Abstract
RNA interference (RNAi) has generated significant interest as a strategy to suppress viral infection, but in some cases antiviral activity of unmodified short-interfering RNA (siRNA) has been attributed to activation of innate immune responses. We hypothesized that immunostimulation by unmodified siRNA could mediate both RNAi as well as innate immune stimulation depending on the mode of drug delivery. We investigated the potential of immunostimulatory RNAs (isRNAs) to suppress influenza A virus in vivo in the mouse lung. Lipidoid 98N12-5(1) formulated with unmodified siRNA targeting the influenza nucleoprotein gene exhibited antiviral activity. Formulations were optimized to increase antiviral activity, but the antiviral activity of lipidoid-delivered siRNA did not depend on sequence homology to the influenza genome as siRNA directed against unrelated targets also suppressed influenza replication in vivo. This activity was primarily attributed to enhancement of innate immune stimulation by lipidoid-mediated delivery, which indicates increased toll-like receptor (TLR) activation by siRNA. Certain chemical modifications to the siRNA backbone, which block TLR7/8 activation but retain in vitro RNAi activity, prevented siRNA-mediated antiviral activity despite enhanced lipidoid-mediated delivery. Here, we demonstrate that innate immune activation caused by unmodified siRNA can have therapeutically relevant effects, and that these non-RNAi effects can be controlled through chemical modifications and drug delivery.
Introduction
RNA interference (RNAi), mediated by short double-stranded RNA (dsRNA) molecules termed short-interfering RNAs (siRNAs), causes degradation of target mRNA in a highly sequence-specific manner.1,2 RNAi holds significant promise as an antiviral therapeutic strategy.3,4 RNA viruses have single- or dsRNA genomes that may be good targets for RNAi,4 and RNAi has been used to inhibit a variety of difficult-to-control viruses such human immunodeficiency virus5,6 and hepatitis C.7 Respiratory tract infections such as respiratory syncytial virus (RSV),8,9,10 parainfluenza virus,8 and severe acute respiratory syndrome coronavirus11 have all been suppressed in vivo following intranasal instillation of naked siRNAs. In the case of naked unmodified siRNAs targeting RSV, an RNAi-mediated mechanism of action has been confirmed by 5′-RACE detection of the RSV mRNA cleavage site.12 Lipid carriers have also been used to deliver siRNAs effective against Ebola virus12 and the DNA virus hepatitis B.13,14
In 2003, Ge et al.15 reported selection of siRNA sequences that efficiently inhibited influenza viral replication in cell culture and chicken eggs. The most promising siRNAs targeting the NP (siNP-1496) and PA genes of Influenza A/PR/8/34 (PR8) were subsequently used by Ge et al.16 to inhibit influenza in vivo in mouse models of infection. They reported inhibition of viral titers in the lungs of mice following intravenous delivery by linear poly(ethylenimine) (PEI). Tompkins et al.17 used the same siRNA sequences, which were delivered by hydrodynamic intravenous injection followed by intranasal instillation of oligofectamine/siRNA complexes, to demonstrate protection against a lethal challenge of multiple different influenza subtypes including H5N1. Thomas et al.18 in 2005 also demonstrated efficient inhibition of influenza in mouse lungs using different forms of PEI as a delivery material for siNP-1496. In all reports, the same green fluorescent protein (GFP) sequence (siGFP-949) was used as a control for nonspecific effects.
Initially it was thought that siRNAs avoided the interferon (IFN) response that was observed with longer dsRNA molecules.19 Induction of intracellular IFN responses was thought to occur only when long dsRNA interacted with cytosolic receptors of RNA such as melanoma differentiation-associated-5 and retinoic acid–inducible gene-I, collectively termed the retinoic acid–inducible gene-I-like helicases,20,21 or activated toll-like receptors (TLRs).22 However, it has recently been appreciated that siRNA, while shorter in length, can in certain instances interact with TLR3 (refs. 23,24), TLR7 (ref. 25), and TLR8 (ref. 26) to induce IFN responses. This interaction of siRNA with TLRs depends greatly on siRNA structure and sequence, and it can be abrogated through sequence selection strategies and/or chemical modification of siRNA.27,28,29,30 Chemical modifications such as 2′-fluoro pyrimidines and 2′-O-methyl purines have been shown to prevent the IFN response.13 2′-O-methyl substitutions to the ribose backbone in particular have been shown to suppress TLR7 and TLR8 interactions with immunostimulatory RNAs (isRNAs).30 It has been hypothesized that these modifications modulate binding of the TLR through structural conformational changes and affect hydrogen bonding in the minor groove of dsRNA.28
In both humans and in mice, the IFN-α-inducing siRNA–TLR7 interaction occurs mainly in the endosome of a specialized cell called the plasmacytoid dendritic cell (pDC); the pDC circulates in the blood, resides in large numbers in the liver, and is the chief producer of IFN-α.21,25,31 Many cell types including myeloid DCs (mDCs), monocytes, and monocyte-derived DCs (mo-DCs) express some functional TLR7 (in mice) or TLR8 (in humans). These cells can also respond to immunostimulatory sequences of siRNA to produce cytokines such as tumor necrosis factor-α (TNF-α) in high amounts.32,33 As such, isRNA is a functional definition, and may be determined not only by base sequence, structure, and chemical modifications, but also by location in the endosome of the cells where sequence- and structure-specific TLR interactions occur.
A systemic IFN response to RNA may confound or mask the gene-specific effects of true RNAi in in vivo models of viral infection. As previously mentioned, an RNAi-mediated mechanism of action has been confirmed by 5′-RACE detection of the RSV mRNA cleavage site following dosing of naked unmodified siRNA.34 However, the influenza virus is highly sensitive to type 1 IFNs, and production of type 1 IFNs is an important component of innate defense against influenza and other viruses.35 Small molecule TLR7/8 agonists36 and the TLR3-agonist poly(I:C-L:C)37 have previously shown potent in vivo antiviral activity against influenza through induction of IFN responses.
Clinically relevant use of siRNAs requires a method of drug delivery,38 but the role of drug delivery in enhancing isRNA activity has not been clearly defined. We hypothesized that, in addition to mediating RNAi, siRNA may have an antiviral effect due to innate immune stimulation, and that this effect may be modulated by method of delivery. We have recently established a combinatorial library of lipid-like materials for delivery of nucleic acids termed “lipidoids” that can efficiently deliver small RNA's across cellular barriers.10 Using high-throughput screening methods, promising novel materials were previously identified that exhibited highly efficient in vitro and in vivo delivery of siRNA molecules to elicit RNAi-mediated downregulation of liver-specific targets. We further investigated the ability of siRNA delivered with one promising candidate material, 98N12-5(1), for its ability to suppress influenza A virus replication in the lung using a murine model of influenza infection. Using a new lipidoid nanoparticle formulation optimized to achieve antiviral effects, we describe the efficient prophylactic inhibition of influenza A in mouse lungs due to innate immune stimulation following the delivery of unmodified siRNA.
Results
98N12-5(1)-siRNA nanoparticles have antiviral activity
Short-interfering RNAs were encapsulated in nanoparticles based upon the 98N12-5(1) lipidoid, a novel lipid-like material that has been used for efficient in vivo siRNA–delivery to the liver10,39,40 (Figure 1). We developed a method for generating stable nanoparticles of lipidoid with cholesterol and polyethylene glycol (PEG)-ceramide in sodium acetate and ethanol. Lipidoid was formulated, extruded at high pressure to normalize particle sizes, and then dialyzed to remove ethanol. These particles consistently exhibit a characteristic size of 70–100 nm as measured by dynamic light scattering with >90% encapsulation of RNA (Supplementary Table S1). We investigated the use of dialyzed lipidoid nanoparticles for inhibition of the PR8 strain influenza A virus in a mouse model of infection. Initial studies with formulated 98N12-5(1) particles complexed with siNP-1496 indicated a dose-dependent response following prophylactic treatment 24 hours before infection with a super-lethal challenge of 12,000 plaque-forming units of PR/8 influenza virus. Up to 13-fold (~93%) reduction in lung viral titer compared to phosphate-buffered saline (PBS) injections was observed at 3 mg/kg siNP-1496 dosing (Figure 2). However, dialyzed lipidoid particles containing siGFP-949 also exhibited a 4.7-fold (~77%) reduction in viral titer at 3 mg/kg. Commercially available PEI, by comparison, demonstrated a 5.8-fold (83%) viral titer reduction at 3 mg/kg dosing of siNP-1496. PEI–siRNA nanoparticles also demonstrated some inhibitory effect at 2 mg/kg dosing (3.6-fold inhibition of viral titer); this effect was similar for both siGFP-949 (data not shown) and siNP-1496.
Figure 1.
Chemical structure of 98N12-5(1) with one internal secondary amine and five tail groups.10
Figure 2.
Dose-dependent and formulation-dependent inhibition of influenza viral replication in mouse lung. Black Swiss mice were injected twice IV with PBS without siRNA, freshly prepared PEI–siRNA nanoparticles, freshly prepared 98N12-5(1)-siRNA nanoparticles, or previously lyophilized 98N12-5(1)-siRNA nanoparticles at 1, 2, or 3 mg/kg siRNA before challenge with 12,000 PFU PR/8 influenza virus. Lung viral titer was determined by quantitative plaque-forming assay and is expressed as relative to control (PBS) viral titer. N = 3–6, mean ± SD shown. IV, intravenous; PBS, phosphate-buffered saline; PEI, poly(ethylenimine); PFU, plaque-forming units; siRNA, short-interfering RNA.
Optimization of formulation to increase antiviral activity
To further optimize particle formulation, we varied our method of particle preparation. Upon lyophilization and reconstitution in PBS, lipidoid nanoparticles increased in size to between 400 and 700 nm (Supplementary Table S1). These larger particles were more efficient than nonlyophilized freshly prepared particles at suppressing influenza viral titer (Supplementary Figure S1). Extrusion of particles through a 400-nm filter resulted in similar sizes as extrusion through a 200-nm filter (data not shown). Extrusion of particles through a 400-nm filter followed by dialysis did not result in viral suppression as efficient as extrusion through a 200-nm filter followed by lyophilization. Compared to freshly prepared nanoparticles, lyophilization of lipidoid-siGFP-949 particles only slightly increased the antiviral effect at 3 mg/kg dose. Lyophilized nanoparticles at 3 mg/kg siRNA dosing reduced viral titer 13.4-fold (92.5%) for siGFP-949 and over 300-fold (99.7%) for siNP-1496 (Figure 2). However, at these highest doses lyophilized nanoparticles caused weight loss of 8% after the first injection and 16% after the second injection. At just 2 mg/kg dosing of siNP-1496, viral titer reduction with lyophilized particles was on average 36-fold (~97%) compared to PBS (Figure 2). This lyophilized formulation and 2 mg/kg dose was selected for the rest of our studies.
Antiviral activity is not due to sequence homology with influenza genome
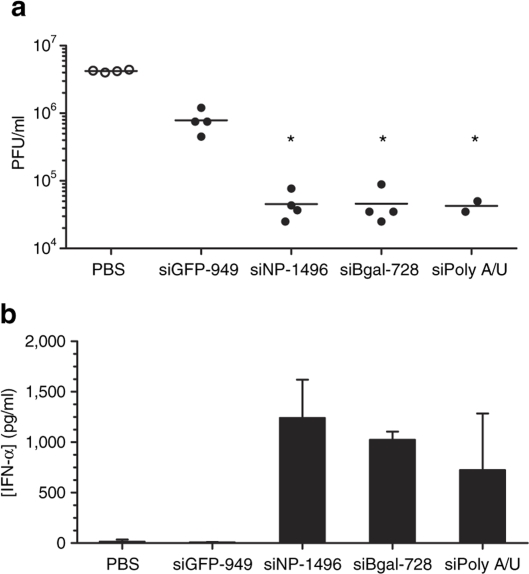
To address issues of sequence-independent inhibition of influenza observed with siGFP-949 particles, we added a scrambled control sequence of siNP-1496 to our experimental set of siRNAs. Lyophilized lipidoid nanoparticles were formulated with siGFP-949, siNP-1496, or scrambled siNP-1496-sc. We also investigated serum IFN responses following dosing with nanoparticles, as some siRNA sequences are known to induce IFN responses through TLR7 in vivo in mice and TLR7/8 in vitro in human cells.25,41 We observed an association between efficient suppression of viral titers (Supplementary Figure S2a) and activation of an innate immune response (Supplementary Figure S2b) using the scrambled siNP-1496 sequence. In addition, viral inhibition in response to an irrelevant siRNA sequence (siβgal-728) known to induce IFN41 was similar in magnitude to that observed with siNP-1496 targeting the flu genome. Similar antiviral activity was observed as well with the sequence siPolyA/U, one strand of which (polyA strand) was reported to not stimulate immune responses when formulated with DOTAP42 (Table 1). Antiviral activity was compared side-by-side with siNP-1496 in lyophilized 98N12-5(1) nanoparticles. Surprisingly, siNP, siβgal, and siPolyA/U lipidoid particles all suppressed influenza reproduction equivalently, achieving over 90-fold reduction in viral titer compared to PBS controls (Figure 3b). Lipidoid nanoparticles containing both siNP and siβgal elicited high levels of serum IFN-α and TNF-α, while neither siGFP-nanoparticles nor PBS injections generated detectable levels of serum IFN-α (Figure 3b).
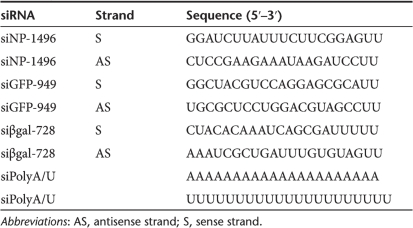
Table 1.
Sequences of unmodified siRNA molecules used throughout this study
Figure 3.
Inhibitory effects of siRNA sequences correlated with induction of systemic type I interferon response. Mice were injected twice IV with lyophilized 98N12-5(1)-siRNA nanoparticles at 2 mg/kg siRNA before (a) infection with 12,000 PFU PR/8 influenza virus, or (b) blood collection by cardiac puncture. (a) Lung viral titer was determined by quantitative plaque-forming assay. Data from individual mice shown; black lines indicate mean for each group. (b) Serum IFN-α concentration was determined by ELISA. Nanoparticles with siNP-1496 or unmatched siβgal-728 sequence elicited high levels of IFN-α that corresponded with significant reductions in viral titer compared to nanoparticles formulated with siGFP-949. *P < 0.05 compared to siGFP-949 group by 5 group 1-way ANOVA followed by Bonferroni's multiple comparison test. ANOVA, analysis of variance; ELISA, enzyme-linked immunosorbent assay; IFN, interferon; GFP, green fluorescent protein; PFU, plaque-forming unit; siRNA, short-interfering RNA.
Antiviral activity is due to immune stimulation
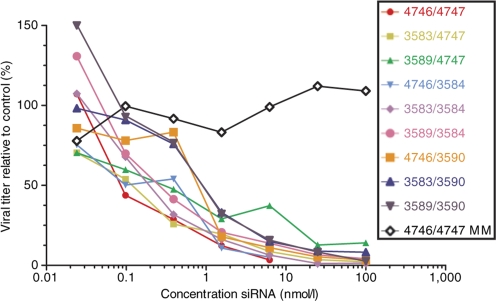
The effect on antiviral activity of chemical modifications to the siRNA backbone was investigated as these modifications can prevent interactions with TLRs.28,29,30 The siNP-1496 sequence was modified with various backbone and nucleic acid substitutions (Table 2). Immune stimulation by modified siRNAs was first investigated in vitro by transfecting modified siRNA into cultured human peripheral blood mononuclear cells (PBMCs) with Gene Porter 2 or DOTAP. IFN-α and TNF-α levels of culture supernatants were measured by enzyme-linked immunosorbent assay 24 hours after transfection (Figure 4). Incorporation of neither deoxythymidine overhangs nor substitution with a phosphorothioate linkage was able to prevent immune stimulation. Substituting the hydroxyl group on the 2′ carbon of ribose with a 2′-O-methyl group prevented immune stimulation when present on either sense or antisense strands in vitro (Figure 4). Just a single base (uridine) modification with 2′-O-methyl could prevent immune stimulation, as the siRNA duplex formed with si4767 (sense) and si3590 (antisense) did not elicit immune stimulation (Figure 4). Chemically modified siRNAs retained the ability to inhibit influenza through RNAi mechanisms in vitro (Figure 5) with the IC50 of all duplexes in the sub-nmol/l range (<0.4 nmol/l). Notably, duplexes with the si3590-modified strand exhibited a slightly reduced potency with an IC50 approaching 1 nmol/l, though siRNA duplexes formed from more heavily 2′-O-methyl substituted RNA strands such as si3589 did not suffer from this slight reduction in activity.
Table 2.
Chemically modified siRNA sequences
Figure 4.
Chemical modification to the siRNA backbone can prevent in vitro induction of innate immune responses in human PBMC cultures. Levels of serum cytokines IFN-α and TNF-α 24 hours after in vitro transfection of human PBMCs with 130 nmol/l unmodified and modified (see Table 1) versions of siNP-1496 complexed with GenePORTER 2 (IFN-α) or DOTAP (TNF-α). Controls are untreated cells, cells with empty transfection agent, or direct incubation with CpG oligo ODN2216 at 500 nmol/l to elicit IFN-α responses through TLR9 interactions. IFN-α is plotted on the left axis (gray) and TNF-α is plotted on the right axis (black). Average of two independent donors is shown. IFN, interferon; PBMC, peripheral blood mononuclear cell; siRNA, short-interfering RNA; TLR, toll-like receptor; TNF, tumor necrosis factor.
Figure 5.
In vitro RNA interference of influenza virus using native and chemically modified siRNAs. Cultured Vero cells were infected with PR/8 influenza virus and transfected with siRNAs. Viral titer is expressed as a percentage relative to nontreated control viral titers. The dose–response RNA interference effects of siRNA were not significantly altered by chemical modifications. The different siRNA duplexes are identified by sense and antisense strand (S/AS) as listed in Table 2. The parent unmodified siRNA is represented by 4746/4747; 4746/4747 MM represents a mismatched version of 4746/4747, while all other duplexes represent introduction of additional modifications into the original 4746/4747 parent siRNA duplex. siRNA, short-interfering RNA.
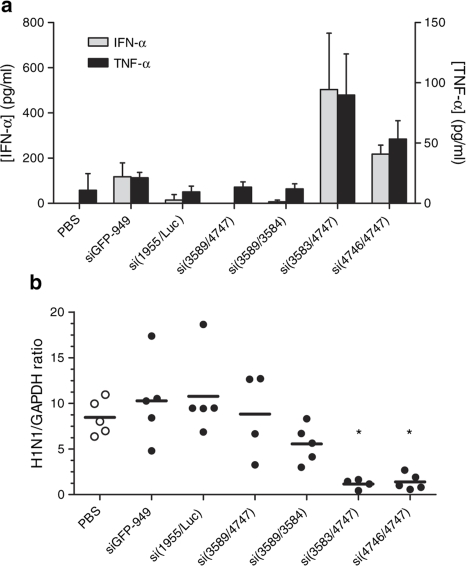
A subset of siNP-1496 siRNA representing unmodified and chemically modified duplexes that retain full in vitro RNAi activity was selected for in vivo investigations of immune stimulation and antiviral activity. These modified siNP-1496 sequences were incorporated into the optimal formulation of lyophilized 98N12-5(1) nanoparticles. We investigated both immune stimulation and suppression of influenza in vivo in a mouse model. Chemical modifications that blocked immune stimulation in human PBMCs in vitro also prevented activation of IFN-α and TNF-α in vivo in Black Swiss mice (Figure 6a).
Figure 6.
Chemical modifications to the siRNA backbone decrease in vivo induction of serum inflammatory cytokines and antiviral responses. Mice were injected twice IV with lyophilized 98N12-5(1) siRNA nanoparticles at 2 mg/kg siRNA before (a) blood collection by cardiac puncture or (b) infection with 12,000 PFU PR8 virus. (a) Serum IFN-α concentration was determined by ELISA. (b) Lung viral RNA level was determined by branched DNA assay and normalized to GAPDH mRNA levels. Data from individual mice shown; black lines indicate mean for each group. *P < 0.05 compared to PBS group by 7-group one-way ANOVA followed by Bonferroni's multiple comparison test. ANOVA, analysis of variance; ELISA, enzyme-linked immunosorbent assay; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IFN, interferon; IV, intravenous; PBS, phosphate-buffered saline; PFU, plaque-forming unit; siRNA, short-interfering RNA.
Plaque assays may be confounded by production of noninfectious viral particles, residual cytokines in the lung samples, or toxicity. Thus, we have established a correlation between functional viral particles, as measured through plaque-forming assay, and levels of viral mRNA (Supplementary Figure S3). Measurement of viral mRNA levels can be normalized to a housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase, to investigate specific inhibition of the siRNA target influenza genome. As measured by branched DNA assay for viral RNA levels, blocking immune stimulation abrogated antiviral responses (Figure 6b). In addition, the chemically modified GL3 si1995/Luc sequence targeting an irrelevant gene, firefly luciferase, formulated in lyophilized 98N12-5(1) particles did not induce either significant immune stimulation or antiviral responses. Delivery of immunostimulatory siRNA's (si3583/3584 and si4746/4747) was associated with tenfold reduction in normalized viral mRNA levels, compared to PBS-treated mice, and exhibited significantly greater antiviral activity than siRNA modified to abolish immunostimulatory activity. These data together provide evidence that replication of the influenza virus was specifically inhibited by the IFN responses and was not due to carrier-mediated toxicity.
Discussion
Previous work15,17,18,43 with the siNP-1496 sequence targeting influenza using only the siGFP-949 sequence as a control did not accurately account for isRNA activity as the siGFP-949 sequence does not seem to activate TLR-mediated IFN responses,44 as we have observed. PEI-mediated delivery of the siNP-1496 sequence was not previously found to induce an IFN response,16,18 although our results indicate that the antiviral activity of PEI was also relatively low. Commonly used and commercially available cationic lipid transfection agents such as DOTAP and Lipofectamine have been used to investigate the biology of isRNA activity in vitro.13,25,28,41 Both lipid- and PEI-based nanoparticles have also been reported to generate isRNA activity in vivo.25,30,45,46 Here, we have demonstrated that the observed antiviral properties of siNP-1496 siRNA are related to delivery-dependent innate immune stimulation rather than an RNAi-mediated antiviral effect. This is the first report to our knowledge comparing isRNA activity amongst different delivery systems and describing the effect of increasing immune stimulation in vivo through systemic delivery of isRNA molecules.
We have developed a novel delivery system that efficiently delivers siRNAs for immune stimulation better than PEI (Figure 2). We report inhibition of viral titers that approaches 90-fold reduction (compared to PBS) at 2 mg/kg RNA dosing and 300-fold reduction at 3 mg/kg dosing using unmodified siRNAs in lyophilized lipidoid nanoparticles. Antiviral activity of lyophilized 98N12-5(1) nanoparticles was superior to PEI nanoparticles encapsulating the same siRNAs (Figure 2), and amplified the difference in isRNA activity between different sequences (Figure 3). Of note, the antiviral activity of optimized lipidoid-siRNA nanoparticles was not limited to just the siNP-1496 sequence. The previously identified isRNA, siβgal-728 (ref. 41), also exhibited antiviral activity. Although the polyA strand of siPolyA/U has been reported to not induce IFN responses when delivered with DOTAP,42 when combined with the polyU strand in the lipidoid nanoparticle this RNA also had significant antiviral activity (Figure 3), likely due to the high uridine content.25,26 Delivery with lyophilized 98N12-5(1) nanoparticles preferentially to IFN-producing cells is likely to increase isRNA potential of any given siRNA sequence including the siGFP-949 sequence. All antiviral activity observed was associated with efficient induction of a type 1 IFN response through induction of IFN-α and a pro-inflammatory cytokine response through the induction of TNF-α (Figures 3 and 6).
The lipidoid material 98N12-5(1) is capable of mediating cytosolic delivery of siRNA molecules and achieving RNAi in vitro and in vivo to the liver.10 Given that efficient TLR7 engagement with siRNA molecules requires endosomal localization,26 our results indicate endosomal uptake as one method of 98N12-5(1) internalization in vivo. PEI, on the other hand, is highly efficient for rapid endosomal escape47,48 and therefore may not release significant RNA in the endosome for interaction with TLRs. Lyophilization of lipidoid nanoparticles increases particle sizes and increases antiviral activity. However, the lyophilization process itself is not sufficient for achieving high antiviral activity because dialysis before lyophilization results in particles with decreased antiviral activity (Supplementary Figure S1). As dialyzed lipidoid-siRNA nanoparticles deliver siRNA to the liver and spleen, albeit without resulting in RNAi in the spleen,10,40 we hypothesize that lyophilizing these particles and increasing the particle size preferentially increases uptake in liver and splenic pDCs.
Understanding and controlling the activation of immune responses is an important step toward using siRNA molecules therapeutically. While we did not observe significant anti-flu effect due to RNAi, large antiviral effect was caused by the immunostimulatory aspect of certain siRNA. The sense strand of the siNP-1496 sequence has been suggested to activate human PBMCs in vitro.30 Our findings are consistent with this report. The siRNA duplexes containing 3′-overhangs with deoxythymidine showed immunostimulatory activity in vitro in human PBMCs (Figure 4) or when formulated and tested in vivo (Figure 6a). Further modification of the 3′-overhangs with a phosphorothioate linkage also had little effect on siRNA-mediated immune activation. Only 2′-O-methyl modifications were able to block IFN responses in vitro and in vivo. Accordingly, only 2′-O-methyl modifications blocked the in vivo antiviral response (Figure 6b). Previous reports have indicated that a minimum of two residues must be modified to prevent immune stimulation.30 We observed blockade of TLR activity with 2′-O-methyl modification of just one central uridine residue (modified strand si3590; Table 2) in the antisense strand of the NP-1496 sequence that efficiently prevented IFN responses in human PBMCs (Figure 4). This central base modification also slightly decreased the potency of RNAi activity, although it was not eliminated (Figure 5), and was not further investigated in vivo. Interestingly, more extensive modification with multiple 2′-O-methyl bases on the sense strand (si3589) molecule did not change RNAi potency. All of the other modifications completely preserved in vitro RNAi activity. While we did not directly test the immunostimulatory activity of our siRNAs in isolated mouse cells in vitro, we have observed similar immunostimulatory activity in vitro with human PBMCs and in vivo in mice consistent with other literature descriptions.25,30,41
Recently, Robbins et al.44 reported on the misinterpretation of anti-influenza effects of the siNP-1496 sequence due to immune stimulation. They showed that the siGFP-949 sequence is uniquely nonstimulatory for unknown reasons, though the low GU-content of the siGFP-949 sequence could be a factor. They also demonstrated that 2′-O-methyl modification of all uridine nucleotides in the sense strand of siNP-1496 can abrogate immune responses and antiviral activity.44 Our results presented here are consistent with the observed differences in immunostimulatory nature of various siRNAs contained in the report by Robbins et al. Although we have demonstrated that minimal modification is necessary to prevent undesired immune stimulation, the exact position and number of chemical modifications required to abolish immunostimulation must be tested and confirmed experimentally on a case-by-case basis.
We have further demonstrated the role that enhanced delivery can have in increasing immunostimulatory siRNA effects. Despite evidence that chemically modified modified siNP-1496 exhibited potent RNAi-mediated antiviral activity in vitro (Figure 5) without significant immune stimulatory activity both in vitro (Figure 4) and in vivo (Figure 6), these siRNA did not reduce virus titer in mice when delivered in lipidoid formulation. However, using a different formulation of 98N12-5(1), we have previously observed RNAi-mediated antiviral activity following direct pulmonary instillation in a mouse model of RSV infection.10 The formulation of 98N12-5(1) described here was optimized for maximum prophylactic effect, which was due to immunostimulatory delivery of RNA. It is possible that the relative contribution of RNAi-mediated gene silencing may be more significant for other delivery methods or other formulations of 98N12-5(1). Stably expressed siNP-1496 is capable of knocking down reporter gene expression (data not shown) and inhibiting influenza virus production in cell culture.16 The lack of observable in vivo RNAi mediated by formulations with siNP-1496 highlights the importance of siRNA delivery in achieving the desired RNAi activity. Thus, additional development is needed to facilitate RNAi-mediated anti-influenza activity of systemically administered siNP-1496 in vivo.
Immunostimulation using siRNA can have potential benefits as well. Immunostimulatory RNA molecules have been used to induce antitumor effects46 and could serve as adjuvants in generating vaccine responses.49 Previously, it was reported that a 13-fold reduction in viral titer in a super-lethal infection model similar to the one described here can lead to a survival benefit; a 63-fold reduction in viral titer approached 100% survival rate.17 We have also investigated a therapeutic model of treating influenza infection with 98N12-5(1) nanoparticles administered after infection with virus. However, no significant benefit was observed with this dosing scheme (Supplementary Figure S4). Still, the prophylactic antiviral effects of 98N12-5(1) nanoparticles encapsulating the siRNAs siNP-1496 or siβgal-726 were robust; we have achieved up to 300-fold reduction in viral titer.
We provide evidence that formulation methods can significantly alter the in vivo activity of unmodified small RNA molecules. As TLR7/8 activity is both cell-specific and compartmentalized to the late endosome, controlled delivery can influence the level of immunostimulatory siRNA activity. As TLR7/8 engagement is important for innate antiviral responses,28,35 avoiding TLRs is important in the development of RNAi therapies in the infectious disease setting. Additionally, excessive activity of TLR7/8 leading to increased type 1 IFN secretion from the pDC has been associated with autoimmunity.50 When siRNAs are administered in vivo they must therefore avoid sustained internalization into the endosome of pDCs and other TLR7/8-responsive cells, or siRNAs must be modified to prevent TLR7/8 activation as we have demonstrated. Nevertheless, an attractive therapeutic potential use of controlled siRNA delivery would be simultaneous RNAi activity and immunostimulatory RNA activity from the same RNA molecule.46 A single RNA with dual functionality may therefore be readily achieved by controlled delivery to a cytosolic location for RNAi activity and an endosomal location in pDCs for innate immune activation. We show that these antiviral IFN and related cytokine responses mediated by certain siRNAs in vivo can be influenced by delivery material and formulation and can be suppressed by chemical modifications to the siRNA. Our work offers a proof of concept of optimizing in vivo drug delivery for isRNA activity, as well as evidence that even a robust delivery system can be rendered immunosilent by certain chemical modifications to siRNA.
Methods
siRNAs. Unmodified siRNA molecules with 3′UU-overhangs were purchased from Dharmacon (Lafayette, CO) with the “in vivo” processing option or the “A4” processing option. The siNP-1496 (ref. 15), siGFP-949 (ref. 15), and siβgal-728 (ref. 41) sequences were as previously reported. Chemically modified siRNAs with the siNP-1496 sequence and GL3 firefly luciferase sequence (si1955/Luc) were synthesized by Alnylam Pharmaceuticals (Cambridge, MA). See Table 1 for sequences and Table 2 for sequences with chemical modifications.
Lipidoid and PEI nanoparticle preparation. Lipidoid 98N12-5(1) (ref. 10) was dissolved to 120 mg/ml in ethanol, cholesterol (Sigma-Aldrich, St Louis, MO) was dissolved to 25 mg/ml in ethanol, and N-palmitoyl-sphingosine-1-[succinyl(methoxypolyethylene glycol)2000] (C16 mPEG 2000 ceramide) (“PEG”) (Avanti Polar Lipids, Alabaster, AL) was dissolved to 100 mg/ml in ethanol. Lipidoid, cholesterol, and PEG were combined at a 15:0.8:7 mass ratio (L:C:P), vortexed briefly, and diluted in a mixture of ethanol and sodium acetate (25 mmol/l with 16.67 mg/ml sucrose) for a final lipidoid concentration of 7.5 mg/ml in 40% ethanol, 60% NaAc. siRNAs were diluted in NaAc, 25 mmol/l, to 500 µg/ml. Diluted lipidoid/cholesterol/PEG were added to diluted siRNA at a 15:1 mass ratio (L:R) and vortexed for 20 minutes to allow complexes to form. Complexed lipidoid-RNA nanoparticles were then extruded 10 passes through a double 200-nm membrane (Whatman, Florham Park, NJ) on a Northern Lipids (British Columbia, Canada) extrusion system at 40 °C. To remove ethanol before injection, nanoparticles were dialyzed in a Slide-A-Lyzer 3500 molecular weight cutoff dialysis cassette (Pierce Biotechnology, Rockford, IL) against PBS. For lyophilization, 10 mg of sucrose was added per ml of extruded complexes before freezing at −80 °C for >2 hours followed by >1 day-lyophilization.
In vivo Jet-PEI was purchased from Polyplus (New York, NY). PEI–siRNA complexes were made according to the manufacturer's protocols immediately before injection and diluted in PBS with 5% glucose.
Lipidoid nanoparticle characterization. For quantification and encapsulation efficiency of siRNA, a 50 µl sample of nanoparticles was diluted 200-fold in Tris-EDTA buffer, mixed with either 50 µl of Tris-EDTA buffer or 50 µl of 0.4% Triton-X-100 in Tris-EDTA, and incubated with 100 µl Quant-It Ribogreen reagent (Invitrogen, Carlsbad, CA) according to the manufacturers protocols for 20 minutes at 37 °C in a 96-well black plate. Fluorescence intensity was determined at 485 nm (ex)/535 nm (em). Total fluorescence in the presence of Triton-X was compared to a standard curve of siRNA diluted in Tris-EDTA to determine total siRNA concentration; siRNA encapsulation efficiency was determined by the ratio of fluorescence signal without Triton-X to signal with Triton-X. Nanoparticle size was assayed by light scattering using a Zeta-PALS instrument (Brookhaven Instruments, Holtsville, NY).
Nanoparticle injections. Male Black Swiss mice were purchased from Taconic Farms (Hudson, NY) and cared for according to the standards of the Massachusetts Institute of Technology under the guidance of the Division of Comparative Medicine. Mice were anesthetized with a mixture of ketamine (10 mg/ml) and xylazine (1.5 mg/ml) in PBS by intraperitoneal injection. Nanoparticles were resuspended or diluted in PBS to 250 µg siRNA/ml immediately before injection. PBS at volumes equivalent to nanoparticle injections was used as a control. Injections were made intravenously into the retroorbital plexus.
Influenza infection and viral titer assays. Mice were dosed with nanoparticles twice at 0 and 20 hours and infected with influenza at 24 hours. Influenza virus A/PR/8/34 (PR8) was diluted to 240,000 plaque-forming units/ml in PBS with 0.3 wt% bovine serum albumin and 100 U/ml penicillin/streptomycin. After anesthetizing mice with the ketamine/xylazine mixture, 50 µL (12,000 plaque-forming units) of diluted PR8 virus was instilled intranasally dropwise. At 48 hours, 24 hours after infection, mice were sacrificed and the lungs removed. Whole lungs were flash frozen in liquid nitrogen in 2 ml of PBS with 0.3 wt% bovine serum albumin and subjected to two freeze–thaw cycles. Lungs were then homogenized by sonication to release virus, centrifuged at 800 relative centrifugal force, 4 °C for 4 minutes, and supernatant samples were frozen at −80 °C until analysis.
To determine viral titer by plaque-forming unit assay, Madin–Darby canine kidney cells were seeded at 0.5 × 106cells/well in 6-well plates in Dulbecco's modified Eagle's medium (with 10 mmol/l HEPES, 10% fetal bovine serum, 100 U/ml penicillin/streptomycin, 2 mmol/l glucose) and allowed to grow to single-layer confluence overnight. Media was aspirated from wells, and 200 µl of virus-containing samples serially diluted tenfold in PBS were added onto cells in triplicate. Following a 1-hour incubation period with periodic shaking to distribute viral particles evenly, cells were covered with 2 ml of semisolid 1% agar/media solution to limit viral particle spread to cell-to-cell contacts. Plaques were counted after 3 days.
To determine viral titer by viral RNA levels, whole lungs were harvested, flash frozen with liquid nitrogen, and then mechanically pulverized. Lung samples were reconstituted in PBS, digested in cell lysis buffer and proteinase K (Epicentre, Madison, WI), and assayed using a branched DNA assay (Panomics, Fremont, CA) with probes specific to H1N1 genomic RNA. Levels of H1N1 genome were measured and compared to glyceraldehyde 3-phosphate dehydrogenase mRNA levels as a housekeeping gene for normalization.
In vivo cytokine assays. Mice were dosed with nanoparticles following a similar schedule as described above without influenza infection. Mice were dosed with nanoparticles twice at 0 and 20 hours. At 28 hours, 8 hours after the second injection, mice were killed and blood was collected by cardiac puncture. Serum was collected following clotting and centrifugation in serum collection tubes (Sarstedt, Newton, NC). Sandwich enzyme-linked immunosorbent assay was used to quantify serum IFN-α levels (PBL Laboratories, Piscataway, NJ) and serum TNF-α levels (eBioscience, San Diego, CA).
In vitro cytokine assays. Human PBMCs were purified from anonymous donor blood buffy coats (Institute for Transfusion Medicine Blood Bank, Suhl, Germany) by density centrifugation in Ficoll-Histopaque 1077 (Sigma-Aldrich, Munich, Germany) and plated in 96-well plates at 106cells/ml. GenePORTER 2 (Genlantis, San Diego, CA) or DOTAP (Roche Applied Science, Indianapolis, IN) were complexed with siRNAs and incubated at 130 nmol/l siRNA with PBMCs. After 24 hours, supernatants were collected and assayed by sandwich enzyme-linked immunosorbent assay for levels of human IFN-α and human TNF-α (BenderMed, Vienna, Austria).
In vitro RNAi assays. Vero cells were seeded at 10,000 cells/well in 96-well plates. After 1-day incubation (80–90% confluence), cells were transfected with siRNA complexed with Lipofectamine (Invitrogen). Cells were washed once with PBS after 24 hours then infected with PR/8 virus. Viral titer was assayed by immuno-staining plaque assay using primary anti-influenza A antibody (Chemicon, Temecula, CA) following fixing and permeabilization of cells.
SUPPLEMENTARY MATERIALTable S1. Physical characteristics of lipidoid-siRNA particles from representative batches.Figure S1. Fold reduction of viral titer due to different methods of lipidoid nanoparticle formulation. Particles were extruded through a 400 nm filter followed by dialysis, a 200 nm filter followed by lyophilization, or a 200 nm filter followed by first dialysis then lyophilization. Mice were injected Twice IV with nanoparticles at 2mg/kg siRNA prior to challenge with 12,000 PFU PR/8 influenza virus. Lung viral titer was determined by quantitative plaque forming assay and is expressed as relative to control (PBS) viral titer. N = 3 to 4, mean + SD shown. * p < 0.05 by two-tailed t-test.Figure S2. Inhibitory effects of mismatched siRNA control sequence correlated with induction of systemic Type I interferon response. Mice were injected Twice IV with lyophilized 98N12-5(1)-siRNA nanoparticles at 2 mg/kg siRNA prior to (a) infection with 12,000 PFU PR/8 influenza virus, or (b) blood collection by cardiac puncture. (a) Lung viral titer was determined by quantitative plaque forming assay. (b) Serum IFN-α concentration was determined by ELISA. Nanoparticles with siNP-1496 or scrambled control sequence both elicited high levels of IFN-α that corresponded with significant reductions in viral titer over nanoparticles formulated with siGFP-949. N = 3 to 4, mean + SD shown.Figure S3. Correlation between plaque forming unit (PFU) assay and branched DNA (bDNA) viral mRNA assay for determining viral load. Mice were instilled with siRNA by intranasal route (IN) 24 hours prior to infection with 400 or 2000 PFU of PR8 virus. After 48 hours post infection, mice were sacrificed and lungs immediately harvested, flash frozen in liquid nitrogen, and ground into powder.Figure S4. Survival following low dose influenza challenge and treatment with lipidoid nanoparticles. Mice were infected with 500 PFU by intranasal route (IN) at day 0. Lipidoid 98N12-5(1) nanoparticles encapsulating siNP-1496 were injected daily on days -2 through 0 (prophylactic) or days -2 through +2 (therapeutic). Body weight was tracked (a), and mice were euthanized when body weight dropped below 30% of original (b). Each group consisted of n=8 animals, except the Not Infected group (n=3). Differences in survival between study groups were not significant by Log-rank (Mantel-Cox) analysis.
Supplementary Material
Physical characteristics of lipidoid-siRNA particles from representative batches.
Fold reduction of viral titer due to different methods of lipidoid nanoparticle formulation. Particles were extruded through a 400 nm filter followed by dialysis, a 200 nm filter followed by lyophilization, or a 200 nm filter followed by first dialysis then lyophilization. Mice were injected Twice IV with nanoparticles at 2mg/kg siRNA prior to challenge with 12,000 PFU PR/8 influenza virus. Lung viral titer was determined by quantitative plaque forming assay and is expressed as relative to control (PBS) viral titer. N = 3 to 4, mean + SD shown. * p < 0.05 by two-tailed t-test.
Inhibitory effects of mismatched siRNA control sequence correlated with induction of systemic Type I interferon response. Mice were injected Twice IV with lyophilized 98N12-5(1)-siRNA nanoparticles at 2 mg/kg siRNA prior to (a) infection with 12,000 PFU PR/8 influenza virus, or (b) blood collection by cardiac puncture. (a) Lung viral titer was determined by quantitative plaque forming assay. (b) Serum IFN-α concentration was determined by ELISA. Nanoparticles with siNP-1496 or scrambled control sequence both elicited high levels of IFN-α that corresponded with significant reductions in viral titer over nanoparticles formulated with siGFP-949. N = 3 to 4, mean + SD shown.
Correlation between plaque forming unit (PFU) assay and branched DNA (bDNA) viral mRNA assay for determining viral load. Mice were instilled with siRNA by intranasal route (IN) 24 hours prior to infection with 400 or 2000 PFU of PR8 virus. After 48 hours post infection, mice were sacrificed and lungs immediately harvested, flash frozen in liquid nitrogen, and ground into powder.
Survival following low dose influenza challenge and treatment with lipidoid nanoparticles. Mice were infected with 500 PFU by intranasal route (IN) at day 0. Lipidoid 98N12-5(1) nanoparticles encapsulating siNP-1496 were injected daily on days -2 through 0 (prophylactic) or days -2 through +2 (therapeutic). Body weight was tracked (a), and mice were euthanized when body weight dropped below 30% of original (b). Each group consisted of n=8 animals, except the Not Infected group (n=3). Differences in survival between study groups were not significant by Log-rank (Mantel-Cox) analysis.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health AI56267 (to J.C.), P50-CA112967 (to J.C.), 1U54 CA119349 (to R.L. and J.C.), EB00244 (to R.L. and D.G.A.), and a grant from Alnylam (to R.L. and D.G.A.). We thank J. Maraganore for helpful comments. A.S., T.N., J.S., S.S.M., and A.d.F are current employees of Alnylam Pharmaceuticals. D.G.A and R.L. report receiving consulting fees from Alnylam Pharmaceuticals.
REFERENCES
- Sharp PA., and , Zamore PD. Molecular biology. RNA interference. Science. 2000;287:2431–2433. doi: 10.1126/science.287.5462.2431. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA., and , Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Leonard JN., and , Schaffer DV. Antiviral RNAi therapy: emerging approaches for hitting a moving target. Gene Ther. 2006;13:532–540. doi: 10.1038/sj.gt.3302645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurgers KB, Sharkey CM, Warfield KL., and , Bavari S. Oligonucleotide antiviral therapeutics: antisense and RNA interference for highly pathogenic RNA viruses. Antiviral Res. 2008;78:26–36. doi: 10.1016/j.antiviral.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Ban HS, Kim SS, Wu H, Pearson T, Greiner DL, et al. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134:577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi JJ, June CH., and , Kohn DB. Genetic therapies against HIV. Nat Biotechnol. 2007;25:1444–1454. doi: 10.1038/nbt1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Umehara T., and , Kohara M. Therapeutic application of RNA interference for hepatitis C virus. Adv Drug Deliv Rev. 2007;59:1263–1276. doi: 10.1016/j.addr.2007.03.022. [DOI] [PubMed] [Google Scholar]
- Bitko V, Musiyenko A, Shulyayeva O., and , Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat Med. 2005;11:50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- DeVincenzo J, Cehelsky JE, Alvarez R, Elbashir S, Harborth J, Toudjarska I, et al. Evaluation of the safety, tolerability and pharmacokinetics of ALN-RSV01, a novel RNAi antiviral therapeutic directed against respiratory syncytial virus (RSV) Antiviral Res. 2008;77:225–231. doi: 10.1016/j.antiviral.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BJ, Tang Q, Cheng D, Qin C, Xie FY, Wei Q, et al. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat Med. 2005;11:944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Hensley LE, Kagan E, Yu EZ, Geisbert JB, Daddario-DiCaprio K, et al. Postexposure protection of guinea pigs against a lethal ebola virus challenge is conferred by RNA interference. J Infect Dis. 2006;193:1650–1657. doi: 10.1086/504267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- Chen Y, Cheng G., and , Mahato RI. RNAi for treating hepatitis B viral infection. Pharm Res. 2008;25:72–86. doi: 10.1007/s11095-007-9504-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Q, McManus MT, Nguyen T, Shen CH, Sharp PA, Eisen HN, et al. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc Natl Acad Sci USA. 2003;100:2718–2723. doi: 10.1073/pnas.0437841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Q, Filip L, Bai A, Nguyen T, Eisen HN., and , Chen J. Inhibition of influenza virus production in virus-infected mice by RNA interference. Proc Natl Acad Sci USA. 2004;101:8676–8681. doi: 10.1073/pnas.0402486101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins SM, Lo CY, Tumpey TM., and , Epstein SL. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc Natl Acad Sci USA. 2004;101:8682–8686. doi: 10.1073/pnas.0402630101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M, Lu JJ, Ge Q, Zhang C, Chen J., and , Klibanov AM. Full deacylation of polyethylenimine dramatically boosts its gene delivery efficiency and specificity to mouse lung. Proc Natl Acad Sci USA. 2005;102:5679–5684. doi: 10.1073/pnas.0502067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG., and , Taylor JM. Small-interfering RNAs in the radar of the interferon system. Nat Cell Biol. 2003;5:771–772. doi: 10.1038/ncb0903-771. [DOI] [PubMed] [Google Scholar]
- Marques JT., and , Williams BR. Activation of the mammalian immune system by siRNAs. Nat Biotechnol. 2005;23:1399–1405. doi: 10.1038/nbt1161. [DOI] [PubMed] [Google Scholar]
- Kumagai Y, Takeuchi O, Kato H, Kumar H, Matsui K, Morii E, et al. Alveolar macrophages are the primary interferon-α producer in pulmonary infection with RNA viruses. Immunity. 2007;27:240–252. doi: 10.1016/j.immuni.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Alexopoulou L, Holt AC, Medzhitov R., and , Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K, Bhuyan P, Capodici J., and , Weissman D. Small interfering RNAs mediate sequence-independent gene suppression and induce immune activation by signaling through toll-like receptor 3. J Immunol. 2004;172:6545–6549. doi: 10.4049/jimmunol.172.11.6545. [DOI] [PubMed] [Google Scholar]
- Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, et al. Sequence-specific potent induction of IFN-α by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- Sioud M. Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J Mol Biol. 2005;348:1079–1090. doi: 10.1016/j.jmb.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Sioud M. RNA interference and innate immunity. Adv Drug Deliv Rev. 2007;59:153–163. doi: 10.1016/j.addr.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Eberle F, Giessler K, Deck C, Heeg K, Peter M, Richert C, et al. Modifications in small interfering RNA that separate immunostimulation from RNA interference. J Immunol. 2008;180:3229–3237. doi: 10.4049/jimmunol.180.5.3229. [DOI] [PubMed] [Google Scholar]
- Zamanian-Daryoush M, Marques JT, Gantier MP, Behlke MA, John M, Rayman P, et al. Determinants of cytokine induction by small interfering RNA in human peripheral blood mononuclear cells. J Interferon Cytokine Res. 2008;28:221–233. doi: 10.1089/jir.2007.0090. [DOI] [PubMed] [Google Scholar]
- Judge AD, Bola G, Lee AC., and , MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Lian ZX, Okada T, He XS, Kita H, Liu YJ, Ansari AA, et al. Heterogeneity of dendritic cells in the mouse liver: identification and characterization of four distinct populations. J Immunol. 2003;170:2323–2330. doi: 10.4049/jimmunol.170.5.2323. [DOI] [PubMed] [Google Scholar]
- Gorden KB, Gorski KS, Gibson SJ, Kedl RM, Kieper WC, Qiu X, et al. Synthetic TLR agonists reveal functional differences between human TLR7 and TLR8. J Immunol. 2005;174:1259–1268. doi: 10.4049/jimmunol.174.3.1259. [DOI] [PubMed] [Google Scholar]
- Vollmer J, Tluk S, Schmitz C, Hamm S, Jurk M, Forsbach A, et al. Immune stimulation mediated by autoantigen binding sites within small nuclear RNAs involves Toll-like receptors 7 and 8. J Exp Med. 2005;202:1575–1585. doi: 10.1084/jem.20051696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez R, Elbashir S, Borland T, Toudjarska I, Hadwiger P, John M, et al. RNAi-mediated silencing of the respiratory syncytial virus nucleocapsid defines a potent anti-viral strategy Antimicrob Agents Chemother 2009(epub ahead of print) [DOI] [PMC free article] [PubMed]
- Diebold SS, Kaisho T, Hemmi H, Akira S., and , Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Hammerbeck DM, Burleson GR, Schuller CJ, Vasilakos JP, Tomai M, Egging E, et al. Administration of a dual toll-like receptor 7 and toll-like receptor 8 agonist protects against influenza in rats. Antiviral Res. 2007;73:1–11. doi: 10.1016/j.antiviral.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Saravolac EG, Sabuda D, Crist C, Blasetti K, Schnell G, Yang H, et al. Immunoprophylactic strategies against respiratory influenza virus infection. Vaccine. 2001;19:2227–2232. doi: 10.1016/s0264-410x(00)00450-3. [DOI] [PubMed] [Google Scholar]
- Whitehead KA, Langer R., and , Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Kamenetsky M, Grefhorst A, Anderson NN, Racie TS, Bramlage B, Akinc A, et al. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc Natl Acad Sci USA. 2008;105:11915–11920. doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinc A, Goldberg M, Qin J, Dorkin JR, Gamba-Vitalo C, Maier M, et al. Development of lipidoid-siRNA formulations for systemic delivery to the liver. Mol Ther. 2009;17:872–879. doi: 10.1038/mt.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judge AD, Sood V, Shaw JR, Fang D, McClintock K., and , MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- Bourquin C, Schmidt L, Hornung V, Wurzenberger C, Anz D, Sandholzer N, et al. Immunostimulatory RNA oligonucleotides trigger an antigen-specific cytotoxic T-cell and IgG2a response. Blood. 2007;109:2953–2960. doi: 10.1182/blood-2006-07-033258. [DOI] [PubMed] [Google Scholar]
- Ge Q, Eisen HN., and , Chen J. Use of siRNAs to prevent and treat influenza virus infection. Virus Res. 2004;102:37–42. doi: 10.1016/j.virusres.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Robbins M, Judge A, Ambegia E, Choi C, Yaworski E, Palmer L, et al. Misinterpreting the therapeutic effects of small interfering RNA caused by immune stimulation. Hum Gene Ther. 2008;19:991–999. doi: 10.1089/hum.2008.131. [DOI] [PubMed] [Google Scholar]
- Robbins M, Judge A, Liang L, McClintock K, Yaworski E., and , MacLachlan I. 2'-O-methyl-modified RNAs act as TLR7 antagonists. Mol Ther. 2007;15:1663–1669. doi: 10.1038/sj.mt.6300240. [DOI] [PubMed] [Google Scholar]
- Poeck H, Besch R, Maihoefer C, Renn M, Tormo D, Morskaya SS, et al. 5'-Triphosphate-siRNA: turning gene silencing and Rig-I activation against melanoma. Nat Med. 2008;14:1256–1263. doi: 10.1038/nm.1887. [DOI] [PubMed] [Google Scholar]
- Akinc A, Thomas M, Klibanov AM., and , Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- Sonawane ND, Szoka FC., and , Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J Biol Chem. 2003;278:44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- Krieg AM. Toll-free vaccines. Nat Biotechnol. 2007;25:303–305. doi: 10.1038/nbt0307-303. [DOI] [PubMed] [Google Scholar]
- Krieg AM. The toll of too much TLR7. Immunity. 2007;27:695–697. doi: 10.1016/j.immuni.2007.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Physical characteristics of lipidoid-siRNA particles from representative batches.
Fold reduction of viral titer due to different methods of lipidoid nanoparticle formulation. Particles were extruded through a 400 nm filter followed by dialysis, a 200 nm filter followed by lyophilization, or a 200 nm filter followed by first dialysis then lyophilization. Mice were injected Twice IV with nanoparticles at 2mg/kg siRNA prior to challenge with 12,000 PFU PR/8 influenza virus. Lung viral titer was determined by quantitative plaque forming assay and is expressed as relative to control (PBS) viral titer. N = 3 to 4, mean + SD shown. * p < 0.05 by two-tailed t-test.
Inhibitory effects of mismatched siRNA control sequence correlated with induction of systemic Type I interferon response. Mice were injected Twice IV with lyophilized 98N12-5(1)-siRNA nanoparticles at 2 mg/kg siRNA prior to (a) infection with 12,000 PFU PR/8 influenza virus, or (b) blood collection by cardiac puncture. (a) Lung viral titer was determined by quantitative plaque forming assay. (b) Serum IFN-α concentration was determined by ELISA. Nanoparticles with siNP-1496 or scrambled control sequence both elicited high levels of IFN-α that corresponded with significant reductions in viral titer over nanoparticles formulated with siGFP-949. N = 3 to 4, mean + SD shown.
Correlation between plaque forming unit (PFU) assay and branched DNA (bDNA) viral mRNA assay for determining viral load. Mice were instilled with siRNA by intranasal route (IN) 24 hours prior to infection with 400 or 2000 PFU of PR8 virus. After 48 hours post infection, mice were sacrificed and lungs immediately harvested, flash frozen in liquid nitrogen, and ground into powder.
Survival following low dose influenza challenge and treatment with lipidoid nanoparticles. Mice were infected with 500 PFU by intranasal route (IN) at day 0. Lipidoid 98N12-5(1) nanoparticles encapsulating siNP-1496 were injected daily on days -2 through 0 (prophylactic) or days -2 through +2 (therapeutic). Body weight was tracked (a), and mice were euthanized when body weight dropped below 30% of original (b). Each group consisted of n=8 animals, except the Not Infected group (n=3). Differences in survival between study groups were not significant by Log-rank (Mantel-Cox) analysis.