Abstract
Boolean logic performs a logical operation on one or more logic input and produces a single logic output. Here, we describe a microfluidic DNA computing processor performing Boolean logic operations for gene expression analysis and gene drug synthesis. Multiple cancer-related genes were used as input molecules. Their expression levels were identified by interacting with the computing related DNA strands, which were designed according to the sequences of cancer-related genes and the suicide gene. When all the expressions of the cancer-related genes fit in with the diagnostic criteria, positive diagnosis would be confirmed and then a complete suicide gene (gene drug) could be synthesized as an output molecule. Microfluidic chip was employed as an effective platform to realize the computing process by integrating multistep biochemical reactions involving hybridization, displacement, denaturalization, and ligation. By combining the specific design of the computing related molecules and the integrated functions of the microfluidics, the microfluidic DNA computing processor is able to analyze the multiple gene expressions simultaneously and realize the corresponding gene drug synthesis with simplicity and fast speed, which demonstrates the potential of this platform for DNA computing in biomedical applications.
INTRODUCTION
DNA computation is a kind of mode to realize molecular computing function, which depends on the information from DNA molecules and the interactions between DNA molecules. In some case of DNA computing, enzymes may be needed for DNA cleavage∕ligation.1 In early time, the study of DNA computation is mainly focused on the implementation of intractable computational problems.2, 3, 4 Recently, the concept of autonomous molecular computer5, 6, 7, 8 has been proposed and it has been investigated to control the gene expressions logically in a test tube.8 However, such kind of molecular computer in test tube is mainly based on the principles of Turing machine, in which the expressions of cancer-related genes can only be computed in sequence. Thus, multifold computation steps should be needed in order to meet the requirement of increased number of cancer-related genes.
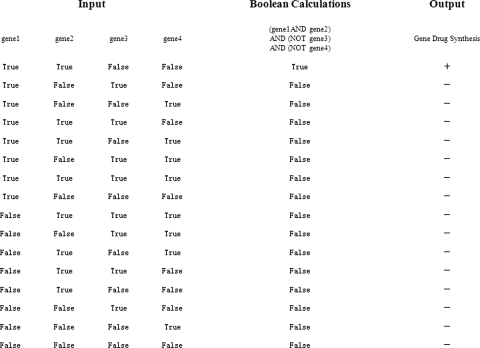
Microfluidics (or laboratory-on-a-chip) is a newly developed technology, which has been used in a wide range of fields including chemical, biology, and medicine due to its integration, miniaturization, automation, and the potential capability for high throughput assay.9, 10, 11, 12, 13 Recently, it has also been investigated for the study of gene analysis14, 15 and DNA computation.16 Here, we report a microfluidic DNA computing processor for gene expression analysis and corresponding gene drug synthesis, in which the DNA computing process performs Boolean logic operations.17, 18 It can be stated as following: Gene 1 and gene 2 and (not gene 3) and (not gene 4).
Multiple cancer-related genes are used as input molecules. Over-expression of a gene is considered as a Boolean true value and under-expression of a gene is considered as a Boolean false value. The diagnosis will be true (i.e., positive diagnosis) for only one combination of gene regulations (gene 1 is true, gene 2 is true, gene 3 is false, and gene 4 is false); all other combinations of gene regulations will be false (i.e., negative diagnosis). Only if the diagnosis is true will a complete gene drug be synthesized as an output molecule (Fig. 1).
Figure 1.
Illustration of gene expression analysis and corresponding gene drug synthesis performing Boolean calculations.
EXPERIMENTS AND RESULTS
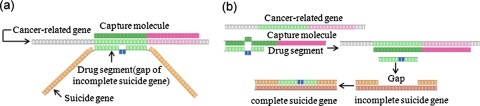
Computation is processed by the interaction between the input molecules (cancer-related genes) and the computing related biomolecules on a microfluidic chip, which is composed of parallel channels. The computing related biomolecules consist of capture molecule, drug segment, and incomplete suicide gene with multiple gaps. These molecules are designed according to the sequences of the cancer-related genes and the suicide gene (HSV-tk).19, 20 The sequences of the suicide gene and the cancer-related gene are contrasted, and the similar sequence between them is selected as the drug segment. The sequence of the gap in the incomplete suicide gene is as same as that of the drug segment. A single-strand DNA is set as the capture molecule, which is complementary to both the input molecule and the drug segment partially [Fig. 2a]. The capture molecule is at least twice as long as the drug segment. Because the complementary sequence to the capture molecule is longer, the cancer-related gene can displace the drug segment from the capture molecule, and the displaced drug segment can be transferred to fill the gap of the incomplete suicide gene [Fig. 2b].
Figure 2.
(a) Schematic illustration of the relationship of cancer-related gene, capture molecule, and drug segment. The drug segment (light green and blue) is the sequence of suicide gene which is most similar to that of cancer-related gene. The capture molecule (dark green and pink) is complementary to both the part of cancer-related gene (light green and purple) and the part of drug segment (light green). (b) Schematic illustration of the computing process at molecular level. Initially, the capture molecule partially hybridizes with the drug segment. When the cancer-related gene appears, the capture molecule is inclined to hybridize with the cancer-related gene and the drug segment will be displaced. The displaced drug segment can be transferred to fill the gap of the incomplete suicide gene.
In this DNA computing processor, only two kinds of breast cancer-related genes (oncogene C-erbB-2 and antioncogene nm23)21, 22 were selected as input molecules to demonstrate how to identify the over-expression and under-expression of the genes. The number of genes that could be used as inputs into the processor is dependent on the number of drug segments corresponding to the target genes (i.e., gaps of the suicide gene) that can be designed. According to our estimation, the sequence of the suicide gene (about 1000 nucleotides) can allow about dozens of drug segments (about 10–20 nucleotides) be designed theoretically. If the sequence of drug segment is too short, it will arouse nonspecific displacement reaction and false positive result. This “error” will limit the number of genes that can be analyzed in the processor.
In this work, we just establish a model to demonstrate the feasibility of the DNA computing processes on a microfluidic platform. So we used the synthetic ssDNA oligonucleotides to represent the cancer-related genes (C-erbB-2 and nm23) and A drug template strand complementary to both drug segments A and B (corresponding to C-erbB-2 and nm23, respectively) to represent the incomplete suicide gene. For the use of microfluidic computing processor in the analysis of real-world sample, pretreatment of genes, involving mRNA extraction, reverse transcript, and amplification unit,23 should be integrated into the microfluidic platform. Drug segments A and B could hybridize with the drug template strand and then be ligated by T4 DNA ligase. The ligated double strands were regarded as complete suicide gene. In order to observe the computing processes clearly, C-erbB-2 and drug segment B were labeled with FAM (green), and nm23 and drug segment A were labeled with TAMRA (red). FAM and TAMRA are two kinds of fluorescence dye, the fluorescence of the former is green and the latter is red. The sequences and labels of all molecules involved in our work were shown in Table 1.
Table 1.
Sequences and labels of the molecules.
| Molecules | Sequences (5′−3′) and labels |
|---|---|
| C-erbB-2 | FAM-GGGCATGGTCCACCACAGGCACCGCAGCTCAT |
| Capture molecule 1 | Acrylamide-ATGAGCTGCGGTGCCTGTGGTGGACCATGCCC |
| Drug segment A | TAMRA-GGGCAGGGTCCAC |
| nm23 | TAMRA-CTGTGATACAGGAACCATGGCCAACTGTGAGC |
| Capture molecule 2 | Acrylamide-GCTCACAGTTGGCCATGGTTCCTGTATCACAG |
| Drug segment B | PO4-AGGAACCATGGCCAAC-FAM |
| Drug template strand | GTTGGCCATGGTTCCTGTGGACCCTGCCC |
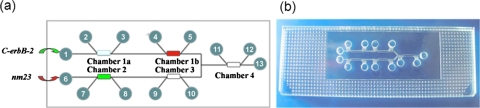
A glass microfluidic chip was designed and used as the platform for DNA computing processor (Fig. 3). The microfluidic chip was fabricated using standard photolithography and wet chemical etching technique, as described elsewhere.24 The channels were 80 μm deep, 200 μm wide, and 30–35 mm long. Each chamber was 80 μm deep, 500 μm wide, and 1.75 mm long. The diameter of the reservoirs was 1.5 mm. The channel wall of the glass chip was treated with Repel silane (2% solution of dimethyl dichlorosilane in chloroform) for 90 min.25 Capture molecule 1 (corresponding to C-erbB-2) at 10 μM was covalently immobilized in the polyacrylamide hydrogel plugs within chambers 1a and 1b.26, 27, 28, 29 Similarly, capture molecule 2 (corresponding to nm23) was immobilized in chamber 2. Saturated drug segments A and B were hybridized with capture molecule 1 in chamber 1b and capture molecule 2 in chamber 2, respectively. In chambers 3 and 4, blank hydrogel microplugs without capture molecules did not participate in computing and only acted as valves.30
Figure 3.
(a) Schematic of the microfluidic chip. (b) The photography of the microfluidic chip.
DNA computing for the expressions of C-erbB-2 and nm23 were performed in two parallel microchannels simultaneously. Continuous biochemical reactions for DNA calculations were accomplished under the action of electric fields. Platinum electrodes were placed in contact with the solution in reservoirs of both sides of the hydrogel plugs and connected to a low-voltage power supply (48 V). 1×TE and 0.5M NaCl was chosen as the buffer because it is a kind of high-salt buffer that can assure the high hybridization rate.26, 27, 28, 29
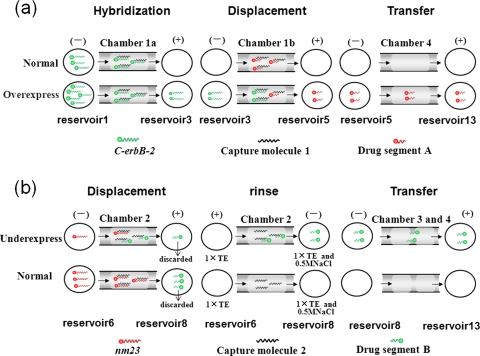
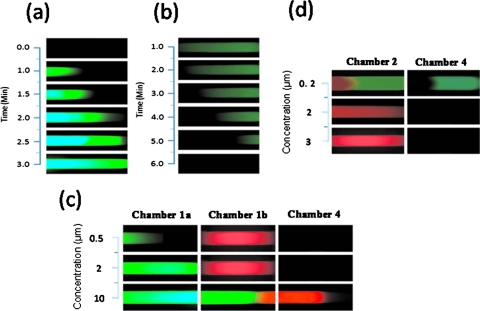
Here, 2 μM was set as the normal concentration of C-erbB-2 and nm23. When 5 μl of C-erbB-2 at 2 μM in 0.5M NaCl and 1×TE buffer was loaded into reservoir 1 and platinum electrodes were placed in reservoirs 3 and 5, C-erbB-2 was electrophoresed into chamber 1a, hybridized and captured by capture molecule 1. The rate of capture was closely related to the concentration of C-erbB-2. When C-erbB-2 was at the normal concentration (2 μM), it could reach the right edge of chamber 1a after 3 min [Figs. 4a, 5a]. When the concentration of C-erbB-2 was above 2 μM, the velocity of capture would be raised.26 After electrophoresed 3 min, the over-expressed C-erbB-2 (10 μM) was moved out of the hydrogel microplug in chamber 1a and then electrophoresed into chamber 1b and displaced some drug segment A (8 min experientially). This part of drug segment A was then moved into reservoir 5 and finally transferred through chamber 4 to reservoir 13 (10 min experientially). Following the computation process above, it was clear that only when C-erbB-2 was over-expressed, drug segment A could be transferred into reservoir 13 [Figs. 4a, 5c].
Figure 4.
(a) Illustration of the computing process of C-erbB-2 over-expression. (b) Illustration of the computing process of nm23 under-expression. Illumination: During the rinse step, when the electrophoresis conditions are changed, both nm23 (red) and drug segment B (green) in chamber 2 can be detached from the capture molecule 2, migrated into reservoir 8, and transferred into reservoir 13 finally. As nm23 is not complementary completely to the drug template strand and cannot ligate with drug segment A to synthesize complete gene drug AB, it has no effect on the final drug synthesis, so it is not shown in the rinse process or the transfer process in Fig. 4b.
Figure 5.
(a) Fluorescent images of 2 μM C-erbB-2 migrating to the right edge of chamber 1a. (b) Fluorescent images of drug segment B in chamber 2 rinsing off when changed the electrophoresis conditions. (c) Fluorescent images of computing process of C-erbB-2 (green) in three concentrations and transferring of drug segment A (red). Drug segment A could be detected in chamber 4 only when C-erbB-2 was over-expressed (10 μM). (d) Fluorescent images of computing process of nm23 (red) in three concentrations and transferring of drug segment B (green). Drug segment B could be detected in chamber 4 only when nm23 was under-expressed (0.2 μM). Because hydrogel has concentrated function to oligonucleotides (Ref. 31), we showed the pictures of drug segments A and B in chamber 4, which were clearer than that in reservoir 13.
At the same time, in the parallel channel, nm23 was also input into reservoir 6 to be analyzed [Fig. 4b]. In the microfluidic DNA computing processor, drug segment B was required only when the expression of antioncogene nm23 below the normal level. When the concentration of nm23 was at the normal level (2 μM), drug segment B in chamber 2 could be displaced by nm23 totally within 3 min, and electrophoresed into reservoir 8. The drug segment B in reservoir 8 was not required and discarded by vacuum pump. In following processes, no drug segment B would be rinsed off or transferred to reservoir 13. When the concentration of nm23 was below the normal level, its displacement velocity would be reduced28 so drug segment B in chamber 2 could not be displaced by nm23 completely, and some would remain in chamber 2. The displaced part of drug segment B would then be electrophoresed into reservoir 8, which was not required and discarded by vacuum pump. Then 5 μl of 0.5M NaCl and 1×TE buffer was set into reservoir 8. By changing eletrophoresis conditions (buffer in reservoir 6 turned to 1×TE and direction of electric field diverted), the remaining part of drug segment B in chamber 2 that was required would be rinsed off from capture molecule 2 [Fig. 5b] and migrated into reservoir 8. Then the drug segment B was migrated from reservoir 8 to 10 through chamber 3, and finally transferred into reservoir 13 through chamber 4 (total 15 min experientially). After the computation process described above, drug segment B could be transferred into reservoir 13 only if nm23 was under-expressed [Figs. 4b, 5d]. Low-salt buffer (1×TE) is selected to denature double DNA strands in the rinse step because low salt concentration leads to less shielding of the negatively charged backbones of hybridized DNA double strands, resulting in a stronger electrostatic repulsion between the strands.32
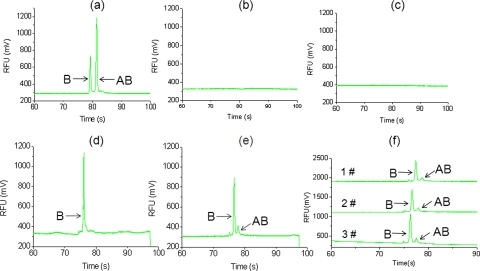
Then the drug template strand and T4 DNA ligase were placed into reservoir 13. In reservoir 13, drug segments A and B could hybridize with the drug template strand and be ligated by T4 DNA ligase to synthesize the complete gene drug at 20 °C for 25 min. The output molecule in chamber 13 was further detected by examining the length of the DNA strands through electrophoresis in another glass chip,33 and the electropherograms were shown in Fig. 6. The detected fluorescence was from drug segment B labeled by FAM, and the fluorescence from drug segment A labeled by TAMRA was filtered in our experiment. “B” represented the drug segment B and “AB” represented the complete suicide gene. It could be observed that drug AB could be synthesized only when the C-erbB-2 was over-expressed and nm23 was under-expressed simultaneously. In Fig. 6e, the completed drug peak AB seemed small compared to the incomplete B peak. This result may be due to the low efficiency of enzyme ligation. We will further optimize the conditions of enzyme ligation, such as time, temperature, enzyme concentration, etc., to increase the output of complete drug AB. Figure 6f showed the reproducibility of three runs of computing on the same microfluidic chip, with the conditions that c-erbB-2 was 10 μM and nm23 was 0.2 μM. We used relative standard deviations (RSDs) to assay the reproducibility of peak B (i.e., drug segment B) and peak AB (i.e., complete drug AB) in height and migration time. The RSDs of the height and migration time of peak B were 17.27% and 21.58%, respectively (n=3). The RSDs of the height and migration time of peak AB were 0.73% and 0.7%, respectively (n=3).
Figure 6.
Electropherograms of the output molecule in reservoir 13. B represents drug segment B and AB represents complete suicide gene (drug AB). (a) Electropherogram of the standard sample of drug segments A, B, and AB. (b) Electropherogram of the output molecule when C-erbB-2 ↑ and nm23 in normal level. (c) Electropherogram of the output molecule when both C-erbB-2 and nm23 in normal level. (d) Electropherogram of the output molecule when C-erbB-2 in normal level and nm23 ↓. (e) Electropherogram of the output molecule when C-erbB-2 ↑ and nm23 ↓. (f) The reproducibility of three runs of computing on the same chip, with the conditions that c-erbB-2 was 10 μM and nm23 was 0.2 μM.
Different capture molecules can be immobilized not only in different channels but also in a same channel.26 The specificity of hybridization between the capture molecules and the target gene would assure that other genes would not affect the computing process and result.26, 29 So only two parallel channels are needed theoretically to analyze the multiple over-expressions and under-expressions of cancer-related genes, respectively.
SUMMARY
A microfluidic DNA computing processor performing Boolean calculations was constructed for gene expression analysis and gene drug synthesis. Two breast cancer-related genes (oncogene C-erbB-2 and antioncogene nm23) were used as input molecules. Their expressions were identified in two parallel microfluidic channels simultaneously by interacting with the computing related DNA strands. Test for over-expression of C-erbB-2, the multistep operations involved hybridization, displacement, and transfer. Test for under-expression of nm23, the multistep operations involved displacement, rinse, and transfer. When the expressions of both genes fit in with the criteria for breast cancer diagnosis, positive diagnosis would be confirmed and a complete suicide gene could be synthesized by DNA ligation as an output molecule. By combining the specific design of the computing related molecules and the integrated functions of the microfluidics, the microfluidic DNA computing processor is able to analyze the multiple gene expressions simultaneously and realize the corresponding gene drug synthesis with simplicity and fast speed, which demonstrates the potential of this platform for DNA computing in biomedical applications
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (Grant Nos. 20635030 and 20575067), the Hi-Tech Research and Development Program of China (863 Program) (Grant No. 2006AA02Z305), and the Ministry of Science and Technology of China (973 Program) (Grant Nos. 2007CB714505 and 2007CB714507). Y. Zhang and H. Yu contributed equally in this work.
References
- Adleman L. M., Science 266, 1021 (1994). 10.1126/science.7973651 [DOI] [PubMed] [Google Scholar]
- Ouyang Q., Kaplan P. D., Liu S. M., and Libchaber A., Science 278, 446 (1997). 10.1126/science.278.5337.446 [DOI] [PubMed] [Google Scholar]
- Seelig G., Soloveichik D., Zhang D. Y., and Winfree E., Science 314, 1585 (2006). 10.1126/science.1132493 [DOI] [PubMed] [Google Scholar]
- Johnson C. R., DNA Computing 4287, 360 (2006). 10.1007/11925903_28 [DOI] [Google Scholar]
- Benenson Y., Paz-Elizur T., Adar R., Keinan E., Livneh Z., and Shapiro E., Nature (London) 414, 430 (2001). 10.1038/35106533 [DOI] [PubMed] [Google Scholar]
- Benenson Y., Adar R., Paz-Elizur T., Livneh Z., and Shapiro E., Proc. Natl. Acad. Sci. U.S.A. 100, 2191 (2003). 10.1073/pnas.0535624100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adar R., Benenson Y., Linshiz G., Rosner A., Tishby N., and Shapiro E., Proc. Natl. Acad. Sci. U.S.A. 101, 9960 (2004). 10.1073/pnas.0400731101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benenson Y., Gil B., Ben-Dor U., Adar R., and Shapiro E., Nature (London) 429, 423 (2004). 10.1038/nature02551 [DOI] [PubMed] [Google Scholar]
- Gao Y., Shen Z., Wang H., Dai Z. P., and Lin B. C., Electrophoresis 26, 4774 (2005). 10.1002/elps.200500283 [DOI] [PubMed] [Google Scholar]
- Kong J., Jiang L., Su X. O., Qin J. H., Du Y. G., and Lin B. C., Lab Chip 9, 1541 (2009). 10.1039/b818430e [DOI] [PubMed] [Google Scholar]
- Shi W. W., Qin J. H., Ye N. N., and Lin B. C., Lab Chip 8, 1432 (2008). 10.1039/b808753a [DOI] [PubMed] [Google Scholar]
- Zhong R. T., Pan X. Y., Jiang L., Dai Z. P., Qin J. H., and Lin B. C., Electrophoresis 30, 1297 (2009). 10.1002/elps.200800491 [DOI] [PubMed] [Google Scholar]
- Ye N. N., Qin J. H., Shi W. W., Liu X., and Lin B. C., Lab Chip 7, 1696 (2007). 10.1039/b711513j [DOI] [PubMed] [Google Scholar]
- Qin J. H., Liu Z. Y., Wu D. P., Zhu N., Zhou X. M., Fung Y. S., and Lin B. C., Electrophoresis 26, 219 (2005). 10.1002/elps.200406158 [DOI] [PubMed] [Google Scholar]
- Liu D. Y., Shi M., Huang H. Q., Long Z. C., Zhou X. M., Qin J. H., and Lin B. C., J. Chromatogr., B: Biomed. Sci. Appl. 844, 32 (2006). 10.1016/j.jchromb.2006.06.041 [DOI] [PubMed] [Google Scholar]
- Grover W. H. and Mathies R. A., Lab Chip 5, 1033 (2005). 10.1039/b505840f [DOI] [PubMed] [Google Scholar]
- Kolpashchikov D. M. and Stojanovic M. N., J. Am. Chem. Soc. 127, 12348 (2005). [DOI] [PubMed] [Google Scholar]
- Rinaudo K., Bleris L., Maddamsetti R., Subramanian S., Weiss R., and Benenson Y., Nat. Biotechnol. 25, 795 (2007). 10.1038/nbt1307 [DOI] [PubMed] [Google Scholar]
- Sandalon Z., Fusenig N. E., McCutcheon J., Taichman L. B., and Garlick J. A., Gene Ther. 8, 232 (2001). 10.1038/sj.gt.3301344 [DOI] [PubMed] [Google Scholar]
- Bonini C., Bondanza A., Perna S. K., Kaneko S., Traversari C., Ciceri F., and Bordignon C., Mol. Ther. 15, 1248 (2007). 10.1038/sj.mt.6300190 [DOI] [PubMed] [Google Scholar]
- Nakopoulou L. L., Tsitsimelis D., Lazaris A. C., Tzonou A., Gakiopoulou H., Dicoglou C. C., and Davaris P. S., Cancer Detect. Prev. 23, 297 (1999). 10.1046/j.1525-1500.1999.99035.x [DOI] [PubMed] [Google Scholar]
- Tommasi S., Fedele V., Crapolicchio A., Bellizzi A., Paradiso A., and Reshkin S. J., Int. J. Mol. Med. 12, 131 (2003). [PubMed] [Google Scholar]
- Zhong R. T., Liu D. Y., Yu L. F., Ye N. N., Dai Z. P., Qin J. H., and Lin B. C., Electrophoresis 28, 2920 (2007). 10.1002/elps.200600604 [DOI] [PubMed] [Google Scholar]
- Zhou Z. M., Liu D. Y., Zhong R. T., Dai Z. P., Wu D. P., Wang H., Du Y. G., Xia Z. N., Zhang L. P., Mei X. D., and Lin B. C., Electrophoresis 25, 3032 (2004). 10.1002/elps.200305966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yershov G., Barsky V., Belgovskiy A., Kirillov E., Kreindlin E., Ivanov I., Parinov S., Guschin D., Drobishev A., Dubiley S., and Mirzabekov A., Proc. Natl. Acad. Sci. U.S.A. 93, 4913 (1996). 10.1073/pnas.93.10.4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen K. G., Ross D. J., and Tarlov M. J., Anal. Chem. 74, 1436 (2002). 10.1021/ac0156969 [DOI] [PubMed] [Google Scholar]
- Zangmeister R. A. and Tarlov M. J., Langmuir 19, 6901 (2003). 10.1021/la034424t [DOI] [Google Scholar]
- Zangmeister R. A. and Tarlov M. J., Anal. Chem. 76, 3655 (2004). 10.1021/ac035238v [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yu H., Dong X. L., Qin J. H., and Lin B. C., Chem. J. Chin. Iniv. 30, 1128 (2009). [Google Scholar]
- Koh C. G., Tan W., Zhao M. Q., Ricco A. J., and Fan Z. H., Anal. Chem. 75, 6379 (2003). 10.1021/ac035124j [DOI] [PubMed] [Google Scholar]
- Dhopeshwarkar R., Li S. A., and Crooks R. M., Lab Chip 5, 1148 (2005). 10.1039/b509063f [DOI] [PubMed] [Google Scholar]
- Petersen J., Poulsen L., Birgens H., and Dufva M., PLoS ONE 4, e4808 (2009). 10.1371/journal.pone.0004808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Li B. W., Zhong R. T., Qin J. H., Zhu Y. S., and Lin B. C., Electrophoresis 29, 4956 (2008). 10.1002/elps.200800490 [DOI] [PubMed] [Google Scholar]