Abstract
Lentiviral vectors enter cells with high efficiency and deliver stable transduction through integration into host chromosomes, but their preference for integration within actively transcribing genes means that insertional mutagenesis following disruption of host proto-oncogenes is a recognized concern. We have addressed this problem by combining the efficient cell and nuclear entry properties of HIV-1–derived lentiviral vectors with the integration profile benefits of Sleeping Beauty (SB) transposase. Importantly, this integration enzyme does not exhibit a preference for integration within active genes. We generated integrase-deficient lentiviral vectors (IDLVs) to carry SB transposon and transposase expression cassettes. IDLVs were able to deliver transient transposase expression to target cells, and episomal lentiviral DNA was found to be a suitable substrate for integration via the SB pathway. The hybrid vector system allows genomic integration of a minimal promoter-transgene cassette flanked by short SB inverted repeats (IRs) but devoid of HIV-1 long terminal repeats (LTRs) or other virus-derived sequences. Importantly, integration site analysis revealed redirection toward a profile mimicking SB-plasmid integration and away from integration within transcriptionally active genes favored by integrase-proficient lentiviral vectors (ILVs).
Introduction
Sleeping Beauty (SB) is a Tc1/mariner-type DNA transposon originally regenerated from extinct transposons found in salmonid fish.1 Wild-type transposons consist of the coding sequence for a 340 amino-acid transposase protein flanked by two nonidentical 230 bp inverted repeat (IR)-direct repeats that contain 32–34 bp transposase binding sites.2 SB has been developed as a vector by substituting the transposase coding sequence with a transgene expression cassette. Following delivery to cells, the transposase protein is provided in trans to mediate cut-and-paste transposition of the transgene into the target cell genome. Cleavage is dependent upon the presence of flanking TA dinucleotides and is enhanced when the transposon is flanked by TATA motifs.3 SB integration occurs exclusively at TA dinucleotides, and DNA repair following integration results in a duplication signature with TA dinucleotides on either side of the transposon.4,5 Importantly, integration occurs within genes at a frequency close to that expected from random integration and is not biased toward actively transcribing genes.6,7 Thus, unlike HIV-1 that has been shown to integrate preferentially within genes (~70% of sites) and is strongly biased toward actively transcribing genes,8 SB integration may be less likely to cause adverse effects. However, as a plasmid-based system, SB lacks the advantages of lentiviral vectors in terms of efficient cell entry and nuclear translocation. Combining transposase-mediated integration with lentiviral delivery could produce highly attractive vectors for gene therapy of mitotic cells that can be stably altered through the transient expression of transposase. We have designed integrase-deficient lentiviral vectors (IDLVs) that incorporate an IR-flanked transgene expression cassette for transposition or express the transposase protein from episomal lentiviral DNA. We provide proof-of-principle data confirming that transposition from IDLVs is achievable and results in a characteristic TA dinucleotide integration signature, and report that transposition is restricted to a defined range of transposase concentrations. The system allows delivery of an IR-flanked expression cassette and avoids genomic integration of HIV-1 long terminal repeats (LTRs) or other virus-derived sequences such as the woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) that is often included in lentiviral vectors to improve vector titer.9 We also show that hybrid vector integration via SB transposition results in a reduced frequency of integration into active genes relative to integrase-proficient lentiviral vectors (ILVs).
Results
IDLVs for transposon delivery and transient transposase expression
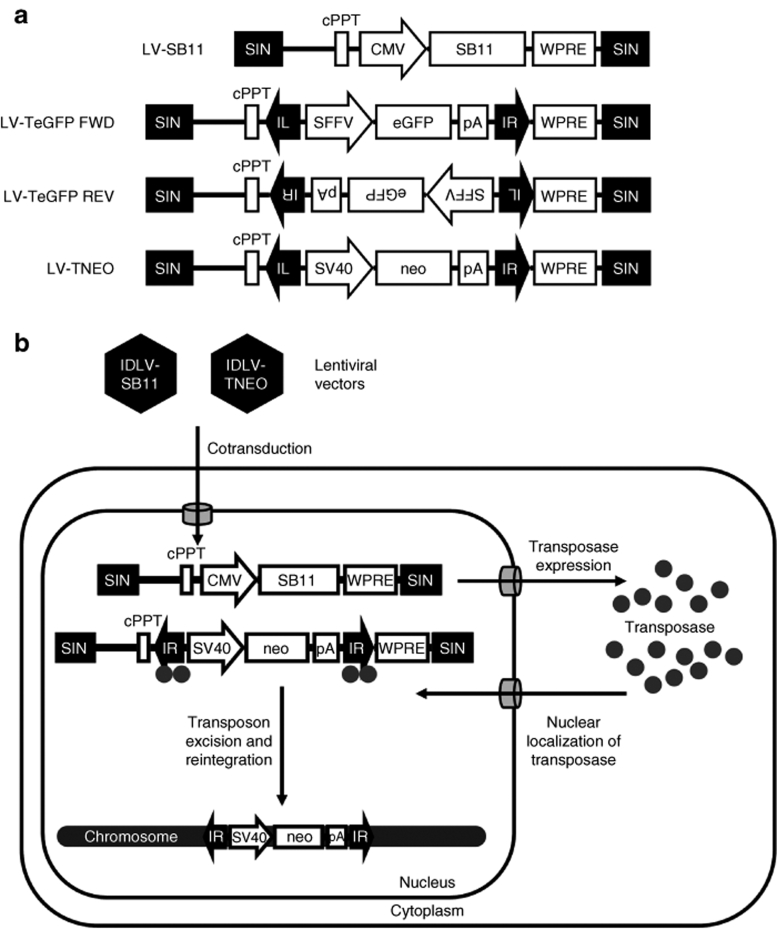
Consideration was given to the configuration of lentiviral constructs incorporating a SB transposon. Incorporation of a transposon into a lentiviral vector backbone has the potential to truncate vector genome transcription in producer cells at a previously identified polyadenylation signal in the right IR of the transposon.10 This could have caused loss of downstream elements, including the 3′-HIV-LTR. To investigate this possibility, we constructed IDLVs in which a transposon (T) containing an enhanced green fluorescent protein (eGFP) expression cassette was inserted in either the forward or reverse orientation with respect to the lentiviral backbone (IDLV-TeGFP forward and IDLV-TeGFP reverse, Figure 1a). Vector titer was determined by transduction of HEK293T cells and quantification of eGFP-positive cells by flow cytometry after 48 hours. Interestingly, vector titers were notably reduced when the transposon was in the reverse orientation (9.9 × 105 transducing units/ml) relative to the lentiviral backbone compared to the forward orientation (3.4 × 108 transducing units/ml), and thus the forward orientated constructs were used in all subsequent experiments.
Figure 1.
Constructs and experimental strategy. (a) Lentiviral vector constructs. LV-SB11 carries a Sleeping Beauty transposase expression cassette. LV-TeGFP FWD and LV-TeGFP REV carry a Sleeping Beauty transposon containing an eGFP expression cassette that is in the sense and antisense orientations respectively. LV-TNEO carries a Sleeping Beauty transposon containing a neomycin phosphotransferase expression cassette in the sense orientation. CMV, immediate early promoter of human cytomegalovirus; cPPT, central polypurine tract; eGFP, enhanced green fluorescent protein; FWD, forward; IL, Sleeping Beauty transposon left inverted repeat; IR, Sleeping Beauty transposon right inverted repeat; LV, lentiviral vector; neo, neomycin-phosphotransferase gene; pA, SV40 polyadenylation signal; REV, reverse; SB11, hyperactive Sleeping Beauty transposase; SFFV; spleen focus forming virus LTR promoter; SIN, self-inactivating (U3-deleted) HIV-1 long terminal repeat; SV40, simian virus 40 promoter; TeGFP, transposon with enhanced green fluorescent protein; TNEO, transposon with neomycin resistance; WPRE, woodchuck posttranscriptional regulatory element. (b) Experimental strategy used in this study. Cells are cotransduced with integrase-deficient lentiviral vectors carrying the Sleeping Beauty transposase and transposon. Transposase protein is expressed and localized to the nucleus where it binds to the transposon inverted repeats and catalyzes excision of the transposon from episomal lentiviral DNA. The excised transposon is mobile and able to subsequently reintegrate elsewhere, for example, into a host cell chromosome.
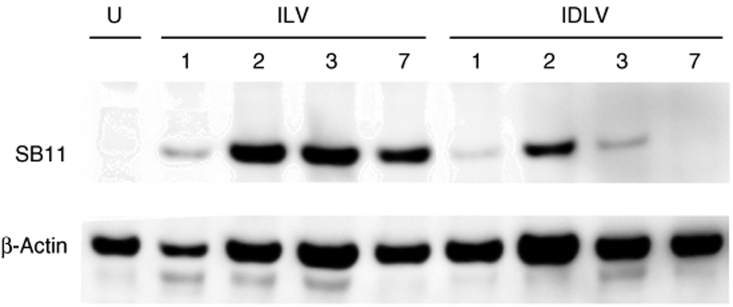
Although incorporation of transposon and transposase elements into the same plasmid vector has been reported,11 we reasoned that transposase expression from a separate, independent lentiviral vector (Figure 1b) would enable optimization of the transposon-transposase ratio in target cells. It was also necessary to demonstrate transience of transposase expression in order to minimize the risk of transposon remobilization. IDLVs are able to mediate transient transgene expression in dividing cell populations through dilution of the episomal vector genome copy number during cell division.12 We generated an ILV and an IDLV for transposase expression, ILV-SB11 and IDLV-SB11, and analyzed transposase expression over time by western blotting of samples from HeLa cells transduced with these vectors (Figure 2). The level of transposase expression mediated by ILV-SB11 was highest after 2 days and as expected remained stable thereafter. When expressed from IDLV-SB11, transposase was readily detectable within 2 days of transduction but subsequently declined, becoming undetectable by day 7. Thus, transient transposase expression can be achieved by IDLV delivery, and this should minimize the risk of subsequent remobilization of the transposon cassette.
Figure 2.
Western blot for expression of transposase from lentiviral vectors. Integrase-proficient ILV-SB11 and integrase-deficient IDLV-SB11 transposase expression vectors were prepared in parallel and concentrated by ultracentrifugation. 106 HeLa cells were transduced with 0.5 µg p24 of vector per well. At 1, 2, 3, and 7 days post-transduction, cells were trypsinized and pellets of equal cell number were frozen for subsequent determination of protein expression by western blot. IDLV, integrase-deficient lentiviral vector; ILV, integrase-proficient lentiviral vector; U, untransduced.
Optimization of transposase expression enhances transposition from a hybrid SB-lentivirus vector
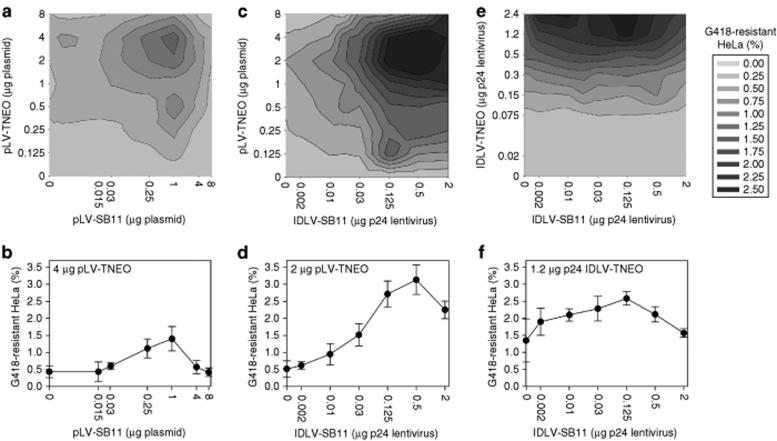
It has been previously reported that the optimal rate of SB transposition from plasmid DNA occurs within a defined transposase concentration range.13 To investigate this in the context of SB-IDLV hybrid vectors, we used an integration assay1 in which a neomycin phosphotransferase cassette for G418 selection replaced the eGFP cassette (pLV-TNEO, Figure 1a). In order to determine the optimal conditions for transposition from an IDLV backbone, double titration experiments were performed in HeLa cells in which both transposon and transposase levels were varied and integration frequency was determined by G418-resistant colony counting (Figure 3). Transfection of both transposon and transposase plasmids resulted in gene marking of 1.4 ± 0.4% at the optimal transposase concentration, with a background (no transposase) rate of 0.4 ± 0.2% (Figure 3a,b). Transposition from transposon plasmid driven by IDLV-SB11 led to optimal gene marking of 3.1 ± 0.4% (background 0.5 ± 0.2%) (Figure 3c,d). When both transposon and transposase were delivered by IDLV transduction, the highest rate of gene marking observed was 2.6 ± 0.2% with background integration of 1.35 ± 0.6% (Figure 3e,f). These data confirm the feasibility of IDLV-mediated delivery of the SB transposon and transposase to target cells. Double titration of transposase and transposon template revealed optimal ranges for transposase activity and suggest that at very high levels, there may be inhibitory or toxic phenomena that reduce transposition.
Figure 3.
Efficiency of chromosomal integration. 105 HeLa cells were transduced or transfected with Sleeping Beauty components in a double titration integration assay. The transposon amounts tested are given by the y-axis labels in a, c, and e, whereas the transposase amounts are given by the x-axis labels. All transposon-transposase combinations were tested, and each combination was tested in triplicate. (a) Plasmid transposon, plasmid transposase; (b) cross-section at transposon plasmid mass was 4 µg; (c) plasmid transposon, IDLV transposase; (d) cross-section at transposon plasmid mass was 2 µg; (e) IDLV transposon, IDLV transposase; (f) cross-section where the SB-IDLV vector dose was 1.2 µg p24. The rate of integration was assessed by the number of G418-resistant colonies formed and is expressed as a percentage of the transduced or transfected cell number assuming 100% plating efficiency. IDLV, integrase-deficient lentiviral vector; SB11, hyperactive Sleeping Beauty transposase.
Characteristic TA signatures confirm SB transposition from IDLVs
G418-resistant HeLa cell colonies were produced by transduction with ILV-TNEO or IDLV-TNEO alone, IDLV-TNEO plus IDLV-SB11, or transfection with plasmids pLV-TNEO plus pLV-SB11 under the previously optimized conditions. Integration sites were recovered from surviving colonies using ligation-mediated PCR. ILV-specific primers in the HIV-1 LTR were used for cells transduced with ILV-TNEO or IDLV-TNEO alone. Transposon-specific primers in the transposon IR were used for cells transfected with the two SB-plasmids or transduced with both SB-IDLV vectors.
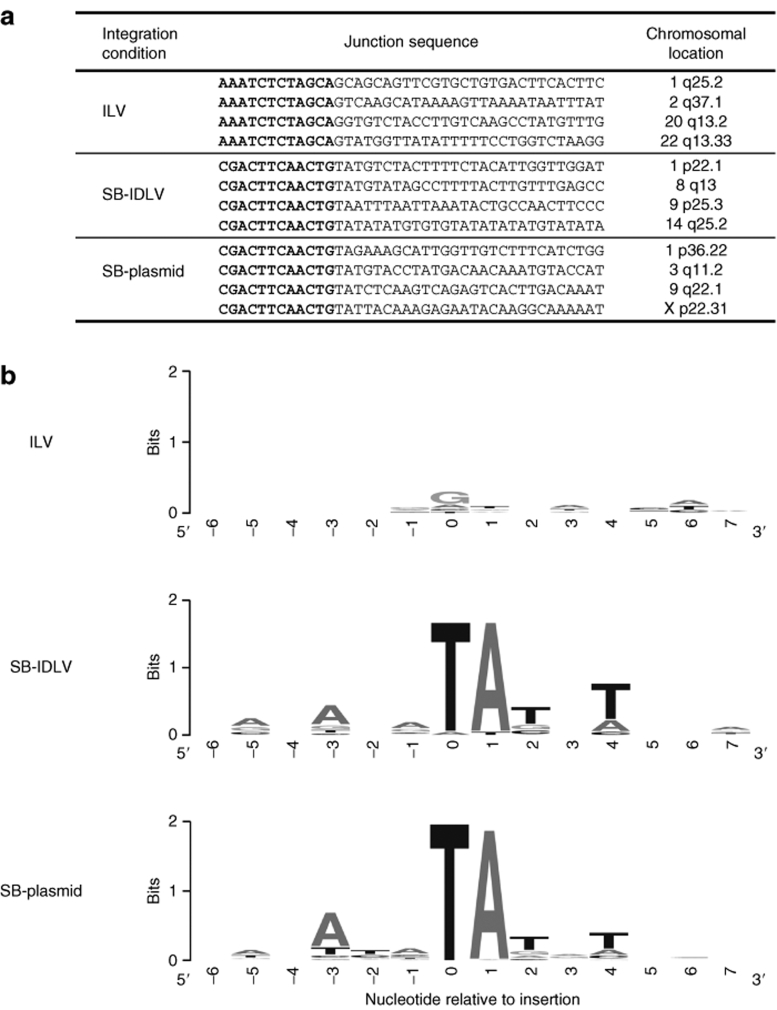
A BLAT search of the recovered sequences against the University of California at Santa Cruz human genome was used to identify junctions between chromosomal DNA and the transposon IR or the viral 3′-LTR. In total, 752 integration sites mapping to unique genomic positions were obtained from cells transfected with the two SB-plasmids (SB-plasmid sites), and transposase-mediated integration was confirmed by the presence of a characteristic TA dinucleotide signature in the chromosomal DNA immediately flanking the integrated transposon (Supplementary Table S1 and representative examples shown in Figure 4a). Similar transposon-chromosome junction signatures were found at 161 sites from cells transduced with both SB-IDLVs (SB-IDLV sites), confirming transposition events. In addition, 976 LTR-chromosome junctions were isolated from cells transduced with ILV-TNEO, and only 10 such sites were recovered from cells transduced with IDLV-TNEO alone. Background integration of IDLVs has been previously shown to result in different junctions to those produced by integrase-mediated integration.14
Figure 4.
Primary sequence at integration sites. (a) Sample junction sequences of integration sites recovered by ligation-mediated PCR. The beginning of the flanking chromosomal sequence is shown for illustration. Virus LTR or transposon IR sequence is shown in bold, and flanking chromosomal DNA is shown in normal type. (b) A sequence logo was generated using the WebLogo tool to identify preferred base pair usage at the three integration site types. Position 0 denotes the first base of the flanking chromosomal sequence 3′ of the integration site. ILV, integrating lentiviral vector; SB-IDLV, hybrid Sleeping Beauty–integrase-deficient lentiviral vector; SB-plasmid, Sleeping Beauty plasmid vector.
The primary DNA sequence immediately flanking each site was scanned for base composition using the WebLogo sequence logo tool15 (Figure 4b). Transposon integration sites recovered from both SB-IDLV and SB-plasmid cells showed, in addition to the ubiquitous flanking TA dinucleotide, a weak palindromic integration site of the form 5′-ANA(TA)TNT-3′ (ref. 6). By contrast, ILV integration sites showed a weak preference for a GT dinucleotide at the first two bases downstream of the integration site, as has been previously reported.16 Overall, these results provide evidence that episomal lentiviral DNA can act as a suitable substrate for SB transposition.
Integration site profile of SB transposition from IDLVs
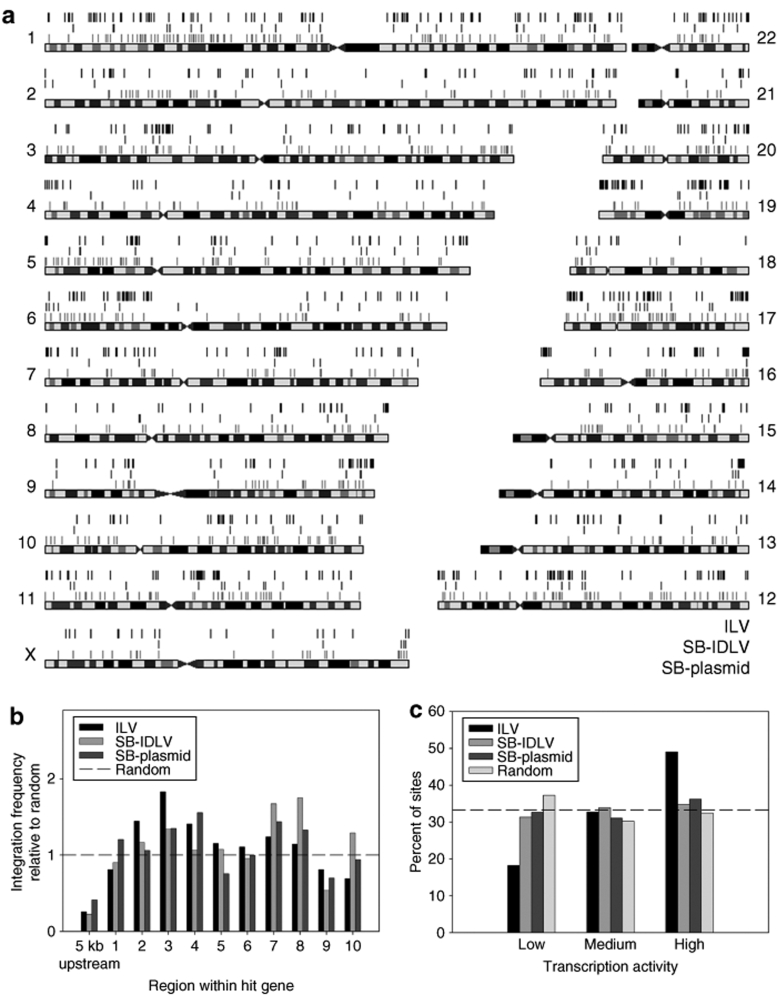
Mapping integration sites with respect to chromosomes demonstrated that all three delivery systems integrated widely across the entire genome (Figure 5a). Bioinformatic comparison of integration sites at the gene level revealed three key features of hybrid vector integration.
Figure 5.
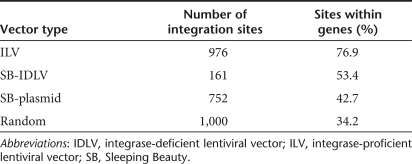
Integration profiles of vector types. 105 HeLa cells per well were transduced with ILV-TNEO at multiplicity of infection 0.01 (ILV sites, top line), transduced with 1.2 µg p24 IDLV-TNEO and 0.13 µg IDLV-SB11 (SB-IDLV sites, middle line), or transfected with 4 µg pLV-TNEO and 1 µg pLV-SB11 (SB-plasmid sites, bottom line). Cells were incubated in nonselective medium for 3 days followed by a two-week incubation in medium containing 1 mg/ml G418. Genomic DNA was extracted and transposon-chromosome or lentivirus-chromosome junctions were recovered by ligation-mediated PCR and sequenced. (a) Sequences were mapped to the University of California at Santa Cruz (UCSC) human genome by BLAT search and integration sites were depicted relative to chromosomes using the UCSC Genome Graphs tool. Note that HeLa cells are karyotypically abnormal. (b) Intragenic position of integration sites within genes. RefSeq genes containing integration sites were divided by length into 10 equally sized regions and a 5 kb upstream region, and the proportion of integration sites within each region was counted. To allow statistical comparison of integration preferences with average genomic content, 1,000 random chromosomal sites were generated by multiplying the total length of the genome by a random number between 0 and 1 and converting this value to a chromosomal coordinate. Vector integration frequencies are expressed relative to the proportion of random sites within each region. (c) Transcriptional activity of genes containing integration sites. All RefSeq genes were scored for transcription in HeLa cells using a published microarray dataset. All genes were then assigned to one of three transcription levels (containing equal numbers of genes) to give low, medium, and highly transcribed genes. Integration sites within genes were then scored according to whether the hit gene was transcribed at a low, medium, or high level. For each vector type, the number of intragenic sites per transcription level is expressed as a percentage of the total number of intragenic sites. A dashed line at 33.3% of sites is included to show theoretically equal distribution of sites between the transcription levels. ILV, integrating lentiviral vector; SB-IDLV, hybrid Sleeping Beauty–integrase-deficient lentiviral vector; TNEO, transposon with neomycin resistance.
1. The proportion of ILV integration sites within RefSeq genes was 77%, much greater than that for SB-IDLV (53%) or SB-plasmid (43%) (P < 0.01). After generating 1,000 random integration sites by bioinformatics, we estimated the expected rate of random integration within genes to be 34% (Table 1). Thus, both SB-plasmid and SB-IDLV integration exhibited a smaller but significant bias (P < 0.01) toward genes, and this phenomenon has been previously described for SB-plasmid systems.6
Table 1.
Integration profiles of vectors relative to RefSeq genes
2. When integrations within genes were mapped relative to their position within the gene or upstream region (Figure 5b), no bias toward transcription start sites was detected and no significant variation in integration pattern was observed along the length of the gene.
3. Lentiviral vector integration occurs preferentially within transcriptionally active genes.17 We estimated the transcription activity of all RefSeq genes in HeLa cells using a published HeLa microarray transcriptome. All genes were then categorized as having low, medium, or high transcription activity (each level containing equal numbers of genes). When considering only genes containing integration sites, ILV integration exhibited a clear preference for genes with high levels of transcriptional activity (P < 0.01), whereas SB-IDLV and SB-plasmid integration showed no bias toward any particular level of transcription, and resembled the profile generated for random integration events (Figure 5c).
Discussion
Integrating vectors for gene therapy can provide sustained transgene expression in target cells, but carry a risk of disrupting host proto-oncogenes or tumor suppressor genes and contributing to cell transformation.18,19 The integration site preferences of most vector types have now been characterized. γ-Retroviral vectors integrate preferentially near transcription start sites, whereas lentiviral vectors show a preference for integration within actively transcribed genes.8,20 Adeno-associated virus vectors integrate preferentially within CpG islands and the first 1 kb of genes.21 By contrast, SB transposon has a near random integration profile with a low preference for genes and transcription start sites.6,7
One approach to addressing the problem of insertional mutagenesis has been the development of IDLVs. In these vectors, mutations within the integrase catalytic DDE domain (including D64V) result in failure of integrase-mediated integration and diversion toward circularization pathways. Transgene expression from IDLV vectors has been demonstrated in a variety of tissues and can be sustained over an extended period in postmitotic cells.12,22 However, in mitotic cells, there is dilution of nonintegrated, nonreplicating viral DNA and thus a gradual loss of gene expression. In this study, we have further developed IDLVs with an approach that would be particularly useful in mitotic cells, which is to replace integration of lentiviral DNA by the HIV-1 integrase with integration by the SB transposase. We hypothesized that the resultant integration pattern would reflect that of the SB transposase and avoid the integration bias of lentiviral vectors toward transcriptionally active, gene dense regions of the genome. In addition, this approach would reduce the length of integrated genetic material to an essential promoter-transgene cassette flanked by the short (230 bp) transposon IRs, preventing chromosomal integration of virus-derived sequences such as the HIV-1 LTR and the WPRE.
The concept of combining the cell entry properties of viruses and the integration machinery of SB has been previously proposed and investigated by Yant et al. using adenoviral vectors.23 These authors reported that transposition from an adenoviral template required precircularization of the vector genome by incorporating a Flp/FRT recombinase system within the viral backbone. In this context, IDLVs have an important advantage inherent to their life cycle. Following cell entry and reverse transcription, double-stranded lentiviral DNA persists in an episomal form, either as linear DNA or as circles produced by homologous recombination or nonhomologous end joining of the viral LTRs. Circular forms account for around 20–30% of recoverable viral genomes, are highly stable and can persist indefinitely in postmitotic cells.14 The concept of transposition from circularized lentiviral-SB hybrid constructs has also recently been investigated by Staunstrup et al. using an IR configuration designed to allow transposition of an entire HIV-LTR flanked circularized proviral DNA.24 Although transposition was demonstrated, this configuration resulted in genomic integration of virus-derived sequences including the HIV-LTRs and WPRE. In contrast, we have demonstrated that the lentiviral backbone tolerated incorporation of a conventional IR-flanked SB transposon and retained high vector titer. Interestingly, orientating the transposon in the sense configuration produced a superior vector titer despite the presence of a polyadenylation signal in the right IR. IDLVs were also used to achieve transient expression of the transposase protein in dividing cells in culture. It has been previously reported that optimization of the transposon-transposase ratio is necessary to maximize the rate of transposase-mediated integration,13 so in this study, the quantities of both components delivered to target cells were titrated in order to identify the optimal conditions for SB transposition from an IDLV backbone. In cells where the level of transposase expression was optimal, transposition was dependent on the amount of transposon DNA available. Increasing the multiplicity of transposon IDLV infection resulted in higher levels of transposition. We also found that there is a restricted window of transposase expression beyond which transposition appears to be inhibited, as has been reported previously.13 It is notable that the level of gene marking observed with the hybrid vector, in the range of 1–3% G418-resistant HeLa cells, was similar to that reported previously with plasmid-based SB systems.13 Strategies to improve the efficiency of the hybrid system might include increasing the multiplicity of IDLV transduction in order to provide higher levels of transposon donor DNA, or increasing the proportion of transposon donor DNA that is in the correct physical conformation for transposition, perhaps through active IDLV circularization by the Flp or Cre recombinases.23 Finally, recently developed hyperactive transposases may increase the rate of stable integration, as has been recently reported.24
We provide proof-of-principle evidence that IDLVs can act as a suitable template for SB transposition, and that IDLV-mediated transposase expression is restricted to a limited time period in mitotic cells. It is clear from our bioinformatic comparison of integration sites that the integration profile of the hybrid vector is significantly different from that of an integrating lentiviral vector and closely resembles that of the plasmid-based SB vector. In theory, this should reduce the risk of insertional mutagenesis compared to conventional integrating lentiviral vectors, but this will need to be further characterized using defined assays for insertional mutagenesis.25,26 In common with other episomal vectors, host DNA repair pathways can mediate low-level background integration of IDLVs, albeit at higher levels than that detected from SB-plasmid vectors. It may be that linearized forms of viral DNA is more efficiently integrated than circularized plasmid DNA.27 Although we do not anticipate that such background integration events will result in any significant risk of transposon mobilization, it must be noted that deliberate ectopic expression of transposase has been used to mediate mutagenic effects as part of cancer gene discovery studies.28,29,30 Safety issues linked to residual background integration could be addressed by linking transposase expression in the IDLV to a suicide gene, as this could enable prodrug killing of cells containing full-length integrants. Further studies will be required to investigate issues such as possible promoter silencing and loss of transgene expression at sites of transposase-mediated integration, as reported in studies using SB-plasmids.31 Ultimately, it is feasible that the transposon and transposase expression cassettes could both be incorporated into a single IDLV vector. Such hybrid vector systems would be well suited for disorders where cells are dividing and gene correction confers a strong survival advantage. Transduction of cells permissive to lentiviral vectors but not plasmid-based systems, such as hematopoietic stem cells, would be particularly attractive.32 One good target disease would be X-linked severe combined immunodeficiency, as stable gene transfer of the interleukin common γ-chain to a relatively small number of hematopoietic stem cells is sufficient to support effective immune reconstitution, and long-term hematopoietic reconstitution in tumor-prone models would be useful for evaluating the risk of insertional mutagenesis following SB transposition compared to existing lentiviral and γ-retroviral vectors.
Materials and Methods
Plasmids. pT2/SVNeo, pT2/HB (ref. 3), and pCMV-SB11 (ref. 13) are enhanced versions of the original SB system as previously described.1 The lentiviral backbone is a previously described variant of pHR containing a self-inactivating 3′-LTR, a central polypurine tract, and the WPRE with a mutated X-protein start codon.9,33 To produce pT2/SFFV-eGFP, the SFFV-eGFP fragment from pHR/SIN-SEW33 was cloned into pT2/HB between the EcoRI and XbaI sites. The BamHI transposon fragment from pT2/SFFV-eGFP was ligated into the BamHI site in an empty lentiviral backbone between the central polypurine tract and the WPRE to produce pLV-TeGFP forward and pLV-TeGFP reverse. Repeating this with the BamHI transposon fragment from pT2/SVNeo gave pLV-TNEO. The CMV-SB11 fragment from pCMV-SB11 was cloned into the lentiviral backbone between the EcoRI and KpnI sites to produce pLV-SB11. Maps and sequences are available upon request.
Cell culture. Human HeLa and HEK293T cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen, Paisley, UK) supplemented with 10% fetal calf serum (Sigma-Aldrich, Poole, UK), 100 units/ml penicillin, and 100 µg/ml streptomycin (Invitrogen).
Preparation and quantification of lentiviral vector. ILVs were produced as previously described33 by transient co-transfection of HEK293T cells using polyethylenimine (Sigma-Aldrich) to deliver the lentiviral vector plasmid, the vesicular stomatitis virus glycoprotein expression plasmid pMD.G2, and the second-generation packaging plasmid pCMVΔ8.74. Supernatant containing lentiviral vector was harvested 48 and 72 hours after transfection and concentrated by ultracentrifugation. IDLVs were prepared in the same way but substituting the packaging plasmid pCMVΔ8.74 D64V in which an amino acid mutation inactivates the integrating ability of the viral integrase.22 The Gag p24 titer of the vectors was quantified with the Retro-Tek HIV-1 p24 Antigen ELISA kit (ZeptoMetrix, Buffalo, NY).
Western blot. 106 HeLa cells were transduced with 0.5 µg p24 of each vector per well. At 1, 2, 3, and 7 days post-transduction, cells were trypsinized and transposase expression analyzed by western blot. Cells were lysed in lysis buffer (1% Nonidet P40, 130 mmol/l NaCl, 20 mmol/l Tris-HCl pH 8.0, 10 mmol/l NaF, 1 mmol/l phenylmethylsulfonyl fluoride, and 2 mmol/l EDTA) at 95 °C for 2 minutes. An equivalent of 105 cells was used for analysis. Samples were resolved using 4–12% Bis-Tris gels and MES buffer (NuPAGE; Invitrogen) and blotted onto polyvinylidene fluoride membranes. Blots were probed with monoclonal mouse anti-SB transposase antibody MAB2798 (R&D Systems, Abingdon, UK) or mouse anti-β-actin antibody A5316 (Sigma-Aldrich, Poole, UK) as a loading control. To visualize the primary antibodies, horseradish peroxidase-conjugated donkey anti-mouse antibody HAF018 (R&D Systems) and the Pierce ECL Western detection system (Amersham Life Sciences, Amersham, UK) were used.
Integration assay. Chromosomal integration was assessed by G418-resistant colony assay.1 Twenty-four hours before plasmid transfection or lentiviral transduction, 105 HeLa cells were seeded into each well of 24-well plates. Plasmids were transfected using Lipofectamine 2000 (Invitrogen). Where cells were both transfected and transduced, transfection was first performed for 4 hours before cells were washed with phosphate-buffered saline and subsequently transduced. Cells were maintained without selection for 3 days, trypsinized and re-seeded into 24-well plates in triplicate at 1:50 dilutions, and maintained in 1 mg/ml G418 (Invitrogen, San Diego, CA) for two weeks. Colonies were fixed with paraformaldehyde, stained with crystal violet, and imaged using a flatbed scanner. Colonies were counted using CellProfiler software (Massachusetts Institute of Technology, Cambridge, MA).34
Ligation-mediated PCR (LM-PCR). To recover integration sites, cells were transfected, transduced, and selected as described above. Single cell colonies were expanded to 106 cells and DNA was extracted by Proteinase K digestion and salting out. 1 µg of DNA was digested with NlaIII to produce vector-chromosome junction fragments and BamHI to destroy transposon-lentivector fragments where transposition did not occur. A linker was produced by annealing oligonucleotide 5′-GTA ATA CGA CTC ACT ATA GGG CTC CGC TTA AGG GAC CGC ATG-3′ and the phosphorylated oligonucleotide 5′-P-CGG TCC CTT AAG CGG AG-3′ as previously described.35 Digested DNA was ligated to excess linker. PCR was performed with GoTaq polymerase (Promega, Madison, WI) using the linker first round primer 5′-GTA ATA CGA CTC ACT ATA GGG C-3′ and either the HIV 3′-LTR first round primer 5′-AGT GCT TCA AGT AGT GTG TGC C-3′ (ref. 36) or the transposon first round primer 5′-CTG GAA TTG TGA TAC AGT GAA TTA TAA GTG-3′ (ref. 37) under conditions 95 °C 2 minutes; 30× (95 °C 30 seconds; 55 °C 30 seconds; 72 °C 1 minute); 72 °C 5 minutes. PCR products were diluted 1:50 and a second round of PCR was performed under the same conditions using the linker second round primer 5′-AGG GCT CCG CTT AAG GGA C-3′ and either the HIV 3′-LTR second round primer 5′-GTC TGT TGT GTG ACT CTG GTA AC-3′ or the transposon second round primer 5′-CTT GTG TCA TGC ACA AAG TAG ATG TCC-3′. PCR products were shotgun-cloned into the Topo-TA vector (Invitrogen, Paisley, UK) and sequenced (Functional Biosciences, Madison, WI) using the second round primers.
Pyrosequencing of LM-PCR products. Additional exponential fusion primer PCR was performed on LM-PCR products, thereby adding GS Flx specific primers (A and B) for amplification and sequencing to the ends of the LM-PCR amplicons. Primers were designed in accordance with the manufacturers' instructions (454 GS Flx; Roche, Branford, CT). Each fusion primer A contained an individual recognition sequence of 6 nucleotides (N) that allowed identification of different samples sequenced in the same sequencing run. Briefly, primer A was joined to a lentiviral vector specific LTR primer (fusion primer A-LTR, sequence: 5′-GCC TCC CTC GCG CCA TCA GNN NNN NTG TGT GAC TCT GGT AAC TAG-3′) or SB IRDR-R specific primer (fusion primer A-IRDRR, sequence: 5′-GCC TCC CTC GCG CCA TCA GNN NNN NGT ATT TGG CTA AGG TGT ATG-3′) and primer B was joined to a linker cassette-specific primer (fusion primer B-LK, sequence: 5′-GCC TTG CCA GCC CGC TCA GAG GGC TCC GCT TAA GGG AC-3′). Forty nanograms of purified LM-PCR products were used as a starting material for the fusion primer PCR. PCR conditions were as follows: initial denaturation for 120 seconds at 95 °C; 12 cycles at 95 °C for 45 seconds, 60 °C for 45 seconds, and 72 °C for 60 seconds; final elongation 300 seconds at 72 °C. Ten microliters of the PCR products were analyzed by gel electrophoresis on a 2% agarose gel.
Bioinformatics and statistics. Raw sequence reads obtained after sequencing were trimmed and aligned to the human genome. Integration sites were considered to be valid if vector-genome junction sequence was present and the flanking genomic region had a unique sequence match of at least 95% after alignment to the human genome (University of California at Santa Cruz, RefSeq genes and RepeatMasker; Alignment March 2006) (ref. 38). Chromosome graphs were generated using the University of California at Santa Cruz Genome Graphs tool. HeLa cell expression data were obtained from the public Gene Expression Omnibus database with accession number GSM157868, a dataset obtained from total RNA using a GeneChip Scanner 3000 (Affymetrix, High Wycombe, UK). Sequence logos were produced with the WebLogo tool.15 Statistical comparisons were performed by χ2 analysis, with level of significance set at P < 0.01.
SUPPLEMENTARY MATERIALTable S1. Integration site data is presented for the three vector systems which achieve stable integration. Integration proficient lentivirus (ILV), Hybrid sleeping beauty and non-integrating lentivirus (NILV-SB) and sleeping beauty plasmid (SB-plasmid). The genomic DNA sequence flanking the lentiviral-LTR or SB inverted repeat (IR) is shown. Sequences were aligned to the human genome, and integration sites were deemed valid if vector-genome junction sequence was detected and at least 95% of the adjacent sequence matched human genome DNA. The orientation of integrants is shown as sense (+) or reverse (-). The RefSeq gene name and ID are shown, along with gene length and where indicated, the distance to transcription start site (TSS).
Supplementary Material
Integration site data is presented for the three vector systems which achieve stable integration. Integration proficient lentivirus (ILV), Hybrid sleeping beauty and non-integrating lentivirus (NILV-SB) and sleeping beauty plasmid (SB-plasmid). The genomic DNA sequence flanking the lentiviral-LTR or SB inverted repeat (IR) is shown. Sequences were aligned to the human genome, and integration sites were deemed valid if vector-genome junction sequence was detected and at least 95% of the adjacent sequence matched human genome DNA. The orientation of integrants is shown as sense (+) or reverse (−). The RefSeq gene name and ID are shown, along with gene length and where indicated, the distance to transcription start site (TSS).
Acknowledgments
Research at the Institute of Child Health and Great Ormond Street Hospital for Children NHS Trust benefits from R&D funding received from the NHS Executive. C.A.V. is the recipient of a Child Health Research Appeal Trust award. A.J.T. is a Senior Wellcome Trust fellow. W.Q. is a Leukaemia Research Fund Senior Clinical Lecturer. This work was also supported by the Deutsche Forschungsgemeinschaft DFG (SPP1230).
REFERENCES
- Ivics Z, Hackett PB, Plasterk RH., and , Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Izsvák Z, Khare D, Behlke J, Heinemann U, Plasterk RH., and , Ivics Z. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J Biol Chem. 2002;277:34581–34588. doi: 10.1074/jbc.M204001200. [DOI] [PubMed] [Google Scholar]
- Cui Z, Geurts AM, Liu G, Kaufman CD., and , Hackett PB. Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J Mol Biol. 2002;318:1221–1235. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- Yant SR., and , Kay MA. Nonhomologous-end-joining factors regulate DNA repair fidelity during Sleeping Beauty element transposition in mammalian cells. Mol Cell Biol. 2003;23:8505–8518. doi: 10.1128/MCB.23.23.8505-8518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izsvák Z, Stüwe EE, Fiedler D, Katzer A, Jeggo PA., and , Ivics Z. Healing the wounds inflicted by sleeping beauty transposition by double-strand break repair in mammalian somatic cells. Mol Cell. 2004;13:279–290. doi: 10.1016/s1097-2765(03)00524-0. [DOI] [PubMed] [Google Scholar]
- Yant SR, Wu X, Huang Y, Garrison B, Burgess SM., and , Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry C, Hannenhalli S, Leipzig J., and , Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder AR, Shinn P, Chen H, Berry C, Ecker JR., and , Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Qasim W, Mackey T, Sinclair J, Chatziandreou I, Kinnon C, Thrasher AJ, et al. Lentiviral vectors for T-cell suicide gene therapy: preservation of T-cell effector function after cytokine-mediated transduction. Mol Ther. 2007;15:355–360. doi: 10.1038/sj.mt.6300042. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Izsvak Z, Minter A., and , Hackett PB. Identification of functional domains and evolution of Tc1-like transposable elements. Proc Natl Acad Sci USA. 1996;93:5008–5013. doi: 10.1073/pnas.93.10.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen JG, Yant SR, Meuse L, Huang Z, Xu H., and , Kay MA. Helper-independent Sleeping Beauty transposon-transposase vectors for efficient nonviral gene delivery and persistent gene expression in vivo. Mol Ther. 2003;8:654–665. doi: 10.1016/s1525-0016(03)00216-8. [DOI] [PubMed] [Google Scholar]
- Apolonia L, Waddington SN, Fernandes C, Ward NJ, Bouma G, Blundell MP, et al. Stable gene transfer to muscle using non-integrating lentiviral vectors. Mol Ther. 2007;15:1947–1954. doi: 10.1038/sj.mt.6300281. [DOI] [PubMed] [Google Scholar]
- Geurts AM, Yang Y, Clark KJ, Liu G, Cui Z, Dupuy AJ, et al. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Nightingale SJ, Hollis RP, Pepper KA, Petersen D, Yu XJ, Yang C, et al. Transient gene expression by nonintegrating lentiviral vectors. Mol Ther. 2006;13:1121–1132. doi: 10.1016/j.ymthe.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM., and , Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B, Burgess SM., and , Munroe DJ. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. J Virol. 2005;79:5211–5214. doi: 10.1128/JVI.79.8.5211-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B., and , Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Miller DG, Trobridge GD, Petek LM, Jacobs MA, Kaul R., and , Russell DW. Large-scale analysis of adeno-associated virus vector integration sites in normal human cells. J Virol. 2005;79:11434–11442. doi: 10.1128/JVI.79.17.11434-11442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez-Muñoz RJ, Balaggan KS, MacNeil A, Howe SJ, Schmidt M, Smith AJ, et al. Effective gene therapy with nonintegrating lentiviral vectors. Nat Med. 2006;12:348–353. doi: 10.1038/nm1365. [DOI] [PubMed] [Google Scholar]
- Yant SR, Ehrhardt A, Mikkelsen JG, Meuse L, Pham T., and , Kay MA. Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nat Biotechnol. 2002;20:999–1005. doi: 10.1038/nbt738. [DOI] [PubMed] [Google Scholar]
- Staunstrup NH, Moldt B, Mátés L, Villesen P, Jakobsen M, Ivics Z, et al. Hybrid lentivirus-transposon vectors with a random integration profile in human cells Mol Ther 2009. epub ahead of print [DOI] [PMC free article] [PubMed]
- Modlich U, Bohne J, Schmidt M, von Kalle C, Knöss S, Schambach A, et al. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108:2545–2553. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Ponzoni M, Bartholomae C, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- Brinster RL, Chen HY, Trumbauer ME, Yagle MK., and , Palmiter RD. Factors affecting the efficiency of introducing foreign DNA into mice by microinjecting eggs. Proc Natl Acad Sci USA. 1985;82:4438–4442. doi: 10.1073/pnas.82.13.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier LS, Carlson CM, Ravimohan S, Dupuy AJ., and , Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- Keng VW, Villanueva A, Chiang DY, Dupuy AJ, Ryan BJ, Matise I, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol. 2009;27:264–274. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison BS, Yant SR, Mikkelsen JG., and , Kay MA. Postintegrative gene silencing within the Sleeping Beauty transposition system. Mol Cell Biol. 2007;27:8824–8833. doi: 10.1128/MCB.00498-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis RP, Nightingale SJ, Wang X, Pepper KA, Yu XJ, Barsky L, et al. Stable gene transfer to human CD34(+) hematopoietic cells using the Sleeping Beauty transposon. Exp Hematol. 2006;34:1333–1343. doi: 10.1016/j.exphem.2006.05.023. [DOI] [PubMed] [Google Scholar]
- Demaison C, Parsley K, Brouns G, Scherr M, Battmer K, Kinnon C, et al. High-level transduction and gene expression in hematopoietic repopulating cells using a human imunodeficiency virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum Gene Ther. 2002;13:803–813. doi: 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
- Lamprecht MR, Sabatini DM., and , Carpenter AE. CellProfiler: free, versatile software for automated biological image analysis. BioTechniques. 2007;42:71–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- Hacker CV, Vink CA, Wardell TW, Lee S, Treasure P, Kingsman SM, et al. The integration profile of EIAV-based vectors. Mol Ther. 2006;14:536–545. doi: 10.1016/j.ymthe.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Wu X., and , Burgess SM. Integration target site selection for retroviruses and transposable elements. Cell Mol Life Sci. 2004;61:2588–2596. doi: 10.1007/s00018-004-4206-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda R, Kokubu C, Yusa K, Keng VW, Horie K., and , Takeda J. Sleeping beauty transposase has an affinity for heterochromatin conformation. Mol Cell Biol. 2007;27:1665–1676. doi: 10.1128/MCB.01500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Integration site data is presented for the three vector systems which achieve stable integration. Integration proficient lentivirus (ILV), Hybrid sleeping beauty and non-integrating lentivirus (NILV-SB) and sleeping beauty plasmid (SB-plasmid). The genomic DNA sequence flanking the lentiviral-LTR or SB inverted repeat (IR) is shown. Sequences were aligned to the human genome, and integration sites were deemed valid if vector-genome junction sequence was detected and at least 95% of the adjacent sequence matched human genome DNA. The orientation of integrants is shown as sense (+) or reverse (−). The RefSeq gene name and ID are shown, along with gene length and where indicated, the distance to transcription start site (TSS).