Summary
Centrioles are cylindrical structures found at the core of the mitotic spindle pole, which also act as basal bodies to nucleate formation of cilia. Centrioles have a complex, nine-fold symmetric structure, and reproduce by an intriguing duplication process. The complexity and apparent self-reproduction of centrioles raises the question of how such a structure could have evolved, making them a favorite topic for theological speculation by "intelligent design" creationists. In fact, centrioles are capable of robust self-assembly and can tolerate dramatic perturbations while still maintaining basic functionality. Far from being irreducibly complex, centrioles appear to be based on a rather minimal underlying core structure requiring only a handful of genes to construct.
Introduction: phylogeny, terminology, and theology
The centrosome (1) is a bipartite structure, consisting of a pair of cylindrical microtubule-based organelles called centrioles (2), embedded in an amorphous network of proteins known collectively as Pericentriolar Material (PCM). The microtubule-nucleating function of the centrosome is carried out by gamma tubulin ring complexes docked on the PCM.
In contrast to the PCM, which has relatively little discernable structure (3), centrioles have a remarkably complex structure (4), which has raised the question of how something so complicated may have evolved. The complexity of the centriole suggests that a large number of genes may be required to build it, which poses a challenge for evolution because lack of any one of those genes would eliminate the functionality of the centriole, so that a fitness benefit would only be accrued once the entire gene set was established. This review will address the question of centriole evolution in light of recent experimental results which suggest that centrioles, while complicated-looking, may be substantially less complex than previously suspected.
Centriole structure and function
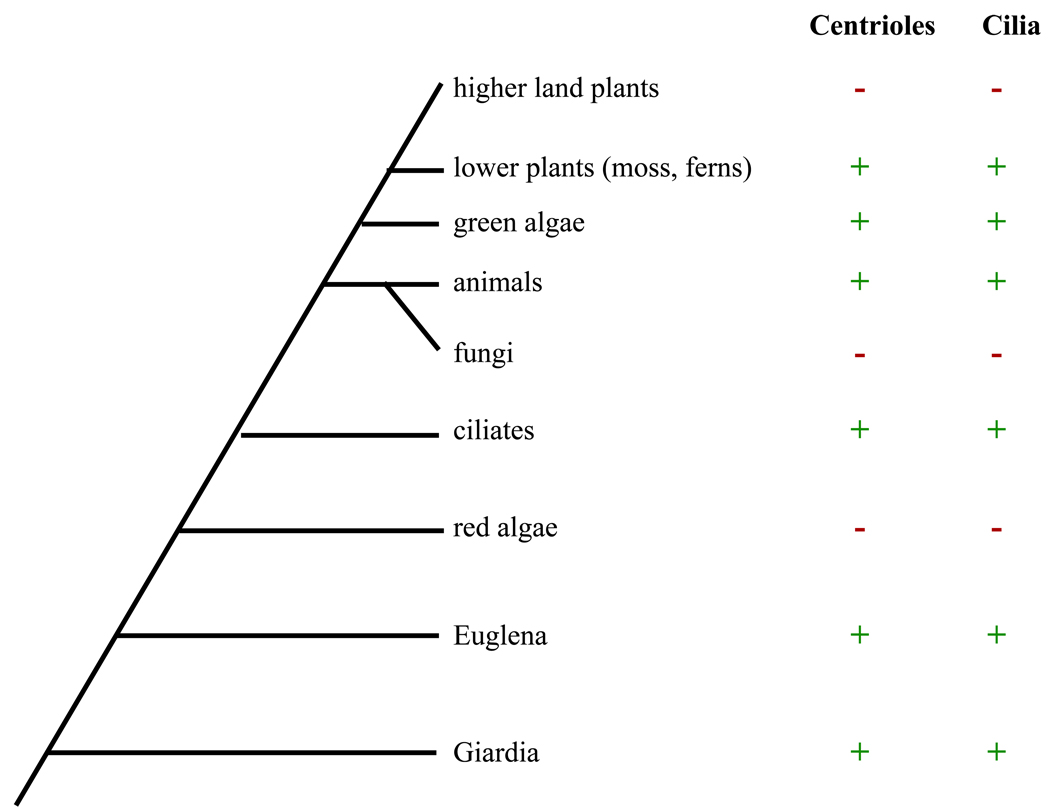
Centrioles consist of nine microtubule triplet blades arranged in a cylinder. At one end of the centriole is a spoke-like arrangement called the cartwheel. A variety of fibers and protrusions extend from the centriole and probably act to anchor it to various elements of the cytoskeleton (5,6). What is all this complex structure for, given that the main microtubule nucleating activity of the centrosome resides in the PCM and not the centriole? Experimental evidence shows that centrioles are required for the formation of a persistent centrosome (7) but when centrioles are removed from cells, bipolar spindles can still form using an alternative self-organization pathway that does not require centrosomes (8). What then is the real function of centrioles, if they aren't needed for mitosis? A look at the phylogeny of eukaryotes (9) shows us the answer (Figure 1). All species, without exception, that have centrioles, always have cilia at some stage of their life cycle, and vice versa. For example, lower plants such as mosses and ferns lack both centrioles and cilia in most cells but suddenly form them both during spermatogenesis. Indeed, centrioles are strictly required for the formation of cilia, and perform multiple functions on the behalf of cilia (10). First of all, the microtubule doublets of cilia grow as a direct outgrowth of the microtubule triplets of the centriole (11, 12). When a centriole forms a cilium, it becomes known as a Basal Body. In addition to directly nucleating ciliary microtubules, basal bodies dictate the orientation and positioning of cilia (13), and act as recruiting centers for molecules involved in ciliogenesis (14). The evolution of centrioles must thus be considered together with the evolution of cilia (15,16), and any fitness benefit that a eukaryotic precursor organism could have attained from the innovation of centrioles must have come from the ability to make cilia.
Figure 1.
Presence of centrioles correlates strictly with presence of cilia throughout eukaryotic phylogeny. (+) and (−) indicate whether centrioles or cilia are present or absent, respectively, in the given phylum. All phyla that currently lack centrioles appear to have descended from ancestors that once contained centrioles.
Self-reproduction of centrioles?
In most cells, centrioles form by an apparent "duplication" process by which each pre-existing centriole gives rise to a new centriole, at right angles to the first one (17). When this happens, the old centriole is called the mother and the new one the daughter. Microsurgical removal of centrioles indicated that cells lacking centrioles cannot form new ones, suggesting that the duplication pathway was the only way that centrioles could form (18). If new centrioles can only form from old ones, how could centrioles ever evolve in the first place within a cell that initially lacked them?
This type of chicken-and-egg paradox was heightened by claims that centrioles contained their own organellar genome (19,20). Indeed, if centrioles contained their own genomes, it would strongly imply that they arose via an endosymbiotic mechanism similar to that which gave rise to mitochondria and chloroplasts (21). Alternatively, it has been proposed that centrioles may have evolved from a virus (16). Requirement of an internal genome would also explain why centrioles undergo duplication and would also potentially make it impossible for centrioles to form de novo. However, a substantial number of experiments have subsequently proven beyond any doubt that centrioles do not contain a DNA-based genome of their own (22–26). It has been clearly shown that centrosomes contain specific RNA molecules associated with them, but these are encoded by genes that, while having an unusual intron-poor structure, are still found within the nucleus (27).
Since centrioles apparently do not contain their own genomes after all, this removes one part of the chicken and egg paradox. The problem posed by centriole duplication is also easily disposed of because, in fact, centrioles can form de novo. De novo formation is a natural occurrence in certain organisms that lack centrioles through parts of their life cycle and then re-acquire them at other stages (28,29). When centrioles are removed even from "normal" cells, new centrioles can form de novo with great efficiency (30,31). Why, then, did initial surgical studies suggest centriole duplication was obligatory? One possibility is the cells were damaged during the procedure. However it has also been shown that experimental removal of centrioles results in cell cycle arrest (32) in G1. De novo centriole formation can only occur in S phase (30,31), so G1 arrest would prevent centriole formation. This G1 arrest appears to be due to an increase in stress-sensitivity, and if the stress response is circumvented, de novo centriole assembly can occur (33).
Thus, the apparent paradox raised by centriole duplication turns out to be a non-issue. Centrioles can form de novo in cells that lack them without any difficulty, thus providing a way for the whole centriolar chain of being to begin.
Centriole complexity is not so irreducible after all
Centrioles look highly complex, but so do snowflakes. Extremely simple self-organizing chemical and physical processes can generate structures of extraordinary complexity, and one must be careful to distinguish between two different types of complexity. One type of complexity is called "informational complexity" which can be measured as the number of bits of information required to explicitly describe the structure in question. For instance, one could recognize image features such as corners and edges and record their positions within the object. Structures that look visually complicated, such as a Persian rug, will have much higher informational complexity than a simple-looking pattern such as a checkerboard. An alternative measure of complexity stems from the fact that simple computer programs can generate complex fractal patterns. Depending on the pattern, a very simple program may suffice, while for other structures, it may take a longer program to generate the pattern. This type of consideration has led to a newer way to describe structures in terms of their "algorithmic complexity", also known as Kolmogorov complexity, which is the size of the smallest computer program (usually measured in bits) sufficient to generate the pattern (34,35). Evolution and genomics is concerned with algorithmic complexity and not informational complexity. No matter how visually complicated a centriole is, its evolvability depends only on how complicated the genomic "program" must be that generates the structure.
We must therefore consider how many different genes are really needed to build a structure that is minimally functional as a centriole. This question has two parts: (a) how much of the structure of the centriole is really critical for its function, and (b) how many genes are necessary to generate the critical core structure. We will tackle the first question first - how much could the structure of a centriole be altered and still work? In this case, "work" would be defined as being able to provide at least some fitness benefit to a cell. Since the main job of centrioles is to form cilia, we can focus on how centriole structure contributes to assembly of cilia. Templating of cilia by centrioles requires is a set of preexisting microtubules, but the canonical arrangement of nine triplets in the centriole and nine doublets in the axoneme is by no means absolutely required for ciliary function, as organisms are known that have other numbers of centriolar or axonemal microtubules (36–39). These cilia and flagella are motile, despite deviating from the canonical ninefold symmetry, hence we can only conclude that there is nothing magical about the number nine. Moreover, it is possible for centrioles having nine triplets to generate cilia having more than nine doublets, indicating that cilia have a degree of self-organization independent of the influence of centrioles (40). Mutants in proteins of the centriole cartwheel can produce centrioles with variable numbers of triplets instead of nine (41,42), and these modified centrioles can still form cilia, demonstrating that ninefold symmetry is not an essential feature of centrioles. It is also important to point out that the existence of a symmetrical array of triplets (be it nine-fold symmetric or with some other symmetry) is not required for the triplets themselves to form. Mutants also exist in which centrioles form asymmetric arrangements of triplet-containing units in variable orientations (43). These studies demonstrate that single triplet subunit-sized chunks of centrioles can form without the overall rotational symmetry being present, so that if a single chunk could perform some useful function, this could be selected for prior to the development of the final nine-fold symmetrical structure.
Although ciliary motility is a highly coordinated process that might be quite sensitive to deviation away from nine-fold symmetry, cilia also play important sensory functions that would require little more than the microtubules and a membrane into which receptors could be localized. Cilia can also drive gliding motility which is independent of normal ciliary motility and might not require strong nine-fold symmetry (44). In one interesting protist, the cilium consists almost entirely of an elongated central pair of microtubules, lacking the nine doublets over most of its length, yet this is able to drive swimming (45). It thus seems reasonable that even a very rudimentary centriole-like precursor could allow formation of a proto-cilium that would give cells a tremendous advantage in terms of either sensory or gliding functions.
The second key question is how many genes would have to evolve in order to generate a centriole. One way to get at this question is to ask how many genes are essential to maintain centrioles. A recent genome-wide RNAi screen in Drosophila has argued that only nine genes are required for centriole duplication (46). The apparent complexity of the centriole proteome, which likely consists of at least 50–100 proteins (47,48), does not contradict the idea that only a small core set of genes are essential for centriole formation, provided the majority of the centriole proteins constitute add-ons, for example fibers that attach centrioles to different cytoskeletal elements.
Combining these considerations, it is apparent that only a small number of molecular innovations would be needed to produce some reduced, fragmentary version of a centriole that could in turn nucleate some sort of microtubule-based cellular extension that would be useful for gliding or sensation.
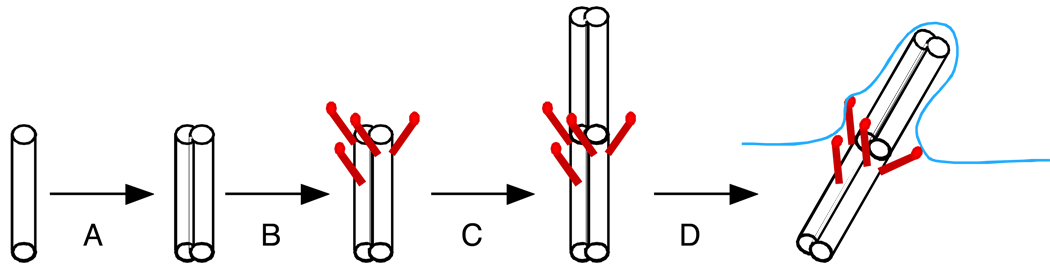
I propose that the original evolutionary precursor to the centriole may have resembled a single triplet blade subunit of the present day centriole, and consisted of a microtubule doublet or triplet structure that was able to extend a rigid proto-cilium consisting of a single doublet surrounded by plasma membrane out into the extracellular environment (Figure 2). Extension of microtubules from the end of the doublet would not require additional evolutionary novelty since it is known that centriole triplets can serve as templates from assembly of purified tubulin (12). The resulting structure, while simple, would have been able to provide basic sensory and gliding motility functions. The molecular requirements for formation of such a structure could be quite minimal: one or more proteins involved in forming the doublet microtubule structure plus a protein capable of linking the base of the structure to the cellular cortex. It is not currently known how microtubule doublets and triplets form, but the tektin family of proteins is thought to be involved in the process (49). Evolution of one or more tektins, plus one or more proteins that can bind microtubule doublets and attach to the cortex, such as ODF2/cenexin (50), might have been sufficient to produce a centriole precursor with basic function.
Figure 2.
Proposed evolution of a proto-centriole capable of nucleating a primitive cilium-like structure. (A) Evolution of tektins allows formation of stable microtubule doublet and triplet structures. (B) Acquisition of one or more appendage proteins allows docking of proto-centriole onto cell cortex. (C) Microtubules can extend from the end of the double structure. (D) If microtubules extend from the end of a docked doublet it would produce a primitive cilium-like structure that could be used for sensory or gliding functions.
Once the original proto-centriole was established, it eventually was modified by addition of further gene products to produce the characteristic nine-fold symmetric array of triplets found today. Genes required for this transition would be recognizable as those which when mutated lead to aberrations in symmetry without affecting assembly of the triplet blade subunits themselves. This phenotype has been seen for mutants in the centriole cartwheel proteins SAS-6 and BLD10 (41, 42) suggesting that these genes may have evolved after the proto-centriole in order to bring about the modern cylindrical symmetric structure. The acquisition of a symmetric cylindrical arrangement might provide a structure with greater mechanical strength to support more powerful motility, and might allow a cilium to project at a right angle to the cell surface by presenting a rotationally symmetric array of attachment appendages. Although as discussed above variations in structure are seen in some lineages, the cylinder of nine triplets is by far the most common arrangement. This relatively invariant centriole architectural plan seen among eukaryotes, combined with the fact that the ninefold triplet architecture is found even in the earliest branching eukaryotic lineages, such as Giardia, suggests that the structure evolved just once and was then inherited throughout the eukaryotes. Modifications to centriole structure, including the reduction of centrioles to discs of singlets in nematodes would thus have occurred by secondary loss events.
Conclusions
How centrioles evolved remains an open question. The lack of an intrinsic centriole genome and the ability of centrioles to form de novo suggests that an endosymbiotic origin is not required. The apparent complexity of centrioles is probably deceptive, as only a handful of genes may really be needed to form a functional centriole precursor.
Acknowledgments
The author acknowledges the support of the Searle Scholars Program, the WM Keck Foundation, and NIH grant R01 GM077004
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ou Y, Zhang M, Rattner JB. The centrosome: The centriole-PCM coalition. Cell Motil. Cytoskel. 2004;57:1–7. doi: 10.1002/cm.10154. [DOI] [PubMed] [Google Scholar]
- 2.Beisson J, Wright M. Basal body/centriole assembly and continuity. Curr. Opin. Cell Biol. 2003;15:96–104. doi: 10.1016/s0955-0674(02)00017-0. [DOI] [PubMed] [Google Scholar]
- 3.Dictenberg JB, Zimmerman W, Sparks CA, Young A, Vidair C, Zheng Y, Carrington W, Fay FS, Doxsey SJ. Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J. Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vorobjev IA, Chentsov YuS. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 1982;93:938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geimer S, Melkonian M. The ultrastructure of the Chlamydomonas reinhardtii basal apparatus: Identification of an early marker of radial asymmetry inherent in the basal body. J. Cell Sci. 2004;117:2663–2674. doi: 10.1242/jcs.01120. [DOI] [PubMed] [Google Scholar]
- 6.Hagiwara H, Ohwada N, Aoki T, Takata K. Ciliogenesis and ciliary abnormalities. Med. Electron Microscop. 2000;33:109–114. doi: 10.1007/s007950000009. [DOI] [PubMed] [Google Scholar]
- 7.Bobinnec Y, Khodjakov A, Mir LM, Rieder CL, Edde B, Bornens M. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J. Cell Biol. 1998;143:1575–1589. doi: 10.1083/jcb.143.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waters JC, Salmon E. Pathways of spindle assembly. Curr. Opin. Cell Biol. 1997;9:37–43. doi: 10.1016/s0955-0674(97)80149-4. [DOI] [PubMed] [Google Scholar]
- 9.Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- 10.Dawe HR, Farr H, Gull K. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J. Cell Sci. 2007;120:7–15. doi: 10.1242/jcs.03305. [DOI] [PubMed] [Google Scholar]
- 11.Ringo DL. Flagellar motion and fine structure of the flagellar apparatus in Chlamydomonas. J. Cell Biol. 1967;33:543–571. doi: 10.1083/jcb.33.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snell WJ, Dentler W, Haimo LT, Binder LI, Rosenbaum JL. Assembly of chick brain tubulin onto isolated basal bodies of Chlamydomonas reinhardtii. Science. 1974;185:33–38. doi: 10.1126/science.185.4148.357. [DOI] [PubMed] [Google Scholar]
- 13.Boisvieux-Ulrich E, Laine MC, Sandoz D. The orientation of ciliary basal bodies in quail oviduct is related to the ciliary beating cycle commencement. Biol Cell. 1985;55:147–150. doi: 10.1111/j.1768-322x.1985.tb00417.x. [DOI] [PubMed] [Google Scholar]
- 14.Deane JA, Cole DG, Seeley ES, Diener DR, Rosenbaum JL. Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr. Biol. 2001;11:1586–1590. doi: 10.1016/s0960-9822(01)00484-5. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell DR. Speculations on the evolution of 9+2 organelles and the role of central pair microtubules. Biol. Cell. 2002;96:691–696. doi: 10.1016/j.biolcel.2004.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Satir P, Guerra C, Bell AJ. Evolution and persistence of the cilium. Cell Motil. Cytoskel. 2007;64:906–913. doi: 10.1002/cm.20238. [DOI] [PubMed] [Google Scholar]
- 17.Azimzadeh J, Bornens M. Structure and duplication of the centrosome. J. Cell Sci. 2007;120:2139–2142. doi: 10.1242/jcs.005231. [DOI] [PubMed] [Google Scholar]
- 18.Maniotis A, Schliwa M. Microsurgical removal of centrosomes blocks cell reproduction and centriole generation in BSC-1 cells. Cell. 1991;67:495–504. doi: 10.1016/0092-8674(91)90524-3. [DOI] [PubMed] [Google Scholar]
- 19.Randall J, Disbrey C. Evidence for the presence of DNA at basal body sites in Tetrahymena pyriformis. Proc. Roy. Soc. B. 1965;162:473–491. doi: 10.1098/rspb.1965.0051. [DOI] [PubMed] [Google Scholar]
- 20.Hall JL, Ramanis Z, Luck DJL. Basal body/centriolar DNA: molecular genetic studies in Chlamydomonas. Cell. 1989;59:121–132. doi: 10.1016/0092-8674(89)90875-1. [DOI] [PubMed] [Google Scholar]
- 21.Chapman MJ, Dolan MF, Margulis L. Centrioles and kinetosomes: form, function, and evolution. Quart. Rev. Biol. 2000;75:409–429. doi: 10.1086/393621. [DOI] [PubMed] [Google Scholar]
- 22.Johnson KA, Rosenbaum JL. The basal bodies of Chlamydomonas reinhardtii do not contain immunologically detectable DNA. Cell. 1990;62:615–619. doi: 10.1016/0092-8674(90)90105-n. [DOI] [PubMed] [Google Scholar]
- 23.Kuroiwa T, Yorihuzi T, Yabe N, Ohta T, Uchida H. Absence of DNA in the basal body of Chlamydomonas reinhardtii by fluorimetry using a video-intensified microscope photon counting system. Protoplasma. 1990;158:155–164. [Google Scholar]
- 24.Holmes JA, Johnson DE, Dutcher SK. Linkage group XIX of Chlamydomonas reinhardtii has a linear map. Genetics. 1993;133:865–874. doi: 10.1093/genetics/133.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall JL, Luck DJL. Basal body-associated DNA: in situ studies in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 1995;92:5129–5133. doi: 10.1073/pnas.92.11.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pyne CK. Sur l'absence d'incorporation de la thymidine trite dans les cinetosomes de Tetrahymena pyriformis (Cilies Holotriches) C.R. Acad. Sci. Paris D. 1968;267:755–757. [Google Scholar]
- 27.Alliegro MC, Alliegro MA. Centrosomal RNA correlates with intron-poor nuclear genes in Spisula oocytes. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6993–6997. doi: 10.1073/pnas.0802293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizukami I, Gall J. Centriole replication II. Sperm formation in the fern, Marsilea, and the cycad, Zamia. J. Cell Biol. 1966;29:97–111. doi: 10.1083/jcb.29.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heath IB, Kaminskyj SG, Bauchop T. Basal body loss during fungal zoospore encystment: evidence against centriole autonomy. J. Cell Sci. 1986;83:135–140. doi: 10.1242/jcs.83.1.135. [DOI] [PubMed] [Google Scholar]
- 30.Marshall WF, Vucica Y, Rosenbaum JL. Kinetics and regulation of de novo centriole assembly. Implications for the mechanism of centriole duplication. Curr. Biol. 2001;11:308–317. doi: 10.1016/s0960-9822(01)00094-x. [DOI] [PubMed] [Google Scholar]
- 31.Khodjakov A, Rieder CL, Sluder G, Cassels G, Sibon O, Wang CL. De novo formation of centrosomes in vertebrate cells arrested during S phase. J. Cell Biol. 2002;158:1171–1181. doi: 10.1083/jcb.200205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hinchcliffe EH, Miller FJ, Cham M, Khodjakov A, Sluder G. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 2001;291:1547–1550. doi: 10.1126/science.1056866. [DOI] [PubMed] [Google Scholar]
- 33. Uetake Y, Loncarek J, Nordberg JJ, English CN, La Terra S, Khodjakov A, Sluder G. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J. Cell Biol. 2007;176:173–182. doi: 10.1083/jcb.200607073. Provides the explanation for cell cycle arrest following centrosome removal - the cells do not arrest because of the lack of centrosomes per se, but because lack of centrosomes makes them highly sensitive to exogenous stress.
- 34.Bennett CH. How to define complexity in physics, and why. In: Zurek WH, editor. Complexity, Entropy, and the Physics of Information, Santa Fe Institute Studies in the sciences of complex systems. volume III. Addison-Wesley; 1990. [Google Scholar]
- 35.Cover TM, Thomas JA. Elements of information theory. Wiley-Interscience; 2006. [Google Scholar]
- 36.Schrevel J, Besse C. Un type flagellaire fonctionnel de base 6+0. J. Cell Biol. 1975;66:492–507. doi: 10.1083/jcb.66.3.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedlaender M, Wahrman J. Giant centrioles in neuropteran meiosis. J. Cell Sci. 1966;1:129–144. doi: 10.1242/jcs.1.1.129. [DOI] [PubMed] [Google Scholar]
- 38.Simpson AGB, Bernard C, Fenchel T, Patterson DJ. The organization of Mastigamoeba schizophrenia n. Sp.: more evidence of ultrastructural idiosyncrasy and simplicity in pelobiont protists. Eur. J. Protistol. 1997;33:87–98. [Google Scholar]
- 39.Mansir A, Justine J. The microtubular system and posttranslationally modified tubulin during spermatogenesis in a parasitic nematode with amoeboid and aflagellate spermatozoa. Mol. Reprod. Dev. 1998;49:150–167. doi: 10.1002/(SICI)1098-2795(199802)49:2<150::AID-MRD6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 40.Raff EC, Hutchsen JA, Hoyle HD, Nielsen MG, Turner FR. Conserved axoneme symmetry altered by a component beta-tubulin. Curr. Biol. 2000;10:1391–1394. doi: 10.1016/s0960-9822(00)00784-3. [DOI] [PubMed] [Google Scholar]
- 41. Hiraki M, Nakazawa Y, Kamiya R, Hirono M. Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centrioles. Curr. Biol. 2007;17:1778–1783. doi: 10.1016/j.cub.2007.09.021. Demonstrates that centrioles can form that have fewer than nine triplets, by rescuing a deletion of a cartwheel spoke protein with truncated versions that produce spokes of reduced length.
- 42. Nakazawa Y, Hiraki M, Kamiya R, Hirono M. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr. Biol. 2007;17:2169–2174. doi: 10.1016/j.cub.2007.11.046. Demonstrates that centrioles can form that have different numbers of triplets besides nine, using a mutation in a centriole cartwheel protein.
- 43. Rodrigues-Martins A, Bettencourt-Dias M, Riparbelli M, Ferreira C, Ferreira I, Callaini G, Glover DM. DSAS-6 organized a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr. Biol. 2007;17:1465–1472. doi: 10.1016/j.cub.2007.07.034. Demonstrates that a centriole can form from triplet units that are facing in the wrong directions and apparently not connected in the usual way, suggesting a high degree of modularity and robustness in centriole assembly, features that could contribute to evolvability.
- 44.Bloodgood RA. Directed movements of ciliary and flagellar membrane components: a review. Biol. Cell. 1992;76:291–301. doi: 10.1016/0248-4900(92)90431-y. [DOI] [PubMed] [Google Scholar]
- 45.Omoto CK, Gibbons IR, Kamiya R, Shingyoji C, Takahashi K, Witman GB. Rotation of the central pair microtubules in eukaryotic flagella. Mol. Biol. Cell. 1999;10:1–4. doi: 10.1091/mbc.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dobbelaere J, Josue F, Suijkerbuijk S, Baum B, Tapon N, Raff J. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 2008;6:e224. doi: 10.1371/journal.pbio.0060224. A screen of almost the whole genome reveals just nine genes essential for centriole duplication.
- 47.Keller LC, Romijn EP, Zamora I, Yates JR, Marshall WF. Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of human ciliary disease genes. Curr. Biol. 2005;15:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 48.Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH, Culver BP, Yates JR, Winey M. New Tetrahymena basal body protein components identify basal body domain structure. J. Cell Biol. 2007;178:905–912. doi: 10.1083/jcb.200703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amos LA. The tektin family of microtubule-stabilizing proteins. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-7-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Donkor Ff, Moennich M, Czirr E, Hollemann T, Hoyer-Fender S. Outer dense fibre protein 2 (ODF2) is a self-interacting centrosomal protein with affinity for microtubules. J. Cell Sci. 2004;117:4643–4651. doi: 10.1242/jcs.01303. [DOI] [PubMed] [Google Scholar]