Abstract
The RNA-binding protein Bicaudal C is an important regulator of embryonic development in C. elegans, Drosophila and Xenopus. In mouse, bicaudal C (Bicc1) mutants are characterized by the formation of fluid-filled cysts in the kidney and by expansion of epithelial ducts in liver and pancreas. This phenotype is reminiscent of human forms of polycystic kidney disease (PKD). Here, we now provide data that Bicc1 functions by modulating the expression of polycystin 2 (Pkd2), a member of the transient receptor potential (TRP) superfamily. Molecular analyses demonstrate that Bicc1 acts as a post-transcriptional regulator upstream of Pkd2. It regulates the stability of Pkd2 mRNA and its translation efficiency. Bicc1 antagonized the repressive activity of the miR-17 microRNA family on the 3′UTR of Pkd2 mRNA. This was substantiated in Xenopus, in which the pronephric defects of bicc1 knockdowns were rescued by reducing miR-17 activity. At the cellular level, Bicc1 protein is localized to cytoplasmic foci that are positive for the P-body markers GW182 and HEDLs. Based on these data, we propose that the kidney phenotype in Bicc1−/− mutant mice is caused by dysregulation of a microRNA-based translational control mechanism.
Keywords: P-bodies, Polycystic kidney disease, microRNA, Pronephros, Translational regulation, Mouse, Xenopus
INTRODUCTION
The vertebrate kidney is essential to conserve water, electrolytes and metabolites, as well as to remove metabolic waste products from the body. It develops through three successive and increasingly complex renal structures: the pronephros, mesonephros and metanephros (Saxén, 1987; Vize et al., 2002). These three kidney forms appear very different, but all rely on the same functional unit, the nephron. The structure and formation of the nephron is evolutionarily conserved (Mobjerg et al., 2000; Zhou and Vize, 2004; Dressler, 2006; Raciti et al., 2008). The kidney as a whole, and the nephron in particular, is an example of a tubular organ (Hogan and Kolodziej, 2002). As such, the kidney has attracted much attention in understanding how mesenchymal cells undergo the mesenchymal-epithelial transition (MET) to form the highly specialized renal tubules. In addition, the integrity of the tubules needs to be maintained throughout life to promote proper kidney function. In humans, the disruption of these processes results in a wide array of genetic diseases. Among these, polycystic kidney diseases (PKDs) are the leading cause of end-stage renal disease in children and adults (Torres and Harris, 2007; Wilson and Goilav, 2007). They are characterized by fluid-filled cysts that result from unregulated expansion of renal epithelial cells. The most frequent form of PKD, autosomal dominant PKD (ADPKD), is caused by mutations in PKD1 and PKD2 (Burn et al., 1995; Hughes et al., 1995; Mochizuki et al., 1996; The International Polycystic Kidney Disease Consortium, 1995). These genes encode polycystin 1 (PKD1; polycystic kidney disease 1), an 11-transmembrane-domain protein, and polycystin 2 (PKD2; polycystic kidney disease 2), a member of the transient receptor potential (TRP) superfamily. PKD1 and PKD2 are present in a cation channel complex that is involved in mechanosensation-triggered Ca2+ influx into cells. The second type of PKD, autosomal recessive PKD (ARPKD), is primarily caused by mutations in a single gene, PKHD1, which encodes a receptor-like membrane protein of unknown function known as polyductin, fibrocystin or tigmin (Hildebrandt et al., 1997; Onuchic et al., 2002; Ward et al., 2002; Xiong et al., 2002). Many animal models of PKD have been developed to better understand the process of cyst formation (Guay-Woodford, 2003; Drummond, 2005). Among these, mice with spontaneous mutations in the bicaudal C (Bicc1) locus are the least understood (Cogswell et al., 2003).
Bicc1 encodes an evolutionarily conserved RNA-binding molecule that consists of five N-terminal KH (hnRNP K homology) RNA-binding domains and a C-terminal protein-protein interaction SAM (sterile alpha motif) domain. Bicaudal C was originally identified in a Drosophila mutagenesis screen as a gene in which heterozygous females produced embryos with ‘double-abdomen’ phenotypes (Mohler and Wieschaus, 1986). Subsequent studies in both Drosophila and C. elegans suggest that Bicaudal C regulates mRNA stability and translation via modulation of mRNA polyadenylation (Mahone et al., 1995; Saffman et al., 1998; Wang et al., 2002; Suh et al., 2006; Chicoine et al., 2007). Previously, we identified the Xenopus and mouse homologs of Bicaudal C (Wessely and De Robertis, 2000; Wessely et al., 2001). Interestingly, bicc1 is expressed in the renal epithelial cells of the Xenopus pronephros and loss of Bicc1 results in a PKD-like phenotype (Tran et al., 2007). Eliminating Bicc1 protein using antisense morpholino oligomers (MOs) impairs the physiological role of the pronephros by interfering with its osmoregulatory function, which leads to edema formation in the Xenopus embryo. At the molecular level, Bicc1 is required for the differentiation of renal cells of the late distal tubule and pronephric duct.
In mouse, Bicc1 is expressed in the meso- and metanephric kidney (Wessely et al., 2001). Here, we now further investigate the biological role of Bicc1 by eliminating the gene using homologous recombination. The Bicc1 mutant mice rarely survived postnatally and developed severe PKD. At birth, cysts were detected along the entire length of the nephron. Interestingly, the loss-of-function phenotype of Bicc1 was remarkably similar to that of Pkd2. Experiments using the Xenopus pronephros, the mouse metanephric kidney and human HEK293T cells showed that both genes actually function in the same pathway, with Bicc1 acting upstream of Pkd2. Moreover, Bicc1 was localized to cytoplasmic foci that contain proteins involved in post-transcriptional regulation of mRNAs, and Bicc1 modulated Pkd2 protein levels by antagonizing microRNA (miRNA)-mediated repression within the 3′UTR of Pkd2 mRNA.
MATERIALS AND METHODS
Gene targeting
The Bicc1 mutation was performed by fusing the β-galactosidase (lacZ) gene with exon 4 of Bicc1 and by deleting part of exon 4 and all of exons 5-8. Correct integration of the targeting construct was confirmed by Southern blot (see Fig. 1A for the position of the 5′ and 3′ probes). Electroporation and injection of the positive clones was performed by the ES Cell and Transgenic Core facilities at UCLA. Mice carrying the Bicc1 mutation were backcrossed into the B6SJLF1/J hybrid strain (Jackson Laboratories) and all the experiments described here used the B6SJLF1/J hybrid.
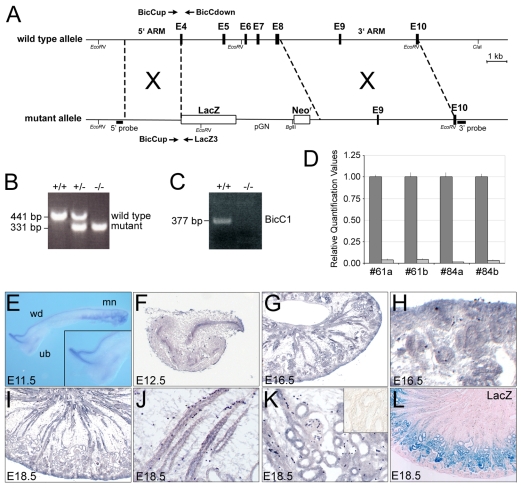
Fig. 1.
Expression analysis and knockout of Bicc1 in mouse. (A) Schematic of Bicc1 loss-of-function strategy. (B) PCR-based genotyping of Bicc1 mice. (C) RT-PCR for Bicc1 in kidneys from Bicc1+/+ and Bicc1−/− mice using primers spanning exons 9 to 11. (D) qPCR analysis of Bicc1 expression using primers covering exons 15 and 16. Each of the four sets represents a comparison of a Bicc1+/+ (dark gray) and a Bicc1−/− mutant (light gray) littermate. (E) Expression of Bicc1 mRNA in the Wolffian duct (wd), mesonephros (mn) and T-stage branching of the metanephros (ureteric bud, ub) at E11.5. Inset is a magnified view of the ureteric bud. (F-K) In situ hybridization on paraplast sections of metanephric kidneys at E12.5 (F), E16.5 (G) and E18.5 (I) and magnified views of the nephrogenic zone at E16.5 (H), and of the collecting ducts (J) and glomeruli and proximal tubules (K) at E18.5. Inset in K shows sense control. (L) lacZ staining of an E18.5 Bicc1+/− kidney section counterstained with Eosin.
Xenopus embryo manipulations
Xenopus embryos obtained by in vitro fertilization were maintained in 0.1× modified Barth medium (Sive et al., 2000) and staged according to Nieuwkoop and Faber (Nieuwkoop and Faber, 1994). The antisense MOs (GeneTools, LLC) used in this study were (5′ to 3′): xBicC-MO1, GGGACAAAGATGCTCATTTTAACAG; xBicC-MO2, GCCACTATCTCTTCAATCATCTCCG; Pkd2-MO, GGTTTGATTCTGCTGGGATTCATCG; and miR-17-MO, ACTACCTGCACTGTAAGCACTTTGA. Unless otherwise indicated, a total of 3.2 pmol of Pkd2-MO, miR-17-MO and Std-MO or a mixture of 3.2 pmol xBicC-MO1 and 3.2 pmol xBicC-MO2 (xBicC-MO1+2) was injected radially at the 2- to 4-cell stage into Xenopus embryos.
For synthetic mRNA, pCS2-xBicC* (Tran et al., 2007), pCS2-xBicC-GFP, pCS2-Pkd2 and pCS2-Pkd2-myc were linearized with NotI and transcribed with SP6 RNA polymerase. pXEXβGal-Pkd2(short_UTR) and pXEXβGal-Pkd2(long_UTR) were linearized with Asp718 and transcribed with T7 RNA polymerase using the mMessage mMachine (Ambion). For the synthetic miR-17 duplex, two oligos (5′-rArCrCrUrGrCrArCrUrGrUrArArGrCrArCrUrUrUrGTT-3′ and 5′-rCrArArArGrUrGrCrUrUrArCrArGrUrGrCrArGrGrUAG-3′) were synthesized and annealed (Integrated DNA Technologies). Rescue experiments and miRNA reporter assays in Xenopus and HEK293T cells were performed as previously described (Tran et al., 2007; Agrawal et al., 2009).
In situ hybridization, immunohistochemistry, lectin staining and histology
Whole-mount and paraplast section in situ hybridizations were performed as previously described (Tran et al., 2007; Agrawal et al., 2009). The sequence of the LNA-modified miR-17 degenerate oligomer is 5′-CTACLNACTGLNACACLNATRTLNADAGLNACACLNATTTLNAG-3′.
For immunohistochemistry and lectin staining, mouse kidneys were fixed in 4% paraformaldehyde and Xenopus embryos were fixed in Dent's fixative. The following antibodies/lectins were used: anti-acetylated α-tubulin (Sigma); peroxidase-conjugated Dolichos biflorus agglutinin (DBA, Sigma); anti-Pkd2 (Millipore); anti-phospho-histone H3 (Millipore); biotinylated Lotus tetragonolobus agglutinin (LTA, Vector Laboratories); anti-Nkcc2 (Slc12a1) (kind gift of Dr Mark Knepper, NIH); and anti-Pcna (DAKO). β-galactosidase staining was performed on cryostat sections or whole-mounts following the protocol described by Ma et al. (Ma et al., 2002) with minor modifications in the fixation timing. For histological staining, tissues were fixed in Bouin's Fixative and stained with Hematoxylin and Eosin (H&E), Periodic Acid Schiff (PAS) or Mallory's Tetrachrome.
RT-PCR and polyadenylation assay
Metanephric kidneys were processed for reverse transcription using standard protocols. Isolation of capped mRNA was performed using the mRNA-ONLY Eukaryotic mRNA Isolation Kit (Epicentre Biotechnologies). Conventional PCR for mouse Bicc1 was performed using primers hBicC1 upper (5′-GGGTTGTCTTCCTCTTGTGT-3′) and hBicC1 lower (5′-AGAGTGAGTTTGGGGTTGTT-3′), which amplify a 377 bp fragment covering exons 9 to 11. Quantitative PCR was performed using the Applied Biosystems 7500 FAST Real-time PCR System and TaqMan Gene Expression Assays.
The length of the poly(A) tail of Pkd2 mRNA was determined using RNA ligation-coupled RT-PCR (RL-PCR) as previously described (Charlesworth et al., 2004) using the Pkd2 primer 5′-CTGGTAGTCTCCCCTCTGT-3′. To visualize the Pkd2-specific amplification products, the PCR reactions were separated on a 1% agarose gel and processed for non-radioactive Southern blotting using a Pkd2-specific probe.
Protein analysis
HEK293T cells were transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Kidneys and HEK293T cells were lysed in T-PER Tissue Protein Extraction Reagent (Thermo Scientific) and processed for western blot analysis using standard protocols. For the subcellular localization studies, transfected HEK293T cells were cultured on Lab-TekII chamber slides (Nalgene Nunc) and processed for immunofluorescence analysis 48 hours post-transfection. Stress granule formation was studied in HeLa cells treated with 20 μM clotrimazole (Sigma) for 45 minutes in serum-free medium.
The following primary antibodies were used: anti-actin (Sigma); anti-calregulin (Santa Cruz); anti-Eif3η (Santa Cruz); anti-Eif4E (Santa Cruz); anti-GM130 (BD Biosciences); anti-GW182 (Abcam and Santa Cruz); anti-Lamp2 (DSHB); anti-Pkd2 (Santa Cruz); anti-Pkd2 (YCC2) (Markowitz et al., 1999); and anti-p70 S6 kinase-α [Rps6kb1; this antibody cross-reacts with HEDLs (Stoecklin et al., 2006); Santa Cruz].
RESULTS
Expression of bicaudal C in the kidney
Bicaudal C (Bicc1) has been shown to be mutated in two mouse models of PKD, bpk and jcpk (Cogswell et al., 2003). bpk mice, in particular, are widely used as a late onset model of PKD (Sweeney et al., 2000; Shillingford et al., 2006). However, the molecular mechanism of cyst formation by impaired Bicc1 function is still poorly understood. To address this, we generated a targeted mutation of Bicc1 in mouse, in which parts of exon 4 were fused in frame to β-galactosidase, replacing exons 4-8 (Fig. 1A). The fusion protein lacks all known functional domains of Bicc1 (the RNA-binding KH domains and the protein-protein interaction SAM domain). Southern blot and RT-PCR analyses confirmed proper targeting of the Bicc1 locus and the absence of alternatively spliced transcripts (Fig. 1B-D; see Fig. S1A in the supplementary material; data not shown), supporting the conclusion that these mice lack a functional Bicc1 protein.
To better understand the kidney phenotype of Bicc1−/− mice, we first analyzed the expression of Bicc1 by in situ hybridization at different stages of kidney development. At embryonic day (E) 11.5, Bicc1 mRNA was detected in the cranial and caudal tubules of the mesonephros, the Wolffian duct and the first branch of the ureteric bud (UB) tree (Fig. 1E; data not shown). Later, Bicc1 was present in the UB tree, but was also found in renal vesicles and in the comma- and S-shaped bodies (Fig. 1F-H; data not shown). At E18.5, Bicc1 mRNA was detected in all epithelial components of the kidney (Fig. 1I-K). lacZ staining faithfully recapitulated the expression of Bicc1 mRNA at early stages of development in the posterior notochord/ventral node and the developing endoderm (see Fig. S1B-C′ in the supplementary material) (Wessely et al., 2001; Blum et al., 2007). However, in the kidney, β-galactosidase activity was mainly observed in the proximal tubules at E18.5 (Fig. 1L). This discrepancy was probably due to reduced stability of the Bicc1-β-galactosidase fusion protein as the lacZ mRNA pattern was indistinguishable from that of Bicc1 mRNA in the kidneys of Bicc1-heterozygous mice (see Fig. S1D,D′ in the supplementary material).
Polycystic kidney disease phenotype in Bicc1−/− mice
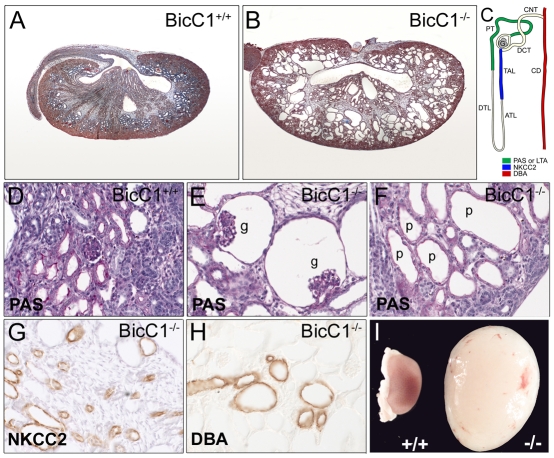
Bicc1 heterozygous intercrosses showed a lower than expected ratio of homozygous mutant progeny (see Table S1 in the supplementary material). The lethality of Bicc1−/− mice increased during development. Although Mendelian ratios were observed in embryos up to E14.5, more than half of the mutant mice died perinatally, for unknown reasons. Similar to bpk and jcpk mice (Nauta et al., 1993; Flaherty et al., 1995), surviving Bicc1−/− mice developed severely enlarged kidneys on both sides, caused by dramatic polycystic malformations (Fig. 2A,B,I). Cells lining the cysts lost their cuboidal shape and adopted a squamous appearance (Fig. 2D,F). Despite the increased size of the mutant kidneys, the nephrogenic zone was reduced (Fig. 2A,B). These mice also developed cysts in the bile ducts of the liver and the excretory ducts of the pancreas (see Fig. S2 in the supplementary material). In PKD, cysts arise from different segments of the nephron. To identify the origin of cysts in Bicc1−/− mice, segment-specific markers were analyzed. As shown in Fig. 2C-H, cysts at birth [post-natal day (P) zero, P0] were derived from glomeruli, proximal tubules and, to a lesser extent, from the thick ascending limb of the loop of Henle and collecting ducts. When Bicc1−/− kidneys were analyzed at earlier stages, cysts were first detected at E15.5. At this stage, cysts were mainly of glomerular origin, but extended towards more distal segments in subsequent stages (see Fig. S3 in the supplementary material). Interestingly, the early phase of cyst formation was not caused by uncontrolled proliferation. No obvious changes in proliferation were detected using antibodies against phospho-histone H3 and proliferating cell nuclear antigen (Pcna) (see Fig. S4 in the supplementary material). In agreement with the observations made in mouse, the PKD-like phenotype of Xenopus embryos lacking Bicc1 protein (Tran et al., 2007) was not due to defects in proliferation (data not shown).
Fig. 2.
Polycystic kidney disease (PKD) phenotype in Bicc1−/− mice. (A,B) Mallory's Tetrachrome staining on Bicc1+/+ (A) and Bicc1−/− (B) littermates at P0. (C) Schematic of a nephron indicating the localization of the segment-specific markers shown in D-H. ATL, ascending thin limb of Henle's loop; CD, collecting duct; CNT, connecting tubule; DCT, distal convoluted tubule; DTL, distal thin limb or Henle's loop; G, glomerulus; PT, proximal tubule; TAL, thick ascending limb of Henle's loop. (D-F) Periodic Acid Schiff (PAS) staining of Bicc1+/+ (D) and Bicc1−/− (E,F) kidneys showing glomerular (g) and proximal tubule (p) cysts at P0. (G,H) Immunohistochemistry on sections of kidneys from P0 mutant mice using an Nkcc2 antibody and Dolichos biflorus agglutinin (DBA). (I) Morphology of a Bicc1−/− cystic kidney compared with a Bicc1+/+ littermate at P21.
Bicc1 regulates Pkd2
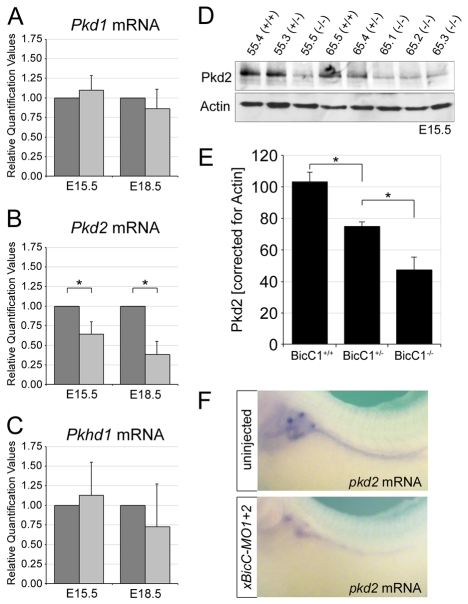
In humans, PKD is caused primarily by mutations in PKD1, PKD2 and PKHD1 (Torres and Harris, 2007; Wilson and Goilav, 2007). To determine whether the loss of Bicc1 affected any of these genes, we performed quantitative PCR (qPCR) analysis using multiple kidneys, comparing Bicc1+/+ with Bicc1−/− littermates at E15.5 and E18.5. As shown in Fig. 3A-C, Pkd2 mRNA was significantly downregulated in Bicc1−/− kidneys, whereas no significant changes were detected for Pkd1 or Pkhd1. This effect was stage dependent, as the downregulation of Pkd2 mRNA was more pronounced at E18.5. Western blot analyses comparing kidneys from Bicc1+/+, Bicc1+/− and Bicc1−/− littermates at E15.5, i.e. at the onset of cyst development, showed a dramatic decrease in Pkd2 protein levels (using two different antibodies; Fig. 3D,E and see Fig. S8A in the supplementary material). Interestingly, this reduction was dose dependent, as heterozygous kidneys also had lower amounts of Pkd2 compared with the wild type.
Fig. 3.
Bicc1 regulates Pkd2. (A-C) qPCR analysis for mouse Pkd1, Pkd2 and Pkhd1 mRNA levels in Bicc1+/+ (dark gray) and Bicc1−/− (light gray) littermates. The averages and s.d. from six kidney pairs at E15.5 and four pairs at E18.5 are shown (*, P<0.05, Student's t-test). (D) Western blot analysis comparing Pkd2 protein levels in E15.5 kidneys from two Bicc1 mouse litters (#55 and #65) of the indicated genotypes using the Pkd2-specific antibody from Santa Cruz. Actin served as a loading control. (E) Quantification of multiple Pkd2 western blot analyses comparing several mouse litters at E15.5 and normalized to actin. Average values and s.d. are indicated (*, P<0.05, Student's t-test). (F) Whole-mount in situ hybridization for Pkd2 mRNA on uninjected and xBicC-MO1+2-injected Xenopus embryos at stage 39.
To explore the significance of this observation, we turned to Xenopus, in which the loss of Bicc1 results in a PKD-like phenotype (Tran et al., 2007). In Xenopus, pkd2 mRNA and protein were expressed in all renal epithelial cells of the pronephros (Fig. 3F; see Fig. S5A-C″ in the supplementary material), a pattern identical to that of bicc1 (Tran et al., 2007). As shown in Fig. 3F, the expression of pkd2 mRNA was significantly reduced in the absence of Bicc1 protein, demonstrating that the regulation of Pkd2 by Bicc1 is evolutionarily conserved.
Epistasis analysis between Bicc1 and Pkd2 in the Xenopus pronephros
In addition to these molecular observations, the phenotype of Bicc1−/− embryos is similar to that of Pkd2 mutants (Wu et al., 2000; Pennekamp et al., 2002). Bicc1−/− mice showed kidney, liver and pancreatic cysts (Fig. 2; see Fig. S2 in the supplementary material), as well as defects in left-right patterning (data not shown). Therefore, we decided to test the interaction between Bicc1 and Pkd2 using the Xenopus pronephric kidney. Knockdown of Xenopus Pkd2 protein with an antisense MO (Pkd2-MO) resulted in a PKD-like phenotype highly reminiscent of that observed in embryos lacking Bicc1 (Tran et al., 2007) (see Figs S5-S7 in the supplementary material). These defects are specific as they were not seen in embryos that were microinjected with a standard control MO (Tran et al., 2007) (data not shown). In addition, the expression of nbc-1 in the late distal tubule was rescued by co-injection of pkd2-myc mRNA (see below, Fig. 4D).
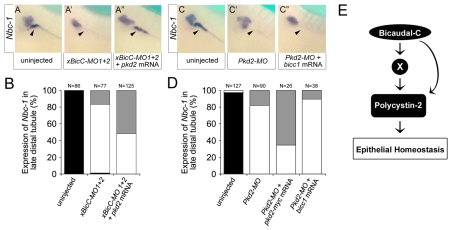
Fig. 4.
Bicc1 is epistatic to Pkd2. (A-A″) Analysis of the expression of nbc-1 in the Xenopus late distal tubule at stage 39 by whole-mount in situ hybridization of uninjected control embryos, and embryos radially injected with xBicC-MO1+2 in the presence or absence of a single injection of 2 ng pkd2 mRNA. (B) Quantification of the expression of nbc-1 in the late distal tubule from the experiments shown in A-A″. Black, bilateral expression; white, no expression; gray, unilateral expression rescued by co-injected mRNA. The number of embryos analyzed is indicated. (C-D) Reciprocal experiments to those in A-B using Xenopus embryos injected with either Pkd2-MO alone or together with pkd2-myc or bicc1 mRNA. Co-injection with pkd2-myc rescued nbc-1 expression, whereas co-injection with bicc1 did not. (E) Flow diagram outlining the proposed mechanism of Bicc1 activity.
Next, we examined the epistatic relationship between Bicc1 and Pkd2. We asked whether pkd2 mRNA could rescue the effects of xBicC-MO1+2 injections and vice versa, i.e. whether bicc1 mRNA could rescue the effects of Pkd2-MO injections. We followed the same strategy described by Tran et al. (Tran et al., 2007), assaying for the expression of nbc-1 in the late distal tubule by in situ hybridization. Although this paradigm represents only part of the PKD-like phenotype, it provides a powerful readout to quantitatively assess Bicc1 and Pkd2 function in the pronephros. We categorized embryos into those with bilateral expression of nbc-1 in the late distal tubule, those with reduced expression and those with unilateral expression rescued by the co-injected mRNA. Xenopus embryos were radially injected at the 2-cell stage with either xBicC-MO1+2 or Pkd2-MO. At the 4-cell stage, a subset of these embryos was then injected with either pkd2 or bicc1 mRNA into a single blastomere. Embryos were grown until stage 39 and processed for whole-mount in situ hybridization for nbc-1 mRNA. As previously observed (Tran et al., 2007), nbc-1 expression in the late distal tubule was lost upon injection of xBicC-MO1+2 (Fig. 4A,A′,B). When pkd2 mRNA was injected, the expression of nbc-1 was restored on the injected side in 52% of cases (Fig. 4A″,B). Interestingly, in the reciprocal experiment, bicc1 mRNA was unable to rescue the effects of the injected Pkd2-MO, whereas a pkd2 mRNA construct (pkd2-myc) that is not targeted by the Pkd2-MO did rescue this expression domain (Fig. 4C-D).
Together, these data suggested that Bicc1 acts upstream of Pkd2 and is necessary for Pkd2 activity. Based on these observations, we propose that either the Pkd2 mRNA itself, or a gene that regulates its expression, is a target for Bicc1 activity (Fig. 4E). In the absence of Bicc1, the spatiotemporal regulation of Pkd2 is disturbed, but not completely disrupted.
Subcellular localization of Bicc1
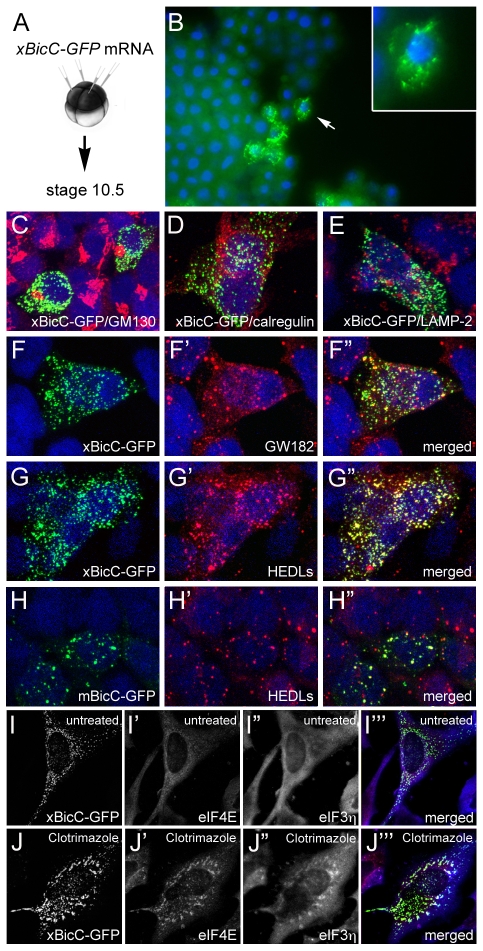
In order to understand the molecular mechanism of Bicc1, we next studied its subcellular localization. We used a Xenopus Bicc1-GFP fusion construct (xBicC-GFP) that has the same biological activity as its untagged counterpart, i.e. it induces ectopic endoderm formation in whole embryos and endodermin mRNA in ectodermal explants (see Fig. S8B in the supplementary material; data not shown) (Wessely and De Robertis, 2000). mRNA encoding xBicC-GFP was microinjected into the animal pole of Xenopus embryos and analyzed by fluorescence microscopy at gastrula stage (Fig. 5A,B). The protein was not nuclear, but was instead detected in cytoplasmic foci of unknown identity. To further investigate this localization, the xBicC-GFP fusion protein was transfected into HEK293T cells and analyzed by immunofluorescence 48 hours later. xBicC-GFP localization was compared with staining by a panel of antibodies that recognize subcellular cytoplasmic compartments: the Golgi apparatus (GM130; Golga2), endoplasmic reticulum (calregulin; calreticulin) and lysosomes (Lamp2). As shown in Fig. 5C-E, xBicC-GFP did not colocalize with any of these proteins, suggesting that it is not part of these structures.
Fig. 5.
Subcellular localization of Bicc1. (A,B) mRNA encoding a Xenopus Bicc1-GFP fusion protein (xBicC-GFP) was injected into the animal region of Xenopus embryos and analyzed at gastrula stage by immunofluorescence microscopy. Inset is a magnified view of a single cell (arrow). (C-H″) HEK293T cells were transfected with plasmids encoding xBicC-GFP (C-G″) or a mouse Bicc1-GFP fusion protein (mBicC-GFP; H-H″) and processed for immunofluorescence with antibodies against GM130 (C), calregulin (D), Lamp2 (E), GW182 (F-F″) and HEDLs (G-H″), using red fluorescent secondary antibodies. Nuclei were counterstained with DAPI (blue). (I-J‴) HeLa cells were transfected with pCS2-xBicC-GFP and were either left untreated (I-I‴) or treated with 20 μM clotrimazole (J-J‴). Stress granule formation was visualized with antibodies against Eif4E (red) and Eif3η (blue). Note that even in the untreated cells, xBicC-GFP is partially colocalized with Eif4E, which is a marker for P-bodies and stress granules.
Another structure that appears as distinct cytoplasmic foci are the so-called ‘processing bodies’ (P-bodies, or GW-bodies) (Parker and Sheth, 2007). Interestingly, immunofluorescence using antibodies against two components of these foci, GW182 (Tnrc6a) and HEDLs (Edc4) (Kedersha and Anderson, 2007), showed overlapping expression with the xBicC-GFP foci (Fig. 5F-G″). This colocalization was confirmed in multiple cell lines (IMCD3, LLC-PK1, MDCK and HeLa), as well as with two additional constructs: Flag-tagged Xenopus Bicc1 and a mouse Bicc1-GFP fusion protein (Fig. 5H-I; data not shown).
P-bodies are closely related to a second class of RNA granules known as stress granules. These are normally not present in cells, but are induced under adverse conditions to halt mRNA metabolism and are dynamically linked to P-bodies (Anderson and Kedersha, 2008). To explore whether Bicc1 is also found in stress granules, HeLa cells were transfected with xBicC-GFP. Stress granule formation was induced by the addition of clotrimazole and visualized by immunofluorescence using antibodies against Eif3η (Eif3B; which stains only stress granules) and Eif4E (which stains P-bodies and stress granules). Under these conditions, xBicC-GFP was found in large aggregates that were positive for these stress granule markers (Fig. 5I-J‴). Based on these results, we conclude that Bicc1 is a component of the cytoplasmic mRNA metabolism machinery.
Bicc1 is a post-transcriptional regulator
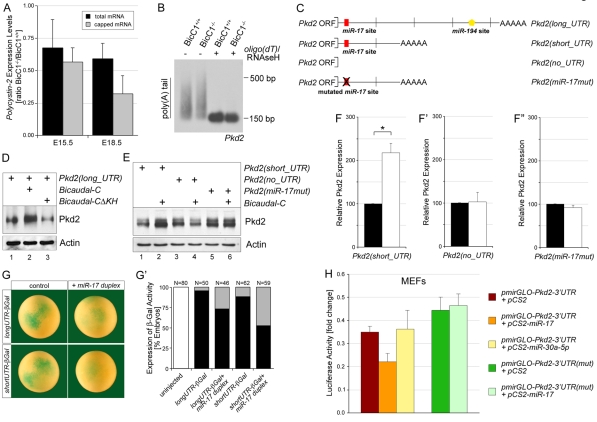
GW182 protein has been shown to be involved in mRNA degradation, stability, polyadenylation and miRNA activity (Parker and Sheth, 2007). To study whether Bicc1 regulates Pkd2 levels via such mechanisms, we first re-evaluated the qPCR and western blot data from the Bicc1 mouse kidneys (Fig. 3B,D,E; data not shown). The average decrease in Pkd2 mRNA levels, comparing six independent pairs of E15.5 kidneys from Bicc1+/+ and Bicc1−/− littermates, was 29% (Fig. 6A). Similar calculations for Pkd2 protein levels from seven independent pairs showed a 54% decrease (Fig. 3E). When analyzed in this manner, Pkd2 protein levels were more substantially decreased than mRNA levels. Post-transcriptional regulation is often accompanied by mRNA degradation and is preceded by changes in the level of translationally active capped mRNA. To distinguish between total and capped mRNA, a 5′-phosphate-dependent exonuclease was used to degrade non-capped RNAs. qPCR analysis of these samples demonstrated that in Bicc1−/− kidneys, the reduction in the percentage of capped Pkd2 mRNA was greater than that in total Pkd2 mRNA (Fig. 6A). As with total mRNA (Fig. 3B, Fig. 6A), the reduction in capped Pkd2 mRNA in Bicc1−/− kidneys was stage dependent. The differences at E15.5 were rather modest, whereas those at E18.5 were very pronounced. This effect was not accompanied by changes in the polyadenylation of Pkd2 mRNA. RNA ligation-coupled RT-PCR (RL-PCR) (Charlesworth et al., 2004) did not detect any obvious differences in the length of the poly(A) tail of Pkd2 mRNA in Bicc1+/+ and Bicc1−/− mouse kidneys at E15.5 (Fig. 6B).
Fig. 6.
Post-transcriptional regulation by Bicc1. (A) qPCR analysis of the reduction in Pkd2 mRNA in the kidneys of Bicc1+/+ and Bicc1−/− mouse littermates, comparing total and capped mRNA at E15.5 and E18.5. (B) RL-PCR to determine the length of the poly(A) tail of Pkd2 mRNA, comparing kidneys of Bicc1+/+ and Bicc1−/− littermates at E15.5. As a control for the specificity of the reaction, oligo(dT) and RNase H were added to remove the poly(A) tail before the RL-PCR, which collapsed the smear seen in the gel into a distinct band. (C) Schematic representation of Pkd2 constructs [Pkd2(long_UTR), Pkd2(short_UTR), Pkd2(no_UTR), Pkd2(miR-17mut)]. Predicted conserved miRNA binding sites are indicated. (D,E) Western blot of Pkd2 expression in HEK293T cells transfected with the four Pkd2 constructs together with an empty vector control, Xenopus bicc1 or a bicc1 construct lacking the RNA-binding KH domains (xBicCΔKH). Equal loading was confirmed by actin. (F-F″) Quantification of three independent western blots showing mean values and s.d. (*, P<0.05). (G,G′) Xenopus embryos were injected with an mRNA containing the lacZ gene fused to the short or long 3′UTR of Pkd2 in the presence or absence of a miR-17 duplex and stained for lacZ expression at stage 10 (G). Multiple experiments were quantified and the number of embryos analyzed is indicated (G′). White, no lacZ staining; black, strong lacZ staining; gray, reduced lacZ staining. (H) Luciferase reporter assay of mouse embryonic fibroblasts (MEFs) transfected with pmirGLO-Pkd2-3′UTR and pmirGLO-Pkd2-3′UTR-mut in the presence of pCS2, pCS2-miR-17 or pCS2-miR-30a-5p. Values were corrected for the expression of Renilla luciferase and calculated as fold change compared with the pCS2 control. Multiple independent experiments were averaged and the s.d. is indicated (P<0.05).
Since neither capping nor polyadenylation of Pkd2 mRNA was significantly changed at E15.5, i.e. at the onset of cyst formation, we next asked whether overexpression of Bicc1 could directly increase the translation of Pkd2 mRNA. Post-transcriptional regulation normally resides within the 3′UTR of a given mRNA. Interestingly, two different variants of the Pkd2 3′UTR could be found in databases: a long UTR corresponding to the published full-length Pkd2 sequence (GenBank NM_008861) and a shorter one that lacks 1.3 kb in the middle of the 3′UTR (GenBank BC062969), presumably owing to alternative splicing (Fig. 6C). Plasmids containing both Pkd2 variants were transfected into HEK293T cells in the presence or absence of Xenopus Bicc1 and processed for western blot analysis. As shown in Fig. 6D-F, Bicc1 significantly increased the amount of Pkd2 protein. This increase required the RNA-binding domain of Bicc1 because a construct lacking the KH domains (xBicCΔKH) did not show such an effect (Fig. 6D; see Fig. S9C in the supplementary material). It also required the 3′UTR of Pkd2 because a construct completely lacking the 3′UTR (Pkd2_noUTR) was resistant to the activity of Bicc1 (Fig. 6E,F′).
Bicc1 protein colocalized with GW182 (Fig. 5F-F″), and GW182 is a crucial component in the regulation of mRNA translation and stability via miRNAs (Liu et al., 2005; Rehwinkel et al., 2005). Thus, it seemed plausible that the regulation of Pkd2 by Bicc1 involved miRNAs. To explore this, the 3′UTR of Pkd2 mRNA was analyzed for potential miRNA binding sites using TargetScan (Version 5.0), PicTar and DIANA microT (version 3.0). As shown in Fig. 6C, two evolutionarily conserved miRNA binding sites, one for the miR-17 family and one for miR-194, were detected. However, the short 3′UTR of Pkd2 only contained a miR-17 binding site. Even though we do not have any evidence that a Pkd2 mRNA with the short 3′UTR exists in vivo, the fact that this construct was still regulated by Bicc1 (Fig. 6E,F) suggests that Bicc1 acts via the miR-17 family. In situ hybridization using a degenerate locked nucleic acid (LNA)-modified oligomer that recognizes three members of the miR-17 family (miR-17, miR-20b and miR-106a) detected expression in the mouse metanephric kidney at E14.5 and P0 and in the Xenopus pronephros (see Fig. S9A-C in the supplementary material). To assess whether this miR-17 binding site in the 3′UTR of Pkd2 was functional, two assays were performed. First, HEK293T cells were transfected with Pkd2(miR-17mut), a construct in which the seed sequence of the miR-17 binding site is mutated so that it does not bind miR-17 (Fig. 6C). This construct, in contrast to its non-mutated counterpart, no longer showed any regulation of Pkd2 protein by Bicc1 (Fig. 6E,F″). Moreover, the mutated construct had a consistently higher baseline expression of Pkd2 (Fig. 6E, compare lanes 1 and 5), arguing that this miR-17 binding site plays a role in the modulation of Pkd2 expression in HEK293T cells.
Secondly, to test whether the putative miR-17 binding site is responsive to miR-17, the long and short 3′UTRs of Pkd2 were fused to a nuclear β-galactosidase gene (nlacZ). Synthetic mRNA of these constructs was injected into the animal region of Xenopus embryos in the absence or presence of a synthetic miR-17 duplex. Embryos were processed for β-galactosidase staining at stage 10. The 3′UTR constructs in the absence of the miR-17 duplex showed strong lacZ staining (Fig. 6G,G′). Co-injection of the miR-17 duplex significantly reduced this staining. To quantify these miR-17 effects, mouse embryonic fibroblasts (MEFs) were transfected with a dual luciferase reporter construct that harbors the 3′UTR of Pkd2 (pmirGLO-Pkd2-3′UTR) in the presence or absence of miR-17, or, as control, miR-30a-5p (pCS2-miR-17, pCS2-miR-30a-5p). As shown in Fig. 6H, miR-17 reduced luciferase expression by 37%, whereas miR-30a-5p did not have any repressive effect. Moreover, this repression was specific, as no effect was observed when the miR-17 binding site was mutated (pmirGLO-Pkd2-3′UTR-mut).
Bicc1 and miR-17
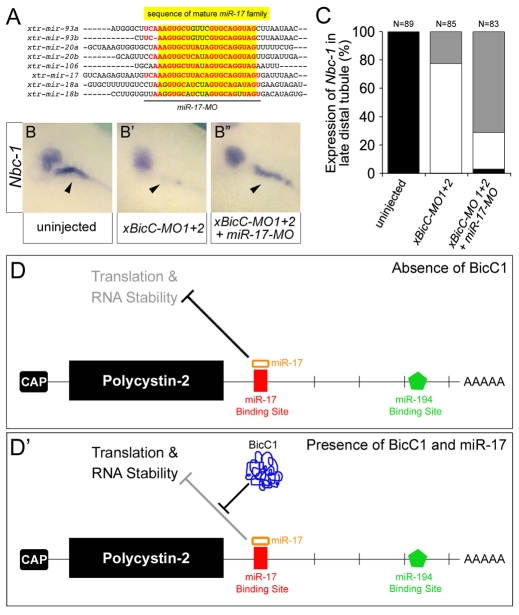
The experiments described above supported the hypothesis that Bicc1 acts as a post-transcriptional regulator of Pkd2 mRNA via its miR-17 binding site. However, the miR-17 duplex, as also seen with other miRNAs (Filipowicz et al., 2008), reduced the expression of the Pkd2 3′UTR reporters (Fig. 6G,G′), whereas Bicc1 increased it (Fig. 6D-F). Thus, a more logical interpretation of our data is that Bicc1 actually releases the repression by miR-17 (Fig. 7D,D′). This was not due to changes in miR-17 levels in Bicc1 mutants. qPCR analyses of representative miR-17 family members (corresponding to the three different primary transcripts) did not detect any differences in expression levels between Bicc1+/+ and Bicc1−/− kidneys (data not shown). This argued that Bicc1 and miR-17 both converge on the Pkd2 transcript. We tested this hypothesis using the PKD-like phenotype in Xenopus as a paradigm. miR-17 activity was knocked down using an antisense MO that targets several members of the miR-17 family (miR-17-MO, Fig. 7A). Xenopus embryos were injected with xBicC-MO1+2 alone, or together with miR-17-MO, and assayed for nbc-1 expression in the late distal tubule by in situ hybridization. bicc1 morphants lost nbc-1 (Fig. 7B,B′,C), whereas miR-17-MO had no effect on nbc-1 expression (data not shown). However, upon simultaneous reduction of miR-17 and Bicc1 activity, nbc-1 mRNA levels were recovered in 50% of the embryos (Fig. 7B″,C). This suggested that the miR-17 family acted either downstream of, or parallel to, Bicc1, further supporting the proposed model (Fig. 7D,D′).
Fig. 7.
Cross-talk between Bicc1 and the miR-17 miRNA family. (A) Alignment of the Xenopus miR-17 family members. Mature forms are highlighted in yellow. The sequence targeted by the miR-17 antisense MO (miR-17-MO) is indicated by the black line. The nucleotides shared between miR-17-MO and the individual members are indicated in red. (B-B″) Analysis of the expression of nbc-1 by whole-mount in situ hybridization of uninjected control embryos, embryos injected with xBicC-MO1+2 alone or with miR-17-MO. Arrowheads indicate the expression of nbc-1 in the Xenopus late distal tubule. Note that this expression domain is rescued upon co-injection of the two antisense MOs. (C) Quantification of the experiments shown in B-B″. Black, bilateral expression; white, reduced or no expression; gray, unilateral, rescued expression in the late distal tubule. (D,D′) Models for the post-transcriptional regulation of Pkd2 mRNA by the miR-17 family in the absence or presence of Bicc1.
Together, the fact that the miR-17 family is expressed in the kidney and that Pkd2, a gene with a miR-17 binding site, is affected by Bicc1, support the hypothesis that Bicc1 is part of a post-transcriptional regulatory complex that is involved in epithelial homeostasis and which, when disrupted, leads to PKD.
DISCUSSION
Bicaudal C has been shown to be an important developmental regulator in Drosophila, Xenopus, C. elegans and mouse (Mahone et al., 1995; Saffman et al., 1998; Wessely and De Robertis, 2000; Wang et al., 2002; Cogswell et al., 2003; Suh et al., 2006; Tran et al., 2007; Maisonneuve et al., 2009). Here, we extend previous studies that showed that Bicc1 is required for the proper development of the mouse metanephric kidney (Cogswell et al., 2003). Loss of Bicc1 resulted in renal cyst formation. These cysts were initially located in the glomerulus and the proximal tubules, but were later found along the entire length of the nephron. This phenotype is reminiscent of human forms of PKD and adds to the existing animal models for PKD (Guay-Woodford, 2003; Torres and Harris, 2007). However, the molecular nature of the cystic phenotype in Bicc1 mutant mice remained obscure. Based on the following observations, we now propose that Bicc1 functions as a post-transcriptional regulator of Pkd2. (1) The Bicc1 and Pkd2 mouse knockout phenotypes both result in the formation of cysts in the kidney, pancreas and liver, as well as in defects in left-right asymmetry (this study, data not shown) (Wu et al., 2000; Pennekamp et al., 2002). (2) Mice lacking a functional Bicc1 protein show reduced expression levels of Pkd2 mRNA and protein. (3) Xenopus embryos lacking Pkd2 develop a PKD-like phenotype that is highly reminiscent of that of embryos lacking Bicc1 (Tran et al., 2007). (4) pkd2 mRNA rescues the effects of bicc1 knockdown in Xenopus, but bicc1 mRNA does not rescue pkd2 morphants. (5) Bicc1 enhances the translation of Pkd2 mRNA in HEK293T cells. (6) Bicc1 protein colocalizes with proteins that are involved in the post-transcriptional regulation of mRNAs. (7) Pkd2 mRNA contains a binding site for the miR-17 miRNA family in its 3′UTR, and knockdown of miR-17 can rescue the phenotype of bicc1 morphants in Xenopus.
One of the most revealing aspects of this study was the realization that Bicc1 is not localized to the nucleus, but acts as a post-transcriptional regulator in the cytoplasm (Fig. 5) (Maisonneuve et al., 2009; Stagner et al., 2009). The protein was localized to cytoplasmic foci that also contain proteins known to be involved in cytoplasmic mRNA turnover and was recruited to stress granules upon treatment with clotrimazole. It is noteworthy that the number of Bicc1-positive foci was much higher than the number of P-bodies detectable in HEK293T cells. Thus, whether Bicc1 is a bona fide P-body protein or whether it is a part of a more specialized subset of the cytoplasmic mRNA maintenance machinery remains to be determined. In particular, it will be important to verify the subcellular localization of the endogenous protein. However, one tempting speculation is that Bicc1 is part of the submicroscopic P-body subcomplex that has recently been described (Franks and Lykke-Andersen, 2007). The implications from the subcellular localization of Bicc1 are in agreement with data concerning its function in invertebrates. In C. elegans, GLD-3, a homolog of Bicaudal C, has been shown to act as a translational regulator by functioning as a specificity subunit for the cytoplasmic poly(A) polymerase GLD-2 (Wang et al., 2002). During germ line development, the GLD-2—GLD-3 complex binds to the 3′UTR of gld-1 mRNA and thereby regulates the rate of translation of GLD-1 protein (Suh et al., 2006). In Drosophila, Bicaudal C mutant flies develop a ‘double-abdomen’ phenotype due to ectopic expression of the posterior determinant Oskar throughout the egg (Mahone et al., 1995; Saffman et al., 1998). Molecular analysis demonstrated that Bicaudal C is required for the correct timing and localized expression of Oskar (Saffman et al., 1998), a process that is probably regulated by the interaction of Bicaudal C with the CCR4-NOT deadenylase (Chicoine et al., 2007).
At the molecular level, we propose that Bicc1 antagonizes the repressive activity of a miR-17-containing silencing complex that is present on the 3′UTR of Pkd2 (Fig. 7D,D′). The nature of this antagonism remains unknown. Bicc1 could directly replace individual proteins of the RNP silencing complex (e.g. argonaute 2; Eif2c2). Alternatively, its binding could convert the RNP complex from an inhibitory to an activating complex, as has recently been shown (Vasudevan et al., 2007). One corollary of this model is that Bicc1 does not function in a one-to-one relationship with one gene (e.g. Pkd2), but instead regulates multiple targets (see below). It probably recognizes its target mRNAs via an RNA interface generated by the interaction between members of the miR-17 miRNA family and the 3′UTR. A similar scenario has recently been described for post-transcriptional regulation via the AU-rich element (ARE), in which a complex between the ARE, fragile X mental retardation 1 (FXR1) and miR369-3 regulates a subset of target genes (Vasudevan and Steitz, 2007; Vasudevan et al., 2007). The proposed mechanism might also explain why, despite being initially described in Drosophila in 1986 (Mohler and Wieschaus, 1986), the function of Bicaudal C has been so difficult to define and why many attempts to identify the targets of its activity have been unsuccessful. For example, the small decrease in Pkd2 mRNA levels in Bicc1−/− mutant mice would be disregarded in a microarray-based approach looking for twofold differences in gene expression.
This model also provided an explanation for one remaining conundrum of our present study: namely, if Pkd2 were the only target of Bicc1, the reduction of Pkd2 protein levels by ~50% should have been sufficient to cause early onset cyst formation. However, heterozygous mice lacking one copy of Pkd2 do not show this early phenotype, but instead develop a limited number of cysts later in life (Wu et al., 2000). Similarly, if Pkd2 were the only target of Bicc1 activity, then one would expect that the complete loss of Pkd2 in Pkd2−/− mice would have a phenotypically stronger effect than the 50% reduction of Pkd2 in the Bicc1−/− mice. Therefore, Bicc1 has to regulate additional genes to cause such a strong cystic phenotype. This notion is not surprising as Bicc1 is an RNA-binding molecule and RNA-binding molecules normally regulate multiple targets. Based on our model, the presence of miR-17 binding sites serves as a predictive tool to identify these additional targets of Bicc1. Indeed, using this bioinformatics approach we detected miR-17 binding sites in several genes involved in PKD (see Fig. S9D in the supplementary material): Pkd1 and Glis3 have a highly conserved binding site, whereas Pkhd1, Nek8, Nphp1, Ift88 (Polaris; Tg737) and Nphp3 have weakly conserved sites, while a third group of genes (Cep290, Hnf1b and Glis2) had sites for miR-17-3p, a miRNA transcribed from the opposite strand of the miR-17 precursor RNA.
Based on the presence of miR-17 binding sites in the 3′UTR of multiple PKD genes, we would also expect to find facets of the phenotype that have still evaded detection. For example, a decrease in Ift88 expression might result in subtle defects in ciliogenesis. However, we have not been able to detect such ciliary defects either in Xenopus (Tran et al., 2007) or mouse (data not shown) by immunofluorescence, but this will need a thorough analysis using electron microscopy. In addition, the miR-17 miRNA family has been implicated in many biologically important processes, including the cell cycle and cancer, and in stem cells (Cloonan et al., 2008; Foshay and Gallicano, 2008; Mendell, 2008). Mice lacking miR-17 family members develop a range of phenotypes, but an analysis of the kidneys is still lacking (Ventura et al., 2008). Interestingly, transgenic mice overexpressing miR-17 show overall growth retardation, yet from all the organs analyzed, the kidneys look the most normal (Shan et al., 2009). The fact that these mice do not develop renal cysts is not surprising, as the present study proposes that one function of Bicc1 is to protect the kidney from superfluous miR-17 activity. Instead, a cystic phenotype may only be observed once the Bicc1 gene dose is reduced.
In conclusion, the present study reveals a novel and provocative aspect of kidney development and disease. The post-transcriptional regulation of genes involved in PKD adds a new level of complexity. Bicc1 is, to our knowledge, the first gene to be genetically linked to this regulatory process. In addition, this study might very well spearhead similar studies in other organ systems, as it is highly unlikely that the kidney is the only organ in which post-transcriptional regulation plays such an important role.
Supplementary Material
Acknowledgements
We thank Drs S. El-Dahr, J. Larraín, T. Obara, M. Oelgeschläger and J. Venuti and all members of the laboratory for critically reviewing the manuscript and for helpful discussions; Susanne Bogusch (University of Hohenheim) for expert technical assistance; Dr P. Vize and the NIBB/NIG/NBRP Xenopus laevis EST project for plasmids; and Drs M. Knepper, S. Somlo and G. Wu for the Nkcc2, Pkd2, GM130 and calregulin antibodies. The anti-Lamp2 monoclonal antibody was obtained from the Developmental Studies Hybridoma Bank (DSHB) developed under the auspices of the NICHD and maintained by The University of Iowa. This work was supported by grants from the DFG to M.B. and from the Polycystic Kidney Disease Foundation (#103a2r) and NIH/NIDDK (5R21DK070671-03, 5R21DK077763-03, 1R01DK080745-01A2) to O.W. E.M.D.R. is an Investigator of the Howard Hughes Medical Institute. Deposited in PMC for release after 6 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.046045/-/DC1
References
- Agrawal R., Tran U., Wessely O. (2009). The miR-30 miRNA family regulates Xenopus pronephros development and targets the transcription factor Xlim1/Lhx1. Development 136, 3927-3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. (2008). Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33, 141-150 [DOI] [PubMed] [Google Scholar]
- Blum M., Andre P., Muders K., Schweickert A., Fischer A., Bitzer E., Bogusch S., Beyer T., van Straaten H. W., Viebahn C. (2007). Ciliation and gene expression distinguish between node and posterior notochord in the mammalian embryo. Differentiation 75, 133-146 [DOI] [PubMed] [Google Scholar]
- Burn T. C., Connors T. D., Dackowski W. R., Petry L. R., Van Raay T. J., Millholland J. M., Venet M., Miller G., Hakim R. M., Landes G. M., et al. (1995). Analysis of the genomic sequence for the autosomal dominant polycystic kidney disease (PKD1) gene predicts the presence of a leucine-rich repeat. The American PKD1 Consortium (APKD1 Consortium). Hum. Mol. Genet. 4, 575-582 [DOI] [PubMed] [Google Scholar]
- Charlesworth A., Cox L. L., MacNicol A. M. (2004). Cytoplasmic polyadenylation element (CPE)- and CPE-binding protein (CPEB)-independent mechanisms regulate early class maternal mRNA translational activation in Xenopus oocytes. J. Biol. Chem. 279, 17650-17659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicoine J., Benoit P., Gamberi C., Paliouras M., Simonelig M., Lasko P. (2007). Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Dev. Cell 13, 691-704 [DOI] [PubMed] [Google Scholar]
- Cloonan N., Brown M. K., Steptoe A. L., Wani S., Chan W. L., Forrest A. R., Kolle G., Gabrielli B., Grimmond S. M. (2008). The miR-17-5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 9, R127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogswell C., Price S. J., Hou X., Guay-Woodford L. M., Flaherty L., Bryda E. C. (2003). Positional cloning of jcpk/bpk locus of the mouse. Mamm. Genome 14, 242-249 [DOI] [PubMed] [Google Scholar]
- Dressler G. R. (2006). The cellular basis of kidney development. Annu. Rev. Cell Dev. Biol. 22, 509-529 [DOI] [PubMed] [Google Scholar]
- Drummond I. A. (2005). Kidney development and disease in the zebrafish. J. Am. Soc. Nephrol. 16, 299-304 [DOI] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9, 102-114 [DOI] [PubMed] [Google Scholar]
- Flaherty L., Bryda E. C., Collins D., Rudofsky U., Montogomery J. C. (1995). New mouse model for polycystic kidney disease with both recessive and dominant gene effects. Kidney Int. 47, 552-558 [DOI] [PubMed] [Google Scholar]
- Foshay K. M., Gallicano G. I. (2008). miR-17 family miRNAs are expressed during early mammalian development and regulate stem cell differentiation. Dev. Biol. 326, 431-443 [DOI] [PubMed] [Google Scholar]
- Franks T. M., Lykke-Andersen J. (2007). TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 21, 719-735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guay-Woodford L. M. (2003). Murine models of polycystic kidney disease: molecular and therapeutic insights. Am. J. Physiol. 285, F1034-F1049 [DOI] [PubMed] [Google Scholar]
- Hildebrandt F., Otto E., Rensing C., Nothwang H. G., Vollmer M., Adolphs J., Hanusch H., Brandis M. (1997). A novel gene encoding an SH3 domain protein is mutated in nephronophthisis type 1. Nat. Genet. 17, 149-153 [DOI] [PubMed] [Google Scholar]
- Hogan B. L., Kolodziej P. A. (2002). Organogenesis: molecular mechanisms of tubulogenesis. Nat. Rev. Genet. 3, 513-523 [DOI] [PubMed] [Google Scholar]
- Hughes J., Ward C. J., Peral B., Aspinwall R., Clark K., San Millan J. L., Gamble V., Harris P. C. (1995). The polycystic kidney disease 1 (PKD1) gene encodes a novel protein with multiple cell recognition domains. Nat. Genet. 10, 151-160 [DOI] [PubMed] [Google Scholar]
- Kedersha N., Anderson P. (2007). Mammalian stress granules and processing bodies. Methods Enzymol. 431, 61-81 [DOI] [PubMed] [Google Scholar]
- Liu J., Rivas F. V., Wohlschlegel J., Yates J. R., 3rd, Parker R., Hannon G. J. (2005). A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 7, 1261-1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Rogers K., Zbar B., Schmidt L. (2002). Effects of different fixatives on beta-galactosidase activity. J. Histochem. Cytochem. 50, 1421-1424 [DOI] [PubMed] [Google Scholar]
- Mahone M., Saffman E. E., Lasko P. F. (1995). Localized Bicaudal-C RNA encodes a protein containing a KH domain, the RNA binding motif of FMR1. EMBO J. 14, 2043-2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve C., Guilleret I., Vick P., Weber T., Andre P., Beyer T., Blum M., Constam D. B. (2009). Bicaudal C, a novel regulator of Dvl signaling abutting RNA-processing bodies, controls cilia orientation and leftward flow. Development 136, 3019-3030 [DOI] [PubMed] [Google Scholar]
- Markowitz G. S., Cai Y., Li L., Wu G., Ward L. C., Somlo S., D'Agati V. D. (1999). Polycystin-2 expression is developmentally regulated. Am. J. Physiol. 277, F17-F25 [DOI] [PubMed] [Google Scholar]
- Mendell J. T. (2008). miRiad roles for the miR-17-92 cluster in development and disease. Cell 133, 217-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobjerg N., Larsen E. H., Jespersen A. (2000). Morphology of the kidney in larvae of Bufo viridis (Amphibia, Anura, Bufonidae). J. Morphol. 245, 177-195 [DOI] [PubMed] [Google Scholar]
- Mochizuki T., Wu G., Hayashi T., Xenophontos S. L., Veldhuisen B., Saris J. J., Reynolds D. M., Cai Y., Gabow P. A., Pierides A., et al. (1996). PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science 272, 1339-1342 [DOI] [PubMed] [Google Scholar]
- Mohler J., Wieschaus E. F. (1986). Dominant maternal-effect mutations of Drosophila melanogaster causing the production of double-abdomen embryos. Genetics 112, 803-822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauta J., Ozawa Y., Sweeney W. E., Jr, Rutledge J. C., Avner E. D. (1993). Renal and biliary abnormalities in a new murine model of autosomal recessive polycystic kidney disease. Pediatr. Nephrol. 7, 163-172 [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P. D., Faber J. (1994). Normal Table of Xenopus laevis New York: Garland Publishing; [Google Scholar]
- Onuchic L. F., Furu L., Nagasawa Y., Hou X., Eggermann T., Ren Z., Bergmann C., Senderek J., Esquivel E., Zeltner R., et al. (2002). PKHD1, the polycystic kidney and hepatic disease 1 gene, encodes a novel large protein containing multiple immunoglobulin-like plexin-transcription-factor domains and parallel beta-helix 1 repeats. Am. J. Hum. Genet. 70, 1305-1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Sheth U. (2007). P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635-646 [DOI] [PubMed] [Google Scholar]
- Pennekamp P., Karcher C., Fischer A., Schweickert A., Skryabin B., Horst J., Blum M., Dworniczak B. (2002). The ion channel polycystin-2 is required for left-right axis determination in mice. Curr. Biol. 12, 938-943 [DOI] [PubMed] [Google Scholar]
- Raciti D., Reggiani L., Geffers L., Jiang Q., Bacchion F., Subrizi A. E., Clements D., Tindal C., Davidson D. R., Kaissling B., et al. (2008). Organization of the pronephric kidney revealed by large-scale gene expression mapping. Genome Biol. 9, R84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J., Behm-Ansmant I., Gatfield D., Izaurralde E. (2005). A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11, 1640-1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffman E. E., Styhler S., Rother K., Li W., Richard S., Lasko P. (1998). Premature translation of oskar in oocytes lacking the RNA-binding protein Bicaudal-C. Mol. Cell. Biol. 18, 4855-4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxén L. (1987). Organogenesis of the Kidney Cambridge, UK: Cambridge University Press; [Google Scholar]
- Shan S. W., Lee D. Y., Deng Z., Shatseva T., Jeyapalan Z., Du W. W., Zhang Y., Xuan J. W., Yee S. P., Siragam V., et al. (2009). MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat. Cell Biol. 11, 1031-1038 [DOI] [PubMed] [Google Scholar]
- Shillingford J. M., Murcia N. S., Larson C. H., Low S. H., Hedgepeth R., Brown N., Flask C. A., Novick A. C., Goldfarb D. A., Kramer-Zucker A., et al. (2006). The mTOR pathway is regulated by polycystin-1, and its inhibition reverses renal cystogenesis in polycystic kidney disease. Proc. Natl. Acad. Sci. USA 103, 5466-5471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive H. L., Grainger R. M., Harland R. M. (2000). Early Development of Xenopus laevis: a Laboratory Manual Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Stagner E. E., Bouvrette D. J., Cheng J., Bryda E. C. (2009). The polycystic kidney disease-related proteins Bicc1 and SamCystin interact. Biochem. Biophys. Res. Commun. 383, 16-21 [DOI] [PubMed] [Google Scholar]
- Stoecklin G., Mayo T., Anderson P. (2006). ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 7, 72-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N., Jedamzik B., Eckmann C. R., Wickens M., Kimble J. (2006). The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proc. Natl. Acad. Sci. USA 103, 15108-11512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney W. E., Chen Y., Nakanishi K., Frost P., Avner E. D. (2000). Treatment of polycystic kidney disease with a novel tyrosine kinase inhibitor. Kidney Int. 57, 33-40 [DOI] [PubMed] [Google Scholar]
- The International Polycystic Kidney Disease Consortium (1995). Polycystic kidney disease: the complete structure of the PKD1 gene and its protein. Cell 81, 289-298 [DOI] [PubMed] [Google Scholar]
- Torres V. E., Harris P. C. (2007). Polycystic kidney disease: genes, proteins, animal models, disease mechanisms and therapeutic opportunities. J. Intern. Med. 261, 17-31 [DOI] [PubMed] [Google Scholar]
- Tran U., Pickney L. M., Ozpolat B. D., Wessely O. (2007). Xenopus Bicaudal-C is required for the differentiation of the amphibian pronephros. Dev. Biol. 307, 152-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S., Steitz J. A. (2007). AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 128, 1105-1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S., Tong Y., Steitz J. A. (2007). Switching from repression to activation: microRNAs can up-regulate translation. Science 318, 1931-1934 [DOI] [PubMed] [Google Scholar]
- Ventura A., Young A. G., Winslow M. M., Lintault L., Meissner A., Erkeland S. J., Newman J., Bronson R. T., Crowley D., Stone J. R., et al. (2008). Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132, 875-886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vize P., Woolf A., Bard J. (2002). The Kidney: From Normal Development to Congenital Diseases San Diego: Academic Press; [Google Scholar]
- Wang L., Eckmann C. R., Kadyk L. C., Wickens M., Kimble J. (2002). A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419, 312-316 [DOI] [PubMed] [Google Scholar]
- Ward C. J., Hogan M. C., Rossetti S., Walker D., Sneddon T., Wang X., Kubly V., Cunningham J. M., Bacallao R., Ishibashi M., et al. (2002). The gene mutated in autosomal recessive polycystic kidney disease encodes a large, receptor-like protein. Nat. Genet. 30, 259-269 [DOI] [PubMed] [Google Scholar]
- Wessely O., De Robertis E. M. (2000). The Xenopus homologue of Bicaudal-C is a localized maternal mRNA that can induce endoderm formation. Development 127, 2053-2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessely O., Tran U., Zakin L., De Robertis E. M. (2001). Identification and expression of the mammalian homologue of Bicaudal-C. Mech. Dev. 101, 267-270 [DOI] [PubMed] [Google Scholar]
- Wilson P. D., Goilav B. (2007). Cystic Disease of the Kidney. Ann. Rev. Path. 2, 341-368 [DOI] [PubMed] [Google Scholar]
- Wu G., Markowitz G. S., Li L., D'Agati V. D., Factor S. M., Geng L., Tibara S., Tuchman J., Cai Y., Park J. H., et al. (2000). Cardiac defects and renal failure in mice with targeted mutations in Pkd2. Nat. Genet. 24, 75-78 [DOI] [PubMed] [Google Scholar]
- Xiong H., Chen Y., Yi Y., Tsuchiya K., Moeckel G., Cheung J., Liang D., Tham K., Xu X., Chen X. Z., et al. (2002). A novel gene encoding a TIG multiple domain protein is a positional candidate for autosomal recessive polycystic kidney disease. Genomics 80, 96-104 [DOI] [PubMed] [Google Scholar]
- Zhou X., Vize P. D. (2004). Proximo-distal specialization of epithelial transport processes within the Xenopus pronephric kidney tubules. Dev. Biol. 271, 322-338 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.