Abstract
We introduce a new caged glutamate, based in a ruthenium bipyridyl core, that undergoes heterolytic cleavage after irradiation with visible light with wavelengths up to 532 nm, yielding free glutamate in less than 50 ns. Glutamate photorelease occurs also efficiently following two-photon (2P) excitation at 800 nm, and has a functional cross section of 0.14 GM.
Introduction
Caged compounds, also called phototriggers, are widely used in biological research. They consist of a chemical entity composed of two parts: the “caged” compound of interest (in our case, a relevant biomolecule) and a “cage” moiety that inhibits its action [1]. Upon irradiation, the compound of interest is freed and can interact with the surrounding media. In physiological research, the caged compound is a biomolecule, and the surrounding media is a cell, a tissue, or the entire organism.
In the mammalian central nervous system, glutamate is the most ubiquitous excitatory neurotransmitter. As a result, the ability to precisely deliver glutamate in space and time is crucial to modulate the activity of neural circuits. An ideal way to achieve this is to use optical photorelease of glutamate. Unfortunately, all the currently available caged glutamate compounds require biologically harmful ultraviolet (UV) light or near UV irradiation, with only a few being useful for two-photon (2P) uncaging [2–4]. Most of these caged compounds are based on nitrobenzyl or nitroindole derivatives as cages. Uncaging occurs through a multi-step reaction, which is relatively slow and difficult to control. Indeed, one of the most used caged glutamates, CNB-glutamate (CNB = γ-carboxynitrobenzyl) [5], has a t1/2 of about 21 μs. Other near-UV caged compounds, such as CDMNB-capsaicin (alpha-carboxy-4,5-dimethoxy-2-nitrobenzyl-capsaicin) [6], and the widely used MNI-Glut have faster kinetics, with MNI-Glut having a t1/2 around 200 ns in water [7].
As an alternative chemical platform to nitrobenzyl or nitroindole derivatives, we introduced the use of ruthenium polypyridine complexes for caging amines, including the neurotransmitter γ-aminobutyric acid (GABA), and demonstrated that this strategy can lead to a full family of caged compounds that are activated with visible light [8,9]. For biological applications this is key, as it avoids the deleterious effects that UV radiation has on living tissue. Moreover, in this class of ruthenium compounds, the photoreaction consists of a single photochemical step, the breaking of a single metal-ligand bond, and allows for very fast uncaging. Ruthenium bipyridines present a strong metal to ligand charge transfer (MLCT) band at the visible region. Absorption at this band populates a triplet state that is thermally activated to a dissociative d-d state, which leads to photoproducts with a very fast kinetics [10].
We have recently introduced the biological use of a new caged glutamate with a ruthenium-bipyridine core, suitable for UV, visible, and IR irradiation [11]. Within the concentration range employed in biological experiments (100s of μM), the ruthenium (II) complex [Ru(bpy)2(PMe3)(Glu)] (bpy = 2,2′ bipyridine and PMe3 = trimethylphosphine) shows no apparent toxicity and can be used for standard physiological experiments, after minutes to hours of incubations. We now present the synthesis and chemical characterization of this novel caged glutamate, describing in particular its fast photorelease.
Experimental
Syntheses
The synthesis took place under filtered light. The acid form of the photoactive complex, cis-[Ru(bpy)2(PMe3)(GluH2)](PF6)2, was obtained as following: 110 mg of [Ru(bpy)2(PMe3)Cl)PF6 was dissolved in 2 mL of acetone. A suspension of 2 mL of water with 200 mg of a chloride-containing anionic exchange resin (DOWEX 2x8) was added, and stirred for 10 min. The resulting [Ru(bpy)2(PMe3)Cl]Cl solution was filtered to remove the resin, 500 mg of monosodium glutamate and 2.4 mL of 1 M NaOH were then added, and the resulting mixture was heated for 3 h. 1 mL of saturated KPF6 was added, and the resulting precipitate was discarded. The solution was then cooled to 0 °C and acidified with the addition of 5M HCl until pH~2. The final compound, [Ru(bpy)2(PMe3)(GluH2)](PF6)2, precipitated upon addition of excess of KPF6. The yellow-orange solid was then washed three times with cold water and dried. Yield: 52%. 1H-NMR (500 MHz, D2O, d = doublet, m = multiplet, t = triplet): δ = 1.07 (d, J=8.8Hz, 18H); 1.56–1.66 (m, 2H); 1.70–1.80 (m, 3H); 2.00–2.08 (m, 2H); 2.16–2.22 (m, 1H); 2.34–2.38 (m, 1H); 2.75–2.79 (m, 1H); 3.54 (t, J=10Hz, 1H,); 3.97 (t, J=10Hz, 1H,); 4.12 (d, J=12Hz, 1H); 4.50 (d, J=12Hz, 1H); 7.10 (t, J=6Hz, 2H); 7.25 (m, 2H); 7.45 (d, J=6Hz, 3H); 7.50 (d, J=6Hz, 1H); 7.68–7.85 (m, 6H); 7.92 (t, J=6Hz, 2H); 8.12–8.26 (m, 6H); 8.33 (d, J=8Hz, 1H); 8.36 (d, J=8Hz, 1H); 8.46 (d, J=10Hz, 2H); 8.49 (d, J=10Hz, 2H); 9.01 (d, J=5Hz, 1H); 9.04 (d, J=5Hz, 1H); 9.10 (d, J=6Hz, 1H) 9.22 (d, J=6Hz, 1H) ppm. C28H34F12N5O4P3Ru (926.23): Calcd. C 36.3, H 3.7, N 7.6, O 6.9; found C 34.7, H 3.2, N 7.9, O 7.2.
The precursor complex [Ru(bpy)2(PMe3)Cl)]PF6 was obtained following this procedure: 520 mg of [Ru(bpy)2Cl2] was suspended in 20 mL of a 1:1 mixture of methanol and water, and refluxed under N2. 1.2 mL of 1M trimethylphosphine in THF (Tetrahydrofuran, Aldrich 324108) was added by syringe. The reaction was followed using UV-Visible (UV-Vis) spectroscopy. In some cases, additional phosphine solution was added. Once the UV-Vis spectrum was stable, methanol and excess phosphine were removed by vacuum distillation. The resulting aqueous solution was filtered to remove any solids, and precipitated by the addition of excess of KPF6 at 0 °C. The dark orange solid was washed three times with cold water and dried. Yield: 93%. 1H-NMR (500 MHz, D2O): δ = 1.07 (d, J = 8.8Hz, 9H); 7.10 (t, J = 8Hz, 1H); 7.27 (t, J = 8Hz, 1H); 7.51 (d, J = 6Hz, 1H); 7.71 (t, J = 8Hz, 1H); 7.75–7.82 (m, 2H); 7.86 (d, J = 5Hz, 1H); 7.94 (t, J = 8Hz, 1H); 8.12 (t, J = 8Hz, 1H); 8.19 (t, J = 8Hz, 1H); 8.29 (d, J = 7.5Hz, 1H); 8.42–8.53 (m, 3H); 9.19 (d, J = 5Hz, 1H); 9.45 (d, J = 5Hz, 1H) ppm. C23H25ClF6N4P2Ru (669.67): Calcd. C 41.2, H 3.8, N 8.4; found C 39.9, H 3.7, N 8.1. (Although the analyses data for “C” are somewhat unsatisfactory, 1H-NMR and COSY spectra reasonably support the formula.)
Emission measurements
Steady state emission spectra were measured with a PTI-Quantamaster spectrofluorometer and corrected for the instruments response function. Time resolved emission were measured with a Horiba Jobin Yvon FluoroCube-11-NL with an LED source (457 nm, 1.6 ns pulse duration).
Electrochemistry and NMR
1H-NMR spectra were obtained with a 500 MHz apparatus (Bruker). Redox potentials were measured in CH3CN/TBAPF6 (0.1 M) using a three-electrode potentiostat based on an operational amplifier (TL071) in a current-to-voltage configuration, with acquisition software written in QB 4.5. A 1 cm Pt wire with a diameter of 500 μm was used as working electrode. An Ag/AgCl electrode was used as reference, and the potentials were obtained using the Ferrocene/Ferricinium redox couple as a reference. The counter electrode was a 10 cm long Pt wire, coiled around the 2 mL cell.
Photolysis
The UV-Vis spectra were measured with a HP8453 diode-array spectrometer. The quantum yield measurements were performed with a Luxeon Star III Royal Blue high-power light-emitting diode (LED) centered at 450 nm with a 20 nm FWHM (Full Width at Half Maximum). The light was collimated and sent through an optical path of 1 cm. Total irradiation was determined by comparison to a reference sample with [Ru(bpy)2(py)2]2+ as photosubstitution quantum yield standard [12]. The fast photolysis measurements were performed using the second harmonic of a Spectra-Physics (Indi-HG) Nd:YAG laser (532nm, 10ns FWHM) as the pump laser and a low power continuous DPDSS Nd:YAG (532nm, 2mW) as the probe laser.
Neuronal tissue preparation and electrophysiology
Three hundred fifty micrometer thick coronal slices from 14-day-old C57BL/6 mouse cortex were prepared using a Leica VT1000-S vibratome with a cutting solution containing (in mM): 27 NaHCO3, 1.5 NaH2PO4, 222 Sucrose, 2.6 KCl, 2 MgSO4, 2 CaCl2. Slices were incubated at 32°C in artificial cerebrospinal fluid (ACSF) for 30 min and then kept at room temperature for at least 30 min before transferring them to the recording chamber. The recording chamber was bathed in room temperature ACSF (pH 7.4) saturated with 95% O2 and 5% CO2, containing (in mM): 126 NaCl, 3 KCl, 2 MgSO4, 2 CaCl2, 1 NaH2PO4, 26 NaHCO3, and 10 glucose. Neurons were either held at their resting membrane potential (−65 mV), or at +40 mV, where both inhibitory and excitatory events can be recorded. Whole-cell patch clamp electrodes (4–7 MΩ) were used.
Two photon glutamate uncaging
An 350 μM of [Ru(bpy)2(PMe3)(Glu)] solution was prepared by dissolving [Ru(bpy)2(PMe3)GluH2](PF6)2 in normal ACSF at physiological pH and directly used. A somatic uncaging point was selected using a custom software, which also triggered the uncaging pulse and controlled the pulse duration [13]. Laser power was modulated by a Pockels cell (Quantum Technology, Lake Mary, FL, USA). For somatic stimulations, each uncaging target consisted of eight subtargets, each of which was illuminated for 8 ms, giving a total duration of around 70 ms at 800 nm. The subtargets themselves were complex, consisting of five very closely spaced beamlets created by multiplexing the laser beam with a diffractive optical element (DOE) [11].
Results and Discussion
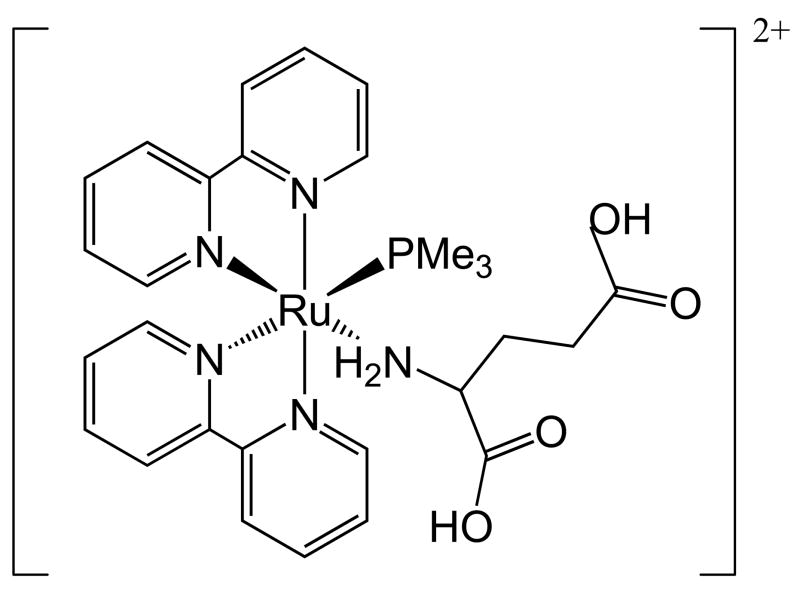
The protonated complex, [Ru(bpy)2(PMe3)(GluH2)](PF6)2,] (see scheme 1) in which the aminoacid is in full protonated glutamic acid form, exhibits a bright orange colour and has a high solubility in water. At physiological pH, the complex exists as the deprotonated glutamate species [Ru(bpy)2(PMe3)(Glu)]. Aqueous solutions present a strong MLCT band centered at 450 nm, characteristic of this family of Rupolypyridines [9]. Cyclic voltammetry of the compound dissolved in acetonitrile shows three redox processes at 0.98, 1.46 and 1.56 V vs. NHE (Normal Hydrogen Electrode), corresponding to the Ru(III)/Ru(II) couple of the original complex, and those of the complexes bearing oxidation products of glutamate, respectively. The second and third couples are not reversible, and some electrocatalytic oxidation processes can occur at such high potentials, which are not expected to happen in physiological conditions (Fig S1, supplementary information).
Scheme 1.
Structure of the protonated form of the complex [Ru(bpy)2(PMe3)(GluH2)]2+
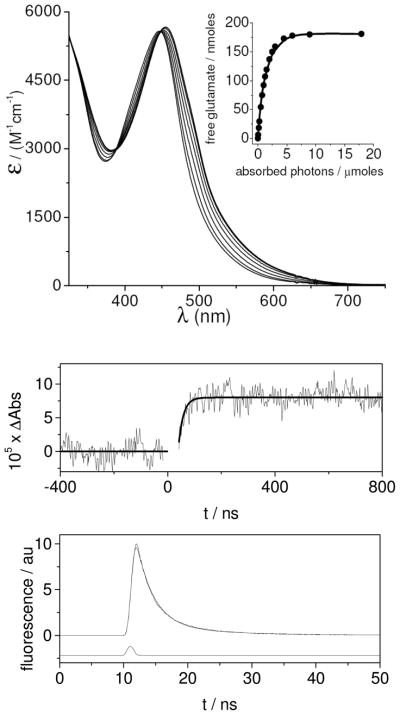
Figure 1 (top) shows the UV-Vis spectrum of the complex while being irradiated with a 450 nm LED in water at pH = 7. The photoreaction proceeds to completion in around 4 min, as indicated by the formation of the aquo-complex. The presence of two isosbestic points, as well as a factor analysis performed on the spectra, indicate that only two colored species are present in the solution, exactly what is expected for the single photoaquation process. The spectrum of the photoproduct was found to be identical to that of the complex [Ru(bpy)2(PMe3)(H2O)]2+, which can be synthesized by refluxing the chloro complex in water. At pH > 12, similar behaviour was observed, although in this case the product is the hydroxo complex [Ru(bpy)2(PMe3)(OH)]+ due to the pKa of the aquo-complex (pKa = 10.3). The complete photolysis at pH = 12 is shown in Fig. S2 as Supplementary information.
Figure 1.
Top: Six successive UV-Vis spectra of [Ru(bpy)2(PMe3)(Glu)] recorded in aqueous solution at pH=7 every 60 s irradiation using 450nm light. Inset: amount of photoreleased glutamate derived from the experimental data (circles); fitting to the theoretical equation (line). Middle: Absorbance changes at 532 nm after flash photolysis of [Ru(bpy)2(PMe3)(Glu)] 1 mM (pump pulse: 532nm, 10 ns FWHM, average of 256 scans). Bottom: Time resolved emission of the same solution. Upper trace: Experimental data (time constant τ = 3.4 ns) Lower trace: excitation pulse.
The inset in Fig. 1 (upper panel) shows the yield of free glutamate generated by photolysis, and was obtained by analysis of the spectra. As mentioned previously, the irradiance of the light source was calibrated using the known efficiency of photolysis of [Ru(bpy)2(py)2]2+ [12]. The yield of photoreleased glutamate was fitted using a two- parameter single-exponential function: y = a[1−exp(−bx)] and the quantum yield (φPC) was calculated as a•b, yielding a value of φPC = 0.13 at pH = 7 and φPC = 0.10 at pH = 12.
To measure the uncaging time, we performed a flash photolysis experiment on a basic aqueous solution of [Ru(bpy)2(PMe3)(Glu)] using the second harmonic of a Nd-YAG laser as the pump source (532 nm, 10 ns FWHM pulses). Absorbance changes recorded at 532 nm by a separate low power CW laser are shown in Fig. 1 (middle panel). The photocleavage occurs within 50 ns, and this is among the fastest reported caged compounds. This timescale is consistent with the measured excited state lifetime of a similar compound containing pyridine instead of glutamate (10–100 ns) [14].
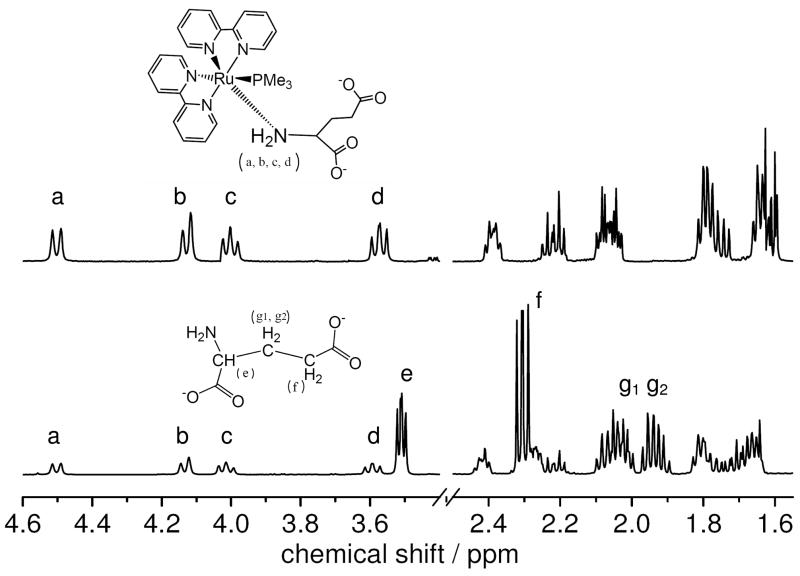
The aqueous solutions of [Ru(bpy)2(PMe3)(Glu)] present a weak emission, with a maximum at 643 nm, following excitation around 450 nm (Fig. S3, supplementary information). The quantum yield of emission was found to be 1.1×10−3 using a degassed solution of [Ru(bpy)3]Cl2 as a standard. The very low quantum yield suggests fast deactivation of the 3MLCT state. The emission lifetime was measured to be ~3.4 ns, much faster than the expected radiative decay for the triplet state, and indicated fast dynamics which quickly populates the dissociative d-d state following irradiation. The glutamate is delivered from the excited complex shortly there after. The photoreaction was also followed by NMR spectroscopy. In the aromatic region of the 1H-NMR spectrum of the complex (Fig S4, supplementary information) it is possible to distinguish 16 signals corresponding to the bipyridine protons. These peaks are found duplicated because the synthesized compound is a roughly 1:1 mixture of diastereomers, resulting from the coordination of l-glutamate with the racemic mixture of Λ and Δ Ru-bpy enantiomers. No effort was made to separate the diastereomers and the chemical and biological tests suggest that both have a similar behavior.
The aliphatic region corresponding to the glutamate moiety is depicted in the upper trace of Fig. 2, where −NH2 protons appear at 4.50, 4.12, 4.00 and 3.57 ppm (a, b, c and d). The continued presence of these signals in a D2O solution is evidence of the absence of isotopic exchange in the complex and clearly indicates that the coordination of glutamate is done via the amine nitrogen, which hampers further protonation. This behavior differs from that of free glutamate which, under the same conditions, exchanges rapidly H+ for D+. Spectra taken after 2 months of storage in D2O, at room temperature, and in the absence of light, show no sign of exchange, also demonstrating that the compound is indeed inert to hydrolysis. After photolysis, many new signals appear in the aromatic region which correspond to the aquo complexes (Fig S4). At the same time, the analysis of the aliphatic region (Fig. 2, bottom trace) provides clear evidence of the appearance of free glutamate. Coordinated −NH2 signals a, b, c and d diminish upon irradiation, and the characteristic signals of the methylene protons of free glutamate (e,f,g1 and g2) appear at 3.50, 2.30 and 2.04 and 1.94 ppm.
Figure 2.
Aliphatic section of the 1H-NMR spectrum of [Ru(bpy)2(PMe3)(Glu)] in D2O before (upper trace) and after (lower trace) photolysis. The signals a, b, c and d correspond to the coordinated amine protons, while e, f, g1 and g2 correspond to free glutamate.
Further addition of pure glutamate (Aldrich, 49621, >98.0% purity) did not produce any additional peaks, but merely increased the intensity of the existing ones e,f,g1 and g2, demonstrating that the aliphatic photoproduct is glutamate. No other signals, besides those of the aquo-complex and free glutamate, appear during the photolysis.
Two-photon (2P) uncaging is becoming increasingly popular for biological experiments, due to its high Z-axis focusing capabilities and the fact that low energy IR photons can easily penetrate living tissues. Recent experiments on similar ruthenium complexes have shown strong two photon absorption, and our own previous studies proved the viability of two photon photocleavage for this class of compounds [15,16]. We therefore investigated the 2P capabilities of our novel caged glutamate. We first incubated acute neocortical mouse brain slices in 350 μM [Ru(bpy)2(PMe3)(Glu)] and carefully monitored the morphology and electrophysiological properties of the neurons and their processes using two-photon microscopy and whole-cell patch recordings. After several hours of incubation, no detectable deleterious effects were observed, and the neurons remained healthy and functioned normally. This indicates that the complex does not modify membrane integrity or has immediate harmful effect on living neurons, and can be used in effectively at these concentrations in biological experiments
We then measured the 2P uncaging efficiency. Because of the difficulties in directly measuring absolute two-photon absorption or uncaging action cross sections, we instead performed a relative “functional cross section” comparing [Ru(bpy)2(PMe3)(Glu)] to MNI-Glut. Uncaging generates free glutamate, which subsequently binds to receptors on the surface of a neuron and opens ion channels. The influx of ions depolarizes the cell, and when the depolarization is sufficiently strong, the cell will fire an action potential. The strength of the initial depolarization, and hence the ability to generate an action potential is dependent on the total amount of glutamate released during the uncaging process. We used this action potential generation measurement as our functional reporter and compared the parameters necessary to reliably generate action potentials in cortical pyramidal neurons in acute mouse brain slices for both the RuBi-Glut and MNI-Glut (N=15 cells for [Ru(bpy)2(PMe3)(Glu)], N=39 cells for MNI-Glut, each with multiple trials).
In the uncaging process, the number of glutamate molecules released is related to the two photon cross section through the following relation:
where σ2 is the two photon absorption cross section, φ the uncaging quantum yield, and C(r,t) and I2(r,t) the spatial and time dependent concentration and light intensity.
While the general expressions for C(r,t) and I(r,t) are complicated [17], we can make some simplifying assumptions for both [Ru(bpy)2(PMe3)(Glu)] and MNI-Glut. Because the total duration of illumination is significantly longer than both the glutamate releasing reactions, and the estimated diffusion times of the compounds in to and out of the excitation volume, we ignore the time dependence of the concentration, and assume a steady state concentration of caged compound. Additionally, we assume that the steady-state concentration is directly proportional to the initial concentration. Using a Gaussian limited focal volume, and measuring the laser pulse width and power on the sample allowed us to relate the intensity to the measured average power. With these simplifications, the functional two photon uncaging action cross section for [Ru(bpy)2(PMe3)(Glu)] is given in relation to MNI-Glut by:
Using the reported uncaging action cross section for MNI-Glut, 0.06 GM at 725 nm (1GM=10−50 cm4 s photon−1), the two photon functional cross section of [Ru(bpy)2(PMe3)(Glu)] at 800 nm is ~0.14 GM.
Conclusions
In conclusion, we have synthesized a new caged compound for glutamate photodelivery with a high uncaging quantum yield at visible wavelengths, as high as 532nm, with very fast photolysis kinetics, and full 2P capabilities. These properties, combined with the ease of synthesis and the lack of apparent toxicity in biological preparations, make the complex [Ru(bpy)2(PMe3)(Glu)] a state-of-the-art tool for physiological research.
Supplementary Material
Acknowledgments
E.M. and R.E. are staff of CONICET. This work was made with funding from CONICET, UBA, NEI, the STV office of Columbia University and the Kavli institute for Brain Science.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mayer G, Heckel A. Angew Chem Int Ed Engl. 2006;45:4900–4921. doi: 10.1002/anie.200600387. [DOI] [PubMed] [Google Scholar]
- 2.Furuta T, Wang SS, Dantzker JL, Dore TM, Bybee WJ, Callaway EM, Denk W, Tsien RY. Proc Natl Acad Sci USA. 1999;96:1193–1200. doi: 10.1073/pnas.96.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morrison J, Wan P, Corrie JE, Papageorgiou G. Photochem Photobiol Sci. 2002;1:960–969. [PubMed] [Google Scholar]
- 4.Rossi FM, Margulis M, Tang CM, Kao JP. J Biol Chem. 1997;272:32933–32939. doi: 10.1074/jbc.272.52.32933. [DOI] [PubMed] [Google Scholar]
- 5.Wieboldt R, Gee KR, Niu L, Ramesh D, Carpenter BK, Hess GP. Proc Natl Acad Sci USA. 1994;91:8752–8756. doi: 10.1073/pnas.91.19.8752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert D, Funk K, Dekowski B, Lechler R, Keller S, Mohrlen F, Frings S, Hagen V. Chembiochem. 2007;8:89–97. doi: 10.1002/cbic.200600437. [DOI] [PubMed] [Google Scholar]
- 7.Cheng Q, Steinmetz MG, Jayaraman V. J Am Chem Soc. 2002;124:7676–7677. doi: 10.1021/ja0259988. [DOI] [PubMed] [Google Scholar]
- 8.Zayat L, Noval MG, Campi J, Calero CI, Calvo DJ, Etchenique R. Chembiochem. 2007;8:2035–2038. doi: 10.1002/cbic.200700354. [DOI] [PubMed] [Google Scholar]
- 9.Zayat L, Salierno M, Etchenique R. Inorg Chem. 2006;45:1728–1731. doi: 10.1021/ic0512983. [DOI] [PubMed] [Google Scholar]
- 10.Campagna S, Puntoriero F, Nastasi F, Bergamini G, Balzani V. Top Curr Chem. 2007;280:117–214. [Google Scholar]
- 11.Fino E, Araya R, Peterka DS, Salierno M, Etchenique R, Yuste Front R. Neural Circuits. 2009;3:2. doi: 10.3389/neuro.04.002.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinnick D, Durham B. Inorg Chem. 1984;23:1440–1445. [Google Scholar]
- 13.Nikolenko V, Poskanzer KE, Yuste R. Nat Methods. 2007;4:943–950. doi: 10.1038/nmeth1105. [DOI] [PubMed] [Google Scholar]
- 14.Durham B, Caspar J, Jeffrey K, Negle K, Meyer T. J Am Chem Soc. 1982;104:4803–4810. [Google Scholar]
- 15.Girardot C, Cao B, Mulatier J, Baldeck PL, Chauvin J, Riehl D, Delaire JA, Andraud C, Lemercier G. ChemPhysChem. 2008;9:1531–1535. doi: 10.1002/cphc.200800186. [DOI] [PubMed] [Google Scholar]
- 16.Nikolenko V, Yuste R, Zayat L, Baraldo LM, Etchenique R. Chem Commun. 2005:1752–1754. doi: 10.1039/b418572b. [DOI] [PubMed] [Google Scholar]
- 17.Kiskin NI, Chillingworth R, McCray JA, Piston D, Ogden D. Eur Biophys J. 2002;30:588–604. doi: 10.1007/s00249-001-0187-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.