Abstract
The ability to control the differentiation of adult hematopoietic stem cells (HSCs) would promote development of new cell-based therapies to treat multiple degenerative diseases. Systemic injection of NaIO3 was used to ablate the retinal pigment epithelial (RPE) layer in C57Bl6 mice and initiate neural retinal degeneration. HSCs infected ex vivo with lentiviral vector expressing the RPE-specific gene RPE65 restored a functional RPE layer, with typical RPE phenotype including coexpression of another RPE-specific marker, CRALBP, and photoreceptor outer segment phagocytosis. Retinal degeneration was prevented and visual function, as measured by electroretinography (ERG), was restored to levels similar to that found in normal animals. None of the controls (no HSCs, HSCs alone and HSCs infected with lentiviral vector expressing LacZ) showed these effects. In vitro gene array studies demonstrated that infection of HSC with RPE65 increased adenylate cyclase mRNA. In vitro exposure of HSCs to a pharmacological agonist of adenylate cyclase also led to in vitro differentiation of HSCs to RPE-like cells expressing pigment granules and the RPE-specific marker, CRALBP. Our data confirm that expression of the cell-specific gene RPE65 promoted fate determination of HSCs toward RPE for targeted tissue repair, and did so in part by activation of adenylate cyclase signaling pathways. Expression by HSCs of single genes unique to a differentiated cell may represent a novel experimental paradigm to influence HSC plasticity, force selective differentiation, and ultimately lead to identification of pharmacological alternatives to viral gene delivery.
Introduction
Stem cells are defined as cells with extensive self-renewal capacity and the ability to differentiate into a wide variety of cell types. Although embryonic stem cells have considerable plasticity and have been shown to be pluripotent, differentiating to all lineages including the germ line, adult stem cells are considered to be already committed to one or a few lineages and to be restricted in their capacity to differentiate. The ability of adult stem cells in a specific organ to generate cells of unrelated types decreases in the more committed progenitors. However, mounting evidence suggests that the initial differentiation into one specific cell type is not as irreversible as originally thought.1 Recent findings in bone marrow stem cells (BMSCs) suggest that the lineage commitment of a stem/progenitor cell is not absolute.2 Furthermore, de-differentiation of fibroblasts to cells with characteristics of embryonic stem cells is also possible.3,4 Much effort has been devoted to deciphering the molecular mechanisms that regulate stem cell plasticity and to use this information to develop clinical therapies. To our knowledge, this is the first report using targeted gene manipulation to specifically program an adult stem cell in order to promote its selective differentiation.
Degenerative diseases of the eye, particularly those involving damage to or loss of the retinal pigment epithelium (RPE) or the retina, are a major health complication associated with aging and diabetes. There is a dire need for methods of repairing damaged RPE. Fortunately, mouse models exist for RPE damage that can be exploited for new experimental therapeutic cell-based strategies. We hypothesized that in hematopoietic stem cells (HSCs), the expression of a gene that is both unique to a terminally differentiated cell type and a transcriptional modulator would shift the balance toward differentiation of HSCs into that unique cell type. Thus, by promoting HSC differentiation more readily into the obligatory cell type, the repair process could be enhanced.
We selected the eye not only because of the increasing number of individuals with vision-threatening conditions, but also because the eye serves as an ideal model system; the retina is highly specialized, allowing precise identification and localization of stem cell–derived tissue. The retina is critically dependent on RPE cells, which help to maintain neural retinal structure and function, and if damaged lead to retinal degeneration and vision loss. One candidate for directing HSC differentiation into RPE is the RPE-specific protein RPE65. RPE65 is critical for the normal formation of 11-cis-retinal, and thus for photoreceptor function. In addition, RPE65 modulates the availability of retinoic acid, a known transcriptional regulator and differentiation inducer.5,6,7,8,9,10 Interestingly, RPE65 has been identified in the yolk sac and has been expressed in nonocular tissues that are rapidly proliferating.11 Furthermore, it is becoming exceedingly evident that proteins critical to a cell's function, such as enzymes and chaperones, have secondary functions to help protect them from mutation.12,13 Thus, we reasoned it was possible that RPE65 may “moonlight” as a transcriptional regulator or have other novel functions that enable it to regulate differentiation of HSCs to RPE cells. We report here that genetic manipulation of HSCs to express RPE65 promotes neuroepithelial cell differentiation, retinal repair and, most importantly, recovery of visual function in an animal model of RPE injury. These observations provide the first demonstration that adult stem cells can be programmed down a particular differentiation pathway by the expression of a protein that dictates cell specificity.
Results
Ex vivo gene transfection of HSCs
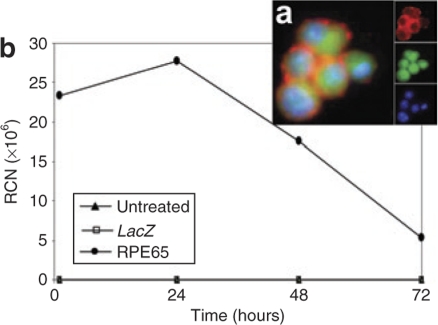
We generated a recombinant lentivirus (LV)–expressing human (h)RPE65 gene under control of a chicken β-actin (CBA) promoter. A companion recombinant virus expressing LacZ was generated as a control. HSCs were infected with LV for 2 hours resulting in an infection efficiency of 65.5 ± 5.5%. Immunohistochemistry at 24 hours (Figure 1a) after infection confirmed expression of RPE65 protein in HSCs and green fluorescent protein (GFP) as cells were isolated from mice expressing GFP. Real-time PCR demonstrated a greater than sevenfold increase in expression of the hRPE65 gene within 1 hour after infection and this dropped to 1.3-fold above baseline at 24 hours after infection (Figure 1b).
Figure 1.
Hematopoietic stem cells (HSCs) infected with RPE65-lentivirus construct for 2 hours resulted in expression of RPE65 mRNA and protein. (a) Immunofluoresence micrograph shows RPE65 protein expression by infected HSCs. The large image is a merge of the separate channels shown in the insets. Blue is nuclear staining from 4′,6-diamidino-2-phenylindole, green is native green fluorescent protein, and red is RPE65 detected by anti-RPE65 antibody followed by rhodamine-conjugated second antibody. (b) Time course of mRNA expression for HSCs infected with RPE65 lentivirus. mRNA was harvested from the infected cells either immediately after the 2-hour infection period ended, 1, 24, 48, and 72 hours after the end of the 2-hour infection period. The mRNA was reverse transcribed using WT-Pico Kit (NuGEN, San Carlos, CA) and amplified using real-time PCR with the human gene–specific primers for RPE65 (TaqMan; ABI). Data were normalized to GSTp1 (detected by GeneChip as having a very low coefficient of variation throughout the experiment) and expressed as relative copy number (RCN). Note that even after 72 hours, the level of expression remained in the 5 × 106 range. For the same time points, the mouse RPE65 was not detectable by TaqMan assay (data not shown).
Programming of HSCs with hRPE65 promotes their recruitment to the subretinal zone
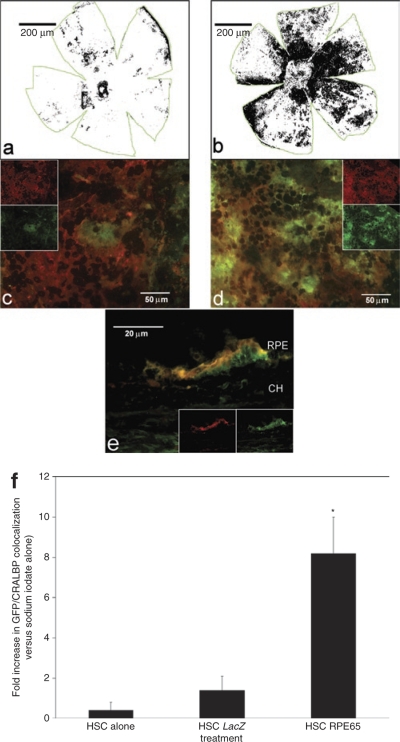
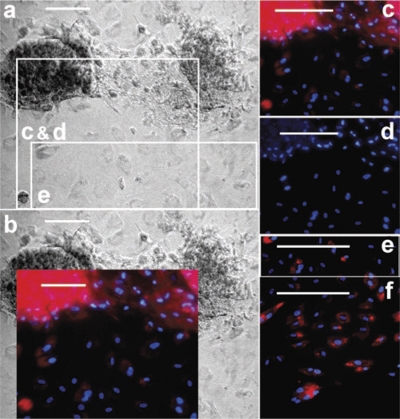
A single dose of 100 mg/kg of sodium iodate given intraperitoneally resulted in essentially total destruction of the RPE layer in the mice not treated with HSCs. Flat-mounted posterior cups from mice treated with HSCs showed varying degrees of GFP localization depending on the experimental group examined (Figure 2 and Supplementary Figure S3). When compared to posterior cup flat mounts of LacZ-infected HSCs (Figure 2a), there was a substantial increase in localized staining in mice injected with RPE65-infected HSCs (Figure 2b). This staining pattern was readily seen at higher magnification, where the amount of GFP staining was considerably greater in mice receiving RPE65-infected HSCs (Figure 2d) than in mice receiving LacZ-infected HSCs (Figure 2c). Staining for the RPE-specific marker CRALBP (red staining) was detected on the apical surface of the RPE layer in cross-section where GFP staining was also observed (Figure 2e). Quantitation of colocalized GFP and CRALBP in the flat-mounted posterior cups confirmed that mice injected with hRPE65-infected HSCs resulted in the highest degree of colocalization for these markers, with a greater than sevenfold increase (P < 0.05) in area positive for both markers compared to other treatment groups (Figure 2f). Additional immunohistochemistry demonstrated that the GFP+ cells also express the RPE-associated developmental protein microophthalmia-associated transcription factor14 and tyrosinase which plays a critical role in RPE melanogenesis15 (Supplementary Figures S4 and S5).
Figure 2.
Immunohistochemistry localized GFP+ cells coexpressing the retinal pigment epithelium (RPE)-specific marker CRALBP to the correct anatomical locale in the sodium iodate–injured eye. All of the images, as well as the quantitative data, were derived from eyes taken 28 days after injury. (a,b) Whole, flat-mounted posterior cups from animals injected with hematopoietic stem cells (HSCs) expressing LacZ or hRPE65, respectively. The images are density slices of colocalized CRALBP and green fluorescent protein (GFP) fluorescence. (c,d) Representative fluorescent micrographs of flat-mounted posterior cups that were stained for CRALBP (red) and GFP (green). Panel c is from a mouse injected with LacZ-infected HSCs whereas d is from a mouse injected with HSCs infected with lentivirus expressing RPE65. Note the significantly greater presence of GFP+ cells in d. The insets in c and d are split red and green channels of the exact image shown in the respective panel, albeit at reduced size, and are included to demonstrate the pattern of localization of each antigen. The cross-section in e shows localized green (GFP+) and red (CRALBP+) cells at the RPE layer. Note the apical localization of the CRALBP staining. (f) Bar form quantitative changes among the treatment groups in areas that are positive for both GFP (HSCs) and CRALBP (RPE). Posterior cups such as those depicted in a and b were used to determine the area of colocalized staining. Again, note the percentage of GFP+ cells that are mature and express CRALBP is much higher in the animals receiving RPE65-expressing HSCs than in any of the other conditions. Each bar represents the mean ± SD of at least four eyes per treatment condition. *P < 0.05 versus control (no cells).
Adoptive transfer of HSCs expressing hRPE65 promotes neural retinal and RPE repair in the sodium iodate ablation model of retinal degeneration
As previously shown, treatment of mice with a single dose of 100 mg/kg of sodium iodate resulted in blebbing, rounding up, detachment, and loss of RPE cells by 7 days after treatment.16,17 Compared to the confluent RPE monolayer in the uninjured eye (Figure 3a), the sodium iodate–treated eye demonstrated a paucity of RPE cells with only very occasional rounded RPE cells remained on the underlying Bruch's membrane (Figure 3b) by day 28 after injection.
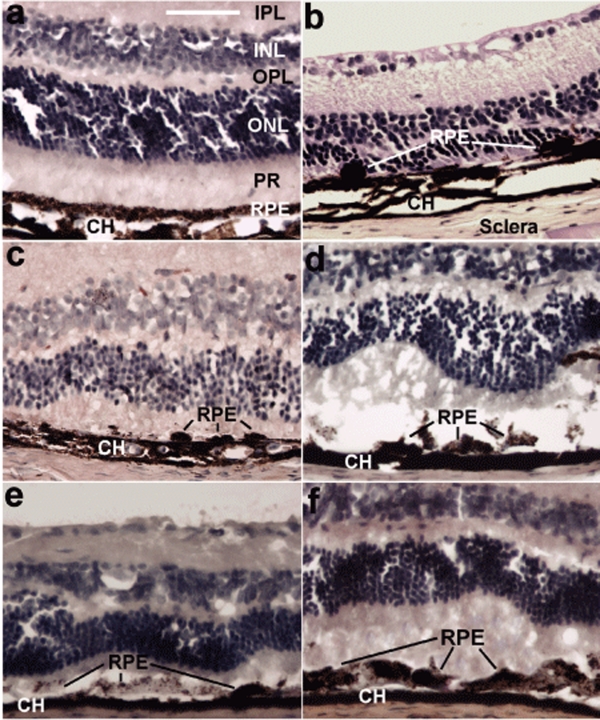
Figure 3.
Hematoxylin and eosin–stained cross-sections showed gross damage from sodium iodate and apparent rescue of retinal pigment epithelium (RPE) and neural retina anatomy by RPE65-transfected hematopoietic stem cells (HSCs). All images are of paraffin-embedded whole globes, cut at 10 µm thickness. (a) A normal uninjured eye with integrity of the RPE layer and photoreceptors. (b) An eye that was injected with 100 mg/kg sodium iodate but was not given any rescuing HSCs. Note the complete absence of the photoreceptor outer segments, near absence of any RPE cells (indicated by lines), and profound decrease in the retinal thickness. (c–f) Cross-sections of eyes from animals that were all given the same dose (100 mg/kg) of sodium iodate, and were also given (c) uninfected HSCs, (d) LacZ-infected HSCs, or (e,f) RPE65-infected HSCs. Animals receiving uninfected HSCs demonstrated only occasional RPE cells on Bruch's membrane at 28 days (c). Animals receiving LacZ-infected HSCs showed a small increase in cells compared to uninfected HSCs but large patches of Bruch's membrane were devoid of cells even at 28 days (d). By contrast, animals receiving RPE65-infected HSCs demonstrated abundant cells on Bruch's membrane. Seven days after treatment, a disorganized and poorly pigmented monolayer of cells was observed (e) and by 28 days (e), a mature highly pigmented epithelial layer had formed on Bruch's membrane. This layer exhibited all the characteristics of native RPE; photoreceptor outer and inner segments were also apparent. Bar = 70 µm. CH, choroid; INL, inner nuclear layer; IPL, inner plexiform layer; ONL, outer nuclear layer; OPL, outer plexiform layer; PR, photoreceptor inner and outer segments.
Animals receiving noninfected HSCs (i.e., naive) did not demonstrate any significant improvement in retinal anatomy at either 7 or 28 days (Figure 3c) and only minimal numbers of GFP+ cells with RPE morphology could be observed, as previously reported by us and others.16,18,19 These cells did not exhibit the morphological or functional characteristics of the cells in the pigmented epithelial layer achieved in the animals that received HSCs expressing hRPE65. Animals receiving cells infected with LV expressing LacZ showed infrequent recruitment of HSCs to the RPE layer and no recruitment to the neural retina by 28 days (Figure 3d).
By contrast, mice receiving HSCs infected with the hRPE65 gene exhibited retinal morphology similar to normal untreated mice by 28 days. Although a rudimentary and poorly pigmented monolayer of epithelial-like cells was observed at day 7 (Figure 3e), by 28 days the RPE layer had developed into a mature highly pigmented epithelial layer on Bruch's membrane (Figure 3f). This layer stained for both GFP at 28 days, but not at 7 days, thus confirming that the cells were HSC-derived and the cells were positive for CRALBP, an RPE-specific marker (Figure 2e).
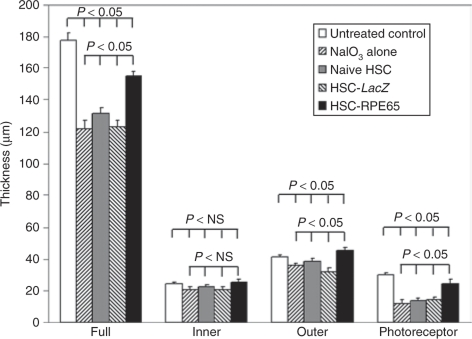
The thickness of the neural retina was also evaluated on cross-sections, and results of those measurements are shown in Figure 4. Twenty-eight days after sodium iodate treatment retinal thickness decreased by 31.3%, from 176.7 ± 19 µm in untreated control mice to 121.4 ± 17 µm in mice injured with sodium iodate (P < 0.05). Thinning of the retina was a combination of a reduction of the inner and outer nuclear layers and an almost complete loss of photoreceptor inner and outer segments. Retinal thickness in the mice injected with HSCs expressing hRPE65 was substantially greater than in mice treated with sodium iodate only, and was reduced by only 12% (154 ± 11 µm) compared to that in untreated controls (P < 0.05), with normal thickness of the inner and outer nuclear layers but a slight reduction remaining in the length of the photoreceptor outer segments (P < 0.05).
Figure 4.
Retinal thickness measurements showed marked reduction in sodium iodate–treated animals and increased thickness in mice injected with RPE65-transfected hematopoietic stem cells (HSCs). Thickness of the full neural retina (full), inner nuclear layer (inner), outer nuclear layer (outer), and photoreceptor inner/outer segment layer (photoreceptor) were taken between 2 and 4 mm temporal to the optic nerve head. Thinning of the retina in the sodium iodate–treated animals was a combination of a reduction in thickness of the inner and outer nuclear layers and an almost complete loss of photoreceptor inner and outer segments. Mice injected with HSC-infected hRPE65 (solid bars) were similar to that in healthy controls (open bars) with normal thickness of the inner and outer nuclear layers but a slight reduction in the length of the photoreceptor outer segments. NS, not significant.
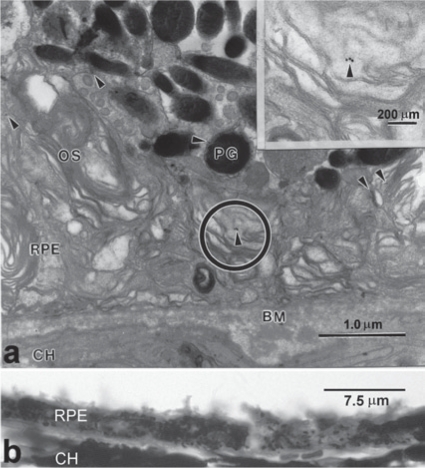
Immunolocalization and transmission electron microscopy studies confirmed that the newly pigmented epithelial layer demonstrated many of the morphological and functional characteristics of native RPE, including polarization through apical immunolocalization of CRALBP (Figure 2e), phagocytosis of rod outer segments, formation of secondary lysosomes, attachment to Bruch's membrane, basal infoldings of the plasma membrane, apical microvilli, and mature and premelanosomes located toward the apical surface (Figure 5a). GFP identified by clusters of immunogold particles (Figure 5a, arrowheads) indicated that these cells, which are morphologically similar to RPE cells, were indeed derived from GFP+ HSCs (Supplementary Figure S1). Figure 5b shows a panoramic view—at high magnification at light level—of the posterior segment of an injured eye from an injured animal 28 days after receiving HSCs expressing RPE65. Note the restoration of the RPE monolayer with near-normal anatomy. Previously using this model, we excluded the possibility of cell fusion using XY fluorescent in situ hybridization. After examining over 150 clearly identified HSC-derived RPE we did not observe fusion.16 While this approach does not completely rule out the possibility of fusion, it strongly indicates that fusion is not the primary mechanism by which the HSC-derived cells are contributing to the RPE layer. To further exclude the possibility of fusion in this study, we used the CRE/loxP LacZ reporter system, wherein the host has a STOP region flanked by loxP sites. The host underwent NaIO3 injury as described but received donor HSCs expressing cyclization recombination protein (CRE). The CRE expressing HSCs were injected systemically and the mice were killed on day 28 and their posterior cups were prepared. In the presence of CRE, the STOP is removed and LacZ is expressed by the cytomegalovirus promoter. The expression of LacZ was monitored by immunohistorchemistry with the chromogenic substrate X-gal. No β-galactosidase activity was detected in the RPE layer of any of these mice (data not shown) further confirming that no fusion occurred.
Figure 5.
Hematopoietic stem cells (HSCs) effect a functional and anatomically correct repair to retinal pigment epithelium (RPE) damaged by sodium iodate. (a) Ultrastructural analysis localized green fluorescent protein (GFP) to functional RPE cells in sodium iodate–damaged eyes. Transmission electron micrograph shows colloidal gold localized GFP within cells in a retina from a mouse treated with sodium iodate and subsequently given RPE65-infected GFP+HSCs. GFP localization is indicated by 18-nm colloidal gold particles (arrowheads) in the RPE cell. The phagocytosed outer segments indicate a functioning RPE. The circle indicated the enlarged area in the inset. BM, Bruch's membrane; CH, choroid; OS, outer segment; PG, melanin pigment granule. (b) Light-level panoramic view at high magnification of a section from the posterior segment of a sodium iodate–injured eye 28 days after the animal received RPE65-infected HSCs. Note the restoration of the RPE monolayer with near normal anatomy.
Gene expression characteristics in vitro
In the in vitro gene array study, HSCs were infected with either LacZ or hRPE65, maintained in culture, and mRNA was isolated at 1, 24, or 72 hours after infection. In this study, there was a significant effect on the genes involved in cellular development at the 1-hour time point in the HSCs infected with hRPE65 compared to the HSCs infected with LacZ (Supplementary Table S1a). Indeed most of the genes identified belong to this category. Top canonical pathways included chemokine signaling and B-cell receptor signaling. Genes showing the greatest increases included pigment epithelium–derived factor, which regulates RPE differentiation.19 At 1 hour after infection in the HSCs infected with hRPE65, gene clusters including those involved in cell signaling and cell interactions (CXCL2, Jag1, Lepr, PPP1r12b) were upregulated. NF-κB signaling cascades as well as expression of a number of immunoglobulin-associated genes were observed in a pattern reminiscent of the multipotent lymphoid myeloid progenitors.20 There was a significant downregulation of clusters of proliferation and cell cycle genes, which appeared to recover at later time points. Thus, this immediate drop is likely due to the effects of viral transfection. There was also a downregulation of negative regulators of proliferation and of endothelial-, erythrocyte-, neural-, and neutrophil-specific differentiation genes suggesting that the cells are not being driven down these differentiation pathways but rather toward other differentiation pathways including RPE (Supplementary Table S1).
Gene expression analysis at 24 hours (Supplementary Table S1b) showed increases in genes of the cell cycle, genes responsible for cellular development, cellular function, and maintenance, genes responsible for RNA trafficking and RNA post-transcriptional modifications, and genes responsible for post-translational modifications. Canonical pathways activated included interferon signaling, chemokine signaling, and epidermal growth factor receptor signaling. Gene expression analysis at 72 hours (Supplementary Table S1c) demonstrated upregulation of cell proliferation genes and top canonical pathways including phototransduction and hematological systems.
One gene that demonstrated increased expression at all three time points was adenylate cyclase. Interestingly, adenylate cyclase has been associated with promoting the transition of de-differentiated fibroblastic RPE to epithelial cells in culture.21 In an effort to identify conditions that induce differentiation of HSCs to RPE in vitro, we tested whether pharmacological activation by adenylate cyclase treatment would have the same effect as expression of RPE65. As shown in Figure 6, using phase contrast microscopy, HSCs maintained in culture and treated for 3 days with forskolin and rolipram exhibited characteristics of RPE pigmentation. Using immunohistochemistry, the cells were examined for the RPE-specific marker CRALBP. Most of the pigmented cells also expressed CRALBP to a high degree as shown in Figure 6c (which is positioned over its corresponding location on the phase contrast image as the inset of Figure 6b). Figure 6d depicts the nuclear staining resulting from 4′,6-diamidino-2-phenylindole. Figure 6e,f shows that some, but not all, cells outside of the large clusters of pigmented cells express CRALBP even though they do not, perhaps yet, have pigment granules.
Figure 6.
Activation of adenylate cyclase promotes differentiation of hematopoietic stem cells (HSCs) to “retinal pigment epithelium–like” cells. (a) Mouse Sca+ lin– HSCs were treated for 3 days with forskolin (10 µmol/l) and rolipram (1 µmol/l) to stimulate adenylate cyclase activity. HSCs exhibited pigmentation after such treatment. These same cells were fixed and stained with primary antibody directed to CRALBP and then exposed to fluorescently labeled secondary antibody. (b) Image from c overlain on the image from a to show the colocalization of intense CRALBP staining with the pigmented cells. (c) CRALBP reactive cells in red and cell nuclei stained in blue with 4′,6-diamidino-2-phenylindole (DAPI) (d shows DAPI only channel). Many of the cells outside of the clumped pigmented cells also expressed CRALBP (e,f), suggesting partial differentiation. Bar = 50 µm.
Gene expression characteristics in vivo
Gene array and real-time PCR analysis of retina/RPE/choroid preparations (posterior segments) demonstrated the absence of hRPE65 expression at day 28 and a reappearance of mouse RPE65 gene, suggesting silencing of the programming hRPE65 and its replacement with the endogenous mouse RPE-specific gene. A number of genes of interest, which met criteria of P < 0.05 and fold change as low as ±1.2, were confirmed by real-time RT-PCR.22 These include genes important in the RPE-dependent retinoid cycle (transthyretin): (i) for degradation of photoreceptors by the RPE (tripeptidyl peptidase, cathepsin D), (ii) for retinal development and enhanced incorporation of HSCs into the retina (colony-stimulating factor 1 receptor),23 and (iii) for transcriptional regulation by retinoic acid (src-like adaptor) were significantly upregulated in eyes 28 days after treatment with hRPE65-infected HSCs compared to animals which received HSCs infected with LacZ. The increased expression of these genes in mice injected with HSCs infected with hRPE65 suggests that there was a functioning RPE-dependent retinoid cycle and lysosomal system by day 28.
In addition, there was regulation of genes involved in cell adhesion, maturation, survival, cell growth, and viability. Top canonical pathways include CXCR4, interleukin-8, and integrin signaling (Supplementary Table S2).
hRPE65-programmed HSCs recover visual function in sodium iodate–treated mice
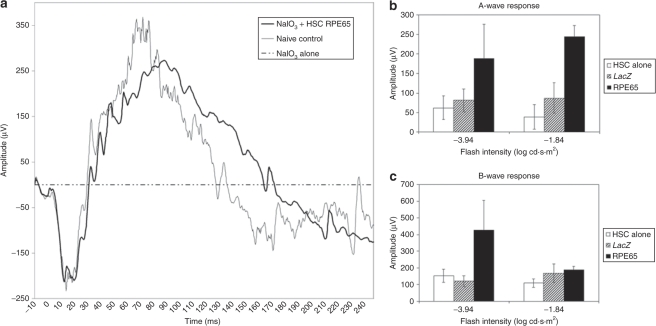
To evaluate whether the morphologic and phenotypic identity of the HSC-derived RPE cells was translated into visual function recovery, electroretinography (ERG) was performed (Figure 7). Mice treated with sodium iodate alone demonstrated an almost complete loss of neural retinal function by 28 days as indicated by a nonresponsive ERG. Of critical note is the scale for the amplitude axes in Figure 7a (RPE65-expressing HSC) as compared to the inset (sodium iodate injury alone). By 28 days in mice treated with HSCs infected with hRPE65, ERGs had returned to levels similar to those observed in normal untreated mice and demonstrated typical scotopic A- and B-wave form (Figure 7a). The A-wave amplitude of the mice treated with HSCs infected with hRPE65 was 57% of the amplitude of the naive controls (Figure 7b), whereas the B-wave amplitude was even higher than the naive controls (127%, Figure 7c). There was no significant increase in ERG signal observed for animals injected with naive HSCs or HSCs expressing LacZ (Figure 7b,c).
Figure 7.
Electroretinography (ERG) response demonstrated the RPE65-transfected HSC-driven rescue of visual function after sodium iodate damage. (a) Typical ERG traces from untreated (naive) control mice and from sodium iodate–injured animals 28 days after receiving HSCs expressing hRPE65. Sodium iodate alone causes a complete loss of normal ERG response. In marked contrast eyes from animals that received HSCs expressing hRPE65 have retinal function that is almost identical to the naive control, in both the A-and the B-wave amplitude. Each of the traces represents the averaged signal from five flashes at a light intensity of −1.84 log cd·s·m2. (b,c) The mean A-wave and B-wave amplitudes, respectively, for low and high flash intensities averaged for n ≥ 5 mice per treatment condition. Animals receiving HSCs expressing hRPE65 after sodium iodate injury show a statistically significant improvement in both A- and B-wave response compared to animals receiving either uninfected HSCs or HSCs expressing LacZ. *P < 0.05.
Discussion
To enhance the reparative capacity of adult HSCs, we approached the process by programming the HSCs for the cellular fate required to repair the specific injured tissue—in this case, the RPE cells of the injured RPE layer. This process requires not only precise differentiation into RPE cells but also sufficient HSC recruitment and proliferation at the site of injury to restore proper tissue function.
Previous studies have shown that BMSCs have the ability for limited differentiation into nonhematopoietic cell types, including glial and neuronal,24,25 skeletal muscle,26 fibroblasts/myofibroblasts,27 hepatocytes and hepatic oval cells,28,29 endothelium,30 and cardiomyocytes.31 We and others have also shown that, in the retina, subpopulations of BMSCs can differentiate into endothelial cells, extramural pericytes, astroglia, and RPE cells.16,30,32,33,34 However, the differentiation of HSCs to endothelial cells occurs with greater frequency.35 Thus, considerable evidence supports the participation of BMSCs in repair of both aberrant and healthy tissue as well as the ability of these cells to differentiate into a variety of cell types.
The physiological significance of this differentiation capacity is still unclear. For most cell types, the contribution of HSCs is quite limited and occurs primarily after severe injury. Although very unlikely, given the abundant presence of the GFP marker in the transplanted retina, this finding leaves open the possibility that the RPE65-infected HSCs are gradually replaced with cells of similar identity derived from the recipient. It is also possible, but again, unlikely, that the donor cells have fused with or were phagocytosed by resident cells.
Previously, using the sodium iodate model and GFP chimeric mice, we have excluded the possibility of fusion by use of fluorescent in situ hybridization for the sex chromosomes (XY fluorescent in situ hybridization).16 In that study, we used a reporter system in which β-galactosidase would have identified HSCs that fused with host RPE cells; however, we did not find evidence of β-galactosidase-positive RPE. Thus, we interpreted the lack of β-galactosidase-positive RPE in our study to support the absence of any HSC fusion with host RPE after sodium iodate injury.
The concept of targeted gene manipulation to specifically promote differentiation has been established for embryonic stem cells.36,37,38 Moreover, developmental studies suggest that a number of critical genes regulate embryonic cell differentiation, e.g., Pax-6, Nanog, and Olig1 and that targeted gene manipulation of embryonic stem cells with specific transcription factors promotes cell-specific differentiation.36,37,38 However, the drivers for adult stem cell differentiation are more elusive but include retinoic acid39 and growth factors.40
Attention has been given to de-differentiation and reprogramming of somatic cells to pluripotent stem cells. Such induced pluripotent stem cells have been generated from mouse fibroblasts by retroviral transduction of four transcription factors: Oct3/4, Sox2, Klf4, and c-Myc4 and from human fibroblasts.3 Unlike our one-step-one-gene protocol in adult HSCs, the use of induced pluripotent stem cells requires a multiple step process: de-differentiation, reprogramming, and differentiation, with each step potentially having inherent difficulties.
Furthermore, the recognition that gene products such as growth factors can act as transcriptional regulators, either directly or via metabolites, implies that proteins unique to a cell phenotype may play a critical role in the terminal differentiation to that particular cell type. RPE65 has secondary functions and important roles in development11 that enable it to serve outside the visual cycle, and as suggested by our data may modulate differentiation of HSCs toward RPE-like cells. Manipulation of differentiation regulators to direct stem cells to a specific phenotype offers a novel therapeutic approach to repair damaged tissues. In particular, neurodegenerative diseases such as Alzheimer's disease, age-related macular degeneration, and amyotrophic lateral sclerosis require replacement of highly specialized cell types. Avenues to achieve this have been largely restricted to allogeneic transplantation of terminally differentiated cells of either embryonic or adult origin, requiring invasive surgical procedures and long-term immunosuppression, even then with limited outcomes.
Systemic delivery of a cell population that has ability to home to injured tissue and differentiate into the correct cell type would be a major advance in therapeutic intervention for chronic degenerative diseases. BMSCs offer just such an approach as they represent an endogenous source of stem cells and can be easily removed and re-administered after pharmacological or gene manipulation, thus allowing for autologous transplantation. Ex vivo pharmacological manipulation of HSCs may be sufficient to direct differentiation and would not be associated with potential risks of viral gene delivery to HSCs. Furthermore, adoptive transfer of these stem cells is minimally invasive. Adult stem cells also have an advantage over embryonic stem cells which, despite their robust ability to proliferate as well as differentiate, have at times resulted in unfavorable outcomes such as development of teratomas and neoplasia.41
Cell type–specific transcriptional regulators initiate cellular differentiation during development. In this study, we have taken this developmental paradigm and applied it to undifferentiated adult cells capable of terminal differentiation. We selected infection of the HSCs with a viral vector containing hRPE65 to test the concept that genetic manipulation by forced expression of a terminal differentiation marker unique to that particular cell type could promote HSC differentiation toward RPE cells and confer RPE cell specificity.
Although we have not identified the mechanism by which RPE65 expression leads to HSC differentiation, we can surmise that RPE65 may directly activate the critical transcription factors needed for this transition. The in vitro array analysis supports this by demonstrating that many transcription factors were upregulated, however, it must be noted that an equal number were downregulated suggesting a unique activation pattern prompted by RPE65. Equally likely, however, is that RPE65 may indirectly activate differentiation by interacting with retinoic acid.8,9,42 Retinoic acid is a potent activator of transcription factors, regulates stem cell division, and serves as a differentiation factor.43 In our array studies, many members of the retinoic acid family were upregulated including retinoic acid receptor and retinoid X receptor. RPE65 may regulate the availability of retinoic acid or may “moonlight” as a transcriptional coactivator. Moreover, forced expression of RPE65 led to increased expression of adenylate cyclase 1, 3, and 5 and exposing HSC to forskolin, an agent that increases intracellular cyclic adenosine monophosphate levels, induced HSC differentiation toward an RPE-like phenotype with pigment granule accumulation and CRALBP expression. This suggests that increasing RPE 65 expression may activate adenylate cyclase, increase cyclic adenosine monophosphate, and RPE differentiation. Our observations corroborate the studies of Neuhuber and co-workers which showed that increasing intracellular cyclic adenosine monophosphate transitioned mesenchymal stem cells toward a “neuronal linage.”44
In the 1 hour gene array study, we observed the downregulation of the transcription factors E2F, which promotes cell proliferation, and Ets, which promotes vascular differentiation and these may help to block certain differentiation pathways. Based on the gene array data, we concluded that expression of hRPE65 induced HSC/hematopoietic precursor cells to differentiate toward the common lymphoid progenitor multipotent progenitor pathway and induced the NF-κB signaling cascade. We also observed expression of a number of immunoglobulin-associated genes in a pattern reminiscent of the multipotent lymphoid myeloid progenitors initially characterized at the single cell level by Cumano et al.20
Morphological and ERG studies showed that the small number of infected HSCs administered was sufficient for amplification and tissue repair. The ultrastructural analyses of the genetically modified HSCs and vision-related functional studies confirmed the phenotype of these cells. Our studies suggest that (i) programming HSCs with RPE65 is essential for successful tissue repair and upregulation of other genes involved in the visual cycle and (ii) mRNA for the human gene remained for <1 month, at which point endogenous mouse RPE65 was expressed, indicating a functional RPE. Others have shown that without ex vivo programming, using even three times the number of c-kit+ or Sca1+c-kit+ cells used in this study, along with facilitating cells, fails to achieve a significant rescue of RPE damage.16,18
Tissue injury with its resultant loss of cellular function and loss of tissue architecture recapitulates aspects of development. Tissue repair re-establishes cellular order and functional specialization much like cellular differentiation does in development. Both tissue repair and development use immature undifferentiated cells that transform under the influence of transcriptional activators in a specific temporal pattern.45 Thus, in our study using the sodium iodate ablation model of retinal degeneration, we observed that gene modulation of adult stem cells provides an efficient source of RPE cells that could potentially be used in treatment of RPE diseases. Furthermore, the same paradigm (well-established in development) can be used to program uncommitted adult stem cells to express cell type–specific genes. Turning on transcriptional activators at specific times by cell type–specific proteins may allow HSCs to be a feasible cellular therapy for a wide variety of diseased tissues.
Materials and Methods
Animals. All animal studies were conducted under an approved protocol by the Institutional Animal Care and Use Committee, and in accordance with guidelines set forth by the National Institutes of Health and the Association for Research in Vision and Ophthalmology. Adult (7–10-week-old) female C57BL6.J mice were purchased from Jackson Laboratories, Bar Harbor, ME.
LV constructs. The hRPE65-LV construct was prepared using a previously reported method.46 Briefly, a DNA expression plasmid containing the hRPE65 gene was kindly provided by William Hauswirth (CBA-RPE65). CBA is a ubiquitous strong promoter composed of a cytomegalovirus immediate-early enhancer (381 bp) and a CBA promoter–exon1–intron1 element (1,352 bp). The CBA-RPE65 plasmid was digested with NotI to remove the hRPE65 gene. The hRPE65 restriction fragment was ligated into pHEF to yield pTVd1.EFehRPE65. The pTFd1EFehRPE65 was digested with XhoI and EcoRI and the restriction fragment was ligated into a XhoI–Eco R1 digested pTY(SIN) plasmid to yield pTY(SIN)hRPE65. 293T cells were cotransfected with pTY(SIN)hRPE65, pHP, and pHEF-VSVG plasmids and virus was concentrated by centrifugation and filtration. Viral titers were determined by immunohistochemical staining for RPE65.
HSC isolation and infection. Ex vivo HSC gene transfer with either retrovirus or LV is typically followed by drug resistant selection in order to ensure that 100% of the infected HSCs are expressing the transgene and to expand the cell population prior to administration to ensure sufficient numbers of cells to repair the damaged tissue. This approach requires cell proliferation without induction of cell differentiation. This is where our protocol differs and offers a potential advantage. The LV infection protocol minimized exposure of the cells to ex vivo culturing conditions. We compensated for the lack of drug selection by infecting the cells with multiplicity of infection that yielded >50% gene transfer efficiency.
Bone marrow from tibiae and femurs of homozygous, in-house colony mice transgenic for GFP were obtained aseptically, and then HSCs were isolated by three-color fluorescence-activated cell sorting using antibodies to Sca1 and c-kit (both from BD Pharmingen, San Diego, CA) as previously described.30,33,47
Specifically, the cells were centrifuged at 300g for 5 minutes, the supernatant removed, and the cells resuspended in Dulbecco's modified Eagle's medium (high glucose), polybreen (10 µg/ml) plus 10% fetal bovine serum to a final cell concentration of 5 × 104 cells/ml. For the infection, the HSCs were split into three 1 × 105 cell aliquots. One control aliquot was not infected with any virus but had 5 µl of phosphate-buffered saline (PBS), pH 7.4 added to the cells. The second aliquot was infected with 5 µl of LacZ recombinant LV (TYF-EZ-LacZ) at a multiplicity of infection of ~50. The third aliquot was infected with 2 µl of hRPE65-LV (TYF-RPE65) for a multiplicity of infection of ~50. Cells were centrifuged at 23 °C at 150g for 2 hours. All experiments that included cell infection with virus were produced in the identical manner. Cells were then washed with PBS 2× by centrifugation and then injected into the mice as described below.
mRNA isolation. After infection, the cells were washed and cultured in Iscove's modified Dulbecco's media (Gibco, Carlsbad, CA) containing 10% fetal bovine serum, 20 ng/ml interleukin-6 (R&D Systems, Minneapolis, MN), 50 ng/ml stem cell factor (R&D Systems), and 20 ng/ml interleukin-3 (R&D Systems). The cells were kept in culture and mRNA was harvested immediately after infection (actually 2 hours after initial exposure to virus, due to the 2-hour infection protocol as described above), 1, and 24 hours after infection (actual 3 and 26 hours after initial exposure to virus, see previous comment). mRNA was harvested using the total aurum RNA kit (Bio-Rad Laboratories, Hercules, CA) per manufacturer's protocol. The mRNA was reverse transcribed and amplified by real-time PCR using gene-specific primers for hRPE65 (Applied Biosystems, Foster City, CA). All samples were normalized using gene-specific primers for TATA-binding protein (Applied Biosystems). For gene array studies, the cells were harvested for mRNA at 1, 24, and 72 hours after infection.
Immunohistochemical analysis of RPE65 protein expression in HSCs. For immunohistochemistry, hRPE65-transfected HSCs were cultured overnight at 37 °C on chamber slides coated with human fibronectin (BD Biosciences, San Jose, CA). The following day, cells were washed with PBS and incubated in fixative solution containing 2% formaldehyde, 0.2% glutaraldehyde in PBS, for 10 minutes at room temperature. After three washes in PBS, cells were permeabilized with 0.1% Triton X-100 for 4 minutes at room temperature. Samples were then washed and blocked by incubation with 10% normal rabbit serum in PBS for 1 hour at room temperature, and incubated with mouse anti-RPE65 (1:200 in blocking solution) (Novus Biologicals, Littleton, CO) overnight at 4 °C. After three washes in PBS, cells were incubated with Alexa Fluor 594 rabbit anti-mouse IgG (1:200 in blocking solution) (Invitrogen, Carlsbad, CA) for 1 hour at room temperature. After extensive washing with PBS, cells were mounted with VectaShield DAPI (Vector Laboratories, Burlingame, CA) for nuclear staining and a coverslip. Negative control samples were processed in the same manner except for the omission of the primary antibody. The cells were examined using an Axiovert 135 fluorescence microscope (Carl Zeiss, Thornwood, NY) with identical settings for laser intensity, gain, etc.
Sodium iodate treatment. The RPE of four groups of C57Bl6/J mice was targeted for ablation by intraperitoneal injection of 100 mg/kg sodium iodate in water.48 Three groups of animals then received 5,000 Sca1+c-kit+GFP+ HSCs [uninfected HSCs (n = 22), LacZ-LV (n = 17), or hRPE65-LV (n = 34)] in 100 µl of sterile saline injected into the left retroorbital sinus 8 hours after administering sodium iodate. A fourth group was given sodium iodate but was not injected with HSCs (n = 14). A fifth group was neither given sodium iodate nor HSCs (n = 7).
Studies to evaluate cell fusion. The RPE of B6.129S4-Gt(ROSA)26Sortm1Sor/J mice was targeted for ablation by intraperitoneal injection of 100 mg/kg sodium iodate.48 Sca1+ and c-kit+ HSCs were isolated from B6.C-Tg (CMV-cre)1Cgn/J mice and HSCs (5,000 cells) were injected into the left retroorbital sinus of the Gt(ROSA) mice 8 hours after administering sodium iodate (n = 10). A second group was given sodium iodate but was not injected with HSCs (n = 10). A third group was neither given sodium iodate nor HSCs (n = 10). All mice were euthanized on day 28. Eyes (n = 60) were enucleated and embedded in optimal cutting temperature compound and then frozen until processed. Ocular tissue was sectioned sagittally (10 µm) and sections were placed on gelatin-coated slides and allowed to air dry. Sections were then fixed for 5 minutes in 2% paraformaldehyde, 0.125% glutaraldehyde in PBS. The fixative was aspirated and slides incubated 3× 1 minute in PBS with 2 mmol/l MgCl2. Slides were then incubated for 2 minutes (3×) in PBS, MgCl2, NP40, desoxycholate, and then incubated 2 minutes with staining buffer without X-gal. X-gal was then added to the staining buffer and incubated at 37 °C for 6 hours in a moist chamber. All sections were examined by light microscope for β-galactosidase-positive cells (blue color). Even in pigmented mice, such as those used in this study, β-galactosidase-positive RPE cells can be identified if present.49
Immunohistology. Twenty-eight days after sodium iodate treatment, the animals were euthanized and the eyes removed. The eyes from each treatment cohort were divided into four groups for further analysis. Some from each treatment cohort were processed for electron microscopy. The second group of eyes was preserved in Trump's fixative for histology using hematoxylin and eosin to observe and compare changes in RPE layer morphology and measure retinal thickness among the different treatment cohorts by light microscopy. Retinal thickness was measured on 7-µm wax-embedded hematoxylin and eosin-stained transverse sections of retina. A total of five sections per eye from four animals were analyzed by a masked observer using a light microscope. Digital photographs were taken between 2 and 4 mm temporal to the optic nerve head and the thickness of the full retina, the inner nuclear layer, the outer nuclear layer, and the photoreceptor inner/outer segment layer were determined morphometrically using Image Pro software.
The third group of eyes was preserved in 4% paraformaldehyde for 2 hours, embedded in optimal cutting temperature medium, then sectioned sagittally for immunohistochemistry to identify and localize GFP+CRALBP+ cells. CRALBP was detected by further reaction with anti-CRALBP (Affinity Bioreagents, Golden, CO) followed by biotinylated goat anti-rabbit IgG (Sigma-Aldrich, St Louis, MO) followed by streptavidin-conjugated rhodamine (Vector Labs, Burlingame, VT).
Two RPE developmental markers were also examined in these sections, and colocalized with GFP. Fluorescein isothiocyanate–conjugated goat anti-GFP (Abcam, Cambridge, MA) was first reacted and washed, followed by antibodies to either microophthalmia-associated transcription factor (rabbit anti-microophthalmia-associated transcription factor; Abcam) or tyrosinase (rabbit anti-tyrosinase; Abcam). The latter two antibodies were detected by secondary reaction to tetramethylrhodamine isothiocyanate–conjugated goat anti-rabbit IgG (Abcam).
The final group of eyes was preserved in 4% paraformaldehyde for 2 hours, dissected to remove the anterior segments and neural retina, and then the posterior cups were reacted with anti-CRALBP, secondary antibody, and streptavidin–rhodamine as above. The latter two groups of eyes were examined by epifluorescence microscopy with identical settings for laser intensity, gain, etc., detecting native GFP in the green channel and CRALBP in the red channel. Colocalization and morphometry were analyzed using ImageJ software (NIH Research Services Branch, Bethesda, MD). At least four posterior cups were examined for each treatment condition. Colocalization was determined using Manders correlation coefficient, and the colocalized area of each posterior cup was determined from calibrated digitized images.50 Mean colocalized area for each treatment was calculated and results reported as fold change relative to control (i.e., sodium iodate–treated mice that received no HSCs).
Transmission electron microscopy. Eyes were enucleated and fixed for 1 hour in cold 4% paraformaldehyde–1% glutaraldehyde in 0.1 mol/l sodium cacodylate–HCl buffer, pH 7.4. Specimens were washed 4× 15 minutes in 0.1 mol/l cacodylate buffer containing 0.1 mol/l glycine. Eyes were postfixed in 1% osmium tetroxide in 0.1 mol/l cacodylate buffer, dehydrated in a methanol series to propylene oxide, and infiltrated and embedded in epoxy resin.
Ultrathin sections on nickel grids were oxidized in 1% (wt/vol) aqueous periodic acid in a Biowave Microwave (Ted Pella, Redding, CA) at 250 W power, 30 °C with a cycle of 2 minutes on, 2 minutes off, 2 minutes on. Grids were washed in PBS, pH 7.4 for 4× 1 minute followed by blocking with 4% (vol/vol) cold water fish gelatin in PBS at 250 W, 30 °C with a cycle of 2 minutes on, 2 minutes off, 2 minutes on. Grids were reacted with polyclonal chicken anti-GFP antibody (Aves Labs, Tigard, OR) at 250 W power, 30 °C with a cycle of 2 minutes on, 2 minutes off, 2 minutes on followed by washing 2× 1 minute with PBS and 2× 1 minute with Tris-buffered saline. Grids were incubated with donkey anti-chicken IgY conjugated to 18-nm colloidal gold (Jackson ImmunoResearch, West Grove, PA) at 250 W, 30 °C with a cycle of 2 minutes on, 2 minutes off, 2 minutes on. Grids were then rinsed with Tris-buffered saline 2× 1 minute followed by 3× 1 minute with deionized water followed by poststaining for 2 minutes with 2% (wt/vol) aqueous uranyl acetate. Controls were omission of the primary antibody and use of secondary antibody alone. Grids were examined and photographed in a JEOL 1200EX transmission electron microscope at an accelerating voltage of 100 kV.
Gene expression and microarray studies. For gene array studies, the cells were harvested at 1, 24, and 72 hours after infection (actually 3, 26, and 74 hours after initial exposure of cells to virus). Total mRNA from cells and retinas was isolated as described. Thirty nanogram of total mRNA were reverse transcribed and amplified using the Ovation RNA Amplification System (NuGEN Technologies, San Carlos, CA) then the cDNA was biotin labeled and fragmented using the FL-Ovation cDNA Biotin Module V2 (NuGEN Technologies). At all steps, the quantity and quality of the preparations were controlled using NanoDrop (NanoDrop Technologies, Wilmington, DE) and the Experion Automated Electrophoresis System (Bio-Rad Laboratories). Mouse Genome 430 2.0 Arrays (Affymetrix, Santa Clara, CA) were hybridized with 3.75 µg cDNA in the hybridization oven 640, washed and stained in the Fluidics Station 400, and scanned with the GeneChip Scanner 3000 (all instruments from Affymetrix). Data acquisition and preliminary analysis were done with the GeneChip Operating Software v1.4.0.036, also from Affymetrix. All experiments were done in triplicate.
For data analysis, the presence call determined by GeneChip Operating Software was used to eliminate those genes that were not present or marginally present in at least two samples from at least one experimental condition. The remaining genes (74–78% for retina samples and 60–66% for HSC samples) were further analyzed using Partek Genomic Suite (Partek, St Louis, MO), using an RMA algorithm. Data distribution was checked for normality (Supplementary Figure S2) and experimental conditions grouping by principal component analysis (Supplementary Figure S2). Two-way analysis of variance was performed, followed by false discovery rate correction for multiple comparisons. The networks and functional analyses were accomplished using the Ingenuity Pathways Analysis (Ingenuity Systems, Redwood City, CA).
Real-time PCR validation of gene array studies. Selected genes (Supplementary Table S3) were validated by real-time PCR. The mRNA was reverse transcribed using WT-Pico Kit (NuGEN, San Carlos, CA) and amplified using real-time PCR with the human gene–specific primers for RPE65 (TaqMan; Applied Biosystems, Foster City, CA). Data were normalized to GSTp1 (detected by GeneChip as having a very low coefficient of variation throughout the experiment) and expressed as relative copy number. We preferred to use relative copy number to measure the level of human RPE65 expression in infected cells because (i) other methods require that the respective gene expression in the control sample be greater than zero (which is not the case in mouse cells) and (ii) we can better examine the dynamics of gene copy numbers over time.51
For endogenous controls, we chose two genes that showed a minimal variation when analyzed by GeneChip: ribosomal protein S17 (GenBank accession no. NM_009092) and glutathione S-transferase pi 1 (GenBank accession no. NM_013541) (Supplementary Figure S2). The detection was done on the 7900HT Fast Real-Time PCR system (Applied Biosystems).
In vitro differentiation of HSC. RPE cells were grown to confluence on 12-well transwell insert plates (Costar, St Louis, MO). Mouse Sca1+ cells were enriched from mouse bone marrow cells using the mouse progenitor and Sca1 enrichment kits (StemCell Technologies, Vancouver, CA) per manufacturer's protocol. The bottom chambers of the transwell plates were coated with fibronectin (Sigma-Aldrich) and 20,000 mouse Sca1+ cells plated. The mouse cells were maintained in Dulbecco's modified Eagle's medium (Mediatech, Herndon, VA), 10% fetal bovine serum (MIDSCI, St Louis, MO), 10 µmol/l forskolin (Sigma-Aldrich), 1 µmol/l rolipram (Sigma-Aldrich), and 100 ng/ml PEDF (Chemicon, Bellercia, MA). The transwell inserts were removed to image the differentiated mouse HSCs using an inverted Zeiss Microscope (Carl Zeiss, Thornwood, NY).
ERG. Scotopic flash ERGs were performed on the recipient mice (n = 6 for each treatment condition of no cells and HSCs infected with LacZ, and n = 17 for the condition of HSCs infected with hRPE65) 28 days after sodium iodate treatment and adoptive transfer of transgenic HSCs as well as on uninjured animals. The animals were adapted to total darkness for 12–18 hours prior to ERG measurements. Mice were anesthetized and the pupils dilated with ophthalmic solutions of atropine and phenylephrine, after which they were subjected to sequences of scotopic flashes of −3.94 and −1.84 log cd·s/m2, with measurements averaged over five flashes. Comparisons were made between treatment conditions for the mean of the maxima for A-wave and B-wave responses for the two sequences of five flashes at each intensity. Significance was calculated by independent samples t-test and checked by Levene's test for equality of variances using SPSS software (SPSS, Chicago, IL).
SUPPLEMENTARY MATERIALFigure S1. Electron micrograph of a section from a normal mouse eye reacted for colloidal gold staining of GFP and serving as control for this reaction. Note the absence of immunogold particles outside of the pigment granules, showing the low non-specific binding of the particles to the specimens. Thus the gold particles evident in Figure 5 show specific GFP localization. BM, Bruch's membrane; PG melanin pigment granule; RPE, retinal pigment epithelial cell.Figure S2. Consistency of endogenous controls across experiments. Note that although these are not the traditional housekeeping genes used as endogenous controls for real-time PCR, they exhibit a remarkable consistency among the in vivo samples, thus validating them as true endogenous controls, both with TaqMan and SYBR Green assays. Samples: (1-3) no treatment; (4-6) NaIO3 only; (7-9) naïve HSC; (11-12) HSC infected with LacZ; (13-15) HSCs infected with hRPE65. The in vitro data (not shown) are identical.Figure S3. Fluorescence (a) and bright-field (b) images of the same 6-µm section of tissue from a normal control (naive) mouse eye that was subsequently incubated with secondary antibody used to detect the primary antibody against CRALBP. The fluorescent image detects autofluorescence in the green channel, as well as autofluorescence and non-specific secondary antibody binding in the red channel. Bright red punctuate fluorescence is due to erythrocytes, as they can be matched with their location in the bright-field image of Panel b.Cell nuclei are stained blue with DAPI. Exposure parameters for this image were increased slightly compared to the image shown in Figure 2c in order to visualize the dim fluorescence; therefore, there is some bleed-over detection between the channels. Scale bar = 50 μm.Figure S4. Immunohistochemical staining of 6 µm sections from mouse eyes injured with NaIO3 and then given HSC expressing RPE65 show microophthalmia-associated transcription factor (MiTF) expression. MiTF is associated with developing RPE and osteoclasts, and was found in these eye tissues only associated with GFP-co-expressing and pigment-containing cells localized to the expected RPE layer. Panel (a) shows the combined bright-field, red channel (MiTF), green channel (GFP) and blue channel (DAPI) digital image captures of the identical section. Panels (b-d) are the separate red, green and blue channels, respectively, that were combined with the bright-field image. Panels (e-h) show a section from a normal control mouse eye demonstrating a lack of non-specific reaction to the MitF and GFP antibodies. MiTF would not be expected to be expressed in fully mature, adult RPE. Scale bars as indicated in Panels (a) and (e) apply to their respective individual channels. The highly red and green fluorescent clumps visible below the RPE are erythrocytes within choroidal vessels.Figure S5. Immunohistochemical staining of 6 µm sections from mouse eyes injured with NaIO3 and then given HSC expressing RPE65 show tyrosinase expression. Tyrosinase is associated with developing RPE and was found in these tissues only associated with GFP-co-expressing and pigment-containing cells localized to the expected RPE layer. Panel (a) shows the combined bright-field, red channel (tyrosinase), green channel (GFP) and blue channel (DAPI) digital image captures of the identical section. Panels (b-d) are the separate red, green and blue channels, respectively, that were combined with the bright-field image. Panels (e-h) show a section from a normal control mouse eye demonstrating a lack of non-specific reaction to the tyrosinase and GFP antibodies. Tyrosinase is not expressed in mature RPE. Scale bars as indicated in Panels (a) and (e) apply to their respective individual channels. As seen also in Supplemental Figure 4, the strongly red and green fluorescent groups of cells visible below the RPE are erythrocytes.Table S1. Table shows the principal genes (and their respective accession numbers) that were either up-regulated or down-regulated 1 hour following infection in HSCs infected with hRPE65 compared to the HSCs infected with LacZ. These genes are the most likely candidates involved in differentiation of the HSCs to RPE. The fold change and p values were calculated by 2-way ANOVA, using Partek Genomic Suite, as described in methods. At the 1 hr time point (3 hr after initial virus exposure, Supplemental Table 1a) in the HSC infected with hRPE65 compared to the HSCs infected with LacZ there was a significant effect on the early response genes, and a significant effect on the genes involved in cellular development including chemokine signaling. The downregulation of clusters of proliferation and cell cycle genes, which appeared to recover at later time points, is probably due to the effects of viral transfection. Downregulation of negative regulators of proliferation and of endothelial, erythrocyte, neural and neutrophil specific differentiation genes was also seen, suggesting that the cells are not being driven down these differentiation pathways. At the 24 hr time point (26 hr after initial exposure to virus, Supplemental Table 1b) increases were seen in genes of the cell cycle, genes responsible for cellular development, cellular function and maintenance, genes responsible for RNA trafficking, RNA-post transcriptional modifications, and genes responsible for post-translational modifications. Canonical pathways activated included interferon signaling, chemokine signaling and epidermal growth factor receptor signaling. Finally, expression analysis at 72 hr (74 hr after initial virus exposure, Supplemental Table 1c) showed upregulation of cell proliferation and phototransduction genes.Table S2. Table shows the principal genes (and their respective accession numbers) that were either up-regulated or down-regulated in posterior segments of mouse eyes 28 days after sodium iodate injury and injection of HSCs infected with hRPE65 compared to the HSCs infected with LacZ. Regulated genes include those involved in cell adhesion, maturation, survival, cell growth and viability. Top canonical pathways include CXCR4, IL-8 and integrin signaling.Table S3. Selected genes and respective primers that were used for real-time PCR validation of GeneChip data. Selected genes and respective primers that were used for real-time PCR validation of GeneChip data. To discriminate between human and mouse RPE65 we used the more specific TaqMan assays. For all the other primers, specificity was ascertained through a dissociation step at the end of the run (“melting curve”).
Supplementary Material
Electron micrograph of a section from a normal mouse eye reacted for colloidal gold staining of GFP and serving as control for this reaction. Note the absence of immunogold particles outside of the pigment granules, showing the low non-specific binding of the particles to the specimens. Thus the gold particles evident in Figure 5 show specific GFP localization. BM, Bruch's membrane; PG melanin pigment granule; RPE, retinal pigment epithelial cell.
Consistency of endogenous controls across experiments. Note that although these are not the traditional housekeeping genes used as endogenous controls for real-time PCR, they exhibit a remarkable consistency among the in vivo samples, thus validating them as true endogenous controls, both with TaqMan and SYBR Green assays. Samples: (1-3) no treatment; (4-6) NaIO3 only; (7-9) naïve HSC; (11-12) HSC infected with LacZ; (13-15) HSCs infected with hRPE65. The in vitro data (not shown) are identical.
Fluorescence (a) and bright-field (b) images of the same 6-µm section of tissue from a normal control (naive) mouse eye that was subsequently incubated with secondary antibody used to detect the primary antibody against CRALBP. The fluorescent image detects autofluorescence in the green channel, as well as autofluorescence and non-specific secondary antibody binding in the red channel. Bright red punctuate fluorescence is due to erythrocytes, as they can be matched with their location in the bright-field image of Panel b.Cell nuclei are stained blue with DAPI. Exposure parameters for this image were increased slightly compared to the image shown in Figure 2c in order to visualize the dim fluorescence; therefore, there is some bleed-over detection between the channels. Scale bar = 50 μm.
Immunohistochemical staining of 6 µm sections from mouse eyes injured with NaIO3 and then given HSC expressing RPE65 show microophthalmia-associated transcription factor (MiTF) expression. MiTF is associated with developing RPE and osteoclasts, and was found in these eye tissues only associated with GFP-co-expressing and pigment-containing cells localized to the expected RPE layer. Panel (a) shows the combined bright-field, red channel (MiTF), green channel (GFP) and blue channel (DAPI) digital image captures of the identical section. Panels (b-d) are the separate red, green and blue channels, respectively, that were combined with the bright-field image. Panels (e-h) show a section from a normal control mouse eye demonstrating a lack of non-specific reaction to the MitF and GFP antibodies. MiTF would not be expected to be expressed in fully mature, adult RPE. Scale bars as indicated in Panels (a) and (e) apply to their respective individual channels. The highly red and green fluorescent clumps visible below the RPE are erythrocytes within choroidal vessels.
Immunohistochemical staining of 6 µm sections from mouse eyes injured with NaIO3 and then given HSC expressing RPE65 show tyrosinase expression. Tyrosinase is associated with developing RPE and was found in these tissues only associated with GFP-co-expressing and pigment-containing cells localized to the expected RPE layer. Panel (a) shows the combined bright-field, red channel (tyrosinase), green channel (GFP) and blue channel (DAPI) digital image captures of the identical section. Panels (b-d) are the separate red, green and blue channels, respectively, that were combined with the bright-field image. Panels (e-h) show a section from a normal control mouse eye demonstrating a lack of non-specific reaction to the tyrosinase and GFP antibodies. Tyrosinase is not expressed in mature RPE. Scale bars as indicated in Panels (a) and (e) apply to their respective individual channels. As seen also in Supplemental Figure 4, the strongly red and green fluorescent groups of cells visible below the RPE are erythrocytes.
Table shows the principal genes (and their respective accession numbers) that were either up-regulated or down-regulated 1 hour following infection in HSCs infected with hRPE65 compared to the HSCs infected with LacZ. These genes are the most likely candidates involved in differentiation of the HSCs to RPE. The fold change and p values were calculated by 2-way ANOVA, using Partek Genomic Suite, as described in methods. At the 1 hr time point (3 hr after initial virus exposure, Supplemental Table 1a) in the HSC infected with hRPE65 compared to the HSCs infected with LacZ there was a significant effect on the early response genes, and a significant effect on the genes involved in cellular development including chemokine signaling. The downregulation of clusters of proliferation and cell cycle genes, which appeared to recover at later time points, is probably due to the effects of viral transfection. Downregulation of negative regulators of proliferation and of endothelial, erythrocyte, neural and neutrophil specific differentiation genes was also seen, suggesting that the cells are not being driven down these differentiation pathways. At the 24 hr time point (26 hr after initial exposure to virus, Supplemental Table 1b) increases were seen in genes of the cell cycle, genes responsible for cellular development, cellular function and maintenance, genes responsible for RNA trafficking, RNA-post transcriptional modifications, and genes responsible for post-translational modifications. Canonical pathways activated included interferon signaling, chemokine signaling and epidermal growth factor receptor signaling. Finally, expression analysis at 72 hr (74 hr after initial virus exposure, Supplemental Table 1c) showed upregulation of cell proliferation and phototransduction genes.
Table shows the principal genes (and their respective accession numbers) that were either up-regulated or down-regulated in posterior segments of mouse eyes 28 days after sodium iodate injury and injection of HSCs infected with hRPE65 compared to the HSCs infected with LacZ. Regulated genes include those involved in cell adhesion, maturation, survival, cell growth and viability. Top canonical pathways include CXCR4, IL-8 and integrin signaling.
Selected genes and respective primers that were used for real-time PCR validation of GeneChip data. Selected genes and respective primers that were used for real-time PCR validation of GeneChip data. To discriminate between human and mouse RPE65 we used the more specific TaqMan assays. For all the other primers, specificity was ascertained through a dissociation step at the end of the run (“melting curve”).
Acknowledgments
We are grateful for the excellent technical assistance provided by Verline Justilien and Shufung (Grace) Zhou of the Electron Microscopy Core at the University of Florida. The microarray experiments were done at the Genetics/Microarray Core in the Davis Heart and Lung Research Institute, The Ohio State University. This work was supported by grants to M.B.G. from the National Eye Institute (EY012601 and EY007739).
REFERENCES
- Poulsom R, Alison MR, Forbes SJ., and , Wright NA. Adult stem cell plasticity. J Pathol. 2002;197:441–456. doi: 10.1002/path.1176. [DOI] [PubMed] [Google Scholar]
- Dawn B., and , Bolli R. Adult bone marrow-derived cells: regenerative potential, plasticity, and tissue commitment. Basic Res Cardiol. 2005;100:494–503. doi: 10.1007/s00395-005-0552-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Lu Y, Lotan D., and , Lotan R. Differential regulation of constitutive and retinoic acid-induced galectin-1 gene transcription in murine embryonal carcinoma and myoblastic cells. Biochim Biophys Acta. 2000;1491:13–19. doi: 10.1016/s0167-4781(00)00055-5. [DOI] [PubMed] [Google Scholar]
- Loudig O, Babichuk C, White J, Abu-Abed S, Mueller C., and , Petkovich M. Cytochrome P450RAI(CYP26) promoter: a distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Mol Endocrinol. 2000;14:1483–1497. doi: 10.1210/mend.14.9.0518. [DOI] [PubMed] [Google Scholar]
- Wang J., and , Yen A. A novel retinoic acid-responsive element regulates retinoic acid-induced BLR1 expression. Mol Cell Biol. 2004;24:2423–2443. doi: 10.1128/MCB.24.6.2423-2443.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollapalli DR., and , Rando RR. The specific binding of retinoic acid to RPE65 and approaches to the treatment of macular degeneration. Proc Natl Acad Sci USA. 2004;101:10030–10035. doi: 10.1073/pnas.0401936101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahng WJ, David C, Nesnas N, Nakanishi K., and , Rando RR. A cleavable affinity biotinylating agent reveals a retinoid binding role for RPE65. Biochemistry. 2003;42:6159–6168. doi: 10.1021/bi034002i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen JJ, Kuhlmann ED, van Vugt AH, Winkens HJ, Janssen BP, Deutman AF, et al. Retinoic acid delays transcription of human retinal pigment neuroepithelium marker genes in ARPE-19 cells. Neuroreport. 2000;11:1571–1579. [PubMed] [Google Scholar]
- Ward SJ., and , Morriss-Kay GM. The functional basis of tissue-specific retinoic acid signalling in embryos. Semin Cell Dev Biol. 1997;8:429–435. doi: 10.1006/scdb.1997.0166. [DOI] [PubMed] [Google Scholar]
- Jeffery CJ. Moonlighting proteins: old proteins learning new tricks. Trends Genet. 2003;19:415–417. doi: 10.1016/S0168-9525(03)00167-7. [DOI] [PubMed] [Google Scholar]
- Markova NG, Pinkas-Sarafova A., and , Simon M. A metabolic enzyme of the short-chain dehydrogenase/reductase superfamily may moonlight in the nucleus as a repressor of promoter activity. J Invest Dermatol. 2006;126:2019–2031. doi: 10.1038/sj.jid.5700347. [DOI] [PubMed] [Google Scholar]
- Tsukiji N, Nishihara D, Yajima I, Takeda K, Shibahara S., and , Yamamoto H. Mitf functions as an in ovo regulator for cell differentiation and proliferation during development of the chick RPE. Dev Biol. 2009;326:335–346. doi: 10.1016/j.ydbio.2008.11.029. [DOI] [PubMed] [Google Scholar]
- Abul-Hassan K, Walmsley R, Tombran-Tink J., and , Boulton M. Regulation of tyrosinase expression and activity in cultured human retinal pigment epithelial cells. Pigment Cell Res. 2000;13:436–441. doi: 10.1034/j.1600-0749.2000.130605.x. [DOI] [PubMed] [Google Scholar]
- Harris JR, Brown GA, Jorgensen M, Kaushal S, Ellis EA, Grant MB, et al. Bone marrow-derived cells home to and regenerate retinal pigment epithelium after injury. Invest Ophthalmol Vis Sci. 2006;47:2108–2113. doi: 10.1167/iovs.05-0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizota A., and , Adachi-Usami E. Functional recovery of retina after sodium iodate injection in mice. Vision Res. 1997;37:1859–1865. doi: 10.1016/s0042-6989(97)00015-1. [DOI] [PubMed] [Google Scholar]
- Atmaca-Sonmez P, Li Y, Yamauchi Y, Schanie CL, Ildstad ST, Kaplan HJ, et al. Systemically transferred hematopoietic stem cells home to the subretinal space and express RPE-65 in a mouse model of retinal pigment epithelium damage. Exp Eye Res. 2006;83:1295–1302. doi: 10.1016/j.exer.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Jablonski MM, Tombran-Tink J, Mrazek DA., and , Iannaccone A. Pigment epithelium-derived factor supports normal development of photoreceptor neurons and opsin expression after retinal pigment epithelium removal. J Neurosci. 2000;20:7149–7157. doi: 10.1523/JNEUROSCI.20-19-07149.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumano A, Paige CJ, Iscove NN., and , Brady G. Bipotential precursors of B cells and macrophages in murine fetal liver. Nature. 1992;356:612–615. doi: 10.1038/356612a0. [DOI] [PubMed] [Google Scholar]
- Kojima A, Nakahama K, Ohno-Matsui K, Shimada N, Mori K, Iseki S, et al. Connexin 43 contributes to differentiation of retinal pigment epithelial cells via cyclic AMP signaling. Biochem Biophys Res Commun. 2008;366:532–538. doi: 10.1016/j.bbrc.2007.11.159. [DOI] [PubMed] [Google Scholar]
- Shi L, Jones WD, Jensen RV, Harris SC, Perkins RG, Goodsaid FM, et al. The balance of reproducibility, sensitivity, and specificity of lists of differentially expressed genes in microarray studies BMC Bioinformatics 20089S10suppl. 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Reca RG, Atmaca-Sonmez P, Ratajczak MZ, Ildstad ST, Kaplan HJ, et al. Retinal pigment epithelium damage enhances expression of chemoattractants and migration of bone marrow-derived stem cells. Invest Ophthalmol Vis Sci. 2006;47:1646–1652. doi: 10.1167/iovs.05-1092. [DOI] [PubMed] [Google Scholar]
- Eglitis MA., and , Mezey E. Hematopoietic cells differentiate into both microglia and macroglia in the brains of adult mice. Proc Natl Acad Sci USA. 1997;94:4080–4085. doi: 10.1073/pnas.94.8.4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezey E, Chandross KJ, Harta G, Maki RA., and , McKercher SR. Turning blood into brain: cells bearing neuronal antigens generated in vivo from bone marrow. Science. 2000;290:1779–1782. doi: 10.1126/science.290.5497.1779. [DOI] [PubMed] [Google Scholar]
- Ferrari G, Cusella-De Angelis G, Coletta M, Paolucci E, Stornaiuolo A, Cossu G, et al. Muscle regeneration by bone marrow-derived myogenic progenitors. Science. 1998;279:1528–1530. doi: 10.1126/science.279.5356.1528. [DOI] [PubMed] [Google Scholar]
- Ogawa M, LaRue AC., and , Drake CJ. Hematopoietic origin of fibroblasts/myofibroblasts: its pathophysiologic implications. Blood. 2006;108:2893–2896. doi: 10.1182/blood-2006-04-016600. [DOI] [PubMed] [Google Scholar]
- Petersen BE, Bowen WC, Patrene KD, Mars WM, Sullivan AK, Murase N, et al. Bone marrow as a potential source of hepatic oval cells. Science. 1999;284:1168–1170. doi: 10.1126/science.284.5417.1168. [DOI] [PubMed] [Google Scholar]
- Theise ND, Badve S, Saxena R, Henegariu O, Sell S, Crawford JM, et al. Derivation of hepatocytes from bone marrow cells in mice after radiation-induced myeloablation. Hepatology. 2000;31:235–240. doi: 10.1002/hep.510310135. [DOI] [PubMed] [Google Scholar]
- Grant MB, May WS, Caballero S, Brown GA, Guthrie SM, Mames RN, et al. Adult hematopoietic stem cells provide functional hemangioblast activity during retinal neovascularization. Nat Med. 2002;8:607–612. doi: 10.1038/nm0602-607. [DOI] [PubMed] [Google Scholar]
- Tomita S, Li RK, Weisel RD, Mickle DA, Kim EJ, Sakai T, et al. Autologous transplantation of bone marrow cells improves damaged heart function Circulation 1999100II247–II256.19 suppl [DOI] [PubMed] [Google Scholar]
- Chan-Ling T, Baxter L, Afzal A, Sengupta N, Caballero S, Rosinova E, et al. Hematopoietic stem cells provide repair functions after laser-induced Bruch's membrane rupture model of choroidal neovascularization. Am J Pathol. 2006;168:1031–1044. doi: 10.2353/ajpath.2006.050697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta N, Caballero S, Mames RN, Butler JM, Scott EW., and , Grant MB. The role of adult bone marrow-derived stem cells in choroidal neovascularization. Invest Ophthalmol Vis Sci. 2003;44:4908–4913. doi: 10.1167/iovs.03-0342. [DOI] [PubMed] [Google Scholar]
- Harris JR, Fisher R, Jorgenssen M, Kaushal S., and , Scott EW. CD133 progenitor cells from the bone marrow contribute to retinal pigment epithelium repair. Stem Cells. 2009;27:457–466. doi: 10.1634/stemcells.2008-0836. [DOI] [PubMed] [Google Scholar]
- Schatteman GC. Adult bone marrow-derived hemangioblasts, endothelial cell progenitors, and EPCs. Curr Top Dev Biol. 2004;64:141–180. doi: 10.1016/S0070-2153(04)64007-5. [DOI] [PubMed] [Google Scholar]
- Carlberg AL, Pucci B, Rallapalli R, Tuan RS., and , Hall DJ. Efficient chondrogenic differentiation of mesenchymal cells in micromass culture by retroviral gene transfer of BMP-2. Differentiation. 2001;67:128–138. doi: 10.1046/j.1432-0436.2001.670405.x. [DOI] [PubMed] [Google Scholar]
- Klug MG, Soonpaa MH, Koh GY., and , Field LJ. Genetically selected cardiomyocytes from differentiating embronic stem cells form stable intracardiac grafts. J Clin Invest. 1996;98:216–224. doi: 10.1172/JCI118769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutsatsos IK, Turgeman G, Zhou S, Kurkalli BG, Pelled G, Tzur L, et al. Exogenously regulated stem cell-mediated gene therapy for bone regeneration. Mol Ther. 2001;3:449–461. doi: 10.1006/mthe.2001.0291. [DOI] [PubMed] [Google Scholar]
- Kruse FE., and , Tseng SC. Retinoic acid regulates clonal growth and differentiation of cultured limbal and peripheral corneal epithelium. Invest Ophthalmol Vis Sci. 1994;35:2405–2420. [PubMed] [Google Scholar]
- Hasuike S, Ido A, Uto H, Moriuchi A, Tahara Y, Numata M, et al. Hepatocyte growth factor accelerates the proliferation of hepatic oval cells and possibly promotes the differentiation in a 2-acetylaminofluorene/partial hepatectomy model in rats. J Gastroenterol Hepatol. 2005;20:1753–1761. doi: 10.1111/j.1440-1746.2005.03922.x. [DOI] [PubMed] [Google Scholar]
- Liu C, Chen Z, Chen Z, Zhang T., and , Lu Y. Multiple tumor types may originate from bone marrow-derived cells. Neoplasia. 2006;8:716–724. doi: 10.1593/neo.06253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomhoff R., and , Blomhoff HK. Overview of retinoid metabolism and function. J Neurobiol. 2006;66:606–630. doi: 10.1002/neu.20242. [DOI] [PubMed] [Google Scholar]
- Mendelsohn C, Ruberte E., and , Chambon P. Retinoid receptors in vertebrate limb development. Dev Biol. 1992;152:50–61. doi: 10.1016/0012-1606(92)90155-a. [DOI] [PubMed] [Google Scholar]
- Neuhuber B, Gallo G, Howard L, Kostura L, Mackay A., and , Fischer I. Reevaluation of in vitro differentiation protocols for bone marrow stromal cells: disruption of actin cytoskeleton induces rapid morphological changes and mimics neuronal phenotype. J Neurosci Res. 2004;77:192–204. doi: 10.1002/jnr.20147. [DOI] [PubMed] [Google Scholar]
- Rosenbauer F., and , Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol. 2007;7:105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- Chang LJ., and , Zaiss AK. Lentiviral vectors. Preparation and use. Methods Mol Med. 2002;69:303–318. doi: 10.1385/1-59259-141-8:303. [DOI] [PubMed] [Google Scholar]
- Sengupta N, Caballero S, Mames RN, Timmers AM, Saban D., and , Grant MB. Preventing stem cell incorporation into choroidal neovascularization by targeting homing and attachment factors. Invest Ophthalmol Vis Sci. 2005;46:343–348. doi: 10.1167/iovs.04-0153. [DOI] [PubMed] [Google Scholar]
- Kiuchi K, Yoshizawa K, Shikata N, Moriguchi K., and , Tsubura A. Morphologic characteristics of retinal degeneration induced by sodium iodate in mice. Curr Eye Res. 2002;25:373–379. doi: 10.1076/ceyr.25.6.373.14227. [DOI] [PubMed] [Google Scholar]
- Kostic C, Chiodini F, Salmon P, Wiznerowicz M, Deglon N, Hornfeld D, et al. Activity analysis of housekeeping promoters using self-inactivating lentiviral vector delivery into the mouse retina. Gene Ther. 2003;10:818–821. doi: 10.1038/sj.gt.3301948. [DOI] [PubMed] [Google Scholar]
- Manders EMM, Verbeek FJ., and , Aten JA. Measurement of co-localisation of objects in dual colour confocal images. J Microsc. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- Gavrilin MA, Bouakl IJ, Knatz NL, Duncan MD, Hall MW, Gunn JS, et al. Internalization and phagosome escape required for Francisella to induce human monocyte IL-1beta processing and release. Proc Natl Acad Sci USA. 2006;103:141–146. doi: 10.1073/pnas.0504271103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electron micrograph of a section from a normal mouse eye reacted for colloidal gold staining of GFP and serving as control for this reaction. Note the absence of immunogold particles outside of the pigment granules, showing the low non-specific binding of the particles to the specimens. Thus the gold particles evident in Figure 5 show specific GFP localization. BM, Bruch's membrane; PG melanin pigment granule; RPE, retinal pigment epithelial cell.
Consistency of endogenous controls across experiments. Note that although these are not the traditional housekeeping genes used as endogenous controls for real-time PCR, they exhibit a remarkable consistency among the in vivo samples, thus validating them as true endogenous controls, both with TaqMan and SYBR Green assays. Samples: (1-3) no treatment; (4-6) NaIO3 only; (7-9) naïve HSC; (11-12) HSC infected with LacZ; (13-15) HSCs infected with hRPE65. The in vitro data (not shown) are identical.
Fluorescence (a) and bright-field (b) images of the same 6-µm section of tissue from a normal control (naive) mouse eye that was subsequently incubated with secondary antibody used to detect the primary antibody against CRALBP. The fluorescent image detects autofluorescence in the green channel, as well as autofluorescence and non-specific secondary antibody binding in the red channel. Bright red punctuate fluorescence is due to erythrocytes, as they can be matched with their location in the bright-field image of Panel b.Cell nuclei are stained blue with DAPI. Exposure parameters for this image were increased slightly compared to the image shown in Figure 2c in order to visualize the dim fluorescence; therefore, there is some bleed-over detection between the channels. Scale bar = 50 μm.
Immunohistochemical staining of 6 µm sections from mouse eyes injured with NaIO3 and then given HSC expressing RPE65 show microophthalmia-associated transcription factor (MiTF) expression. MiTF is associated with developing RPE and osteoclasts, and was found in these eye tissues only associated with GFP-co-expressing and pigment-containing cells localized to the expected RPE layer. Panel (a) shows the combined bright-field, red channel (MiTF), green channel (GFP) and blue channel (DAPI) digital image captures of the identical section. Panels (b-d) are the separate red, green and blue channels, respectively, that were combined with the bright-field image. Panels (e-h) show a section from a normal control mouse eye demonstrating a lack of non-specific reaction to the MitF and GFP antibodies. MiTF would not be expected to be expressed in fully mature, adult RPE. Scale bars as indicated in Panels (a) and (e) apply to their respective individual channels. The highly red and green fluorescent clumps visible below the RPE are erythrocytes within choroidal vessels.
Immunohistochemical staining of 6 µm sections from mouse eyes injured with NaIO3 and then given HSC expressing RPE65 show tyrosinase expression. Tyrosinase is associated with developing RPE and was found in these tissues only associated with GFP-co-expressing and pigment-containing cells localized to the expected RPE layer. Panel (a) shows the combined bright-field, red channel (tyrosinase), green channel (GFP) and blue channel (DAPI) digital image captures of the identical section. Panels (b-d) are the separate red, green and blue channels, respectively, that were combined with the bright-field image. Panels (e-h) show a section from a normal control mouse eye demonstrating a lack of non-specific reaction to the tyrosinase and GFP antibodies. Tyrosinase is not expressed in mature RPE. Scale bars as indicated in Panels (a) and (e) apply to their respective individual channels. As seen also in Supplemental Figure 4, the strongly red and green fluorescent groups of cells visible below the RPE are erythrocytes.
Table shows the principal genes (and their respective accession numbers) that were either up-regulated or down-regulated 1 hour following infection in HSCs infected with hRPE65 compared to the HSCs infected with LacZ. These genes are the most likely candidates involved in differentiation of the HSCs to RPE. The fold change and p values were calculated by 2-way ANOVA, using Partek Genomic Suite, as described in methods. At the 1 hr time point (3 hr after initial virus exposure, Supplemental Table 1a) in the HSC infected with hRPE65 compared to the HSCs infected with LacZ there was a significant effect on the early response genes, and a significant effect on the genes involved in cellular development including chemokine signaling. The downregulation of clusters of proliferation and cell cycle genes, which appeared to recover at later time points, is probably due to the effects of viral transfection. Downregulation of negative regulators of proliferation and of endothelial, erythrocyte, neural and neutrophil specific differentiation genes was also seen, suggesting that the cells are not being driven down these differentiation pathways. At the 24 hr time point (26 hr after initial exposure to virus, Supplemental Table 1b) increases were seen in genes of the cell cycle, genes responsible for cellular development, cellular function and maintenance, genes responsible for RNA trafficking, RNA-post transcriptional modifications, and genes responsible for post-translational modifications. Canonical pathways activated included interferon signaling, chemokine signaling and epidermal growth factor receptor signaling. Finally, expression analysis at 72 hr (74 hr after initial virus exposure, Supplemental Table 1c) showed upregulation of cell proliferation and phototransduction genes.
Table shows the principal genes (and their respective accession numbers) that were either up-regulated or down-regulated in posterior segments of mouse eyes 28 days after sodium iodate injury and injection of HSCs infected with hRPE65 compared to the HSCs infected with LacZ. Regulated genes include those involved in cell adhesion, maturation, survival, cell growth and viability. Top canonical pathways include CXCR4, IL-8 and integrin signaling.
Selected genes and respective primers that were used for real-time PCR validation of GeneChip data. Selected genes and respective primers that were used for real-time PCR validation of GeneChip data. To discriminate between human and mouse RPE65 we used the more specific TaqMan assays. For all the other primers, specificity was ascertained through a dissociation step at the end of the run (“melting curve”).