lymphoid tissues, with no detectable expression in other tissues in mammals.44 On this basis, the use of the HMGB2 promoter for transcriptional targeting has potential to develop efficient vectors for local treatment of solid tumors.
Materials And Methods
Cell culture and tissue samples. Cells used in this study were human U87 glioblastoma cell line from American Type Culture Collection (ATCC, Manassas, VA), U251 glioblastoma cell line from the Chinese Academy of Science (Shanghai, China), H1299 human lung cancer cell line (ATCC), HeLa human cervical cancer cell line (ATCC), normal human astrocytes from Clonetics Primary Cell Systems (Lonza, Basel, Switzerland), and HES-1 (NIH code: ES01) and HES-3 (NIH code: ES03) human embryonic stem cells from ES Cell International (Singapore). U87-luciferase cells that stably express luciferase were established as described elsewhere.10 Tumor cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37 °C, 5% CO2. Normal human astrocytes were cultured in special Astrocyte Basal Medium supplemented with the AGM SingleQuots (Lonza). Both HES-1 and HES-3 human embryonic stem cells were amplified on mitotically inactivated mouse embryonic fibroblasts from CF-1 mouse strain (ATCC) and human embryonic stem-derived neural stem cells and neurons were generated as described previously.45,46
Primary glioblastoma tissue samples were obtained from Asterand (Detroit, MI), including 65482A1(OCT embedded, malignant glioma), 77696A2 (fresh-frozen glioblastoma) and 73856A (OCT-embedded glioblastoma), and Capital Biosciences (Gaithersburg, MD), including formalin fixed paraffin embedded tissue samples Glioblastoma FFPE-133727 and Glioblastoma FFPE-126038-41.
Complementary DNA microarray analysis. Total RNA isolated using Trizol (Invitrogen, Carlsbad, CA) and further cleaned up using RNeasy mini kit (Qiagen, Hilden, Germany) was used to prepare labeled complementary RNA using Affymetrix's One-Cycle Target Labeling and Control Reagents (Affymetrix, Santa Clara, CA). Hybridization mixes with 15 µg of fragmented complementary RNA previously checked on the Bioanalyzer were prepared according to Affymetrix's GeneChip Hybridization, Wash, and Stain Kit's instruction and used to hybridize the human GeneChip Human Genome U133 Plus 2.0. After being washed and stained with phycoerythrin, chips were scanned using GeneChip Scanner 3000 and data acquired using the GCOS software. Data obtained from GCOS was exported using the Data Transfer Tool and transferred to GeneSpring and Genedata software, respectively. The GC-RMA preprocessor file was used to reprocess imported CEL files and to normalize the data at the probe level. Replicates (at least three per experiment) were grouped and data normalized using the default setting of Genespring.
Real-time reverse-transcriptase PCR analyses. Total RNA was extracted using RNeasy Mini Kits (Qiagen) and 500 ng of total RNA was used in reverse transcription reactions to generate complementary DNA. Real-time quantitative PCR was performed in a 25-µl reaction volume using 1:100 diluted complementary DNA, Universal PCR Master Mix kit, SYBR green, and 300–900 nmol/l of each primer. Primer pairs of HMGB2 were obtained after searching Quantitative PCR Primer Database primer set 10099, which amplify an 86-bp fragment for HMGB2. β-Actin was used as an endogenous control and the primers were custom made from Superarray (Frederick, MD). Relative quantization of target mRNA expression, normalized to an endogenous control and relative to a calibrator, was calculated using the mathematical expression for fold change 2−ΔΔCt (fold change) as described.47
Cloning of the HMGB2 promoter. PCR was used to clone a 2-kb region of nucleotides upstream of the transcription start site of the HMGB2 gene from U251 glioblastoma and normal human astrocytes (Lonza). The transcription start site of the HMGB2 gene was predicted by online database for transcription start sites. Genomic DNA from 2 × 105 cells was isolated using a Qiagen kit (Qiagen). One hundred nanograms of genomic DNA was amplified using forward primer 5′-AAATTTTTGAGGTCACAGGCTATGACTTTTCATCTT-3′ and reverse primer 5′-ATCCCCACTAATCTGATTGGTTCTGACGATTATT-3′. PCR was performed using high fidelity PCR super mix (Invitrogen): initial denaturation at 94 °C for 3 minutes, then 29 cycles of 94 °C for 1 minute, 30 seconds at 50.5 °C for 30 seconds, 68 °C and 2 minutes 30 seconds. The PCR products extracted from agarose gel were cloned into TOPO 2.1 vector (Invitrogen) first and then subcloned into Kpn1 and Xho1 sites of pGL4.11 luciferase reporter vector (Promega, Madison, WI). To obtain the truncated versions of the HMGB2 promoter, forward primers amplifying 1.5, 1, 0.5, and 0.2 kb regions were designed and amplified by PCR with a common reverse primer from the pGL4. The sequences used for the primers are as follows: 1.5 kb, 5′-ATGCGCTAGCGGCATTAAGTACATTCA-3′; 0.5 kb, 5′-ATGCGAGCTCGACAGACAGGACCTAA-3′; 0.2 kb, 5′-ATGCGCTAGCATCAGGATCGAGCAGT-3′. One kilobyte HMGB2 promoter was obtained by digestion of the two-kilobyte promoter with Sac1 and Xho1.
Transfection, baculoviral transduction, and cell viability assay. One day before transfection, cells (1 × 105) were plated in 24-well plates with growth medium without antibiotics. After cells reached 85–90% confluence, they were transfected using 0.8 µg of pGL4.11 vectors with a luciferase gene under the control of a testing promoter construct. The promoterless basic pGL4.11 vector containing the luciferase gene was used as a control for transfection efficiency. Lipofectamine 2000 (Invitrogen) was used for transfection with a ratio of DNA: Lipofectamine at 2.5:1 throughout the study. Twenty-four hours later, the transfected cells were freeze-thawed with 100 µl of reporter cell lysis buffer (Promega) and 10 µl of the cell extract was used for luciferase activity assays. Measurements were made in a single-tube luminometer (Berthold Lumat LB 9507, Bad Wildbad, Germany). The relative light units were normalized to the protein concentration measured with a protein assay kit from Bio-Rad (Richmond, CA).
To construct baculoviral vectors, we first replaced the luciferase gene in the above plasmid vector accommodating the 0.5-kb HMGB2 promoter with an HSVtk gene obtained from pORF-Tk (Invivogen) using PCR (forward primer, 5′-ATGCCTCGAGTCTTTCCTACAGCTGAGATCAC-3′ and reverse primer, 5′-ATGCTCTAGAAATGTATCTTATCATGTCGAGCTAGC-3′). The promoter and the HSVtk gene were then subcloned into the pFastBac Dual vector (Invitrogen) between BamH1 and Xba1. A fragment with the HSVtk gene alone without the promoter was subcloned into Not1 and Xba1 sites of pFastBac Dual vector to produce a promoterless vector as the negative control. As a reporter, emerald green fluorescent protein driven by the cytomegalovirus promoter from Block-iT pol II miR expression vector (Invitrogen) was subcloned into the pFastBac Dual vector at Kpn1 and Xho1 sites in a second multiple cloning site. The two expression cassettes, the HMGB2/HSVtk cassette and the cytomegalovirus/emerald green fluorescent protein cassette, are separated by two insect cell promoters, the baculovirus late polyhedrin promoter PPH and the very late p10 gene promoter PP10. Both of them are inactive in mammalian cells. The Bac-to-Bac baculovirus expression system (Invitrogen) was then used to generate bacmid DNA. Recombinant baculoviruses were produced and propagated in Spodoptera frugiperda (Sf9) insect cells preadapted to Sf-900 II serum-free medium (Invitrogen). After the viral titer was determined by plaque assay, the viruses were used for cell transduction as specified in the text.
Cell survival rates were measured by MTS assay (Promega) as per manufacturer's instructions. Cells were seeded onto 96-well plates at 1 × 104 cells per well in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and infected with baculoviruses. The spectrophotometric assay for cell proliferation was performed 24 and 48 hours later by measuring OD at 490 nm.
In vivo animal studies. Ten adult female BALB/c athymic, immunoincompetent nude mice (weighing 20 g, 6–8 weeks old) were used. For tumor inoculation, 0.5 × 106 human glioma U87-luciferase cells were injected into the right side of the striatum of anesthetized animals. One week later, in vivo imaging was performed to evaluate tumor growth (designated as day 0) and the animals were then distributed into two groups. On the same day, 1 × 108 plaque-forming units of baculovirus particles in 10 µl was injected into tumor xenografts, followed by daily intraperitoneal injection of GCV (50 mg/kg body weight) for 2 weeks starting 1 day after virus injection. To monitor bioluminescent singles of U87-luciferase cells, anesthetized animals were injected intraperitoneally with D-luciferin (Promega) at 100 mg/kg in phosphate-buffered saline. Bioluminescent imaging was performed with the IVIS imaging system coupled with cool CCD camera (Xenogen, Alameda, CA) at 20 minutes after Luciferin injection. Images acquisition and bioluminescent signals were measured by subtracting the background signal from the ROI signal and the photon intensity was analyzed with the Xenogen living imaging software v2.5. The protocol of the current animal experiment was approved by the local Institutional Animal Care and Use Committee.
For immunohistochemical and western blot analysis of HSVtk expression in the mouse brain, tumor inoculation in BALB/c nude mice and baculoviral vector injection were performed as described earlier. Three days post virus injection, brain tissues were collected. For immunohistochemical analysis, the mice were perfused with 4% formaldehyde in phosphate-buffered saline. The collected brains were post-fixed in 4% paraformaldehyde overnight, followed by cryoprotection in 30% sucrose overnight. The tissues were embedded in OCT and cut into 25 µm thick sections. The sections were blocked in 10% bovine serum albumin in phosphate-buffered saline, followed by overnight incubation with rabbit polyclonal anti-HSVtk antibodies (1:100 dilution, kindly provided by William Summers, Yale University). As the secondary antibody, an antirabbit TRITC-labeled antibody (1:100, Dako, Glostrup, Denmark) was used. For western blotting analysis, the brain region with the tumor xenograft was dissected out from the whole brain and homogenized on ice in 750 µl of a lysis buffer containing a protease inhibitor cocktail. The supernatants of the cell lysates with 50 µg protein were separated on 4–12% Bis-Tris gradient gel (Invitrogen) under a denaturing condition and transferred on a nitrocellulose membrane using the iBlot dry blotting system. The membranes were probed with the rabbit anti-HSVtk antibody (1:1000) and a monoclonal antibody against mouse β-actin (Sigma, St Louis, MO).
Statistical analysis. All data are represented as mean ± SD. The statistical significance of differences was determined by the two-factor analysis of variance with replication followed by Tukey post hoc analysis or unpaired Student's t-test. The statistical analysis of survival data was performed using the log rank test. A P < 0.05 was considered to be statistically significant.
Acknowledgments
The work was supported by Institute of Bioengineering and Nanotechnology, Biomedical Research Council, Agency for Science, Technology and Research (A*STAR) in Singapore.
REFERENCES
- Pulkkanen KJ., and , Yla-Herttuala S. Gene therapy for malignant glioma: current clinical status. Mol Ther. 2005;12:585–598. doi: 10.1016/j.ymthe.2005.07.357. [DOI] [PubMed] [Google Scholar]
- Harrington KJ, Linardakis E., and , Vile RG. Transcriptional control: an essential component of cancer gene therapy strategies. Adv Drug Deliv Rev. 2000;44:167–184. doi: 10.1016/s0169-409x(00)00093-4. [DOI] [PubMed] [Google Scholar]
- Robson T., and , Hirst DG. Transcriptional targeting in cancer gene therapy. J Biomed Biotechnol. 2003;2003:110–137. doi: 10.1155/S1110724303209074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GD, Curtin JF, Candolfi M, Kroeger K, Lowenstein PR., and , Castro MG. Gene therapy and targeted toxins for glioma. Curr Gene Ther. 2005;5:535–557. doi: 10.2174/156652305774964631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi H., and , Hitt MM. Transcriptionally targeted adenovirus vectors. Curr Gene Ther. 2005;5:411–427. doi: 10.2174/1566523054546189. [DOI] [PubMed] [Google Scholar]
- Chen MH, Yang WK, Whang-Peng J, Lee LS., and , Huang TS. Differential inducibilities of GFAP expression, cytostasis and apoptosis in primary cultures of human astrocytic tumours. Apoptosis. 1998;3:171–182. doi: 10.1023/a:1009698822305. [DOI] [PubMed] [Google Scholar]
- McKie EA, Graham DI., and , Brown SM. Selective astrocytic transgene expression in vitro and in vivo from the GFAP promoter in a HSV RL1 null mutant vector—potential glioblastoma targeting. Gene Ther. 1998;5:440–450. doi: 10.1038/sj.gt.3300621. [DOI] [PubMed] [Google Scholar]
- Vandier D, Rixe O, Brenner M, Gouyette A., and , Besnard F. Selective killing of glioma cell lines using an astrocyte-specific expression of the herpes simplex virus-thymidine kinase gene. Cancer Res. 1998;58:4577–4580. [PubMed] [Google Scholar]
- Zamorano A, Mellstrom B, Vergara P, Naranjo JR., and , Segovia J. Glial-specific retrovirally mediated gas1 gene expression induces glioma cell apoptosis and inhibits tumor growth in vivo. Neurobiol Dis. 2004;15:483–491. doi: 10.1016/j.nbd.2003.11.022. [DOI] [PubMed] [Google Scholar]
- Wang CY, Li F, Yang Y, Guo HY, Wu CX., and , Wang S. Recombinant baculovirus containing the diphtheria toxin A gene for malignant glioma therapy. Cancer Res. 2006;66:5798–5806. doi: 10.1158/0008-5472.CAN-05-4514. [DOI] [PubMed] [Google Scholar]
- Horst M, Brouwer E, Verwijnen S, Rodijk M, de Jong M, Hoeben R, et al. Targeting malignant gliomas with a glial fibrillary acidic protein (GFAP)-selective oncolytic adenovirus. J Gene Med. 2007;9:1071–1079. doi: 10.1002/jgm.1110. [DOI] [PubMed] [Google Scholar]
- Bush TG, Savidge TC, Freeman TC, Cox HJ, Campbell EA, Mucke L, et al. Fulminant jejuno-ileitis following ablation of enteric glia in adult transgenic mice. Cell. 1998;93:189–201. doi: 10.1016/s0092-8674(00)81571-8. [DOI] [PubMed] [Google Scholar]
- Delaney CL, Brenner M., and , Messing A. Conditional ablation of cerebellar astrocytes in postnatal transgenic mice. J Neurosci. 1996;16:6908–6918. doi: 10.1523/JNEUROSCI.16-21-06908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush TG, Puvanachandra N, Horner CH, Polito A, Ostenfeld T, Svendsen CN, et al. Leukocyte infiltration, neuronal degeneration, and neurite outgrowth after ablation of scar-forming, reactive astrocytes in adult transgenic mice. Neuron. 1999;23:297–308. doi: 10.1016/s0896-6273(00)80781-3. [DOI] [PubMed] [Google Scholar]
- Faulkner JR, Herrmann JE, Woo MJ, Tansey KE, Doan NB., and , Sofroniew MV. Reactive astrocytes protect tissue and preserve function after spinal cord injury. J Neurosci. 2004;24:2143–2155. doi: 10.1523/JNEUROSCI.3547-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci USA. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronfani L, Ferraguti M, Croci L, Ovitt CE, Scholer HR, Consalez GG, et al. Reduced fertility and spermatogenesis defects in mice lacking chromosomal protein Hmgb2. Development. 2001;128:1265–1273. doi: 10.1242/dev.128.8.1265. [DOI] [PubMed] [Google Scholar]
- Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, et al. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S., and , Pelus LM. Survivin, a cancer target with an emerging role in normal adult tissues. Mol Cancer Ther. 2006;5:1087–1098. doi: 10.1158/1535-7163.MCT-05-0375. [DOI] [PubMed] [Google Scholar]
- Alavi JB., and , Eck SL. Gene therapy for high grade gliomas. Expert Opin Biol Ther. 2001;1:239–252. doi: 10.1517/14712598.1.2.239. [DOI] [PubMed] [Google Scholar]
- Sarkis C, Serguera C, Petres S, Buchet D, Ridet JL, Edelman L, et al. Efficient transduction of neural cells in vitro and in vivo by a baculovirus-derived vector. Proc Natl Acad Sci USA. 2000;97:14638–14643. doi: 10.1073/pnas.260472897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtolainen P, Tyynela K, Kannasto J, Airenne KJ., and , Yla-Herttuala S. Baculoviruses exhibit restricted cell type specificity in rat brain: a comparison of baculovirus- and adenovirus-mediated intracerebral gene transfer in vivo. Gene Ther. 2002;9:1693–1699. doi: 10.1038/sj.gt.3301854. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang X, Guo H., and , Wang S. Axonal transport of recombinant baculovirus vectors. Mol Ther. 2004;10:1121–1129. doi: 10.1016/j.ymthe.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Wang CY., and , Wang S. Astrocytic expression of transgene in the rat brain mediated by baculovirus vectors containing an astrocyte-specific promoter. Gene Ther. 2006;13:1447–1456. doi: 10.1038/sj.gt.3302771. [DOI] [PubMed] [Google Scholar]
- Thomas JO. HMG1 and 2: architectural DNA-binding proteins. Biochem Soc Trans. 2001;29:395–401. doi: 10.1042/bst0290395. [DOI] [PubMed] [Google Scholar]
- Koon N, Schneider-Stock R, Sarlomo-Rikala M, Lasota J, Smolkin M, Petroni G, et al. Molecular targets for tumour progression in gastrointestinal stromal tumours. Gut. 2004;53:235–240. doi: 10.1136/gut.2003.021238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma RR, Hector SM, Clark K, Greco WR, Hawthorn L., and , Pendyala L. Gene expression profiling of a clonal isolate of oxaliplatin-resistant ovarian carcinoma cell line A2780/C10. Oncol Rep. 2005;14:925–932. [PubMed] [Google Scholar]
- Xing Z, Conway EM, Kang C., and , Winoto A. Essential role of survivin, an inhibitor of apoptosis protein, in T cell development, maturation, and homeostasis. J Exp Med. 2004;199:69–80. doi: 10.1084/jem.20031588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D, Zeng YX, Wang HJ, Wen JM, Tao Y, Sham JS, et al. Expression of cytoplasmic and nuclear Survivin in primary and secondary human glioblastoma. Br J Cancer. 2006;94:108–114. doi: 10.1038/sj.bjc.6602904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt WJ, Haviv YS, Lu B, Wang M, Rivera AA, Ulasov IV, et al. The human survivin promoter: a novel transcriptional targeting strategy for treatment of glioma. J Neurosurg. 2006;104:583–592. doi: 10.3171/jns.2006.104.4.583. [DOI] [PubMed] [Google Scholar]
- Ulasov IV, Zhu ZB, Tyler MA, Han Y, Rivera AA, Khramtsov A, et al. Survivin-driven and fiber-modified oncolytic adenovirus exhibits potent antitumor activity in established intracranial glioma. Hum Gene Ther. 2007;18:589–602. doi: 10.1089/hum.2007.002. [DOI] [PubMed] [Google Scholar]
- Zhu ZB, Makhija SK, Lu B, Wang M, Rivera AA, Kim-Park S, et al. Incorporating the survivin promoter in an infectivity enhanced CRAd-analysis of oncolysis and anti-tumor effects in vitro and in vivo. Int J Oncol. 2005;27:237–246. [PubMed] [Google Scholar]
- Sasaki T, Lopes MB, Hankins GR., and , Helm GA. Expression of survivin, an inhibitor of apoptosis protein, in tumors of the nervous system. Acta Neuropathol. 2002;104:105–109. doi: 10.1007/s00401-002-0532-x. [DOI] [PubMed] [Google Scholar]
- Pennartz S, Belvindrah R, Tomiuk S, Zimmer C, Hofmann K, Conradt M, et al. Purification of neuronal precursors from the adult mouse brain: comprehensive gene expression analysis provides new insights into the control of cell migration, differentiation, and homeostasis. Mol Cell Neurosci. 2004;25:692–706. doi: 10.1016/j.mcn.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Jiang Y, de Bruin A, Caldas H, Fangusaro J, Hayes J, Conway EM, et al. Essential role for survivin in early brain development. J Neurosci. 2005;25:6962–6970. doi: 10.1523/JNEUROSCI.1446-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulasov IV, Rivera AA, Sonabend AM, Rivera LB, Wang M, Zhu ZB, et al. Comparative evaluation of survivin, midkine and CXCR4 promoters for transcriptional targeting of glioma gene therapy. Cancer Biol Ther. 2007;6:679–685. doi: 10.4161/cbt.6.5.3957. [DOI] [PubMed] [Google Scholar]
- Herraiz M, Beraza N, Solano A, Sangro B, Montoya J, Qian C, et al. Liver failure caused by herpes simplex virus thymidine kinase plus ganciclovir therapy is associated with mitochondrial dysfunction and mitochondrial DNA depletion. Hum Gene Ther. 2003;14:463–472. doi: 10.1089/104303403321467225. [DOI] [PubMed] [Google Scholar]
- van der Eb MM, Geutskens SB, van Kuilenburg AB, van Lenthe H, van Dierendonck JH, Kuppen PJ, et al. Ganciclovir nucleotides accumulate in mitochondria of rat liver cells expressing the herpes simplex virus thymidine kinase gene. J Gene Med. 2003;5:1018–1027. doi: 10.1002/jgm.450. [DOI] [PubMed] [Google Scholar]
- Brand K, Arnold W, Bartels T, Lieber A, Kay MA, Strauss M, et al. Liver-associated toxicity of the HSV-tk/GCV approach and adenoviral vectors. Cancer Gene Ther. 1997;4:9–16. [PubMed] [Google Scholar]
- van der Eb MM, Cramer SJ, Vergouwe Y, Schagen FH, van Krieken JH, van der Eb AJ, et al. Severe hepatic dysfunction after adenovirus-mediated transfer of the herpes simplex virus thymidine kinase gene and ganciclovir administration. Gene Ther. 1998;5:451–458. doi: 10.1038/sj.gt.3300637. [DOI] [PubMed] [Google Scholar]
- Thust R, Tomicic M, Klocking R, Wutzler P., and , Kaina B. Cytogenetic genotoxicity of anti-herpes purine nucleoside analogues in CHO cells expressing the thymidine kinase gene of herpes simplex virus type 1: comparison of ganciclovir, penciclovir and aciclovir. Mutagenesis. 2000;15:177–184. doi: 10.1093/mutage/15.2.177. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Huang SZ, Wang S., and , Zeng YT. Development of an HSV-tk transgenic mouse model for study of liver damage. FEBS J. 2005;272:2207–2215. doi: 10.1111/j.1742-4658.2005.04644.x. [DOI] [PubMed] [Google Scholar]
- Wallace H, Clarke AR, Harrison DJ, Hooper ML., and , Bishop JO. Ganciclovir-induced ablation non-proliferating thyrocytes expressing herpesvirus thymidine kinase occurs by p53-independent apoptosis. Oncogene. 1996;13:55–61. [PubMed] [Google Scholar]
- Muller S, Ronfani L., and , Bianchi ME. Regulated expression and subcellular localization of HMGB1, a chromatin protein with a cytokine function. J Intern Med. 2004;255:332–343. doi: 10.1111/j.1365-2796.2003.01296.x. [DOI] [PubMed] [Google Scholar]
- Zeng J, Du J, Zhao Y, Palanisamy N., and , Wang S. Baculoviral vector-mediated transient and stable transgene expression in human embryonic stem cells. Stem Cells. 2007;25:1055–1061. doi: 10.1634/stemcells.2006-0616. [DOI] [PubMed] [Google Scholar]
- Reubinoff BE, Itsykson P, Turetsky T, Pera MF, Reinhartz E, Itzik A, et al. Neural progenitors from human embryonic stem cells. Nat Biotechnol. 2001;19:1134–1140. doi: 10.1038/nbt1201-1134. [DOI] [PubMed] [Google Scholar]
- Livak KJ., and , Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]