Abstract
In gene therapeutic approaches targeting hematopoietic cells, insertional mutagenesis may provoke clonal dominance with potential progress to overt leukemia. To investigate the contribution of cell-intrinsic features and determine the frequency of insertional proto-oncogene activation, we sorted hematopoietic subpopulations before transduction with replication-deficient γ-retroviral vectors and studied the clonal repertoire in transplanted C57BL/6J mice. Progressive clonal dominance only developed in the progeny of populations with intrinsic stem cell potential, where expanding clones with insertional upregulation of proto-oncogenes such as Evi1 were retrieved with a frequency of ~10−4. Longitudinal studies by high-throughput sequencing and locus-specific quantitative PCR showed clones with >50-fold expansion between weeks 5 and 31 after transplantation. In contrast, insertional events in proto-oncogenes did not endow the progeny of multipotent or myeloid-restricted progenitors with the potential for clonal dominance (risk <10−6). Transducing sorted hematopoietic stem cells (HSCs) with self-inactivating (SIN) lentiviral vectors in short-term cultures improved chimerism, and although clonal dominance developed, there was no evidence for insertional events in the vicinity of proto-oncogenes as the underlying cause. We conclude that cell-intrinsic properties cooperate with vector-related features to determine the incidence and consequences of insertional mutagenesis. Furthermore, our study offers perspectives for refinement of animal experiments in the assessment of vector-related genotoxicity.
Introduction
Multipotent stem cells support organ development in utero and form a regenerative reserve in the ageing organism. In most organs with a high cell turnover, somatic stem cells represent a rare cell population that is characterized by a largely open chromatin structure allowing the execution of multiple genetic programmes, active maintenance of chromosome telomeres over multiple cell divisions, and the capacity to repopulate specialized niches that support self-renewal.1,2,3 Organ-resident multipotent cells such as hematopoietic stem cells (HSCs) are a preferred resource for novel approaches in regenerative medicine. Gene-modified HSCs can lead to sustained therapeutic effects in patients suffering from severe genetic disorders of blood cell function.4,5
However, transformation of hematopoietic cells by semi-randomly integrating gene vectors has been identified as a dose-limiting toxicity of gene therapy in both animal studies and clinical trials.4,6,7,8,9 Similar to the situation observed in murine and avian tumors caused by replicating retroviruses, upregulation of crucial proto-oncogenes by semi-random vector insertion in their chromosomal neighborhood has been identified as a causal event. The risk of insertional cell transformation by nonreplicating gene vectors depends on the number of insertions acquired per cell, the insertion properties, and the cargo of the vector.10,11,12 The impact of the target cell type is less well understood, although this question is of key interest to identify the parameters that explain the context-dependence of insertional transformation. Underlining this issue, it was recently demonstrated that mature T-lymphocytes are largely refractory to malignant transformation even following retroviral vector–mediated expression of potent oncogenes.13
Current protocols used in model organisms and clinical trials typically target a mixed population composed of a minority of repopulating HSCs (<1%) and a majority (>99%) of more mature hematopoietic progenitor cells (HPCs) that lack the potential for long-term repopulation. To obtain deeper insight into the extent of clonal skewing resulting from insertional mutagenesis, recently several groups have studied insertion profiles in the progeny of CD34+ cells isolated before infusion and compared the results with insertion profiles after long-term repopulation in clinical studies.14,15,16 This approach assumes that insertional mutagenesis may trigger the acquisition of long-term repopulation potential in HPC, which represent the great majority of the CD34+ population.
Following a similar concept, targeting highly purified HSCs has been proposed to prevent insertional adverse events.17 Several studies provided evidence that HPCs, although originally lacking the potential for long-term engraftment and self-renewal, can be transformed by retroviral gene transfer of certain oncogenes, further supporting the hypothesis that progenitor cells may contribute to the origin of transformed clones after insertional mutagenesis.18,19,20 Here, we addressed this important question in a murine model of competitive bone marrow (BM) transplantation using typical transduction conditions for γ-retroviral vectors but sorted cell populations as the starting material. In the same model, we also used serial clonal tracking to investigate the potential induction of clonal restriction following lentiviral vector transduction.
Results
Experimental strategy
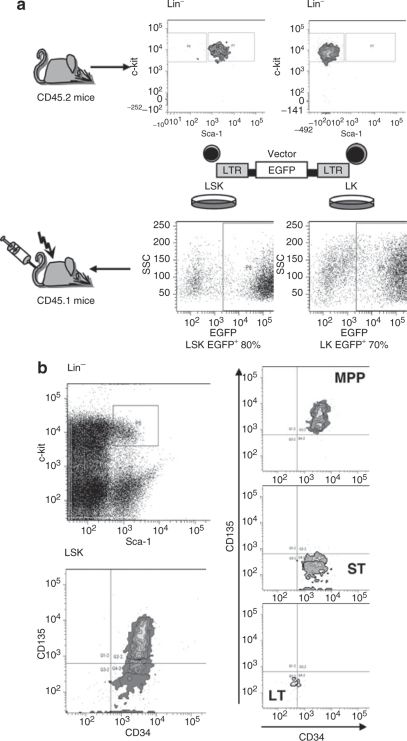
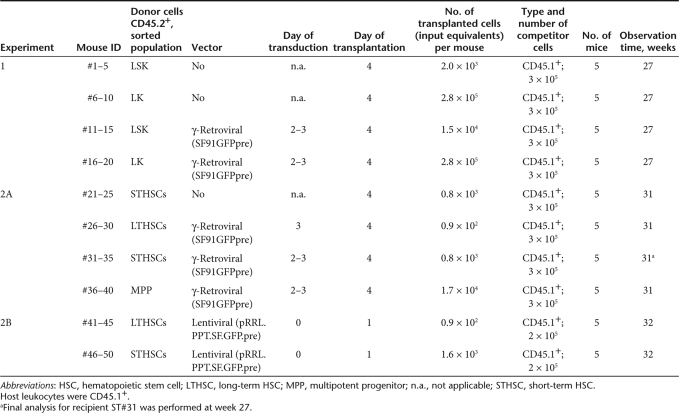
To identify the origin of insertional mutants, we used syngenic C57BL/6J mice in which donor and host cells are distinguished by a chimerism in the common leukocyte antigen (CD45.2+ donor cells). We enriched repopulating hematopoietic cells, including the most primitive HSCs, in the LSK population: lineage marker negative, Sca1+, and c-Kit+.21,22 In parallel, we purified lineage marker negative, Sca1−, and c-Kit+ HPCs, hereafter referred to as LK cells (Figure 1a). The purity of the sorted subpopulations was >94%. Sorted cells were cultured in serum-free media supplemented with recombinant cytokines and transduced with γ-retroviral vectors encoding enhanced green fluorescent protein (EGFP) under control of retroviral long terminal repeat (LTR) enhancer–promoters (SF91EGFPpre*) (Table 1). This vector type is known to trigger insertional dominance or leukemias in murine models and in a clinical trial.7,8
Figure 1.
Scheme of experiment and cell sorting conditions. (a) Sorted LSK and LK cells from bone marrow of untreated CD45.2 mice were prestimulated in serum-free conditions and transduced with γ-retroviral vector expressing EGFP fluorescent protein. Untransduced controls were included for both LSK and LK groups. On day 4 cells were transplanted into lethally irradiated congenic CD45.1 mice along with fresh CD45.1 competitor cells. Mice were observed for 7 months with regular analysis every 5 weeks of transgene expression and clonal dominance status. (b) The sorting procedure for LT and ST fractions of HSC and MPP followed established conditions.23 EGFP, enhanced green fluorescent protein; LSK, lineage marker negative, Sca1+, and c-Kit+; LTR, long terminal repeat; MPP, multipotent progenitor; SSC, side scatter.
Table 1.
Overview of experiments
After preparatory experiments to establish transplant doses and transduction conditions, we achieved similar gene marking rates in LSK and LK cells (80 and 70% EGFP+, respectively) (Figure 1a). Reflecting the initial frequency of the populations, we transplanted 19× more EGFP+ LK than LSK cells (280,000 vs. 15,000 per recipient). The subpopulations were thus used in quantities that match standard experimental conditions in studies of insertional transformation and clonal dominance. Control animals received untransduced donor and host cells following the same scheme (experiment 1, Table 1).
In a second experiment (experiment 2A, Table 1), we further split the LSK population into long-term repopulating HSCs (LTHSCs), short-term repopulating HSC (STHSCs), and multipotent progenitor (MPP) cells following marker definitions established for steady-state BM cells (Figure 1b).23 We transduced all three fractions with the γ-retroviral vector (Supplementary Figure S1a). In another arm of this experiment (experiment 2B, Table 1), we transduced LTHSCs and STHSCs with a lentiviral vector (Supplementary Figure S1b). Therefore, a short culture time (<24 hours) in the presence of only two cytokines was used (Materials and Methods, Table 1).
To monitor long-term repopulation, mice of all groups were prospectively observed for 7 months, with regular analyses of peripheral blood (PB) every 5–6 weeks and detailed final analysis of PB, BM, spleen, and in some cases liver.
Chimerism studies provide no evidence for insertional dominance of HPCs
To detect insertional dominance, we monitored donor chimerism, EGFP expression in PB leukocytes and analyzed the “integrome” of transduced cells using a ligation-mediated PCR (LMPCR) procedure that focuses on dominant bands and neglects weak amplicons; previous studies have indicated that dominant bands correlate with dominant clones.24 Hereafter, we refer to this approach as dominant band insertion site analysis (DBISA).
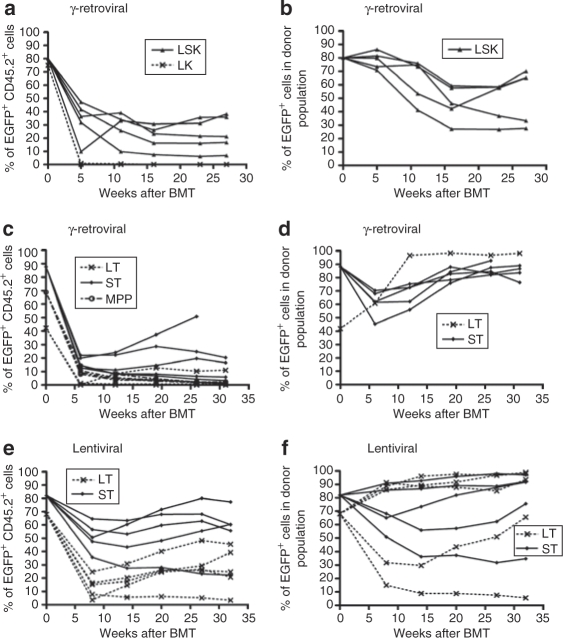
Following the known rules of hematopoietic reconstitution by sorted cell fractions, we expected that LK cells were not capable of long-term reconstitution unless being transformed by insertional mutagenesis. We found that neither transduced (EGFP+) nor untransduced (EGFP−) LK cells gave rise to significant levels of long-term repopulation (kinetics in Figure 2a; representative dot plots in Supplementary Figure S2). At early time points (PB, 5 weeks), EGFP+ LK progeny contributed only 0.31 ± 0.27% (mean ± 95% confidence interval) of the entire population with a further continuous decline until week 27 (0.04 ± 0.04%). Donor chimerism in PB and BM fell to 0.3 ± 0.1 and 0.8 ± 0.3%, respectively (Supplementary Table S1).
Figure 2.
Contribution of EGFP+ donor cells to peripheral blood leukocytes and gene marking within the donor population. (a,b) LSK and LK, γ-retroviral transduction. (c,d) STHSC, LTHSC, MPP, γ-retroviral transduction. (e,f) STHSC and LTHSC, lentiviral transduction. Values at week 0 for STHSC and LTHSC correspond to FACS analysis of cultured aliquots performed 7–18 days after transduction. BM, bone marrow; EGFP, enhanced green fluorescent protein; FACS, fluorescence-activated cell sorter; HSC, hematopoietic stem cell; LSK, lineage marker negative, Sca1+, and c-Kit+; LTHSC, long-term repopulating HSC; MPP, multipotent progenitor; PB, peripheral blood; Spl, spleen; STHSC, short-term repopulating HSC.
MPP progeny (experiment 2A) showed somewhat better survival long-term, although it was also not capable of establishing significant levels of hematopoiesis. EGFP+ MPP progeny contributed 10.38 ± 2.11% of PB leukocytes at early time points (6 weeks post-transplantation) with a decline to 1.48 ± 0.62% at final analysis (31 weeks, Figure 2c). Donor chimerism analysis in PB, BM, and spleen reached only 3.3 ± 1.7, 1.7 ± 0.4, and 2.4 ± 0.3%, respectively (Supplementary Table S1). While γ-retroviral transduction was efficient (~70%, Supplementary Figure S1a), no evidence was obtained for preferential survival of EGFP+ cells within the MPP progeny (Supplementary Figure S2).
LMPCR is more sensitive to detect transduced cells than flow cytometry but preferentially amplifies insertions from dominant clones.24 The pattern of γ-retroviral vector insertion sites (VISs) became reproducible when the starting material contained ~5 × 104 cells, corresponding to ~50 ng of clonal genomic DNA (Supplementary Figure S3a,b). In the LK progeny, authentic insertion sites could be recovered at early time points (week 11). In the MPP progeny, an oligoclonal pattern persisted until late time points (e.g., week 26, Supplementary Figure S4). DBISA revealed that one VIS in LK progeny and 7 out of 38 (18.4%) VISs in MPP progeny marked overt or suggested proto-oncogenes (Stat5b in LK; Fos, Bach2, D16Ertd472e, Notch1, Il4ra, Cd47, and BC031781 in MPP, Supplementary Tables S2 and S3). Despite such “suspicious” hits, some clones disappeared at later time points and none outcompeted competitor cells to contribute to >2% of hematopoiesis. Thus, we obtained no evidence that LK cells or MPP, or their transduced progeny, contribute to systemically relevant levels of clonal imbalance after retroviral vector–mediated insertional mutagenesis.
Dominant clones originate from cultured HSCs
Although transplanting almost 20-fold lower cell numbers in comparison with the LK conditions, the LSK progeny efficiently repopulated hosts in competition with cotransplanted, freshly isolated host-type cells (Figure 2a; engraftment analyses of individual animals in Supplementary Figure S2). γ-Retroviral marking rates for LSK EGFP+ progeny at week 27 in PB were 7–40%, accounting for 30–70% of donor-derived hematopoiesis (Figure 2a,b). Three mice showed a slight increase of the proportion of EGFP+ cells within the donor population after 16 weeks. Similar results were obtained in the progeny of γ-retrovirally transduced STHSCs, in marked contrast to the LK and MPP progeny (Figure 2c,d, Supplementary Figure S2). The progeny of STHSCs persisted at relatively high levels long-term even in the absence of γ-retroviral transduction (untransduced control group in Supplementary Figure S2). We cannot exclude that the culture conditions may have contributed to the long-term repopulation potential of STHSCs (see Discussion).
Interestingly, γ-retroviral transduction of purified LTHSCs was inefficient with our experimental conditions and these cells showed a substantial loss of their competitive repopulation potential (Figure 2c,d and Supplementary Figure S2). Control experiments with lentiviral transduction conditions revealed that this was not due to an experimental artefact in the sorting of LTHSCs.
Efficient marking of STHSCs and LTHSCs after overnight lentiviral transduction
To explore hematopoietic reconstitution after transducing STHSCs and LTHSCs using human immunodeficiency virus-1-based lentiviral vectors with their improved ability to transduce nondividing cells, we devised a short-term transduction protocol in a minimal cytokine cocktail, similar to earlier reports (see Material and methods and Table 1 for details).17 Of note, the lentiviral vector contained the same retroviral enhancer–promoter as the γ-retroviral vector, albeit as a single element located between self-inactivating (SIN) LTR. Considering that SIN vectors with an internal retroviral enhancer–promoter may trigger insertional transformation,12 this vector was chosen to allow for potential upregulation of a proto-oncogene in case of a semi-random insertion event in its vicinity. However, the lentiviral vector design is expected to lower the risk of insertional gene activation because the number of active enhancers is reduced to one, and direct gene activation by the 3′-LTR is not possible.
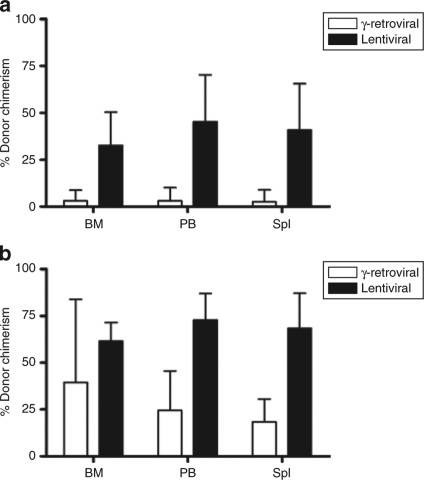
Compared to the results achieved with the prolonged γ-retroviral transduction protocol, we observed a significantly better engraftment of transduced cells with the lentiviral conditions (Wilcoxon rank sum test P < 0.05), although interanimal variability was still pronounced (Figures 2e,f and 3a,b; Supplementary Figure S2 and Supplementary Table S1). Surprisingly, STHSCs again showed superior long-term repopulation than LTHSCs (see Discussion). Within individual animals, marking levels were relatively stable, suggesting no major silencing of expression. Some animals tended to increase marking levels after week 12 (Figure 2e,f), as previously observed using the γ-retroviral conditions (Figure 2 b,d).
Figure 3.
Donor chimerism (CD45.2+ cells) analysis in recipients transplanted with progeny of γ-retrovirally or lentivirally transduced sorted populations of HSCs. (a) Donor chimerism final analysis in BM, PB, and Spl of recipients transplanted with progeny of γ-retrovirally or lentivirally transduced long-term repopulating fraction of HSCs (LTHSCs). Average donor chimerism calculated for five mice of each experimental group. Error bars indicate 95% confidence intervals. (b) Donor chimerism final analysis in BM, PB, and Spl of recipients transplanted with progeny of γ-retrovirally or lentivirally transduced short-term repopulating fraction of HSCs (STHSCs). Error bars indicate 95% confidence intervals. BM, bone marrow; HSC, hematopoietic stem cell; LTHSC, long-term repopulating HSC; PB, peripheral blood; Spl, spleen; STHSC, short-term repopulating HSC.
Insertion site analysis reveals preferential γ-retroviral hits in proto-oncogenes
To address whether a bias toward insertions in proto-oncogenes or related signaling genes had occurred in the progeny of sorted cell fractions, we analyzed hematopoietic samples of all 50 mice (overview in Table 1) every 5–6 weeks by DBISA (band pattern of the five recipients of transduced LSK cells in Figure 4a). Clonal fluctuations occurred in all mice, with oligoclonal stabilization in four of the five recipients (#12–15, Figure 4a). STHSC progeny generated the same oligoclonal pattern with progressive reduction of diversity and long-term persistence (representative data in Figure 4b). The poor results achieved with γ-retroviral transduction of purified LTHSCs indicate that this pattern cannot be attributed to the potential contamination of STHSC cultures with a priori LTHSCs.
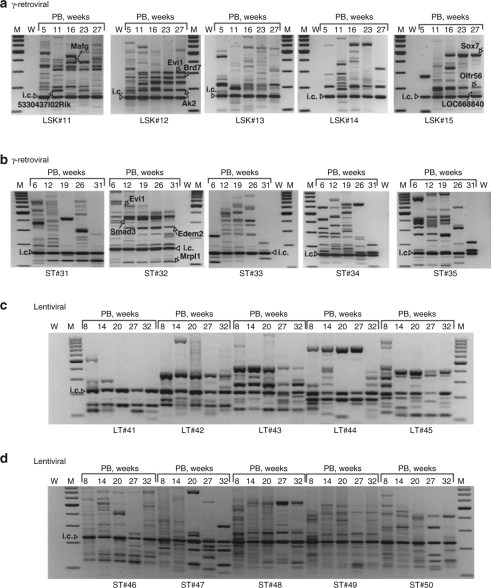
Figure 4.
Insertion sites analysis at different time points after BMT. (a) γ-Retroviral VIS analysis in PB samples of individual LSK recipients #11–15 obtained by LMPCR. Arrows indicate selected insertion sites. (b) γ-Retroviral VIS analysis in PB samples of individual STHSC recipients ST#31–35. (c) Lentiviral VIS analysis in PB samples of individual LTHSC recipients LT#41–45. (d) Lentiviral VIS analysis in PB samples of STHSC recipients ST#46–50. BMT, bone marrow transplantation; i.c., internal control of LMPCR reaction; LSK, lineage marker negative, Sca1+, and c-Kit+; M, marker; LMPCR, ligation-mediated PCR; LTHSC, long-term repopulating hematopoietic stem cell; PB, peripheral blood; STHSC, short-term repopulating hematopoietic stem cell; VIS, vector insertion site; W, water; weeks, weeks after transplantation.
As in our previous studies using DBISA,7,10,24 we excised the most prominent amplicons, likely to reflect dominant clones,24 for sequence analysis. To address whether the pattern observed in the PB reflected the primary site of hematopoiesis, we analyzed samples from BM and spleen. In all cases the dominant bands chosen for insertion site sequencing were also found in at least one of these two major hematopoietic organs (Supplementary Figure S4b,c).
The complete list of “dominant amplicons” recovered in LSK progeny (n = 52) and STHSC progeny (n = 49) by DBISA is provided in Supplementary Tables S2 and S3. Very similar to our earlier studies targeting a bulk population of lineage-negative cells,24 we found an over-representation of insertions within 150 kb of genes listed as common insertion sites in the retrovirally tagged cancer gene database (Supplementary Table S4).25 We found no difference in the distribution around the transcriptional start site between the IDDb, data sets derived from primitive cell populations (LSK and STHSCs) and data sets originating from cell populations that lack the potential for clonal dominance (LK and MPP) (P = 0.432, Wilcoxon rank sum test).
Clonal restriction in the progeny of lentivirally transduced STHSCs and LTHSCs
Insertional pattern of cultured (untransplanted) γ-retrovirally transduced MPP and STHSCs revealed highly polyclonal situations (data not shown), as reported earlier.24 Analysis of cultured cells also demonstrated the ability of γ-retroviral vectors to transduce the progeny of LTHSCs (data not shown). The lentiviral LMPCR protocol demonstrated an initial polyclonal pattern especially in the mice receiving transduced STHSCs, whereas mice receiving transduced LTHSCs showed a more oligoclonal pattern (Figure 4c,d). Based on the number of observed integrations, we estimate that about 10% of the initial LTHSC input (~90 cells per animal, Table 1) manifested as dominant clones. This suggests that many LTHSCs may have lost their homing and engraftment properties due to the induction of cell cycle activity in response to cytokines,26 and that others did not become dominant as a result of the known cell-intrinsic heterogeneity. Towards the final analysis, a trend to oligoclonal dominance was also observed in the STHSC group, with consistent clonal representations in PB, BM, and spleen (Figure 4d; Supplementary Figure S5). Remarkably, this had no impact on overall marking levels and chimerism (Figure 2e,f; Supplementary Figure S1). In contrast, the LTHSC group showed a similar clonality pattern at the final analysis as at week 8 (Figure 4c), revealing the correctness of our conditions for cell sorting.
To address whether the clonality pattern observed in mice receiving lentivirally transduced HSCs was the result of insertional mutagenesis, we sequenced the dominant amplicons, using the same criteria as above (Supplementary Table S5). Dominant amplicons of lentiviral insertions were not enriched for locations close to proto-oncogenes or other signaling genes (Supplementary Table S4) and instead showed a much more frequent insertion in repeat regions than γ-retroviral insertions. Thus, although the vector contained a strong internal retroviral promoter that is capable of long-distance enhancer interactions and proto-oncogene upregulation from a SIN vector,27 “integrome” studies gave no evidence for clonal imbalance as a result of insertional hits in proto-oncogenes by lentiviral vectors. The absence of “suspicious” insertion events reveals a strong cell-intrinsic component in the induction of the observed clonal dominance.
Validation of DBISA by high-throughput sequencing, locus-specific quantitative PCR, and quantitative RT-PCR
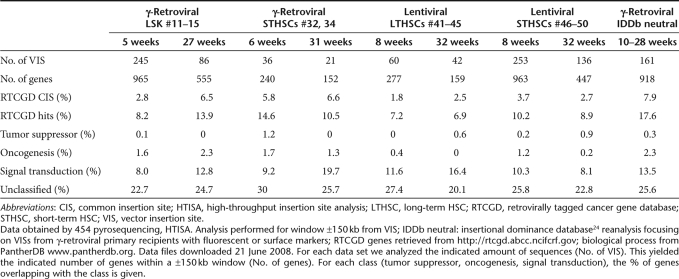
It could be argued that the selection of dominant amplicons for sequencing studies, as performed in DBISA, represents an arbitrary choice. We thus implemented a novel approach of high-throughput insertion site analysis (HTISA) using the 454 sequencing method.28 Using barcoded primers29 for the second exponential PCR step of the LMPCR protocol, we were able to run multiple samples in a single HTISA. In γ-retrovirally transduced LSK and STHSCs, HTISA mapped 3.6× more insertion sites than the “manual” approach (388 vs. 107), and the HTISA sequences were significantly less frequently associated with proto-oncogenes (4.6% of 388 vs. 8% of 107; P < 0.001 in Fisher exact, two-sided) (Table 2, Supplementary Table S4). VISs obtained by HTISA are available on request. Dominant amplicons from DBISA led to a significantly higher number of sequence reads in HTISA, whereas weak amplicons that we had neglected in DBISA gave rise to relatively rare sequence reads in HTISA. This confirmed that VISs close to proto-oncogenes increased the likelihood of clonal dominance, manifesting in dominant PCR products.
Table 2.
VIS distribution according to gene classes (±150 kb), HTISA
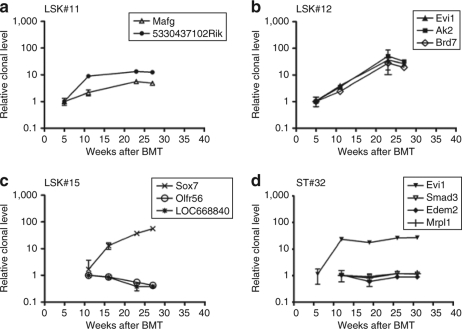
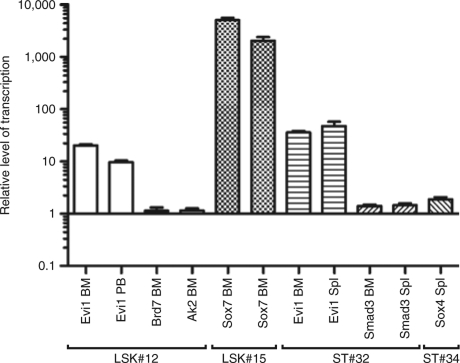
To better quantify the kinetics of clonal dominance, we studied selected clones from four recipients by longitudinal locus-specific quantitative PCR. For 12 retroviral VISs, we designed amplicons with internal probes to detect a fusion sequence between the retroviral vector and the neighboring cellular DNA. These data not only further support the validity of DBISA (compare Figure 5 and Figure 4a,b), they furthermore showed that individual clones amplified between fivefold to 56-fold from week 5 until week 31 (Figure 5). In many recipients analyzed in this way, at least one of the expanding clones had a VIS next to a putative proto-oncogene or developmental gene [Mafg in LSK#11 (Figure 5a); Evi1 and Brd7 in LSK#12 (Figure 5b); Sox7 in LSK#15 (Figure 5c); and again Evi1 in ST#32 (Figure 5d)]. High levels of transcriptional activation were observed for Sox7, a gene involved in Wnt signaling30 (5,098 ± 455x in LSK#15, Figure 6), and also for the transcription factor Evi1, which is essential for HSC's self-renewal31 (>tenfold upregulation in recipients LSK#12 and ST#32, Figure 6).
Figure 5.
Locus-specific qPCR analysis of selected clones in γ-retroviral LSK and STHSC progeny. (a–c) Locus-specific qPCR analysis of selected clones in individual recipients of LSK cells. Relative quantification of a target gene amplicon was estimated in comparison to amplicon at early time points, corresponding week 5 for (a) LSK#11, (b) LSK#12, and week 11 for (c) LSK#15. (d) Locus-specific qPCR analysis of selected clones in recipient ST#32. Relative quantification of Evi1 amplicon was estimated in comparison to amplicon at time point 5 weeks after BMT; for Smad3, Edem2, Mrpl1 amplicons in comparison to 11 weeks. Each analysis was performed in triplicate. Error bars indicate standard deviations. BMT, bone marrow transplantation; LSK, lineage marker negative, Sca1+, and c-Kit+; qPCR, quantitative PCR; STHSC, short-term repopulating hematopoietic stem cell.
Figure 6.
Transcriptional dysregulation of loci targeted by γ-retroviral insertional mutagenesis. Real-time RT-PCR shows dysregulation of selected targeted alleles in BM, PB, Spl of selected mice (LSK#12, LSK#15, ST#32, and ST#34). The expression level of the control mouse CD45.2 (BM, PB) or CD45.1 (Spl) was set to 1. Mean values of at least three measurements. Error bars indicate 95% confidence intervals. BM, bone marrow; LSK, lineage marker negative, Sca1+, and c-Kit+; PB, peripheral blood; RT, reverse transcription; Spl, spleen.
Having validated the consequences of VIS in proto-oncogenes (gene upregulation coupled with clonal expansion), the DBISA data reveal a frequency of functionally relevant insertions in Evi1 in the order of ~1 in 75,000 LSK-derived cells, and 1 in 4,000 STHSC-derived cells (reflecting the numbers for transplanted cells and engrafted mice in Table 1). In contrast, no such event was detected in 1,400,000 transplanted γ-retrovirally transduced LK progeny, 85,000 γ-retrovirally transduced MPP progeny, or 8,000 lentivirally transduced STHSC progeny. In line with this finding, lentiviral insertions in the progeny of enriched HSCs were significantly less likely to occur close to proto-oncogenes and did not show a time-dependent enrichment for such events under the experimental conditions chosen in this study, in contrast to the results obtained with γ-retroviral vectors (Table 2). We thus conclude that cell-intrinsic and vector-related properties cooperate to determine the incidence and consequences of insertional mutagenesis.
Discussion
This study reveals that the cultured progeny of HSCs, but not HPCs, is the primary source of insertional mutants after γ-retroviral transduction. Insertional dominance of LSK-derived cells, and of the STHSCs contained therein, was demonstrated based on progressive clonal restriction with over-representation of hits in potential or established proto-oncogenes, upregulation of the affected genes, and clonal expansion as validated by insertion site–specific quantitative PCR. In contrast, despite the transplantation of almost 20-fold more transduced cells and the relatively frequent detection of insertions in proto-oncogenes, the progeny of MPP and more mature HPC (LK cells) was unable to give rise to dominant clones. These data reveal that the pre-existing intrinsic repopulation potential dominates the susceptibility of hematopoietic cells to insertional mutagenesis.
A surprising finding made in both the γ-retroviral and the lentiviral arms of our study was that STHSCs provided a better chimerism long-term than LTHSCs. This can be attributed to a combined effect of higher cell numbers (reflecting STHSCs abundance in vivo) and a culture-dependent “enhancement” of STHSC function. Of note, the phenotype of STHSCs and LTHSCs has been established using freshly isolated cells.21,22 In our experimental conditions, the sorted cell populations are exposed to cytokines and may thus change their fate in vitro before receiving a potential transforming hit by a retroviral vector. Even relatively short culture periods of >8 hours have been shown to alter the functional properties of HSCs.32 Irrespective of the potential effects of the cell-culture conditions, the absence of sustained clonal dominance in the progeny of MPP or LK cells underlines the existence of hierarchically organized cell-intrinsic features as a confounding factor of insertional transformation.
Our study allows a more accurate determination of the risk associated with γ-retroviral vector insertions. The chance to induce clonal dominance by insertion of a vector with strong enhancer–promoter in the LTR was found to be around 1 in 10,000 repopulating HSCs, at least two orders of magnitude higher than in HPCs. These results strongly suggest that previous γ-retroviral tracking studies performed in animals and clinical trials after transplanting mixed populations of HSCs and HPCs primarily reflected insertional events occurring in the minor fraction of a priori HSCs or their progeny arising during cell culture before transplantation. When attempting to define insertional repertoires before transplantation, it may thus be better to focus on (the progeny of) populations that are highly enriched for HSCs. It will be important to determine whether in genetic diseases that may affect early stages of hematopoiesis (such as X-linked severe combined immunodeficiency), the origin of insertional mutants might extend into more committed populations.33
As γ-retroviral vectors preferentially target open chromatin and active genes,34 the epigenetic status of HSCs and their daughter cells arising during ex vivo culture may contribute to their increased risk by providing better access to the transforming genes. Cell-intrinsic factors that regulate engraftment after transplantation35 play a role in the frequency with which different cell types can lead to dominance, and could be affected by dysregulation of engraftment genes. However, the progeny of cultured LK cells or MPP cells that exhibited limited engraftment was unable to expand to clonal dominance even when containing insertions in proto-oncogenes. In the murine setting studied here, telomere insufficiency appears to be an unlikely explanation for the resistance of more mature progenitor cells to insertional transformation.36 Similarly, transduced mature T-cells were able to engraft recipient mice but were unable to induce uncontrolled expansion even when transduced with retroviral vectors expressing overt oncogenes.13 This suggests that besides potential alterations of engraftment, the primary limitation controlling the emergence of insertional mutants is associated with proliferation potential and sustained self-renewal, which is considered to be the hallmark of long-term repopulating HSCs.37 Of note, even short-term repopulating HSCs are estimated to produce a clonal progeny of >1012 blood cells.37 This is several orders of magnitude above the numbers required to detect insertional mutants in the recently established replating assays,12,38 and suggests that cell-culture assays targeting primary hematopoietic BM cells are not necessarily scoring the transformation of true HSCs.
In contrast to previous studies addressing the oncogenic potential of purified murine HPCs,18,19,20 the vectors used here did not encode leukemogenic oncogenes. Our data imply that insertional upregulation of a single cellular gene, in contrast to the expression of more complex oncogenic fusion proteins such as MLL-ENL18 or MLL-AF9,20 is not sufficient to transform HPCs. The ability to elicit leukemia by genetic modification of HPCs has indeed been shown to depend upon the nature of the oncogene expressed.19 We hypothesize that similar restrictions operate in other organs, greatly reducing the risk of insertional mutagenesis especially in tissues with a high frequency of cells lacking the potential for extensive proliferation.
Initial insight into the signals evoking a self-renewal program with sustained proliferation in transformed HPCs has been obtained.20,37 Some of the underlying genes (such as homeobox genes) may well be hit by insertional mutagenesis.7,24,39 Insertional dysregulation of more than one gene may thus required to convey the potential for clonal dominance to cells lacking stem cell potential. Such a scenario may be possible when dose-escalated gene transfer takes place,10 and is typically encountered in tumors originating from replicating MLV in vivo.25 In contrast, upregulating a single proto-oncogene such as Evi1 or the related gene Prdm16 in HSCs may suffice to induce leukemia.6,27
Our data also indicate that a better purification of stem cells may not necessarily reduce the genotoxic risk of gene transfer, unless the number of transduced HSCs is restricted. Interestingly, we found that lentiviral transduction of a limited number of purified HSCs reduces the risk of inducing clonal imbalance by insertional events close to proto-oncogenes, although the overall efficiency of gene transfer into HSCs was significantly increased compared to the γ-retroviral conditions. Thus, the differences that were previously identified between lentiviral and γ-retroviral gene targets in a study of cultured CD34+ cells, with evidence reported of a significantly higher risk of γ-retroviral vectors to integrate in the vicinity of proto-oncogenes,40 appear to have major functional consequences. However, in the clinical setting, lentiviral insertions in proto-oncogenes may still occur more frequently than observed here, considering the considerably larger cell number typically treated in humans. This may explain why in a comparison of lentiviral and γ-retroviral insertion sites performed in repopulating cells from nonhuman primates, both vector types exhibited a similar over-representation of insertions close to proto-oncogenes.41 Therefore, developing vectors with safer expression cassettes that are less likely to activate proto-oncogenes in stem cells remains an important way to reduce the risk of insertional mutagenesis.12,42,43 A more controversial issue is whether a limitation of target cell numbers, as performed here and suggested by others earlier,17 may be beneficial. When transplanting a limited number of gene-modified HSCs in a myeloablative setting, the increased hematopoietic stress resulting from the need for strong expansion may antagonize the potentially increased safety associated with the smaller insertional repertoire. Conversely, removing STHSCs from LTHSCs (once appropriate markers are available in the human system) might reduce the potential for malignancy in submyeloablative transplant situations where STHSCs are not required to accelerate engraftment.
Finally, the kinetics and insertion profile of the lentivirally transduced HSC populations suggest that clonal dominance occurring after transplantation of gene-modified HSCs is not necessarily caused by insertional mutagenesis but may rather reflect the known cell-intrinsic heterogeneity.32,44,45,46 We would expect that in humans clonal restriction may require longer follow-ups than in the murine model. Bioinformatical analyses of individual genes or gene networks associated with clonal dominance thus need to be coupled with functional studies before expanding cell clones can be addressed as transformed mutants. In this regard, this study indicates that murine experiments performed to address the relative safety of different vectors and target cells on insertional biosafety in the hematopoietic system can be refined to relatively small group sizes, observation times <25 weeks and an “integrome” rather than a tumor endpoint.
Materials and Methods
Cell sorting and flow cytometry. LSK (Linneg/loIL7RnegSca1highckithigh) and LK (Linneg/loIL7RnegSca1negckithigh) hematopoietic subpopulations were isolated by staining of freshly prepared CD45.2 BM cells with PerCPCy5.5-labeled anti-Sca1, APC-labeled anti-c-kit, and a cocktail of PE-labeled monoclonal antibodies (mAbs) directed against lineage markers (CD11b, Gr1, Ter119, CD3, CD4, CD8, B220, IL7R). MPP (LSK CD34+ Flt3+), STHSCs (LSK CD34+ Flt3−), and LTHSCs (LSK CD34− Flt3−) were sorted as described,23 using c-kit-APC, Sca1-PerCPCy5.5, Flk2-PE (CD135), CD34-FITC, and biotinylated antibodies against lineage markers in combination with Streptavidin-PE-Cy. Erythrocytes, debris, and dead cells were excluded by forward scatter, side scatter, and 4,6-diamidino-2-phenylindole gating, and cell aggregates by forward-scatter area vs. pulse width gating. Gates were set according to control samples using the Fluorescence Minus One approach. FACSAria, LSRII, and FACS-Calibur instruments (Becton Dickinson, Heidelberg, Germany) were used for cell sorting and analysis. Data were analyzed using FlowJo software (Tree Star, Ashland, OR). Antibodies were purchased from BDPharmingen or eBiocience (San Diego, CA).
Retroviral constructs and retroviral transduction. The γ-retroviral vector pRSF91GFPpre* contains the LTR derived from spleen focus-forming virus, the primer binding site and leader sequences derived from the murine embryonic stem cell virus, EGFP as the transgene, and the woodchuck hepatitis posttranscriptional regulatory element.47 High titer ecotropic vector stocks were produced following established protocols.47 pRRL.PPT.SF.GFP.pre, a SIN third generation human immunodeficiency virus-1 lentiviral vector,48 was pseudotyped with the glycoprotein of vesicular stomatitis virus. Viral particles were concentrated by low-speed centrifugation and resuspended in serum-free StemSpan medium (Stem Cell Technologies, Vancouver, Canada). For γ-retroviral transduction, cells were cultivated in serum-free medium supplemented with murine interleukin-3 (20 ng/ml), murine stem cell factor (50 ng/ml), human FMS-like tyrosine kinase-3 (50 ng/ml), and human interleukin-11 (50 ng/ml; all cytokines from Peprotech, Hamburg, Germany). Untransduced control cells were kept in the same cytokine conditions until the day of BM transplantation. On the second day LSK and LK cells were γ-retrovirally transduced with a multiplicity of infection (MOI) 5 and 10, respectively, and on day 3 with MOI 10 for both LSK and LK cells. STHSCs and MPP cells were transduced with MOI 10 at days 2 and 3. Given the limited cell number, LTHSCs were γ-retrovirally transduced only once with MOI 20 on day 3. To determine transduction efficiency, aliquots of STHSCs and LTHSCs were kept in vitro in the presence of murine interleukin-3 (20 ng/ml), murine stem cell factor (50 ng/ml), human FMS-like tyrosine kinase-3 (50 ng/ml), human interleukin-11 (50 ng/ml) for additional 3 days (day 7 in vitro) and 10 days (day 18 in vitro), respectively. For lentiviral transduction of LTHSCs and STHSCs, an MOI of 30 was used in serum-free medium in the presence of murine stem cell factor (50 ng/ml), human interleukin-11 (50 ng/ml), and protamine sulphate (8 µg/ml), and cells were cultivated for 20 hours in Retronectin coated plates.
Mice and transplantation conditions. Mice purchased from Charles River (Sulzfeld, Germany) Germany were kept in microisolators in the specific pathogen-free animal facility of Hannover Medical School. C57BL/6J (CD45.2) female mice served as cell donors, congeneic female CD45.1 mice (B6.SJL-PtprcaPep3b/BoyJ) as recipients. Experiments approved by the local ethical committee were performed according to their guidelines. Before BM transplantation (<24 hours), recipients ≥12 weeks of age were conditioned by myeloablative irradiation (10 Gy). Donor cells were cotransplanted intravenously with freshly isolated unfractionated BM cells for radioprotection, as indicated (Table 1). Transplanted mice received ciprofloxacin at 100 mg/ml in drinking water for the first 2 weeks.
Chimerism analysis. Please refer to Supplementary Materials and Methods.
Vector integration site identification, bioinformatical analyses, and real-time PCR of neighboring genes. Please refer to Supplementary Materials and Methods.
Pyrosequencing (HTISA). LMPCR was performed on PB of retrovirally transduced LSK cells (25 samples, 5 mice, 5 time points), PB of retrovirally transduced STHSCs (10 samples, 2 mice, 5 time points), PB and BM of lentivirally transduced STHSCs (20 samples, 5 mice, 3 time points and 1 BM sample), and PB and BM of lentivirally transduced LTHSCs (20 samples, 5 mice, 3 time points and 1 BM sample) as described.49 After exponential PCR, products were digested with Sac1 or HindIII for lentiviral or γ-retroviral amplicons, respectively. A nested PCR with primer containing a 20-bp 454 adapter and an 8-bp DNA barcode29 was performed. The primers were:
γ-retrovirus LTR: 5′-GCC TCC CTC GCG CCA TCA G[barcode]CC ATG CCT TGC AAA ATG GC-3′; lentivirus LTR: 5′-GCC TCC CTC GCG CCA TCA G[barcode]AGT AGT GTG TGC CCG TCT GT-3′; common linker primer: 5′ GCC TTG CCA GCC CGC TCA G-AGT GGC ACA GCA GTT AGG-3′.
DNA concentrations of the nested PCR products were determined and equal amounts (1.7 ng/sample) of DNA were mixed for further emulsion PCR and 454 pyrosequencing (GATC, Konstanz, Germany). The resulting sequences were clustered using CD-HIT software,50 aligned using BLAST and annotated using in-house designed software.
SUPPLEMENTARY MATERIALFigure S1. Transduction efficiency of sorted cells as analyzed by flow cytometry.Figure S2. Engraftment analysis performed on bone marrow of recipients from different experimental groups.Figure S3. LMPCR sensitivity validation in two independent experiments.Figure S4. γ-retroviral vector insertion site analysis in different organs.Figure S5. Lentiviral vector insertion site analysis in different organs.Table S1. Donor chimerism (CD45.2+ cells) final analysis in peripheral blood, bone marrow, and spleen of primary recipients.Table S2. Insertion sites recovered from γ-retroviral LSK and LK EGFP progeny (DBISA).Table S3. Insertion sites recovered from γ-retroviral LTHSCs, STHSCs, and MPP EGFP progeny (DBISA).Table S4. Vector insertion sites distribution according to gene classes (±150 kb), DBISA.Table S5. Insertion sites recovered from lentiviral LTHSC and STHSC EGFP progeny (DBISA).Materials and Methods.
Supplementary Material
Transduction efficiency of sorted cells as analyzed by flow cytometry.
Engraftment analysis performed on bone marrow of recipients from different experimental groups.
LMPCR sensitivity validation in two independent experiments.
γ-retroviral vector insertion site analysis in different organs.
Lentiviral vector insertion site analysis in different organs.
Donor chimerism (CD45.2+ cells) final analysis in peripheral blood, bone marrow, and spleen of primary recipients.
Insertion sites recovered from γ-retroviral LSK and LK EGFP progeny (DBISA).
Insertion sites recovered from γ-retroviral LTHSCs, STHSCs, and MPP EGFP progeny (DBISA).
Vector insertion sites distribution according to gene classes (±150 kb), DBISA.
Insertion sites recovered from lentiviral LTHSC and STHSC EGFP progeny (DBISA).
Acknowledgments
We are grateful for technical assistance in ligation-mediated PCR and flow cytometry by Yvonne Fernandez-Munoz. We thank Thomas Neumann, Cindy Elfers, Martin Hapke, Niels Heinz, and Mathias Rhein for assistance with mouse monitoring and data acquisition, Natalia Bogdanova and Tilo Dörk for help with sequencing, Matthias Ballmaier for sorting procedures, Axel Schambach for providing the vector. Financial support was provided by the Deutsche Forschungsgemeinschaft (DFG SPP1230 and excellence cluster REBIRTH to C.B. and Z.L.), and the Bundesministerium für Bildung und Forschung (TreatID to C.B.). The authors declare that they have no conflicts of interest.
REFERENCES
- Niwa H. Open conformation chromatin and pluripotency. Genes Dev. 2007;21:2671–2676. doi: 10.1101/gad.1615707. [DOI] [PubMed] [Google Scholar]
- Lansdorp PM. Role of telomerase in hematopoietic stem cells. Ann N Y Acad Sci. 2005;1044:220–227. doi: 10.1196/annals.1349.027. [DOI] [PubMed] [Google Scholar]
- Martinez-Agosto JA, Mikkola HK, Hartenstein V., and , Banerjee U. The hematopoietic stem cell and its niche: a comparative view. Genes Dev. 2007;21:3044–3060. doi: 10.1101/gad.1602607. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiuti A, Cattaneo F, Galimberti S, Benninghoff U, Cassani B, Callegaro L, et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med. 2009;360:447–458. doi: 10.1056/NEJMoa0805817. [DOI] [PubMed] [Google Scholar]
- Li Z, Dullmann J, Schiedlmeier B, Schmidt M, von Kalle C, Meyer J, et al. Murine leukemia induced by retroviral gene marking. Science. 2002;296:497. doi: 10.1126/science.1068893. [DOI] [PubMed] [Google Scholar]
- Kustikova O, Fehse B, Modlich U, Yang M, Düllmann J, Kamino K, et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Seggewiss R, Pittaluga S, Adler RL, Guenaga FJ, Ferguson C, Pilz IH, et al. Acute myeloid leukemia is associated with retroviral gene transfer to hematopoietic progenitor cells in a rhesus macaque. Blood. 2006;107:3865–3867. doi: 10.1182/blood-2005-10-4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlich U, Kustikova OS, Schmidt M, Rudolph C, Meyer J, Li Z, et al. Leukemias following retroviral transfer of multidrug resistance 1 (MDR1) are driven by combinatorial insertional mutagenesis. Blood. 2005;105:4235–4246. doi: 10.1182/blood-2004-11-4535. [DOI] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Ponzoni M, Bartholomae C, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- Zychlinski D, Schambach A, Modlich U, Maetzig T, Meyer J, Grassman E, et al. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol Ther. 2008;16:718–725. doi: 10.1038/mt.2008.5. [DOI] [PubMed] [Google Scholar]
- Newrzela S, Cornils K, Li Z, Baum C, Brugman MH, Hartmann M, et al. Resistance of mature T cells to oncogene transformation. Blood. 2008;112:2278–2286. doi: 10.1182/blood-2007-12-128751. [DOI] [PubMed] [Google Scholar]
- Deichmann A, Hacein-Bey-Abina S, Schmidt M, Garrigue A, Brugman MH, Hu J, et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J Clin Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzwaelder K, Howe SJ, Schmidt M, Brugman MH, Deichmann A, Glimm H, et al. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo. J Clin Invest. 2007;117:2241–2249. doi: 10.1172/JCI31661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiuti A, Cassani B, Andolfi G, Mirolo M, Biasco L, Recchia A, et al. Multilineage hematopoietic reconstitution without clonal selection in ADA-SCID patients treated with stem cell gene therapy. J Clin Invest. 2007;117:2233–2240. doi: 10.1172/JCI31666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky G, Kotton DN, Fabian AJ, Gray JT, Lee JS., and , Mulligan RC. Efficiency of transduction of highly purified murine hematopoietic stem cells by lentiviral and oncoretroviral vectors under conditions of minimal in vitro manipulation. Mol Ther. 2005;11:932–940. doi: 10.1016/j.ymthe.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Cozzio A, Passegué E, Ayton PM, Karsunky H, Cleary ML., and , Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntly BJ, Shigematsu H, Deguchi K, Lee BH, Mizuno S, Duclos N, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6:587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Okada S, Nakauchi H, Nagayoshi K, Nishikawa S, Miura Y., and , Suda T. In vivo and in vitro stem cell function of c-kit- and Sca-1-positive murine hematopoietic cells. Blood. 1992;80:3044–3050. [PubMed] [Google Scholar]
- Osawa M, Nakamura K, Nishi N, Takahasi N, Tokuomoto Y, Inoue H, et al. In vivo self-renewal of c-Kit+ Sca-1+ Lin(low/−) hemopoietic stem cells. J Immunol. 1996;156:3207–3214. [PubMed] [Google Scholar]
- Yang L, Bryder D, Adolfsson J, Nygren J, Månsson R, Sigvardsson M, et al. Identification of Lin−Sca1+kit+CD34+Flt3- short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood. 2005;105:2717–2723. doi: 10.1182/blood-2004-06-2159. [DOI] [PubMed] [Google Scholar]
- Kustikova OS, Geiger H, Li Z, Brugman MH, Chambers SM, Shaw CA, et al. Retroviral vector insertion sites associated with dominant hematopoietic clones mark “stemness” pathways. Blood. 2007;109:1897–1907. doi: 10.1182/blood-2006-08-044156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi K, Suzuki T, Stephens RM, Jenkins NA., and , Copeland NG. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 2004;32:D523–D527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie MB, McKnight KD, Kent DG, McCaffrey L, Hoodless PA., and , Eaves CJ. Hematopoietic stem cells proliferate until after birth and show a reversible phase-specific engraftment defect. J Clin Invest. 2006;116:2808–2816. doi: 10.1172/JCI28310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlich U, Schambach A, Brugman MH, Wicke DC, Knoess S, Li Z, et al. Leukemia induction after a single retroviral vector insertion in Evi1 or Prdm16. Leukemia. 2008;22:1519–1528. doi: 10.1038/leu.2008.118. [DOI] [PubMed] [Google Scholar]
- Wang GP, Garrigue A, Ciuffi A, Ronen K, Leipzig J, Berry C, et al. DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer. Nucleic Acids Res. 2008;36:e49. doi: 10.1093/nar/gkn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ., and , Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takash W, Cañizares J, Bonneaud N, Poulat F, Mattéi MG, Jay P, et al. SOX7 transcription factor: sequence, chromosomal localisation, expression, transactivation and interference with Wnt signalling. Nucleic Acids Res. 2001;29:4274–4283. doi: 10.1093/nar/29.21.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyama S, Yamamoto G, Shimabe M, Sato T, Ichikawa M, Ogawa S, et al. Evi-1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell Stem Cell. 2008;3:207–220. doi: 10.1016/j.stem.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Kent DG, Dykstra BJ, Cheyne J, Ma E., and , Eaves CJ. Steel factor coordinately regulates the molecular signature and biologic function of hematopoietic stem cells. Blood. 2008;112:560–567. doi: 10.1182/blood-2007-10-117820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou Y, Ma Z, Lu T., and , Sorrentino BP. Unique risk factors for insertional mutagenesis in a mouse model of XSCID gene therapy. Proc Natl Acad Sci USA. 2006;103:11730–11735. doi: 10.1073/pnas.0603635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinski MK., and , Bushman FD. Retroviral DNA integration–mechanism and consequences. Adv Genet. 2005;55:147–181. doi: 10.1016/S0065-2660(05)55005-3. [DOI] [PubMed] [Google Scholar]
- Lapidot T, Dar A., and , Kollet O. How do stem cells find their way home. Blood. 2005;106:1901–1910. doi: 10.1182/blood-2005-04-1417. [DOI] [PubMed] [Google Scholar]
- Samper E, Fernández P, Eguía R, Martín-Rivera L, Bernad A, Blasco MA, et al. Long-term repopulating ability of telomerase-deficient murine hematopoietic stem cells. Blood. 2002;99:2767–2775. doi: 10.1182/blood.v99.8.2767. [DOI] [PubMed] [Google Scholar]
- Faubert A, Chagraoui J, Mayotte N, Frechette M, Iscove NN, Humphries RK.Complementary and independent function for hoxb4 and bmi2 in HSC activity Cold Spring Harbor Symposia Quant Biol 200873555–564.et al [DOI] [PubMed] [Google Scholar]
- Modlich U, Bohne J, Schmidt M, von Kalle C, Knöss S, Schambach A, et al. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108:2545–2553. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball CR, Pilz IH, Schmidt M, Fessler S, Williams DA, von Kalle C, et al. Stable differentiation and clonality of murine long-term hematopoiesis after extended reduced-intensity selection for MGMT P140K transgene expression. Blood. 2007;110:1779–1787. doi: 10.1182/blood-2006-11-053710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoglio C, Facchini G, Sartori D, Antonelli A, Miccio A, Cassani B, et al. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood. 2007;110:1770–1778. doi: 10.1182/blood-2007-01-068759. [DOI] [PubMed] [Google Scholar]
- Beard BC, Dickerson D, Beebe K, Gooch C, Fletcher J, Okbinoglu T, et al. Comparison of HIV-derived lentiviral and MLV-based gammaretroviral vector integration sites in primate repopulating cells. Mol Ther. 2007;15:1356–1365. doi: 10.1038/sj.mt.6300159. [DOI] [PubMed] [Google Scholar]
- Ryu BY, Evans-Galea MV, Gray JT, Bodine DM, Persons DA., and , Nienhuis AW. An experimental system for the evaluation of retroviral vector design to diminish the risk for proto-oncogene activation. Blood. 2008;111:1866–1875. doi: 10.1182/blood-2007-04-085506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove PW, Kepes S, Hanawa H, Obenauer JC, Pei D, Cheng C, et al. Globin lentiviral vector insertions can perturb the expression of endogenous genes in β-thalassemic hematopoietic cells. Mol Ther. 2008;16:525–533. doi: 10.1038/sj.mt.6300394. [DOI] [PubMed] [Google Scholar]
- Mazurier F, Gan OI, McKenzie JL, Doedens M., and , Dick JE. Lentivector-mediated clonal tracking reveals intrinsic heterogeneity in the human hematopoietic stem cell compartment and culture-induced stem cell impairment. Blood. 2004;103:545–552. doi: 10.1182/blood-2003-05-1558. [DOI] [PubMed] [Google Scholar]
- Ema H, Sudo K, Seita J, Matsubara A, Morita Y, Osawa M, et al. Quantification of self-renewal capacity in single hematopoietic stem cells from normal and Lnk-deficient mice. Dev Cell. 2005;8:907–914. doi: 10.1016/j.devcel.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Murillo A, Lozano ML, Montini E, Bueren JA., and , Guenechea G. Unaltered repopulation properties of mouse hematopoietic stem cells transduced with lentiviral vectors. Blood. 2008;112:3138–3147. doi: 10.1182/blood-2008-03-142661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schambach A, Mueller D, Galla M, Verstegen MM, Wagemaker G, Loew R, et al. Overcoming promoter competition in packaging cells improves production of self-inactivating retroviral vectors. Gene Ther. 2006;13:1524–1533. doi: 10.1038/sj.gt.3302807. [DOI] [PubMed] [Google Scholar]
- Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustikova OS, Modlich U., and , Fehse B. Retroviral insertion site analysis in dominant haematopoietic clones. Methods Mol Biol. 2009;506:373–390. doi: 10.1007/978-1-59745-409-4_25. [DOI] [PubMed] [Google Scholar]
- Li W., and , Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transduction efficiency of sorted cells as analyzed by flow cytometry.
Engraftment analysis performed on bone marrow of recipients from different experimental groups.
LMPCR sensitivity validation in two independent experiments.
γ-retroviral vector insertion site analysis in different organs.
Lentiviral vector insertion site analysis in different organs.
Donor chimerism (CD45.2+ cells) final analysis in peripheral blood, bone marrow, and spleen of primary recipients.
Insertion sites recovered from γ-retroviral LSK and LK EGFP progeny (DBISA).
Insertion sites recovered from γ-retroviral LTHSCs, STHSCs, and MPP EGFP progeny (DBISA).
Vector insertion sites distribution according to gene classes (±150 kb), DBISA.
Insertion sites recovered from lentiviral LTHSC and STHSC EGFP progeny (DBISA).