Abstract
The Sleeping Beauty (SB) transposon system is a nonviral vector that directs transgene integration into vertebrate genomes. We hydrodynamically delivered SB transposon plasmids encoding human α-L-iduronidase (hIDUA) at two DNA doses, with and without an SB transposase gene, to NOD.129(B6)-Prkdcscid IDUAtm1Clk/J mice. In transposon-treated, nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice with mucopolysaccharidosis type I (MPS I), plasma IDUA persisted for 18 weeks at levels up to several hundred–fold wild-type (WT) activity, depending on DNA dose and gender. IDUA activity was present in all examined somatic organs, as well as in the brain, and correlated with both glycosaminoglycan (GAG) reduction in these organs and level of expression in the liver, the target of transposon delivery. IDUA activity was higher in the treated males than in females. In females, omission of transposase source resulted in significantly lower IDUA levels and incomplete GAG reduction in some organs, confirming the positive effect of transposition on long-term IDUA expression and correction of the disease. The SB transposon system proved efficacious in correcting several clinical manifestations of MPS I in mice, including thickening of the zygomatic arch, hepatomegaly, and accumulation of foamy macrophages in bone marrow and synovium, implying potential effectiveness of this approach in treatment of human MPS I.
Introduction
Mucopolysaccharidosis type I (MPS I), caused by deficiency of α-L-iduronidase (IDUA; EC3.2.1.76), is one of the most common MPS diseases in humans.1 MPS I is currently treated by bone marrow transplantation and/or enzyme replacement therapy.2,3 However, there are significant concerns associated with these treatments that include limited positive outcomes and high costs due to the need for life-long therapy.4,5 Gene therapy represents a potential alternative approach, albeit one with its own attendant concerns.3,6,7 Considerable progress has been made over the past few years in developing methods of gene transfer to treat MPS I using viral vectors.8,9,10,11,12,13,14 However, there are complications with viruses as gene-delivery vectors including their integration-site preferences that may increase chances of adverse effects.15,16 Moreover, the need for stringent purification and quality control to prevent replication-competent virus makes the use of these vectors relatively expensive.3,17 Advantages of nonviral vectors include the ease and relatively low cost of producing sufficient amounts required to meet the entire patient population, stability during storage, and lack of immunogenicity.3,18 There are two major problems with nonviral, plasmid-based, gene therapy approaches: (i) expression of the transgene from most plasmids in most cell types is brief due to lack of integration and epigenetic responses;19 and (ii) directing DNA molecules to a specific organ or cell type is inefficient. The first problem can be surmounted by the use of the Sleeping Beauty (SB) transposon system,20,21 a nonviral vector that combines the advantages of viruses and naked DNA. The second problem can be solved using the rapid high-volume, hydrodynamic delivery that specifically targets the liver.22,23 The efficiency of hydrodynamic delivery in mice is ~10% of that reported for retroviral delivery.24,25 However, if the therapeutic expression cassette employs a powerful promoter, a low percentage of genetically modified cells in even a single organ can provide therapeutic levels of a secreted gene product sufficient to treat an entire animal.18,24,26
We have focused on treating MPS I in mice as a model for developing a therapy for humans. The available MPS I mouse models27,28 exhibit many of the clinical manifestations of the respective human disease, including organomegaly, skeletal dysplasia, and neurologic deficits. Recently, we demonstrated adequate delivery of therapeutic transgenes to mouse liver for treatment of MPS I mice using the SB transposon system.24 However, as have others,29 we found that immune responses interfered with long-term IDUA expression.24 In our earlier study, only about one-third of the treated mice responded to immunomodulation and the levels of IDUA activity varied in a wide range. In this study focusing on the effectiveness of SB transposons in restoring normal phenotypes in the absence of immune responses, we used the MPS I mouse model bred onto an immunodeficient background (NOD.129(B6)-Prkdcscid IDUAtm1Clk/J).30 This approach has been useful in gene therapy studies of lysosomal storage diseases particularly when either the cytomegalovirus promoter or the cytomegalovirus-hybrid CAGGS promoter are used for high-level expression of therapeutic gene.31,32,33
Results
Correction of IDUA deficiency by hydrodynamic injection of IDUA-SB transposons
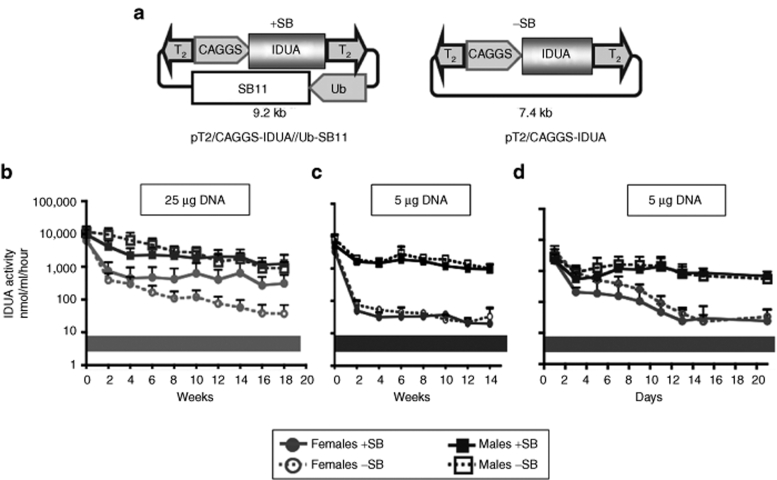
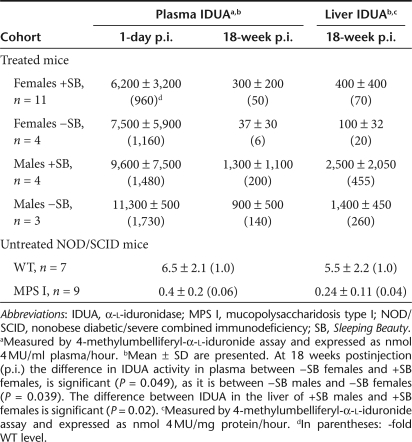
The pT2/CAGGS-IDUA//Ub-SB11 hyperactive T2 transposon has (i) the human IDUA (hIDUA) gene behind the CAGGS promoter and (ii) an ubiquitin-regulated SB transposase sequence on the same plasmid; control plasmids lacked the SB expression cassette (Figure 1a). We hydrodynamically injected nonobese diabetic/severe combined immunodeficiency (NOD/SCID) MPS I mice with either of the two plasmids; littermates of plasmid-treated mice served as untreated controls. Following injection of 25 µg transposon DNA, IDUA activity in plasma was high in all treatment groups for 18 weeks (Figure 1b, Table 1). By 4 months IDUA was about tenfold lower in −SB females than in +SB females (*P = 0.049) whereas in −SB males they were only ~70% that of +SB males. Expression was consistently higher in males than in females. The gender-related difference was especially striking when the injected DNA dose was reduced from 25 to 5 µg (Figure 1c) even though gene delivery, as measured by initial IDUA activities, was about the same in all treatment groups. Within 2 weeks, plasma IDUA levels were ~30-fold lower in females than in males (Figure 1d). There was no statistical difference in plasma IDUA activities between mice treated with or without SB transposase for either gender with 5 µg DNA (Supplementary Table S1). Thus, we observed two unexpected effects from injections into NOD/SCID mice: (i) a major gender effect that was most pronounced at lower doses of the transgene, and (ii) lack of a need for SB transposase to achieve sustained gene expression. The simplest explanation for the similar expression from injections with and without transposase was that prolonged expression of the transgenic IDUA gene came from unintegrated transposons that remained episomal.
Figure 1.
Sleeping Beauty (SB) transposon vectors and plasma levels of IDUA activity in mice following hydrodynamic delivery. (a) pT2/CAGGS-IDUA//Ub-SB11 (+SB) and pT2/CAGGS-IDUA (−SB) vectors contain T2 transposon48 (inverted arrowheads) that carries the human (h)IDUA cDNA inserted between two EcoRI sites of the mini(m)CAGGS promoter. The integrating vector (+SB) contains an SB transposase expression cassette outside the transposon. Expression of the SB11 hyperactive transposase gene49 is regulated by a shortened version of the intermediate-strength ubiquitin (Ub) promoter.24 (b) Expression of transgenic IDUA in NOD/SCID MPS I mice injected with 25 µg of either the +SB or −SB plasmid. Mice aged 4–13 weeks were treated in groups of 10 for +SB females, 4 −SB females, 4 +SB males and 3 −SB males. Mice were euthanized 18 weeks postinjection (p.i.); (c and d) Expression of transgenic IDUA in NOD/SCID MPS I mice following injection with 5 µg of either of the two plasmids: (c) biweekly time-course; the values are pooled from two independent experiments with the duration of 6 and 14 weeks. The values on day 1 through 6 weeks represent 11 +SB females, 11 −SB females, 8 +SB males and 8 −SB males; the values at 8–14 weeks represent 5 +SB females, 5 −SB females, 2 +SB males and 2 −SB males; (d) IDUA expression in males and females in the first 3 weeks following treatment. Values represent six mice/gender/treatment. Plasma IDUA levels in untreated controls were: 6.5 ± 0.7 nmol 4 MU/ml/hour, n = 7 (WT, indicated by the gray bars in panels b–d) and 0.4 ± 0.05 nmol 4 MU/ml/hour, n = 9 (MPS I). Symbols: +SB females, filled gray circles and solid lines; −SB females, open gray circles and dotted lines; +SB males, filled dark squares and solid lines; −SB males, open dark squares and dotted lines. Each point represents the mean IDUA activity ± SD. IDUA, α-L-iduronidase; MPS-I, mucopolysaccharidosis type I; NOD/SCID, nonobese diabetic/severe combined immunodeficiency; WT, wild type.
Table 1.
IDUA activity in plasma and liver of mice treated with 25 µg DNA
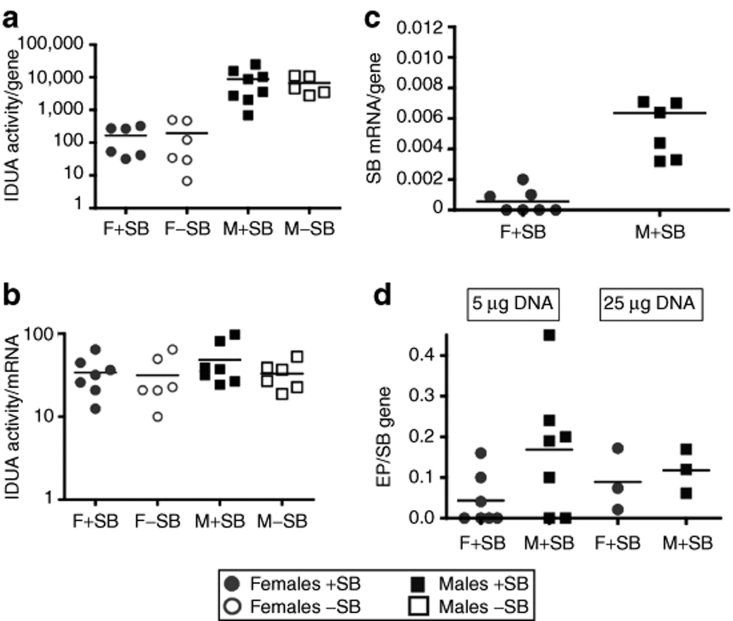
We and others have observed24,34 that plasma IDUA activity directly correlates with IDUA activity in the liver (Table 1). At the 25 µg DNA dose, plasma IDUA activity normalized per gene copy number in liver cells was about the same in +SB mice for both males and females (Supplementary Table S1). However, in −SB female mice treated with 25 µg DNA, normalized IDUA activity was ~90-fold lower than in +SB females, whereas the difference was only about threefold in respective male groups. In contrast, with 5 µg DNA, IDUA activity per gene did not differ ±SB within either sex although it was ~30-fold higher in males than in females (Figure 2a). The differences in IDUA levels in the two sexes could have been due to transcriptional or post-transcriptional regulation. Consequently, we determined the ratio of IDUA activity to IDUA mRNA (Figure 2b) using reverse transcription–PCR. The ratios of IDUA activity to respective IDUA mRNA relative copy number were on average statistically the same (Figure 2b) but transcription was depressed 30–50-fold in females. These data are consistent with transcriptional repression in females compared to in males rather than differences in either translational regulation or stability of the IDUA enzyme.
Figure 2.
Molecular analyses of expression of IDUA and SB transposase genes, and transposition measured by excision product (EP) formation following injection of transposon constructs. (a) IDUA enzyme activity relative to the number of IDUA genes, from pT2/CAGGS-IDUA//Ub-SB11 or pT2/CAGGS-IDUA, respectively, quantified by PCR; (b) IDUA enzyme activity relative to the level of IDUA mRNA quantified by reverse transcription (RT)-PCR of IDUA. There is no statistical difference between the mean values for any of the cohorts; (c) SB mRNA levels, quantified by real-time RT-PCR, relative to SB gene copy number; four of seven samples had mRNA levels below minimal detection; (d) EP formation as a function of SB gene number. Note that four of the female mice injected with 5 µg plasmid had no detectable EPs, which is consistent with the lack of transcription from the SB gene in these mice (c). DNA dose is 5 µg (a–c) and 5 and 25 µg (d). IDUA, α-L-iduronidase; SB, Sleeping Beauty.
If this conclusion was true, then we would not expect to see differences in transposition activity unless there was transcriptional repression of the Ub promoter. Hence, we examined the livers of mice for evidence of transposition using the PCR-based excision product (EP) assay wherein the EP copy number is measured as a function of input plasmids, i.e., the number of SB genes.24,34 Although the EP/SB ratio was four times lower in 5 µg–treated females than in males (Figure 2d), the difference was not statistically significant. Notably, transcription of the SB gene in about half of the female livers was below our limit of detection (Figure 2c), which accounted for half of the difference in SB expression seen between males and females. These data suggest that transcription of SB, when allowed, occurred at about the same rate in males and in females.
If transposition had occurred from ~0.1 to 1% of the plasmids taken up into hepatocytes,35 then as expression from episomal plasmids slowly decreased, we would expect that over time stable expression of IDUA from integrated plasmids would be evident. Accordingly, we calculated the half-lives of plasma IDUA in treated males and females. Because the decay kinetics following hydrodynamic injection were biphasic (Figure 1b,c), we calculated the half-lives of plasma IDUA from the second week postinjection, until killing. Following injection of 25 µg DNA, the average half-life of plasma IDUA was ~3.5-fold lower in +SB compared to −SB mice (90 versus 26 days) for both genders (Figure 1b), consistent with our previous results in C57BL/6 MPS I mice in which transposition led to prolonged higher levels of gene expression.24 However, at the 5 µg DNA dose there was no difference in the average half-life for plasma IDUA in either gender, ~100 days in males and 60 days in females. Thus, at the higher DNA doses there was a significant effect of transposition on the stability of IDUA expression. This conclusion was validated by our observations that on the C57BL/6 background, where the gender differences in IDUA activity were only about threefold, the decay rates in +SB mice were approximately twofold slower than in −SB mice for both sexes over the first 12 days, after which a rapid decline of IDUA activity occurred presumably due to adaptive immune responses35 (data not shown).
Biochemical and metabolic effects of treatment
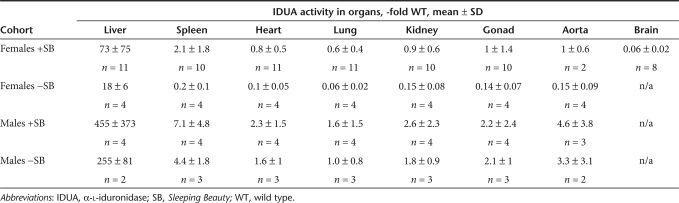
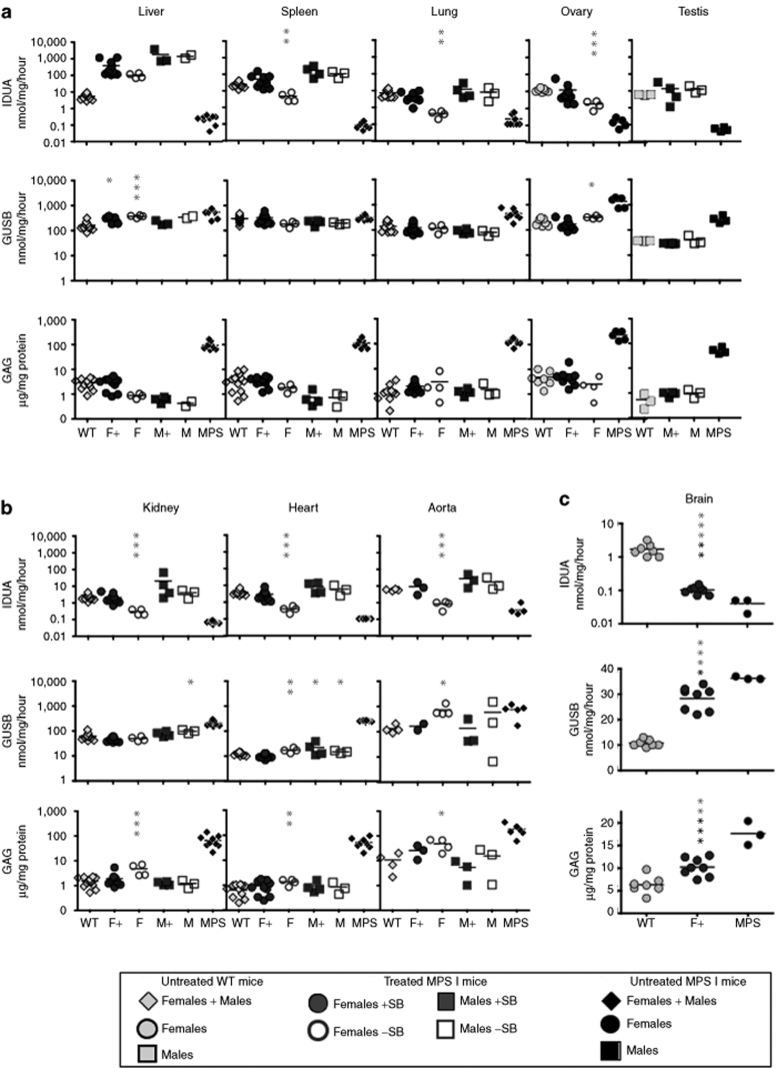
Mice injected with 25 µg transposon DNA were analyzed for therapeutic effects of SB-mediated IDUA delivery (Tables 1 and 2). In the liver, IDUA activity was more than tenfold wild-type (WT) levels in all females, >100-fold WT in all males, and on average about fourfold higher in +SB females than in −SB females but only 40% higher in +SB males than in −SB males. IDUA activity was detected in all examined organs in all treatment groups (Figure 3a–c). There was a decrease of the respective levels of β-glucuronidase (GUSB), another lysosomal enzyme involved in glycosaminoglycan (GAG) catabolism, with the exception of the spleen. Marked clearance of GAG accumulation was observed in all examined tissues of mice from all treatment groups. Restoration of IDUA activity in somatic organs correlated with IDUA expression in the liver (Tables 1 and 2). Liver IDUA in the +SB and −SB males was ~460-fold and 260-fold WT levels, respectively. IDUA levels in the spleen, heart, lung, kidney, gonads, and aorta were up to several-fold WT levels with complete correction of GAG storage. In +SB-treated females, liver IDUA activity was on average 70-fold WT activity and IDUA enzyme was near WT levels in somatic organs. However, in −SB females we observed only ~10% WT IDUA activities in somatic organs and incomplete GAG correction in the kidney, heart, and aorta (Figure 3b). Nevertheless, GAG levels in +SB females were reduced to normal in all organs examined (Figure 3 a,b), demonstrating the positive effect of transposition on correction of MPS I disease.
Table 2.
Restoration of IDUA activity in mouse organs 18 weeks after injection of 25 µg DNA
Figure 3.
Correction of metabolic disease in organs of NOD/SCID MPS I mice 18 weeks following injection of 25 µg DNA. Female and male mice were treated with 25 µg of transposon plasmid +SB (respectively, F+ and M+) or −SB (respectively, F− and M−). Clarified homogenates of the indicated organs were assayed for IDUA and GUSB enzyme activity and soluble GAG levels. (a) Organs apparently cleared of GAG storage: liver, spleen, lung, ovary, and gonads showed complete GAG correction in all treatment groups. (b) Organs incompletely cleared of GAG storage: kidney, heart, and aorta had complete correction in males and females receiving the full SB system (SB transposon + SB transposase), but incomplete GAG correction in −SB females. (c) Effects of treatment on the brain. Values are compared to the respective values in untreated NOD/SCID MPS I mice (black asterisks) and to WT values (gray asterisks). *P < 0.05; **P < 0.01; ***P < 0.001. IDUA activity was assayed with 4-methylumbelliferyl-α-L-iduronide and expressed as nmol 4 MU/mg protein/hour; GUSB activity (nmol 4 MU/mg protein/hour); GAG levels (µg GAG/mg protein). Individual values for each mouse are shown. Black horizontal dash indicates the mean level in each group. Filled dark circles, F+; open circles, F−; filled dark squares, M+ and open squares, M−. Diamonds: mixed group (males + females); gray, untreated WT NOD/SCID; black, untreated MPS I NOD/SCID mice. GAG, glycosaminoglycan; GUSB, β-glucuronidase; IDUA, α-L-iduronidase; MPS I, mucopolysaccharidosis type I; NOD/SCID, nonobese diabetic/severe combined immunodeficiency; SB, Sleeping Beauty; WT, wild type.
Remarkably, IDUA levels in the brain (Figure 3c, Supplementary Table S2) of treated MPS I mice were significantly above those in untreated mice (P = 0.0048) resulting in a reduction in GAG levels (P = 0.0007) and a reduction in GUSB levels (P = 0.03) in comparison to untreated affected mice. Treatment reduced the excess GAG level in the brain by ~66%, a level that was significantly different from respective values in unaffected mice (IDUA and GUSB, P < 0.0001; GAG, P = 0.002). The cellular source of IDUA activity in brain was not pursued.
Clinical effects of treatment
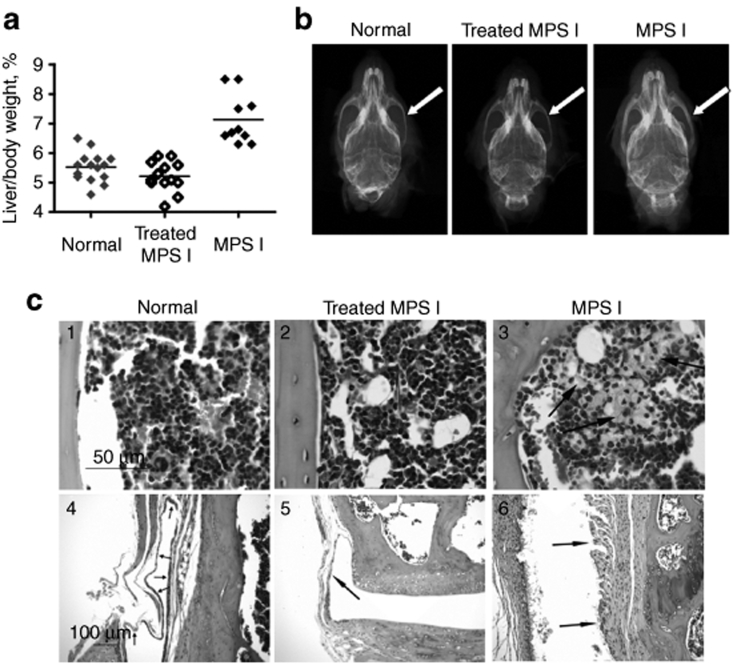
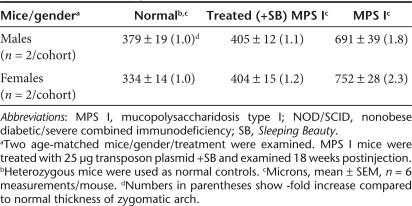
The ratio of liver weight to total body weight in untreated MPS I mice was significantly elevated compared with both normal and 25 µg plasmid-treated MPS I mice (Figure 4a). These results demonstrate that IDUA gene expression from SB transposons led to the correction of hepatomegaly in treated animals. Moreover, radiographic analysis showed correction of the thickening of the bone of the zygomatic arch, an abnormality in untreated NOD/SCID MPS I mice.30 Using digitized radiographs of isolated skulls, we found that untreated NOD/SCID MPS I mice had nearly twice the bone thickness compared with normal (heterozygous) animals and transposon-treated NOD/SCID MPS I mice (Figure 4b, Table 3). Longitudinal sections of femur and tibia revealed no consistent growth plate abnormalities among treatment groups. We did not observe consistent treatment differences in mid-diaphyseal bone areas, cortical thickness or diameters (measured in histological sections), although other investigators have reported partial correction in diaphyseal width increases (measured radiographically) in adult MPS I mice13,14 and almost complete correction in mice following neonatal gene therapy.11 We also did not observe osteocytes with unusually high lysosomal accumulations in our NOD/SCID MPS I mice, as was observed in immunocompetent MPS I mice.11 However, striking accumulations of cells with distended cytoplasms, presumed to represent accumulation of swollen lysosomes, were present within the bone marrow, synovium, periosteum, and skeletal muscle in sections of femur and tibia from the animals in the untreated NOD/SCID MPS I group in this study (Figure 4c). These were completely absent in sections from the treated MPS I and normal mice, strongly suggesting that the IDUA transposon treatment was successful in eliminating this pathological change.
Figure 4.
Correction of phenotypic characteristics in NOD/SCID MPS I mice. Mice treated with 25 µg transposon plasmids +SB were evaluated 18 weeks postinjection and compared to untreated, age-matched controls from the same mouse colony. (a) Correction of hepatomegaly in treated NOD/SCID MPS I mice. For evaluation of hepatomegaly, the entire carcass and the intact liver were weighed. The ratio of liver weight to total body weight in normal controls was 5.7 ± 0.2% (in females, n = 9) and 5.2 ± 0.3% (in males, n = 3). In MPS I mice the respective ratios were: 6.6 ± 0.1% (in females, n = 3) and 7.2 ± 0.3% (in males). Treated MPS I female and male mice had respective ratios of 5.1 ± 0.3% (n = 6) and 5.3 ± 0.1% (n = 7), which is significantly lower than in MPS I mice (P = 0.007 and P = 0.0002 in females and males, respectively). The values for males and females in each group were in the same range and therefore were pooled. Gray diamonds, untreated, normal (WT and heterozygous) controls; black open diamonds, treated NOD/SCID MPS I mice; black filled diamonds, untreated NOD/SCID MPS I mice. (b) Representative skull microradiographs from male mice. Untreated normal (heterozygous) control (left), treated MPS I (center), and untreated MPS I (right). The white arrows indicate increases in thickness of cranial zygomatic arches in the untreated MPS I mouse. Measurements are given in Table 3. (c) Representative sections of bone marrow (1–3) and synovium (4–6) from untreated normal (WT and heterozygous) control (left), treated MPS I (center), and untreated MPS I (right) animals. Arrows in panel 3 indicate clusters of foamy macrophages in the untreated MPS I mouse; these cells were absent in mice in the other two groups. Arrows in panels 4–6 indicate synovial membrane, which is normal in untreated normal (left) and treated MPS I (center) animals and is expanded by the presence of large numbers of foamy macrophages in the untreated MPS I animal (right). Hematoxylin and eosin stain. Bar = 50 µm (sections 1–3) and 100 µm (sections 4–6). MPS I, mucopolysaccharidosis type I; NOD/SCID, nonobese diabetic/severe combined immunodeficiency; SB, Sleeping Beauty; WT, wild-type.
Table 3.
Thickness of zygomatic arch in NOD/SCID mice
Discussion
Our study demonstrates that the SB transposon system is effective in ameliorating clinical manifestations of MPS I disease in an immunodeficient mouse model. Although >99% of transgene expression comes from the liver following hydrodynamic delivery,25,32 restoration of IDUA activity in somatic organs occurred through cross-correction that was dose-dependent on IDUA activity in the liver. Complete correction of GAG levels in tissues occurred in all +SB-treated animals and in −SB males. In contrast, the 20-fold WT IDUA activity in the liver of −SB females resulted in only ~10% WT IDUA activities in somatic organs and as a consequence incomplete GAG correction in kidney, heart, and aorta. These data demonstrate the positive effect of transposition on prolonged IDUA expression and correction of the disease in females at the dose of 25 µg/injection as well as the need to attain IDUA activity in the liver in the range of ~70 times WT levels to restore enzymatic activity in other organs. Gross necropsy of liver specimens revealed no adverse effects on liver tissue and treated mice appeared healthy and their life spans were not shortened. Detailed studies of overexpression of two other lysosomal enzymes, β-glucurunidase36 and arylsulfatase A,37 demonstrated that therapeutic levels of overexpression of these enzymes can be safely achieved, which suggests that there are no a priori reasons to suspect adverse effects following elevation of IDUA in the liver.
The reduction of GAG accumulation in the brain was consistent with the presence of low-level IDUA and indicated that the IDUA activity was not due to a contamination. This finding is important because IDUA enzyme levels as low as 2% of normal can be therapeutic.38 Although enzyme replacement therapy suggests IDUA will not traverse the blood–brain barrier in mice older than 2 weeks,39 our results support studies reporting IDUA activity in the brain in adult mice following liver-directed retroviral gene therapy13,14 and the breaching of the blood–brain barrier when brain capillaries are exposed to prolonged high levels of circulating enzyme.39,40
The NOD/SCID MPS I mice have a relatively short lifespan due to susceptibility to thymic lymphoma and other diseases (http://jaxmice.jax.org/strain/004083_3.html). Consequently, our “long-term” experiments did not extend beyond 4.5 months, which may be insufficient for full development of MPS I disease. As a result, we were not able to find differences in cardiac function between untreated normal mice and MPS I controls (data not shown). The milder MPS I phenotype that we observed in NOD/SCID mice may be due to attenuated inflammatory responses, which would be consistent with the hypothesis41 that the central consequence of GAG storage in MPS animals is inflammation. In addition, all mice produced by breeding NOD.129(B6)-Prkdcscid IDUAtm1Clk/J were profoundly deaf (data not shown), preventing the evaluation of auditory function.
Gender influence on transgene expression has been reported.42,43,44,45 Higher transgene expression levels in males were noted in mice treated with retroviral11 and AAV33 vectors, which led to different clinical outcomes for males and females. In our study, efficacious treatment of females required the complete transposon system for systemic metabolic correction (Figure 3a,b). We observed gender-related differences of IDUA expression that were striking at the 5 µg DNA dose. Our data demonstrating direct correlation of IDUA activity and transcriptional levels (Figure 2a,b) are most consistent with regulation of the CAGGS promoter being the cause of our unexpected findings. The recombinant enhancer motifs of the CAGGS promoter have been reported to have similar gender-specific attributes in rat liver.46 The gender effect may not be restricted to the CAGGS promoter. The expression from ubiquitin promoter also was reduced in some female mice (Figure 2c), resulting in insignificantly higher transposition rates in males than in females at both doses of DNA (Figure 2d), suggesting that the gender difference in IDUA expression was not due to reduced transposition in females.
The duration of gene expression from unintegrated plasmids in our NOD/SCID mice continued at high levels in contrast to in C57BL/6 mice where unintegrated gene expression drops 2–3 orders of magnitude within 2 weeks.24 Because there is no known mechanism that would lead to enhanced integration of plasmids into chromatin in NOD/SCID animals, we hypothesize that the prolonged transgene expression is coming from unintegrated plasmids, which is consistent with prolonged gene expression from unintegrated DNA in skeletal muscle of SCID mice.47 However, at the 25 µg DNA dose in NOD/SCID MPS I mice that received SB transposase, expression is better sustained following integration, suggesting that the dose of transgenic DNA may be even a larger issue than most researchers previously thought32 and that there may be unknown interactions that link dosage and gender effects.
Despite the limitations of immunodeficient MPS I mice, our study demonstrates the attractiveness of using the nonviral SB integrating vector to achieve IDUA levels and sustainability of enzymatic activity comparable to those observed with viral vectors and, as a consequence, of significantly ameliorating clinical manifestations of the disease.
Materials and Methods
Plasmids. The SB transposon plasmid for co-delivery of IDUA and SB genes, pT2/IDUA//Ub-SB11, has been described.24 In short, an expression cassette with the full-length hIDUA cDNA, regulated by the mini(m)CAGGS promoter, was cloned into a T248 transposon. The hyperactive SB11 transposase gene49 regulated by a UbC (ubiquitin C) promoter (pUB6/V5-His; Invitrogen, Grand Island, NY) was placed into the same plasmid as the transposon, a cis configuration. A negative control for transposition consisted of a plasmid containing the IDUA transposon, but lacking the SB transposase component (pT2/IDUA).
Animals. Immune-deficient heterozygous MPS I mice (NOD.129(B6)-Prkdcscid IDUAtm1Clk/J) were purchased from Jackson Laboratories (Bar Harbor, ME) and maintained as a homozygous breeding colony. All mice were housed under specific pathogen-free conditions in AAALAC-accredited facilities. The Institutional Animal Care and Use Committee of the University of Minnesota approved all animal studies. Plasmids were prepared commercially (Aldevron, Fargo, ND) and injected into mice using the hydrodynamics-based procedure as described.25 Blood was collected by retro-orbital phlebotomy under anesthesia. Animals were euthanized by carbon dioxide inhalation and perfused with saline. Selected organs and carcasses were harvested and preserved for analyses as specified below. Because NOD.129(B6)-Prkdcscid IDUAtm1Clk/J mice tend to develop thymic lymphomas (http://jaxmice.jax.org/strain/004083_3.html) that might interfere with interpretation of the experimental results, mice with tumors in the location of the thymus that were identified at the time of necropsy were excluded from the study (~15% mice including untreated MPS I and normal controls). No mice died due to the injection procedure; however, data were not used from mice in which the injection time exceeded 8 seconds.
Lysosomal enzyme quantification. Organs and plasma specimens were stored at −80 °C until used. Whole livers were pulverized by freezing in liquid nitrogen and grinding with mortar and pestle. Portions of each sample were extracted to assay for enzymatic activities and for gene copy number by quantitative PCR. IDUA and GUSB activities were measured in clarified supernatants of tissue homogenates and plasma using fluorometric assays as previously described,8 with 4-MU-α-L-iduronide (Glycosynth, Cheshire, England) and 4-MU-β-D-glucuronide (Sigma, St Louis, MO) as substrates. Protein concentration was determined using the Coomassie Blue assay as formulated by Bio-Rad (Bio-Rad Laboratories, Hercules, CA).
Quantification of tissue GAGs. Tissue GAGs were measured in the same clarified tissue homogenates as those used for enzyme assays, as described.30 In brief, after protein removal by digestion with Proteinase K and nucleic acid digestion with DNase and RNase, GAG concentration was determined using the Blyscan Sulfated Glycosaminoglycan Assay (Accurate Chemical and Scientific, Westbury, NY) and expressed as µg GAG/mg protein in each sample.
PCR analyses. DNA and RNA were isolated from pulverized whole-liver specimens. DNA was isolated from ~50 mg tissue by phenol–chloroform extraction (http://www.molecularcloning.com).
DNA copy number was determined by real-time PCR using an iCycler (Bio-Rad), as previously described.24 The mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) sequence, which served as an internal control of genomic DNA content, was amplified in a separate reaction. A GAPDH standard curve was generated from serial dilutions of mouse genomic DNA and a standard curve for SB transgene was prepared by serial dilutions of genomic DNA extracted from tissues of a Rosa 26-SB11 transgenic mouse harboring a single copy of the SB gene.50 Standard curves for IDUA gene and plasmid “EPs”34 were obtained as described.24 The mockEP used for preparation of EP standard curve was derived from the pT2/BH//spacer plasmid in which the transposon was deleted between the two flanking BamHI sites. The PCR primers were as described,24 with the exception of IDUA primers: 5′-AGGAGATACATCGGTAGG-3′ and 5′-TGTCAAAGTCGTGGTGGT-3′.
Total RNA was isolated using the TRIzol isolation system (Invitrogen Life Technologies, Carlsbad, CA); DNA-treated and purified using Turbo DNA-free kit (Ambion, Austin, TX). A one-step real-time reverse transcription–PCR was performed using EXPRESS One-Step SYBR GreenER Universal (Invitrogen) in a 20-µl reaction mix according to manufacturer's instructions. Reverse transcription–PCR conditions were: 50 °C for 5 minutes followed by 2 minutes at 95 °C and 40 cycles of 95 °C for 15 seconds then 58° C for 30 seconds and a melting curve was performed to verify specificity of amplification. The sense IDUA primer, specific to hIDUA, was 5′-GAAGGACTTGGTCTCCAG-3′; the anti-sense IDUA, SB and GAPDH primers were the same as above. Relative copy numbers of hIDUA and SB were calculated by correlating Ct values to standard curve produced by serial dilutions of a specimen with the highest IDUA activity and Rosa 26-SB11 mouse RNA, respectively, and normalized to murine GAPDH.
Pathology evaluations. For evaluation of the presence or absence of hepatomegaly, the entire carcass was weighed using a laboratory scale and the intact liver was weighed after removal from the carcass. Twelve NOD/SCID mice, two males and two females/cohort (treated with 25 µg DNA +SB and killed 18 weeks postinjection; age-matched untreated MPS I and heterozygote controls) were evaluated for bone changes. After removal of the internal organs, the carcasses were fixed in 10% neutral buffered formalin and were radiographed using anterio-posterior and lateral views with a cabinet radiography unit (Faxitron series; Hewlett Packard, McMinnville, OR) and high-detail film (X-Omat TL; Eastman Kodak, Rochester, NY). The radiographic technique included 45 kilovolt (peak) exposures, 3 milliamperes continuous for 30 seconds, and a film-to-source distance of 61 cm. After review of the films by a veterinary radiologist, the skull, one fore-, one hindlimb and the paws from these limbs were removed. These tissues (skulls, forelimbs, hindlimbs, and paws) were radiographed separately (dorsoventral views) for a more detailed assessment of these sites. All radiographs were then digitized.
Because there appeared to be treatment-related differences in the thickness of the zygomatic arches in these mice based on evaluation of the radiographs, measurements of these sites were made. In each digitized dorsoventral skull radiograph, one measurement of the width of each zygomatic arch (right and left) was made at three different locations (six measurements/mouse; three measurements/zygomatic arch): the midpoint of the arch, one rostral measurement and one caudal measurement. The latter two measurements were made at a distance of ~1.5 mm from the midpoint measurement; each was made at the point where the zygomatic arch began to widen to attach to either the maxilla or the base of the temporal bone. Measurements were made at high magnification using SPOT imaging and morphometry software (Diagnostic Instruments, Sterling Heights, MI).
To evaluate bone diameters, areas, and cortical thicknesses of long bones, one femur and humerus from each mouse (n = 12 mice; 24 long bones) were decalcified in EDTA and a mid-diaphyseal transverse section from each bone was embedded in paraffin, sectioned at 6 µm and stained with hematoxylin and eosin. Measurements of these sections were taken using a ×4 objective and SPOT imaging and morphometry software. The outer cortical perimeter and inner endosteal perimeter was measured in each section; cortical bone area was determined by subtracting the area of the marrow cavity from the total area. Cortical thicknesses (shortest distance from endosteum to periosteum) were measured in 8–10 equidistant sites that included the entire circumference of the bone in each section. Finally, cortical diameter was measured at its greatest width (periosteal surface to opposite periosteal surface) in each section.
For the evaluation of articular cartilage and growth plates, the opposite femur and tibia from six of these same mice (one male and one female from each treatment group) were decalcified in EDTA, processed routinely, and embedded longitudinally in paraffin. Sections were cut at 6 µm, stained with hematoxylin and eosin, and examined by a veterinary pathologist (C.S.C.).
Statistical analysis. Data were analyzed using GraphPad Prizm 4.0 software (GraphPad Software, San Diego, CA). The significance of differences between groups was determined based on exact two-tailed P values obtained with the nonparametric Mann–Whitney test. A value of P < 0.05 was considered statistically significant.
SUPPLEMENTARY MATERIALTable S1. IDUA enzymatic activity and transgene and excision product (EP) copy numbers in livers of treated NOD/SCID MPS I mice.Table S2. Biochemical effects of treatment in the brain of female mice 18 weeks following administration of 25 µg plasmid +SB.
Supplementary Material
IDUA enzymatic activity and transgene and excision product (EP) copy numbers in livers of treated NOD/SCID MPS I mice.
Biochemical effects of treatment in the brain of female mice 18 weeks following administration of 25 µg plasmid +SB.
Acknowledgments
We thank Elizabeth A. Braunlin for testing cardiac function and Vladimir Tsuprun and Sebahattin Cureoglu for testing auditory function of mice; Walter C. Low, and Ilze Matise for helpful discussions; Dan Wolf, Kelly Podetz-Pedersen, and Mayra Garcia-Rivera for assistance with experiments, and the Masonic Cancer Center Comparative Pathology Shared Resource for assistance with pathology assessments (University of Minnesota). This project was supported by grant P01 HD32652 from the National Institutes of Health (NIH) and other funding from Beckman Center for Transposon Research at the University of Minnesota. R.S.M. and P.B.H. have equity in a small biotech company that receives funding from the NIH to explore the use of the SB Transposon System for gene therapy.
REFERENCES
- Neufeld EF., and , Muenzer J.The mucopolysaccharidoses The Metabolic and Molecular Bases of Inherited Disease 20018th edn., vol. III. McGraw-Hill: New York; 3427–3436.In: Scriver, CR, Beaudet, AL, Sly, WS, Valle, D, Childs, B, Kinzler, KW et aleds [Google Scholar]
- Whitley CB, Belani KG, Chang PN, Summers CG, Blazar BR, Tsai MY, et al. Long-term outcome of Hurler syndrome following bone marrow transplantation. Am J Med Genet. 1993;46:209–218. doi: 10.1002/ajmg.1320460222. [DOI] [PubMed] [Google Scholar]
- Hodges BL., and , Cheng SH. Cell and gene-based therapies for the lysosomal storage diseases. Curr Gene Ther. 2006;6:227–241. doi: 10.2174/156652306776359522. [DOI] [PubMed] [Google Scholar]
- Wraith JE. Limitations of enzyme replacement therapy: current and future. J Inherit Metab Dis. 2006;29:442–447. doi: 10.1007/s10545-006-0239-6. [DOI] [PubMed] [Google Scholar]
- Ponder KP., and , Haskins ME. Gene therapy for mucopolysaccharidosis. Expert Opin Biol Ther. 2007;7:1333–1345. doi: 10.1517/14712598.7.9.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sands MS., and , Davidson BL. Gene therapy for lysosomal storage diseases. Mol Ther. 2006;13:839–849. doi: 10.1016/j.ymthe.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Beck M. New therapeutic options for lysosomal storage disorders: enzyme replacement, small molecules and gene therapy. Hum Genet. 2007;121:1–22. doi: 10.1007/s00439-006-0280-4. [DOI] [PubMed] [Google Scholar]
- Hartung SD, Frandsen JL, Pan D, Koniar BL, Graupman P, Gunther R, et al. Correction of metabolic, craniofacial, and neurologic abnormalities in MPS I mice treated at birth with adeno-associated virus vector transducing the human α-L-iduronidase gene. Mol Ther. 2004;9:866–875. doi: 10.1016/j.ymthe.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Di Domenico C, Di Napoli D, Gonzalez Y Reyero E, Lombardo A, Naldini L., and , Di Natale P. Limited transgene immune response and long-term expression of human α-L-iduronidase in young adult mice with mucopolysaccharidosis type I by liver-directed gene therapy. Hum Gene Ther. 2006;17:1112–1121. doi: 10.1089/hum.2006.17.1112. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Carbonaro D, Pepper K, Petersen D, Ge S, Jackson H, et al. Neonatal gene therapy of MPS I mice by intravenous injection of a lentiviral vector. Mol Ther. 2005;11:776–789. doi: 10.1016/j.ymthe.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Liu Y, Xu L, Hennig AK, Kovacs A, Fu A, Chung S, et al. Liver-directed neonatal gene therapy prevents cardiac, bone, ear, and eye disease in mucopolysaccharidosis I mice. Mol Ther. 2005;11:35–47. doi: 10.1016/j.ymthe.2004.08.027. [DOI] [PubMed] [Google Scholar]
- Watson G, Bastacky J, Belichenko P, Buddhikot M, Jungles S, Vellard M, et al. Intrathecal administration of AAV vectors for the treatment of lysosomal storage in the brains of MPS I mice. Gene Ther. 2006;13:917–925. doi: 10.1038/sj.gt.3302735. [DOI] [PubMed] [Google Scholar]
- Ma X, Liu Y, Tittiger M, Hennig A, Kovacs A, Popelka S, et al. Improvements in mucopolysaccharidosis I mice after adult retroviral vector-mediated gene therapy with immunomodulation. Mol Ther. 2007;15:889–902. doi: 10.1038/sj.mt.6300112. [DOI] [PubMed] [Google Scholar]
- Herati RS, Ma X, Tittiger M, Ohlemiller KK, Kovacs A., and , Ponder KP. Improved retroviral vector design results in sustained expression after adult gene therapy in mucopolysaccharidosis I mice. J Gene Med. 2008;10:972–982. doi: 10.1002/jgm.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn DB, Sadelain M., and , Glorioso JC. Occurrence of leukaemia following gene therapy of X-linked SCID. Nat Rev Cancer. 2003;3:477–488. doi: 10.1038/nrc1122. [DOI] [PubMed] [Google Scholar]
- Hackett CS, Geurts AM., and , Hackett PB. Predicting preferential DNA vector insertion sites: implications for functional genomics and gene therapy. Genome Biol. 2007;8 Suppl 1:S12. doi: 10.1186/gb-2007-8-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Reeves L, Sanburn N, Croop J, Williams DA., and , Cornetta K. Packaging cell line DNA contamination of vector supernatants: implication for laboratory and clinical research. Virology. 2001;282:186–197. doi: 10.1006/viro.2001.0826. [DOI] [PubMed] [Google Scholar]
- Essner JJ, McIvor RS., and , Hackett PB. Awakening gene therapy with Sleeping Beauty transposons. Curr Opin Pharmacol. 2005;5:513–519. doi: 10.1016/j.coph.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Hackett PB, Ekker SC, Largaespada DA., and , McIvor RS. Sleeping beauty transposon-mediated gene therapy for prolonged expression. Adv Genet. 2005;54:189–232. doi: 10.1016/S0065-2660(05)54009-4. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Hackett PB, Plasterk RH., and , Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Hackett PB. Integrating DNA vectors for gene therapy. Mol Ther. 2007;15:10–12. doi: 10.1038/sj.mt.6300065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Song Y., and , Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- Zhang G, Budker V., and , Wolff JA. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther. 1999;10:1735–1737. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- Aronovich EL, Bell JB, Belur LR, Gunther R, Koniar B, Erickson DC, et al. Prolonged expression of a lysosomal enzyme in mouse liver after Sleeping Beauty transposon-mediated gene delivery: implications for non-viral gene therapy of mucopolysaccharidoses. J Gene Med. 2007;9:403–415. doi: 10.1002/jgm.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JB, Podetz-Pedersen KM, Aronovich EL, Belur LR, McIvor RS., and , Hackett PB. Preferential delivery of the Sleeping Beauty transposon system to livers of mice by hydrodynamic injection. Nat Protoc. 2007;2:3153–3165. doi: 10.1038/nprot.2007.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izsvák Z., and , Ivics Z. Sleeping beauty transposition: biology and applications for molecular therapy. Mol Ther. 2004;9:147–156. doi: 10.1016/j.ymthe.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Clarke LA, Russell CS, Pownall S, Warrington CL, Borowski A, Dimmick JE, et al. Murine mucopolysaccharidosis type I: targeted disruption of the murine α-L-iduronidase gene. Hum Mol Genet. 1997;6:503–511. doi: 10.1093/hmg/6.4.503. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Rozengurt N, Ryazantsev S, Kohn DB, Satake N., and , Neufeld EF. Treatment of the mouse model of mucopolysaccharidosis I with retrovirally transduced bone marrow. Mol Genet Metab. 2003;79:233–244. doi: 10.1016/s1096-7192(03)00116-1. [DOI] [PubMed] [Google Scholar]
- Ponder KP. Immunology of neonatal gene transfer. Curr Gene Ther. 2007;7:403–410. doi: 10.2174/156652307782151434. [DOI] [PubMed] [Google Scholar]
- Garcia-Rivera MF, Colvin-Wanshura LE, Nelson MS, Nan Z, Khan SA, Rogers TB, et al. Characterization of an immunodeficient mouse model of mucopolysaccharidosis type I suitable for preclinical testing of human stem cell and gene therapy. Brain Res Bull. 2007;74:429–438. doi: 10.1016/j.brainresbull.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Chen YT, Bird A, Xu F, Hou YX, Amalfitano A, et al. Packaging of an AAV vector encoding human acid α-glucosidase for gene therapy in glycogen storage disease type II with a modified hybrid adenovirus-AAV vector. Mol Ther. 2003;7:467–477. doi: 10.1016/s1525-0016(03)00022-4. [DOI] [PubMed] [Google Scholar]
- Rossmanith W, Chabicovsky M, Herkner K., and , Schulte-Hermann R. Cellular gene dose and kinetics of gene expression in mouse livers transfected by high-volume tail-vein injection of naked DNA. DNA Cell Biol. 2002;21:847–853. doi: 10.1089/104454902320908496. [DOI] [PubMed] [Google Scholar]
- Sun B, Zhang H, Franco LM, Young SP, Schneider A, Bird A, et al. Efficacy of an adeno-associated virus 8-pseudotyped vector in glycogen storage disease type II. Mol Ther. 2005;11:57–65. doi: 10.1016/j.ymthe.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Liu G, Aronovich EL, Cui Z, Whitley CB., and , Hackett PB. Excision of Sleeping Beauty transposons: parameters and applications to gene therapy. J Gene Med. 2004;6:574–583. doi: 10.1002/jgm.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman RR, Pirrotta V., and , Geyer PK. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 1993;12:435–442. doi: 10.1002/j.1460-2075.1993.tb05675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler C, Galvin N, Levy B, Grubb J, Jiang J, Zhou XY, et al. Transgene produces massive overexpression of human β-glucuronidase in mice, lysosomal storage of enzyme, and strain-dependent tumors. Proc Natl Acad Sci USA. 2003;100:2669–2673. doi: 10.1073/pnas.0437941100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capotondo A, Cesani M, Pepe S, Fasano S, Gregori S, Tononi L, et al. Safety of arylsulfatase A overexpression for gene therapy of metachromatic leukodystrophy. Hum Gene Ther. 2007;18:821–836. doi: 10.1089/hum.2007.048. [DOI] [PubMed] [Google Scholar]
- Donsante A, Levy B, Vogler C., and , Sands MS. Clinical response to persistent, low-level β-glucuronidase expression in the murine model of mucopolysaccharidosis type VII. J Inherit Metab Dis. 2007;30:227–238. doi: 10.1007/s10545-007-0483-4. [DOI] [PubMed] [Google Scholar]
- Vogler C, Levy B, Grubb JH, Galvin N, Tan Y, Kakkis E, et al. Overcoming the blood-brain barrier with high-dose enzyme replacement therapy in murine mucopolysaccharidosis VII. Proc Natl Acad Sci USA. 2005;102:14777–14782. doi: 10.1073/pnas.0506892102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb JH, Vogler C, Levy B, Galvin N, Tan Y., and , Sly WS. Chemically modified β-glucuronidase crosses blood-brain barrier and clears neuronal storage in murine mucopolysaccharidosis VII. Proc Natl Acad Sci USA. 2008;105:2616–2621. doi: 10.1073/pnas.0712147105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonaro CM, D'Angelo M, He X, Eliyahu E, Shtraizent N, Haskins ME, et al. Mechanism of glycosaminoglycan-mediated bone and joint disease: implications for the mucopolysaccharidoses and other connective tissue diseases. Am J Pathol. 2008;172:112–122. doi: 10.2353/ajpath.2008.070564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidoff AM, Ng CY, Zhou J, Spence Y., and , Nathwani AC. Sex significantly influences transduction of murine liver by recombinant adeno-associated viral vectors through an androgen-dependent pathway. Blood. 2003;102:480–488. doi: 10.1182/blood-2002-09-2889. [DOI] [PubMed] [Google Scholar]
- Chen L, Thung SN., and , Woo SL. Metabolic basis of sexual dimorphism in PKU mice after genome-targeted PAH gene therapy. Mol Ther. 2007;15:1079–1085. doi: 10.1038/sj.mt.6300137. [DOI] [PubMed] [Google Scholar]
- Nathwani AC, Cochrane M, McIntosh J, Ng CY, Zhou J, Gray JT, et al. Enhancing transduction of the liver by adeno-associated viral vectors. Gene Ther. 2009;16:60–69. doi: 10.1038/gt.2008.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober C, Loisel DA., and , Gilad Y. Sex-specific genetic architecture of human disease. Nat Rev Genet. 2008;9:911–922. doi: 10.1038/nrg2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sereemaspun A, Takeuchi K, Sato Y, Iwamoto S, Inakagi T, Ookawara S, et al. Testosterone-dependent transgene expression in the liver of the CAG-lacZ transgenic rat. Gene Expr. 2005;12:305–313. doi: 10.3727/000000005783992016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Laipis PJ, Berns KI., and , Flotte TR. Effect of DNA-dependent protein kinase on the molecular fate of the rAAV2 genome in skeletal muscle. Proc Natl Acad Sci USA. 2001;98:4084–4088. doi: 10.1073/pnas.061014598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z, Geurts AM, Liu G, Kaufman CD., and , Hackett PB. Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J Mol Biol. 2002;318:1221–1235. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- Geurts AM, Yang Y, Clark KJ, Liu G, Cui Z, Dupuy AJ, et al. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Dupuy AJ, Akagi K, Largaespada DA, Copeland NG., and , Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
IDUA enzymatic activity and transgene and excision product (EP) copy numbers in livers of treated NOD/SCID MPS I mice.
Biochemical effects of treatment in the brain of female mice 18 weeks following administration of 25 µg plasmid +SB.