Abstract
Duchenne muscular dystrophy (DMD) is a fatal muscle wasting disorder caused by mutations in the dystrophin gene. In most cases, the open-reading frame is disrupted which results in the absence of functional protein. Antisense-mediated exon skipping is one of the most promising approaches for the treatment of DMD and has recently been shown to correct the reading frame and restore dystrophin expression in vitro and in vivo. Specific exon skipping can be achieved using synthetic oligonucleotides or viral vectors encoding modified small nuclear RNAs (snRNAs), by masking important splicing sites. In this study, we demonstrate that enhanced exon skipping can be induced by a U7 snRNA carrying binding sites for the heterogeneous ribonucleoprotein A1 (hnRNPA1). In DMD patient cells, bifunctional U7 snRNAs harboring silencer motifs induce complete skipping of exon 51, and thus restore dystrophin expression to near wild-type levels. Furthermore, we show the efficacy of these constructs in vivo in transgenic mice carrying the entire human DMD locus after intramuscular injection of adeno-associated virus (AAV) vectors encoding the bifunctional U7 snRNA. These new constructs are very promising for the optimization of therapeutic exon skipping for DMD, but also offer powerful and versatile tools to modulate pre-mRNA splicing in a wide range of applications.
Introduction
Duchenne muscular dystrophy (DMD) is a severe neuromuscular disorder caused by mutations in the dystrophin gene that result in the absence of functional protein. The majority of mutations causing DMD disrupts the open-reading frame and gives rise to prematurely truncated proteins, thus leading to progressive muscle wasting. There is currently no effective treatment for DMD. One of the most promising strategies aims to convert an out-of-frame mutation into an in-frame mutation, which would give rise to internally deleted, but still functional dystrophin.1,2 This can be achieved by using antisense oligonucleotides (AONs) that interfere with splice sites or regulatory elements within the exon and thus induce the skipping of specific exons at the pre-mRNA level.3,4 Alternatively, the antisense sequence can be delivered to cells using viral vectors carrying a gene from which the antisense sequence can be transcribed, such as modified small nuclear RNA (snRNA) genes.5,6 U7 snRNA is normally involved in histone pre-mRNA 3′-end processing, but can be converted into a versatile tool for splicing modulation by a small change in the binding site for Sm/Lsm proteins.7 The antisense sequence embedded into a snRNP particle is therefore protected from degradation and accumulates in the nucleus where splicing occurs. We previously demonstrated that a single treatment with an adeno-associated virus (AAV)2/1 vector containing appropriately modified U7 snRNA induces widespread exon-skipping of the dystrophin pre-mRNA and results in the sustained correction of muscular dystrophy in both the mdx mouse8 and the golden retriever muscular dystrophy dog (A. Vulin, A. Goyenvalle, I. Barthélémy, F. Leturcq, J.C. Kaplan, J. Chelly et al., unpublished results). This long-term restoration of dystrophin represents a strong advantage of this approach over synthetic AONs by eliminating the need for repeated injections.
Considering the diversity of mutations among DMD patients, the translation of this strategy to human will require specific tools adapted to different human dystrophin exons. As with the synthetic AON approach, choice of the antisense sequence is one of the most important parameters to ensure efficient exon skipping. The first targets usually considered to induce exon skipping are the donor splice (DS) and acceptor splice (AS) sites, together with the branch point, and these sites have indeed been successfully targeted on the DMD gene.9,10 Exon skipping can also be achieved by targeting exon-internal sites and in particular exonic splicing enhancers (ESEs),11 but these ESE motifs are only loosely defined and their identification is still complex.12,13,14 Therefore, even after several years of research in the exon-skipping field, there are still no clear rules to guide investigators in the design of their antisense sequences.
Depending on the exon, the most effective antisense sequence will not necessarily target the same sites,15 which implies steps of optimization in each case. Alternatively, it has been demonstrated that tailed AONs carrying a splicing silencer sequence can induce splicing modulation even more efficiently than oligonucleotides acting through duplex formation only.16 We therefore asked in this study whether, based on the idea of these bifunctional oligonucleotides, U7 snRNA can be equipped with splicing silencer sequences to promote the efficient skipping of a particular exon. We have thus designed tailed U7 snRNA constructs carrying canonical binding sites for the heterogeneous ribonucleoprotein A1 (hnRNPA1) targeting the exon 51 of the human dystrophin gene and inserted them into lentiviral vectors to ensure stable expression in human myoblasts. We demonstrate here that bifunctional U7 snRNA constructs can achieve efficient exon 51 skipping in human myoblasts and restore dystrophin expression in cells from DMD patients. Moreover, we show the efficacy of these U7 snRNA constructs in vivo after inserting them into AAV2/1 vectors which were subsequently injected into the tibialis anterior muscle of a mouse model transgenic for the entire human dystrophin locus.17,18
Results
U7 snRNA carrying hnRNPA1 binding sites induces enhanced exon 51 skipping on the dystrophin pre-mRNA of normal myoblasts
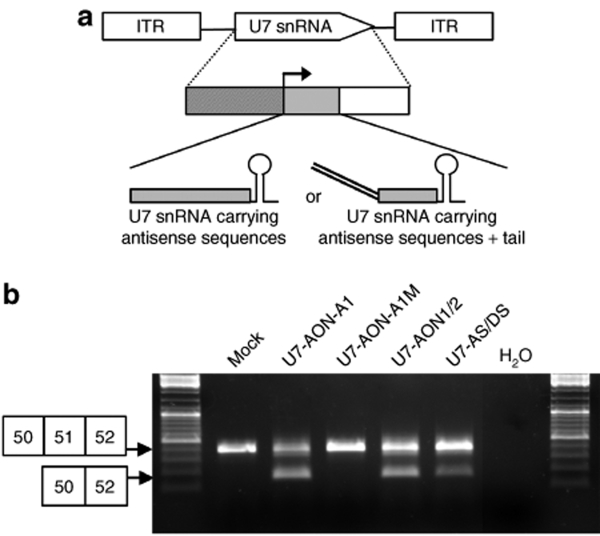
In order to compare the bifunctional approach mediated by U7 snRNA carrying a free tail with splicing silencer motifs with the more commonly used antisense approach to induce exon skipping, we created several engineered U7 snRNA constructs. First, we designed U7 snRNA constructs carrying exclusively antisense sequences to the exon 51, targeting either the AS and DS sites, as previously described by De Angelis et al.5 (Figure 1a, U7-AS/DS) or two exon-internal sequences previously described19 (Figure 1a, U7-AON1/2). We then engineered a U7 snRNA construct carrying 20 nucleotides complementary to exon 51 (corresponding to the h51AON1 sequence inserted in U7-AON1/2) and a free tail harboring high-affinity binding sites for the hnRNPA1 (Figure 1a, U7-AON-A1).20 To distinguish between the effect of the annealing sequence and the tail harboring the splicing silencer motifs, we also created a control U7 snRNA construct with the same antisense sequence but a mutated tail unable to recruit hnRNPA1 (Figure 1, U7-AON-A1M).16 All these different engineered U7 snRNAs were cloned separately into lentiviral vector constructs (Figure 1b) and viral particles were subsequently produced. Immortalized human normal myoblasts21 were transduced with the different lentiviral vectors. Transduction conditions were optimized to induce >95% efficiency using a lentiviral vector encoding green fluorescent protein (data not shown) so that the myoblasts do not have to be sorted after infection. Cells were transduced with the different lentiviral vectors and subsequently amplified for further analysis. Total RNA was then extracted from the transduced cells and analyzed by reverse transcriptase (RT)–PCR, the results of which are presented in Figure 2 (representative of results obtained from at least three independent transductions). Interestingly, the U7-AS/DS targeting the exon 51 splice sites appeared mainly ineffective at inducing skipping of this exon because only a very faint band corresponding to the exon 51–skipped product could be detected. In contrast, the U7-AON1/2 targeting exon-internal sequences was capable of provoking efficient skipping (Figure 2a) suggesting that masking exonic sequences is more effective than masking splice sites, as previously reported for this particular exon.22 The precise skipping of exon 51 was confirmed by direct sequencing of the lower band (Figure 2b). The bifunctional U7-AON-A1 appeared very effective at inducing exon 51 skipping, whereas the control U7-AON-A1M targeting the same sequence on exon 51 but carrying a mutated tail did not induce any skipping. Comparison of these two constructs suggests that skipping efficiency was mainly due to the tail harboring motifs for hnRNPA1 binding and not solely the antisense sequence. The addition of the tail A1 to the AON1 antisense sequence therefore seems to achieve a level of exon 51 skipping comparable to that observed with U7-AON1/2 which could be considered as a positive control for exon 51 skipping, as it results from previous optimization.19
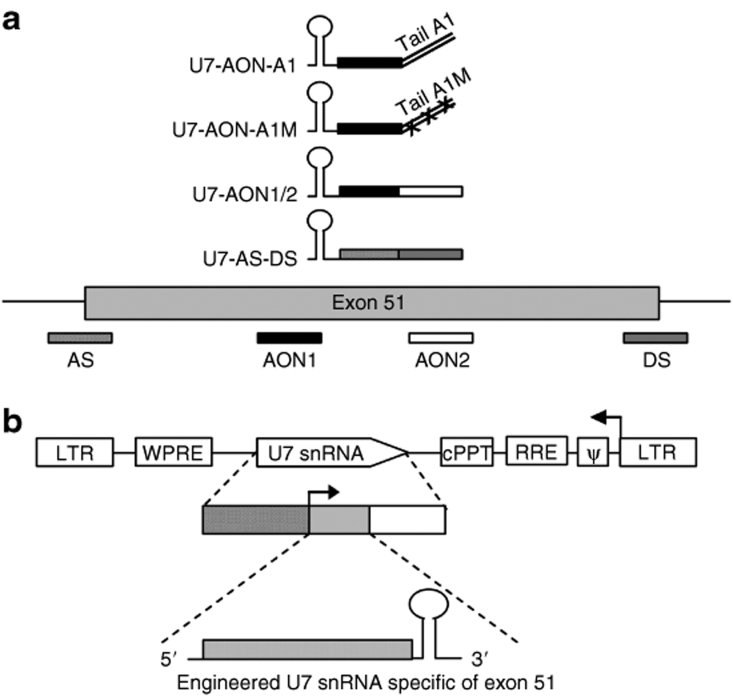
Figure 1.
Scheme depicting the different U7 snRNA constructs and the lentiviral vector used in this work. (a) Representation of the different U7 snRNAs and their target on the dystrophin pre-mRNA. Antisense sequences are represented by rectangular boxes. The U7-AS/DS construct targets the acceptor splice (AS) and donor splice (DS) sites of the exon 51 (represented by hatched boxes) and the U7-AON1/2 construct targets two exonic splicing enhancer (ESE) (represented by black and white boxes). The bifunctional U7-AON-A1 construct carries a complementary sequence to the first ESE (AON1) and a tail harboring two canonical binding sites (TATGATAGGGACTTAGGGTG) for the heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1).16 The control construct U7-AON-A1M targets the same sequence on the exon 51 but carries a mutated tail where the binding sites for hnRNPA1 have been mutated to TACGCT (represented by two lines with crosses). (b) Top: Schematic map of the U7 snRNA lentiviral vectors. The two long-terminal repeats (LTRs) enclose the encapsidation signal (ψ), the Rev-responsive element (RRE), the central polypurine tract (cPPT), the woodchuck hepatitis virus responsive element (WPRE), and the U7 snRNA cassette inversely inserted. Middle: The U7 snRNA cassette is constituted of the engineered U7 snRNA sequence (gray box) carrying different antisense sequences with or without tail as previously described (bottom), placed under the control of its natural U7 promoter (hatched box) and 3′ downstream elements (open box). AON, antisense oligonucleotide; snRNA, small nuclear RNA.
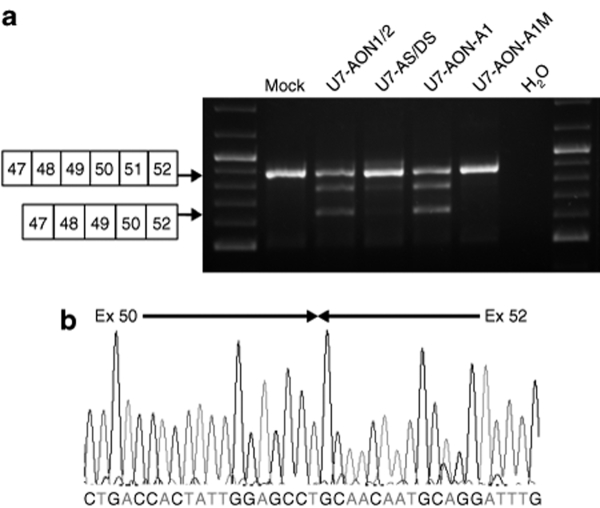
Figure 2.
Comparison of exon 51 skipping in human wild-type myoblasts transduced by different lentiviral vectors. (a) Normal human myoblasts were transduced with lentiviral vectors encoding different U7 snRNA constructs and exon 51 skipping is assessed by nested reverse transcriptase–PCR. (b) Bands of the expected size were analyzed by sequencing, which confirmed the precise skipping of exon 51, joining exons 50 and 52. Note that additional bands between the nonskipped and skipped products are visible in some analyses. This is due to heteroduplex formation and has been described previously.34 AON, antisense oligonucleotide; AS, acceptor splice; DS, donor splice.
Efficient exon-skipping and dystrophin expression induced by engineered U7 snRNA specific of exon 51 in DMD cells
To check that these U7 constructs were able to restore human dystrophin expression, the same experiments were repeated in immortalized myoblasts from a DMD patient carrying a deletion of exons 49 and 50. This particular mutation induces a frame-shift (represented by the black circle in Figure 3a) resulting in the absence of functional dystrophin. Skipping of exon 51 is expected to create an in-frame transcript which should restore dystrophin expression in these cells. Analysis of transduced DMD myoblasts by RT-PCR revealed an apparent higher level of exon 51 skipping induced by all four U7 constructs, compared to the level observed in normal myoblasts, even though viral infections were performed under exactly the same conditions (Figure 3b). This time, U7-AS/DS was capable of inducing a detectable level of exon 51 skipping, even though this was still much lower than that induced by U7-AON1/2. Remarkably, the bifunctional U7-AON-A1 induced complete skipping of exon 51, and the band corresponding to the unskipped product was no longer detectable. The mutated control U7-AON-A1M induced minimal exon 51 skipping, suggesting that the AON1 antisense sequence masking exon-internal sequences had an additional effect on the tail carrying silencer motifs. Sequencing of the lower RT-PCR product showed a precise junction of exons 48 to exon 52, confirming exon 51 skipping (Figure 3c). Protein samples were isolated from the same transduced DMD myoblasts and analyzed by western blotting (Figure 3d). The U7-AON1/2 and the bifunctional U7-AON-A1 construct induced restoration of dystrophin to a level comparable with that detected in normal myoblasts. No dystrophin was detectable in protein extracts of myoblasts treated with U7-AS/DS and U7-AON-A1M, presumably because the level of exon 51 skipping was too low in these cells.
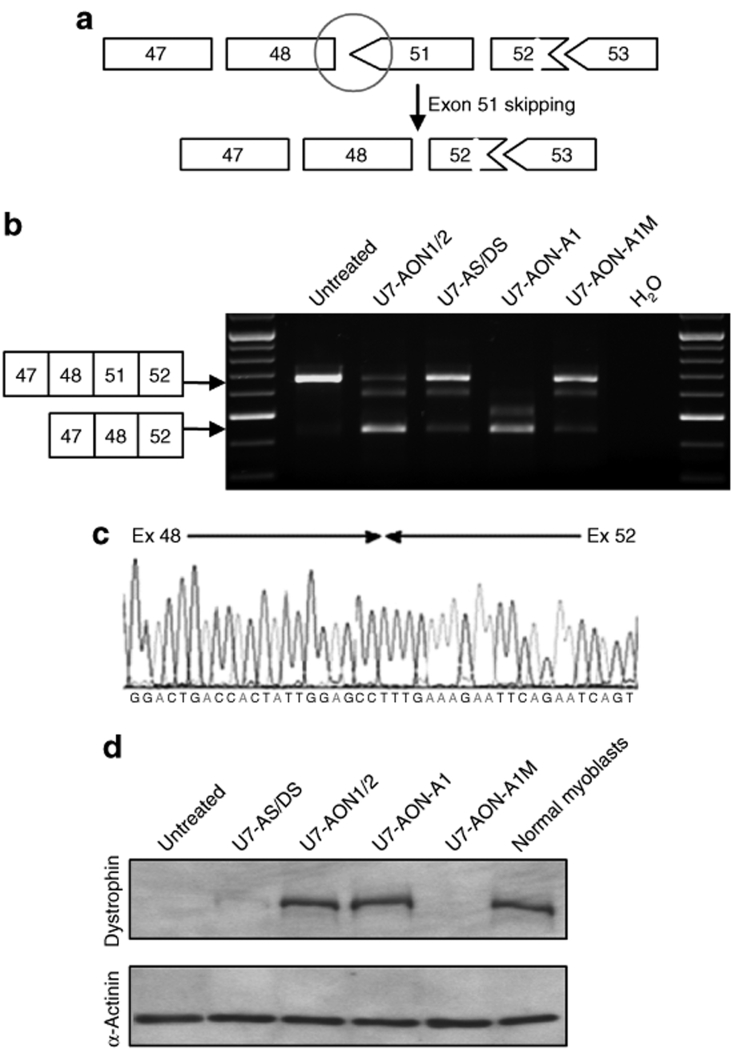
Figure 3.
Comparison of exon 51 skipping in DMD myoblasts and subsequent dystrophin rescue. (a) Schematic representation of the Δ49–50 DMD deletion, which creates a codon de-phasing between the exons 48 and 51 (black circle) and the expected link generated by exon 51 skipping. (b) DMD myoblasts carrying the Δ49–50 deletion were transduced with lentiviral vectors encoding the different U7 snRNA constructs. Exon 51 skipping was assessed by nested reverse transcriptase–PCR and a fragment of the expected size was detected in each case. Additional bands due to heteroduplex formation and activation of a cryptic splice site within the exon 51 (previously described34) are visible in same analyses. (c) Bands were analyzed by sequencing confirming the precise skipping of exon 51, joining exons 48 and 52. (d) Western blot probed with dystrophin antibody (top gel) and α-actinin antibody (bottom gel). Note that the dystrophin detected corresponds to an internally deleted protein missing exons 49, 50, and 51. AON, antisense oligonucleotide; AS, acceptor splice; DMD, Duchenne muscular dystrophy; DS, donor splice.
Tailed U7 snRNA can induce exon-skipping wherever the targeted exonic region is and induce dystrophin rescue
Encouraged by the results obtained with the U7-AON-A1 construct, we wanted to investigate further the potential of the bifunctional approach and especially check if tailed U7 snRNAs could induce exon skipping when targeting various regions within the exon, as long as they carry the silencer motifs. For that purpose, new U7 snRNA constructs were engineered which carried the same free tail containing hnRNPA1 binding sites but targeted either the beginning (U7-5′-A1 construct) or the end (U7-3′-A1 construct) of the exon (Figure 4a). In order to evaluate the contribution of the tail without (as much as possible) any additional effect due to the antisense sequence, we decided to avoid directly masking the AS and DS sites. Therefore, we arbitrarily chose to target nt +5 to +25 of exon 51 for the U7-5′-A1 construct and nt +209 to +229 for the U7-3′-A1 construct (which corresponds to 5 nt before the end of the exon). For both constructs, a control U7 was also created, targeting exactly the same sequence but carrying a mutated tail (U7-5′-A1M and U7-3′-A1M constructs). Lentiviral vectors encoding these different engineered U7 constructs were produced and used to transduce immortalized DMD myoblasts as before. The RT-PCR results presented in Figure 4b showed a strong band which corresponds to an exon 51–skipped product in all cells treated with the U7 snRNA constructs carrying the hnRNPA1 binding motifs (U7-5′-A1, U7-AON-A1 and U7-3′-A1). Low levels of exon 51 skipping were also detected in myoblasts treated with control constructs U7-5′-A1M and U7-AON-A1M carrying a mutated tail, suggesting a slight effect of the antisense sequence. Protein samples isolated from the same transduced myoblasts showed dystrophin restoration (Figure 4c), the levels of which correlated with the degree of exon skipping observed by RT-PCR. A high level of dystrophin expression could be detected in myoblasts treated with either U7-5′-A1, U7-AON-A1, or U7-3′-A1 constructs, whereas no detectable band could be observed in myoblasts treated with the control constructs U7-5′-A1M, U7-AON-A1M, or U7-3′-A1M. The addition of the tail A1 to these antisense sequences therefore seems to significantly increase their exon-skipping potential which would be almost negligible without it. However, the most efficient exon 51 skipping and the highest restoration of dystrophin is achieved by U7-AON-A1 where the tail A1 is combined with a previously optimized antisense sequence (AON1).19
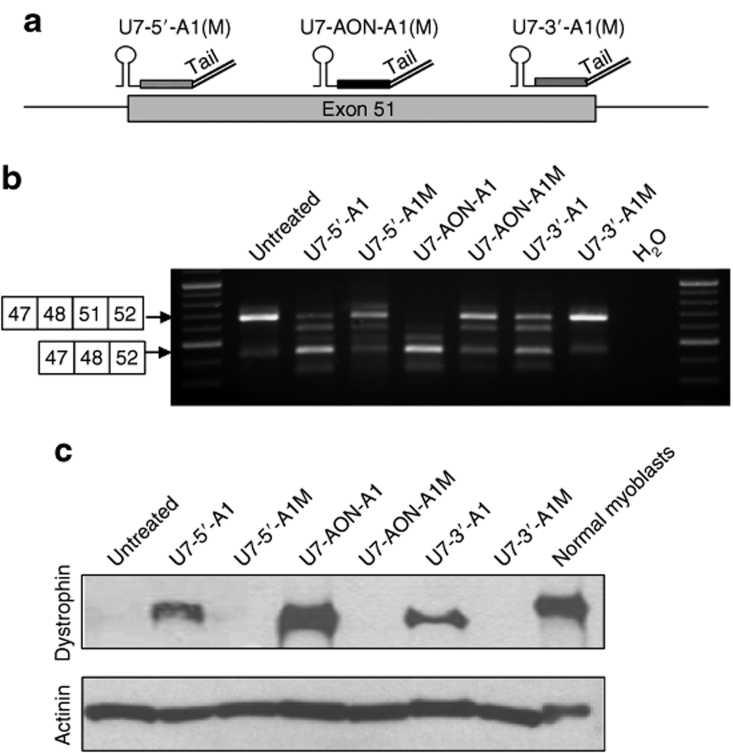
Figure 4.
Comparison of exon 51 skipping in DMD myoblasts induced by bifunctional U7 snRNA depending on their targeted sequence on the exon. (a) Schematic representation of the hybridization of the different U7 snRNA constructs on the exon 51 depending on their antisense sequence. The U7-5′-A1 and U7-5′-A1M carry a complementary sequence to the 5′ of the exon 51 (nt 5–25), whereas the U7-3′-A1 and U7-3′-A1M carry a complementary sequence to the 3′ of the exon 51 (nt 209–229 which corresponds to 5 nt before the end of the exon). The U7-AON-A1 and U7-AON-A1M carry a complementary sequence to a middle exonic splicing enhancer (AON1). (b) DMD myoblasts carrying the Δ49–50 deletion were transduced with lentiviral vectors encoding the different U7 snRNA constructs and exon 51 skipping was assessed by nested reverse transcriptase–PCR. Additional bands due to heteroduplex formation and activation of a cryptic splice site within the exon 51 (previously described34) are visible in same analyses. (c) Western blot probed with dystrophin antibody (top gel) and α-actinin antibody (bottom gel). AON, antisense oligonucleotide; DMD, Duchenne muscular dystrophy.
AAV vectors carrying engineered U7 snRNA induce efficient exon 51 skipping in hDMD mice
The hDMD mouse offers the possibility to evaluate in vivo the efficiency of constructs specifically engineered to target the human dystrophin gene and therefore represents an excellent preclinical model. In order to investigate the effectiveness of the bifunctional approach in vivo, we cloned the different engineered U7 snRNA constructs into an AAV2-based vector (Figure 5a) and subsequently produced AAV1 pseudotyped vectors (AAV2/1) encoding the various U7 snRNA constructs. AAV-U7-AON-A1, AAV-U7-AON-A1M, AAV-U7-AON1/2, and AAV-U7-AS/DS vectors were injected into the tibialis anterior muscles of hDMD mice and RNA from treated muscles were isolated 4 weeks after the injection. RT-PCR results presented in Figure 5b illustrate a similar hierarchy of efficiency as that demonstrated in vitro in immortalized myoblasts. The highest level of exon 51 skipping was obtained in muscles treated with the AAV vector encoding the U7-AON-A1 construct, thus confirming the efficiency of the bifunctional approach in vivo.
Figure 5.
Comparison of exon 51 skipping in vivo in hDMD mice induced by AAV vectors encoding different engineered U7 snRNA. (a) Top: Structure of the AAV vectors encoding the different U7 snRNA cassettes. The U7 snRNA cassette is inserted between two AAV2 inverted terminal repeats (ITRs) and is constituted of the engineered U7 snRNA sequence (gray box) carrying different antisense sequences with or without tail as previously described (bottom), placed under the control of its natural U7 promoter (hatched box) and 3′ downstream elements (open box). (b) AAV2/1 vectors encoding different engineered U7 snRNA were injected in the tibialis anterior of adult hDMD mice. Four weeks after the injection, muscles were harvested and analyzed by nested reverse transcriptase–PCR for exon 51 skipping. AAV, adeno-associated virus; AON, antisense oligonucleotide; AS, acceptor splice; DS, donor splice; snRNA, small nuclear RNA.
Discussion
DMD is a severe, progressive neuromuscular disorder that affects 1 in 3,500 newborn boys23 and generally leads to death between the ages of 20–35 years. There is currently no effective treatment for this myopathy but several strategies have been considered since the discovery of the dystrophin gene in the late 1980s. Among these, the antisense-mediated exon skipping aiming at restoring the open-reading frame has recently gained increasing attention. In this study, we investigated the possibility of inducing effective exon skipping mediated by a splicing silencer sequence, in order to overcome the optimization required for every new targeted exon. We have therefore designed bifunctional U7 snRNA carrying a nonhybridizing tail harboring high-affinity binding sites for the hnRNPA1 protein, as well as U7 snRNAs carrying exclusively antisense sequences. We demonstrated enhanced exon 51 skipping induced by the bifunctional U7 snRNA construct combining the tail A1 and a previously described antisense sequence, compared to those simply masking important splice sites. Interestingly, the U7-AS/DS construct targeting both the AS and DS sites appeared much less efficient than the U7-AON1/2 targeting exon-internal sequences, suggesting that either an ESE or a proper internal secondary RNA structure, or a combination of the two, must play an important role in the correct splicing of exon 51. Previous work using oligonucleotides on this exon has also shown that targeting exonic sequences induced a more effective skipping of exon 51 than targeting AS and DS sites.22 This clearly illustrates the careful selection of targets required in order to achieve efficient skipping depending on the exon. When tested in myoblasts from a DMD patient carrying a deletion of exons 49–50, all U7 snRNA constructs appeared more effective even though viral infections were performed under exactly the same conditions as in normal myoblasts. This observation has previously been reported by others19,24 and could be explained by the nonsense-mediated mRNA decay, that protects the cell from potentially detrimental dominant negative truncated proteins.25 Thus, in human control cells, the skipping of exon 51 results in frame-shifted transcript that contains a stop codon and will be targeted for nonsense-mediated mRNA decay. In patient myoblasts however, the original transcripts are subjected to degradation while in frame mRNA resulting from the skipping of exon 51 are not. Therefore, skipped products which are not subjected to nonsense mediated decay, could appear enriched.
Remarkably, the bifunctional U7-AON-A1 induces complete exon 51 skipping in patient cells, which is associated with a restoration of dystrophin expression to almost wild-type level. By comparison with the U7-AON-A1M construct targeting the same exonic sequence but carrying a mutated tail unable to recruit the hnRNPA1 protein, we were able to distinguish between the effect of the antisense sequence and the repressor tail. The U7-AON-A1M indeed induced a low level of exon 51 skipping, highlighting the effect of the antisense sequence, as previously reported by Aartsma-Rus et al. using 2′-O-methyl oligonucleotides targeting the same sequence.19 The complete exon 51 skipping observed with U7-AON-A1 is then the result of additive effects: the antisense sequence presumably masking some ESE, combined with the repressor tail which allows the recruitment of hnRNPA1, silencing even more the normal splicing process.
We next investigated whether the effect of the repressor tail depends on the targeted region and therefore designed new tailed U7 snRNAs targeting either the beginning or the end of the exon. In order to avoid as much as possible any additional effect due to the antisense sequence, we did not directly target the AS and DS sites and as a result, control constructs U7-5′-A1M and U7-3′-A1M only induced a marginal exon 51 skipping. In contrast, U7-5′-A1 and U7-3′-A1 constructs carrying the high-affinity binding sites for hnRNPA1, were able to induce some level of exon 51 skipping and restoration of dystrophin expression in DMD myoblasts. These observations confirm that the skipping efficacy can be mainly attributed to the silencing tail. Even though levels of exon 51 were not as high as those obtained with U7-AON-A1 or even U7-AON1/2, these results show that significant skipping can be induced even when U7 snRNAs target nonessential splicing signals, as long as they carry the silencing tail.
The splicing interference is likely to be the result of competition between hnRNPA1 and the different splicing factors. Wherever the bifunctional U7 snRNA targets the exon, hnRNPA1 will presumably inhibit the fixation of essential factors, like U2AF when hnRNPA1 is recruited in proximity to the AS site, or U1 snRNP when recruited near the DS site. The presence of hnRNPA1 will also impair the recruitment of SR proteins by any ESE motifs, which normally facilitate the fixation of the other factors, therefore inhibiting the normal splicing process.
The tail used in this study carries two high-affinity binding sites for hnRNPA1 identified by Burd et al., and has been previously shown to recruit hnRNPA1 very efficiently,16,20 thus giving hnRNPA1 an advantage when competing with other splicing factors. Because hnRNPA1 is very abundant in the nucleus, introducing high-affinity binding sites via the bifunctional U7 snRNAs, which are expressed at quite low levels via the U7 natural promoter, must have a negligible impact on normal hnRNPA1-mediated events.
We also demonstrated in this study the efficacy of these new tailed U7 snRNA constructs in vivo in a mouse model transgenic for the entire human dystrophin locus following intramuscular injection of AAV vectors encoding the bifunctional constructs. AAV vectors have recently emerged as very promising gene transfer vehicles in the treatment of neuromuscular disorders because they enable extremely stable in vivo expression in the vast majority of the musculature. The vectors introduce multiple extrachromosomal copies of the transgene in cells and display a variety of tissue tropisms, depending on the type of capsid used in the packaging system. Vectors packaged with AAV1, 6, 8, and 9 capsids are of particular interest for gene transfer into muscle26 and represent very efficient tools for the delivery of U7 snRNA, as we previously demonstrated in the mdx mouse.8
Although AAV vectors appear to be the vehicle of choice to achieve U7 snRNA–mediated exon skipping in DMD patients, lentiviral vectors used in the in vitro part of this study represent a molecular tool to induce high and stable levels of expression of different U7 snRNA constructs in human myoblasts, which cannot be efficiently transfected by plasmids (as opposed to oligomer transfection). However, the lentiviral vectors encoding U7 snRNAs that we designed here can also be considered as promising clinical tools especially for stem cell–mediated approaches for DMD. Benchaouir et al. have indeed recently demonstrated the delivery of CD133+ patient cells corrected by U7 snRNA–mediated exon-skipping into mice, suggesting a new potential avenue for autologous cell therapy.27
The new tailed U7 snRNAs constructs designed in this study offer an efficient alternative to the optimization of antisense sequence normally required to achieve effective exon skipping. Although it is evident that the antisense sequence has a major influence, the presence of the repressor tail attached to the U7 snRNAs appears to be sufficient to achieve significant exon skipping. This will prove to be a very useful tool when considering that this strategy has to be extended to many exons in the case of DMD.
In conclusion, the strategy based on bifunctional U7 snRNAs presented in this study is extremely promising for future preclinical and clinical work on DMD. We demonstrate here the efficiency of the approach in two different models taking advantage of patient cells with the appropriate deletion, providing us with the possibility to confirm dystrophin restoration, together with the hDMD mouse model, which enables us to assess the effect of the AAV-U7 snRNAs vectors in vivo. Additionally, as antisense-mediated exon skipping can be implemented in numerous other therapeutic interventions or developmental studies, these hnRNPA1-tailed U7 snRNAs represent very versatile and powerful tools to modulate pre-mRNA splicing in an even wider context.
Materials and Methods
U7 snRNA constructs design. The different U7 snRNA constructs specific to exon 51 were engineered from the previously described U7smOPT-SD23/BP22 (modified murine U7 snRNA gene).8 Antisense sequences targeting the mouse exon 23 were replaced by antisense sequences targeting the human exon 51 of dystrophin mRNA. The construct U7-AON1/2 carries antisense sequences targeting two internal splicing enhancers (ESE) of exon 51: h51AON1: 5′-TCAAGGAAGATGGCATTTCT-3′ and h51AON2: 5′-CCTCTGTGATTTTATAACTTGAT-3′.19 The construct U7-AS/DS carries antisense sequences targeting the AS and DS sites of exon 51: AS: 5′-CCCAAAATATTTTAGCTCCTACTCAGAC-3′ and DS: 5′-ATCAAGCAGAAGGTATGAGAAAAA-3′.5 The choice of targeting two sequences when designing U7-AON1/2 and U7-AS/DS was directed by the previous observation that combination of antisense sequences in U7 snRNA seems to increase its efficiency28 and also to maintain the same size as the tailed U7 constructs. The tailed U7 snRNA constructs carry complementary sequence to either the 5′ region of exon 51 (5′-TACTCAGACTGTTACTCTGGTGACA-3′) (construct U7-5′-A1), the first ESE—h51AON1—(construct U7-AON-A1) or the 3′ region of exon (5′-ATATCAACGAGATGATCATCAAGCA-3′) (construct U7-3′-A1), and a 16-nt long nonhybridizing tail carrying high-affinity binding sites for the hnRNP A1 protein (TATGATAGGGACTTAGGGTG).20 For each tailed U7 snRNA constructed, a corresponding control targeting the same sequence and carrying a mutated version of the tail A1 has been engineered (GGG to CGC mutations) (U7 snRNA-A1M). The resulting U7 snRNA fragments were then introduced either in a lentiviral vector construct for further lentiviral production or into an AAV vector construct for AAV production.
Viral vector production. All the lentiviral vectors were based on the pRRL-cPPT-hPGK-eGFP-WPRE constructs29 where the hPGK-GFP cassette was removed and replaced with the U7 snRNA construct. For a large tropism, lentiviral vectors were pseudotyped with the vesicular stomatosis virus-G protein, and were generated by transfection into 293T cells of a packaging construct, pCMVΔR8.74, a plasmid producing the vesicular stomatosis virus-G envelope (pMD.G) and the vector itself as previously described.30 Viral titers (infectious particles, ip) were determined by transduction of 105 NIH3T3 cells with serial dilutions of the vector preparation in a 12-well plate. Seventy-two hours later, genomic DNA from transduced cells was extracted using a genomic DNA purification kit (Qiagen, Crawley, UK). The infectious particles titer (ip/ml) was determined by quantitative real-time PCR as described elsewhere.31
For subsequent AAV vector production, the different U7 snRNA fragments were introduced at the XbaI site of the pSMD2 AAV2 vector.32 AAV2/1 pseudotyped vectors were prepared by co-transfection in 293 cells of pAAV2-U7 snRNA, pXX6 encoding adenovirus helper functions and pAAV1pITRCO2 that contains the AAV2 rep and AAV1 cap genes. Vector particles were purified on cesium chloride gradients from cell lysates obtained 48 hours after transfection and titers were measured by quantitative real-time PCR.33
Cell culture and lentiviral transduction. Human myoblasts (courtesy of V. Mouly; Institut de Myologie, Paris, France) have been immortalized as described in ref. 21. Immortalized normal myoblasts were grown in F10 medium (Invitrogen, Paisley, UK) supplemented with 20% fetal bovine serum (Invitrogen), 100 U/ml penicillin and 100 U/ml streptomycin. Immortalized DMD myoblasts carrying a deletion of exons 49 and 50 were grown in medium [four parts Dulbecco's modified Eagle's medium (4.5 mg/ml glucose) to one part medium 199] supplemented with 20% fetal bovine serum, 100 U/ml penicillin and 100 U/ml streptomycin. For lentiviral transduction, 1 × 105 myoblasts were seeded in each well of a 12-well plate. After 24 hours, cells were incubated with 106 to 107 ip of lentiviral vector in a total volume of 500 µl of their respective medium mentioned above. After 4 hours of incubation, medium was replaced. Cells were subsequently amplified for further analysis (RNA and protein analysis). All analysis was repeated on cells amplified from at least three independent transductions for each U7 construct. In all cases, copy number of integrated lentiviral vector has been measured by quantitative real-time PCR and shown to be directly comparable.
RNA isolation and RT-PCR analysis. Total RNA was isolated from the human myoblasts using the RNeasy kit according to the manufacturer's instructions (Qiagen). Aliquots of 200 ng of total RNA were used for RT-PCR analysis using the Access RT-PCR System (Promega, Southampton, UK) in a 50 µl reaction using the external primers Ex 46F (5′-AGGAAGCAGATACATTGCT-3′) and Ex 53R (5′-TTTCATTGAACTGTTGCCTC-3′). The cDNA synthesis was carried out at 45 °C for 45 minutes, directly followed by the primary PCR of 30 cycles of 94 °C (30 seconds), 58 °C (1 minute) and 72 °C (2 minutes). Two microlitres of these reactions were then reamplified in nested PCRs by 28 cycles of 94 °C (30 seconds), 58 °C (1 minute) and 72 °C (2 minutes) using the internal primers Ex 47Fi (5′-TTACTGGTGGAAGAGTTGCC-3′) and Ex 52Ri (5′-TGATTGTTCTAGCCTCTTGA-3′). For in vivo experiments in hDMD mice, total RNA was isolated from the injected muscles using TRIzol reagent according to the manufacturer's instructions (Invitrogen). 200 ng of total RNA were used for RT-PCR analysis using the Access RT-PCR System (Promega) in a 50 µl reaction using the external primers Hex 49F2 (5′-AAACTGAAATAGCAGTTCAAGC-3′) and Hex 53R2 (5′-TTGCCTCCGGTTCTGAAGG-3′). The cDNA synthesis was carried out at 45 °C for 45 minutes, directly followed by the primary PCR of 20 cycles of 94 °C (40 seconds), 60 °C (40 seconds) and 72 °C (40 seconds). Two microlitres of these reactions were then reamplified in nested PCRs by 30 cycles of 94 °C (40 seconds), 60 °C (40 seconds) and 72 °C (40 seconds) using the internal primers Hex 50F (5′-AGGAAGTTAGAAGATCTGAGC-3′) and Hex 52R2 (5′-TTCTTCCAACTGGGGACGC-3′). PCR products were analyzed on 2% agarose gels.
Western blot analysis. Protein extracts were obtained from pelleted myoblasts treated with Newcastle buffer (3.8% sodium dodecyl sulfate, 75 mmol/l Tris-HCl pH 6.7, 4 mol/l urea, 10% β-mercaptoethanol, 10% glycerol, 0.001% bromophenol blue). To determine the total protein amount in samples, the bicinchoninic acid protein assay kit and protocol was used (Perbio Science, Northumberland, UK). Samples were denatured at 95 °C for 5 minutes before 50 µg of protein was loaded in a 5% polyacrylamide gel with a 4% stacking gel. Gels were electrophoresed for 4–5 hours at 100 V and blotted to a polyvinylidene fluoride membrane overnight at 40 V. Blots were blocked for 1 hour with 10% nonfat milk in phosphate buffered saline–Tween buffer. Dystrophin and α-actinin proteins were detected by probing the membrane with 1:100 dilution of NCL-DYS1 primary antibody (monoclonal antibody to dystrophin R8 repeat; NovoCastra, Newcastle upon Tyne, UK) and 1:200 dilution of α-actinin primary antibody (Santa Cruz Biotechnology, Santa Cruz, CA) respectively. An incubation with a mouse horseradish peroxidase–conjugated secondary antibody (1:2,000) or goat horseradish peroxidase–conjugated secondary antibody (1:160,000) allowed visualization using ECL Analysis System (GE Healthcare, Chalfont St Giles, UK).
Transgenic human DMD mice injection. Transgenic human DMD mice were imported from the Leiden University Medical Center.17,18 All animal experiments were performed according to the guidelines and protocols approved by the Home Office. Adult hDMD mice (between 6 and 8 weeks of age) were anesthetized under isofluorane and injected with 1011 vg of AAV2/1 vector, encoding the different U7 snRNA constructs, into the tibialis anterior muscles. Mice were killed 4 weeks after the injection and TA muscles were isolated and snap frozen in liquid nitrogen-cooled isopentane.
Acknowledgments
We are grateful to Vincent Mouly (Institut de Myologie, Paris) for providing the immortalized myoblasts used in this study. We also thank Annemieke Aarstma-Rus and Johan T. den Dunnen (Leiden University Medical Center, the Netherlands) for providing the transgenic human DMD mice. This work was supported by Action Duchenne, the Association Monegasque contre les myopathies and Duchenne Parent Project de France. A.G. was supported by an EMBO long-term postdoctoral fellowship.
REFERENCES
- Koenig M, Beggs AH, Moyer M, Scherpf S, Heindrich K, Bettecken T, et al. The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am J Hum Genet. 1989;45:498–506. [PMC free article] [PubMed] [Google Scholar]
- Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H., and , Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Janson AA, Heemskerk JA, De Winter CL, Van Ommen GJ., and , Van Deutekom JC. Therapeutic modulation of DMD splicing by blocking exonic splicing enhancer sites with antisense oligonucleotides. Ann NY Acad Sci. 2006;1082:74–76. doi: 10.1196/annals.1348.058. [DOI] [PubMed] [Google Scholar]
- Mann CJ, Honeyman K, Cheng AJ, Ly T, Lloyd F, Fletcher S, et al. Antisense-induced exon skipping and synthesis of dystrophin in the mdx mouse. Proc Natl Acad Sci USA. 2001;98:42–47. doi: 10.1073/pnas.011408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis FG, Sthandier O, Berarducci B, Toso S, Galluzzi G, Ricci E, et al. Chimeric snRNA molecules carrying antisense sequences against the splice junctions of exon 51 of the dystrophin pre-mRNA induce exon skipping and restoration of a dystrophin synthesis in Delta 48-50 DMD cells. Proc Natl Acad Sci USA. 2002;99:9456–9461. doi: 10.1073/pnas.142302299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun C, Suter D, Pauli C, Dunant P, Lochmüller H, Burgunder JM, et al. U7 snRNAs induce correction of mutated dystrophin pre-mRNA by exon skipping. Cell Mol Life Sci. 2003;60:557–566. doi: 10.1007/s000180300047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schümperli D., and , Pillai RS. The special Sm core structure of the U7 snRNP: far-reaching significance of a small nuclear ribonucleoprotein. Cell Mol Life Sci. 2004;61:2560–2570. doi: 10.1007/s00018-004-4190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyenvalle A, Vulin A, Fougerousse F, Leturcq F, Kaplan JC, Garcia L, et al. Rescue of dystrophic muscle through U7 snRNA-mediated exon skipping. Science. 2004;306:1796–1799. doi: 10.1126/science.1104297. [DOI] [PubMed] [Google Scholar]
- Dunckley MG, Manoharan M, Villiet P, Eperon IC., and , Dickson G. Modification of splicing in the dystrophin gene in cultured Mdx muscle cells by antisense oligoribonucleotides. Hum Mol Genet. 1998;7:1083–1090. doi: 10.1093/hmg/7.7.1083. [DOI] [PubMed] [Google Scholar]
- Mann CJ, Honeyman K, McClorey G, Fletcher S., and , Wilton SD. Improved antisense oligonucleotide induced exon skipping in the mdx mouse model of muscular dystrophy. J Gene Med. 2002;4:644–654. doi: 10.1002/jgm.295. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, De Winter CL, Janson AA, Kaman WE, Van Ommen GJ, Den Dunnen JT, et al. Functional analysis of 114 exon-internal AONs for targeted DMD exon skipping: indication for steric hindrance of SR protein binding sites. Oligonucleotides. 2005;15:284–297. doi: 10.1089/oli.2005.15.284. [DOI] [PubMed] [Google Scholar]
- Fairbrother WG, Yeh RF, Sharp PA., and , Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- Cartegni L., and , Krainer AR. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat Struct Biol. 2003;10:120–125. doi: 10.1038/nsb887. [DOI] [PubMed] [Google Scholar]
- Zhang XH., and , Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton SD, Fall AM, Harding PL, McClorey G, Coleman C., and , Fletcher S. Antisense oligonucleotide-induced exon skipping across the human dystrophin gene transcript. Mol Ther. 2007;15:1288–1296. doi: 10.1038/sj.mt.6300095. [DOI] [PubMed] [Google Scholar]
- Villemaire J, Dion I, Elela SA., and , Chabot B. Reprogramming alternative pre-messenger RNA splicing through the use of protein-binding antisense oligonucleotides. J Biol Chem. 2003;278:50031–50039. doi: 10.1074/jbc.M308897200. [DOI] [PubMed] [Google Scholar]
- Bremmer-Bout M, Aartsma-Rus A, de Meijer EJ, Kaman WE, Janson AA, Vossen RH, et al. Targeted exon skipping in transgenic hDMD mice: A model for direct preclinical screening of human-specific antisense oligonucleotides. Mol Ther. 2004;10:232–240. doi: 10.1016/j.ymthe.2004.05.031. [DOI] [PubMed] [Google Scholar]
- 't Hoen PA, de Meijer EJ, Boer JM, Vossen RH, Turk R, Maatman RG, et al. Generation and characterization of transgenic mice with the full-length human DMD gene. J Biol Chem. 2008;283:5899–5907. doi: 10.1074/jbc.M709410200. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Bremmer-Bout M, Janson AA, den Dunnen JT, van Ommen GJ., and , van Deutekom JC.Targeted exon skipping as a potential gene correction therapy for Duchenne muscular dystrophy Neuromuscul Disord 200212S71–S77.suppl. 1 [DOI] [PubMed] [Google Scholar]
- Burd CG., and , Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu CH, Mouly V, Cooper RN, Mamchaoui K, Bigot A, Shay JW, et al. Cellular senescence in human myoblasts is overcome by human telomerase reverse transcriptase and cyclin-dependent kinase 4: consequences in aging muscle and therapeutic strategies for muscular dystrophies. Aging Cell. 2007;6:515–523. doi: 10.1111/j.1474-9726.2007.00306.x. [DOI] [PubMed] [Google Scholar]
- Arechavala-Gomeza V, Graham IR, Popplewell LJ, Adams AM, Aartsma-Rus A, Kinali M, et al. Comparative analysis of antisense oligonucleotide sequences for targeted skipping of exon 51 during dystrophin pre-mRNA splicing in human muscle. Hum Gene Ther. 2007;18:798–810. doi: 10.1089/hum.2006.061. [DOI] [PubMed] [Google Scholar]
- Moser H. Duchenne muscular dystrophy: pathogenetic aspects and genetic prevention. Hum Genet. 1984;66:17–40. doi: 10.1007/BF00275183. [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Bremmer-Bout M, Janson AA, Ginjaar IB, Baas F, den Dunnen JT, et al. Antisense-induced exon skipping restores dystrophin expression in DMD patient derived muscle cells. Hum Mol Genet. 2001;10:1547–1554. doi: 10.1093/hmg/10.15.1547. [DOI] [PubMed] [Google Scholar]
- Maquat LE. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- Blankinship MJ, Gregorevic P., and , Chamberlain JS. Gene therapy strategies for Duchenne muscular dystrophy utilizing recombinant adeno-associated virus vectors. Mol Ther. 2006;13:241–249. doi: 10.1016/j.ymthe.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Benchaouir R, Meregalli M, Farini A, D'Antona G, Belicchi M, Goyenvalle A, et al. Restoration of human dystrophin following transplantation of exon-skipping-engineered DMD patient stem cells into dystrophic mice. Cell Stem Cell. 2007;1:646–657. doi: 10.1016/j.stem.2007.09.016. [DOI] [PubMed] [Google Scholar]
- Suter D, Tomasini R, Reber U, Gorman L, Kole R., and , Schümperli D. Double-target antisense U7 snRNAs promote efficient skipping of an aberrant exon in three human beta-thalassemic mutations. Hum Mol Genet. 1999;8:2415–2423. doi: 10.1093/hmg/8.13.2415. [DOI] [PubMed] [Google Scholar]
- VandenDriessche T, Naldini L, Collen D., and , Chuah MK. Oncoretroviral and lentiviral vector-mediated gene therapy. Meth Enzymol. 2002;346:573–589. doi: 10.1016/s0076-6879(02)46078-8. [DOI] [PubMed] [Google Scholar]
- Zufferey R, Nagy D, Mandel RJ, Naldini L., and , Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- Charrier S, Stockholm D, Seye K, Opolon P, Taveau M, Gross DA, et al. A lentiviral vector encoding the human Wiskott-Aldrich syndrome protein corrects immune and cytoskeletal defects in WASP knockout mice. Gene Ther. 2005;12:597–606. doi: 10.1038/sj.gt.3302440. [DOI] [PubMed] [Google Scholar]
- Snyder RO, Spratt SK, Lagarde C, Bohl D, Kaspar B, Sloan B, et al. Efficient and stable adeno-associated virus-mediated transduction in the skeletal muscle of adult immunocompetent mice. Hum Gene Ther. 1997;8:1891–1900. doi: 10.1089/hum.1997.8.16-1891. [DOI] [PubMed] [Google Scholar]
- Douar AM, Poulard K, Stockholm D., and , Danos O. Intracellular trafficking of adeno-associated virus vectors: routing to the late endosomal compartment and proteasome degradation. J Virol. 2001;75:1824–1833. doi: 10.1128/JVI.75.4.1824-1833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aartsma-Rus A, Janson AA, Kaman WE, Bremmer-Bout M, den Dunnen JT, Baas F, et al. Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum Mol Genet. 2003;12:907–914. doi: 10.1093/hmg/ddg100. [DOI] [PubMed] [Google Scholar]