Abstract
Background
Microarray technology offers a new opportunity to gain insight into global gene and protein expression profiles in asthma. To identify novel factors produced in the asthmatic airway, we analyzed sputum samples by using a membrane-based human cytokine microarray technology in patients with bronchial asthma (BA).
Methods
Induced sputum was obtained from 28 BA subjects, 20 nonasthmatic atopic control (AC) subjects, and 38 nonasthmatic nonatopic normal control (NC) subjects. The microarray samples of subjects were randomly selected from nine BA subjects, three AC subjects, and six NC subjects. Sputum supernatants were analyzed using a custom human cytokine array (RayBio Custom Human Cytokine Array; RayBiotech; Norcross, GA) designed to analyze 79 specific cytokines simultaneously. The levels of growth-regulated oncogene (GRO)-α, eotaxin-2, and pulmonary and activation-regulated chemokine (PARC)/CCL18 were measured by sandwich enzyme-linked immunosorbent assays (ELISAs), and eosinophil-derived neurotoxin (EDN) was measured by radioimmunoassay.
Results
By microarray, the signal intensities for GRO-α, eotaxin-2, and PARC were significantly higher in BA subjects than in AC and NC subjects (p = 0.036, p = 0.042, and p = 0.033, respectively). By ELISA, the sputum PARC protein levels were significantly higher in BA subjects than in AC and NC subjects (p < 0.0001). Furthermore, PARC levels correlated significantly with sputum eosinophil percentages (r = 0.570, p < 0.0001) and the levels of EDN(r = 0.633, p < 0.0001), the regulated upon activation, normal T cell expressed and secreted cytokine (r = 0.440, p < 0.001), interleukin-4 (r = 0.415, p < 0.01), and interferon-γ (r = 0.491, p < 0.001).
Conclusions
By a nonbiased screening approach, a chemokine, PARC, is elevated in sputum specimens from patients with asthma. PARC may play important roles in development of airway eosinophilic inflammation in asthma.
Keywords: asthma, cytokine microarray analysis, eosinophilic inflammation, pulmonary and activation-regulated chemokine/CCL18
Sputum parameters of airway inflammation are considered to be valid, reliable, and responsive to change.1–3 Numerous inflammatory mediators are produced and detected in the fluid phase of sputum, including cytokines, chemokines, growth factors, angiogenic factors, proteases, and others. The three main types of methods used for the measurement of sputum fluid-phase mediators are bioassays, enzyme assays, and immunoassays. However, these assay methods require identification or prediction of target molecules by investigators, which may allow bias for or against certain molecules. Thus, molecules beyond the radar screen of investigators are not measured, even if they might play important roles in disease pathophysiology. Furthermore, these methods generally do not allow simultaneous detection of multiple cytokines or chemokines.
Microarray technology offers a new opportunity to gain insight into global gene or protein expression profiles in asthma, leading to the identification of asthma-associated molecules. A protein-detecting microarray consists of numerous affinity reagents arrayed on a solid surface. A complex mixture of proteins then binds specific target proteins to unique locations on the array, and bound proteins are subsequently detected and quantified.4 These microarrays may be used to detect multiple proteins, such as cytokines and chemokines, from one sample in an accurate, a reproducible, and a rapid manner.
To identify novel factors produced in the airways of patients with asthma, we analyzed sputum samples from those individuals by using membrane-based human cytokine microarray technology. Among 79 molecules tested, we found that pulmonary and activation-regulated chemokine (PARC [systemic name CCL18]) as well as growth-regulated oncogene (GRO)-α and eotaxin-2 are specifically elevated in sputum from patients with asthma, in comparison to levels in atopic control (AC) subjects and normal control (NC) subjects. Furthermore, the PARC level correlated significantly with the magnitude of airway eosinophilic inflammation. Thus, PARC may play important roles in the pathophysiology of asthma.
MATERIALS AND METHODS
Study Subjects
Induced sputum and peripheral blood specimens were obtained from 28 subjects with bronchial asthma (BA), 20 AC subjects without asthma, and 38 nonatopic NC subjects without asthma. The samples for microarray were selected randomly from nine BA subjects, three AC subjects, and six NC subjects. All of the samples were used for cytokine enzyme-linked immunosorbent assays (ELISAs), and eosinophil-derived neurotoxin (EDN) radioimmunoassay. All subjects were nonsmokers and had not taken oral glucocorticoids for at least 8 weeks prior to the study. Subjects were recruited from a pool of patients or healthy individuals seen at the Mayo Clinic, Rochester, MN. The Mayo Clinic Institutional Review Board reviewed and approved this study, and patients provided informed consent to participate.
Patients with asthma had a history of recurrent wheezing. They had a > 15% increase in FEV1 after inhaling 180 µg of albuterol and/or a positive methacholine challenge result as defined by a 20% reduction from baseline in FEV1 following inhalation of methacholine. At the time of the study, patients with asthma were stable, with no acute asthma symptoms for at least 8 weeks and no evidence of chest infection. Thirteen of 28 BA subjects were receiving regular inhaled corticosteroid therapy.
Atopy was defined by the presence of at least one positive skin-prick test result (> 3 mm wheal diameter) in response to a battery of 15 common aeroallergens. All of the 28 BA subjects were atopic. Fifteen of 20 AC subjects had a history of allergic rhinitis without bronchial hyperresponsiveness. Nonatopic subjects had negative to the skin-prick test to the battery of 15 common aeroallergens, and negative responses to the radioallergosorbent test (Pharmacia Diagnostics; Uppsala, Sweden) to the same common aeroallergens.
Sputum Induction
All subjects underwent sputum induction during the winter. The subjects were premedicated with two puffs of albuterol, total dose 180 µg, via spacer. Sputum induction was performed with aerosolized 3% hypertonic saline solution generated by an ultrasonic nebulizer (DeVilbiss Ultra-Neb 99; DeVilbiss Co; Somerset, PA), as described by Fahy et al,5 with modifications. Prior to sputum production, subjects were asked to rinse their mouth with water, swallow, and blow the nose to minimize contamination of sputum with saliva and postnasal drip fluid. Every 2 min for up to 12 min, they were instructed to cough in order to produce sputum into a sterile container. For all subjects, FEV1 was monitored 15 min post-albuterol therapy and at 4-min intervals thereafter throughout the test for safety. If the FEV1 decreased by ≥20% from the post-albuterol therapy baseline, the sputum induction was discontinued.
Sputum Processing
The collected sputum was processed, as described by Pizzichini et al,1 with modifications. Sputum was processed immediately within 2 h of sample collection. Equal volumes of sputum were placed into two 15-mL polypropylene tubes. One aliquot was treated with an equal volume of saline solution containing 0.1% dithiothreitol (DTT) [1:10 diluted DTT] (Sputolysin 1%; Calbiochem Corp; San Diego, CA) [“with DTT” aliquot]. The other aliquot was mixed with two times the volume of saline solution without DTT (“without DTT” aliquot). Both mixtures were aspirated repeatedly with a Pasteur pipette, then vortexed for 15 s, and placed on a bench rocker for 20 min. With DTT aliquots were filtered through 40-µm strainers (Becton Dickinson; Franklin Lakes, NJ), and then both aliquots were centrifuged at 833 g for 10 min at 4°C. The supernatants from each sample were collected and frozen at −70°C for later assay. Cell pellets from the original aliquot with DTT were resuspended in saline solution and stained with trypan blue, and a total nonsquamous cell count was performed with a hemocytometer. Nonsquamous cell viability was then calculated. Cytospin preparations were adjusted to equal approximately 1.0 × 106/mL and then stained (Diff-Quik stain; Dade Behring Inc; Newark, DE). Four hundred differential nonsquamous cells were counted. The specimen was considered adequate if morphologic identification of total and differential cell counts could be obtained.
Membrane-Based Human Cytokine Microarray Assay
A custom human cytokine array kit (RayBio; RayBiotech; Norcross, GA) was purchased. The array spotted the membrane with 79 specific antibodies toward cytokines and chemokines per our request. For microarray assay, we followed the directions precisely as they were stated by the manufacturer. Briefly, membranes were placed in an eight-well tissue culture tray and incubated with 2 mL of a 1 × blocking buffer at room temperature for 30 min to block membranes. After decanting the blocking buffer from each container, 1 mL of sputum supernatant from the without DTT samples were added and incubated overnight at 4°C. After decanting the samples, all membranes were washed three times with 2 mL of a 1 × wash buffer I at room temperature with shaking, 5 min per wash, followed by washing two times with 1 × wash buffer II at room temperature with shaking, 5 min per wash. One milliliter of 1:250 diluted biotin-conjugated antibodies was prepared and incubated for 2 h at room temperature, and the washing steps were repeated as before. Two mL of 1:1,000 diluted horse radish peroxidase-conjugated streptavidin was added, and membranes were incubated for 1 h at room temperature, followed by additional washing. Spots were visualized using detection buffer C and D (Pierce Biotechnology; Rockford, IL). A total of 500 µL of a 1 × detection buffer C and 500 µL of a 1 × detection buffer D were mixed, loaded onto the membranes to cover the entire surface, and incubated for 5 min. Membranes were wrapped in plastic wrap and exposed to radiographic film (Kodak X-Omat; Kodak; Rochester, NY) for 20 min, and the signal was detected using film developer. Each film was scanned (CanoScan D1230U; Canon Inc; Tokyo, Japan) into an image-processing and analysis program (NIH Image for Macintosh, version 1.63; National Institutes of Health; Bethesda, MD), and spots were digitized into pixel densities. The densities were exported into spreadsheet software (Excel; Microsoft Corp; Redmond, WA), and the background intensity was subtracted before analysis. The data were normalized to the values for positive control subjects provided by the manufacturer as 100%.
Cytokine and Chemokine Quantitation
The levels of interleukin (IL)-4, IL-5, interferon (IFN)-γ (Endogen; Woburn, MA), the regulated upon activation, normal T cell expressed and secreted cytokine (RANTES), and PARC (R&D Systems; Minneapolis, MN) were measured in the sputum supernatants from without DTT samples using sandwich ELISAs according to the respective manufacturers’ protocols. The levels of PARC were also measured in the serum specimens. The sensitivities for IL-4 and IL-5 were < 2.0 pg/mL, and those for IFN-γ, RANTES, and PARC were < 1, 2.5, and 15.6 pg/mL, respectively. The levels of EDN in sputum supernatants were determined using a specific and sensitive double-antibody competitive radioimmunoassay kit (Amersham Pharmacia Biotech; Uppsala, Sweden).6 The sensitivity for EDN was < 1 ng/mL. The result was adjusted for the dilution (without DTT aliquot was diluted threefold). When the cytokine and chemokine levels were below the sensitivity, a level of 0 was used for the comparisons of the levels, and one half the level of the sensitivity was used for the correlations.
Statistical Analysis
The microarray results were shown as mean ± SEM, and ELISA results were shown as median and range. The microarray signal intensities were analyzed only when the molecules were detected in > 50% of BA subjects (ie, five of nine BA subjects). Because most of the data were not normally distributed, differences among the three groups were screened with the nonparametric Kruskal-Wallis test. When this test indicated a significant difference, the difference between two groups was tested for significance with a Mann-Whitney U test. Correlations were analyzed by the nonparametric Spearman correlation test. A p value of < 0.05 was considered statistically significant.
RESULTS
Protein Microarray Analysis
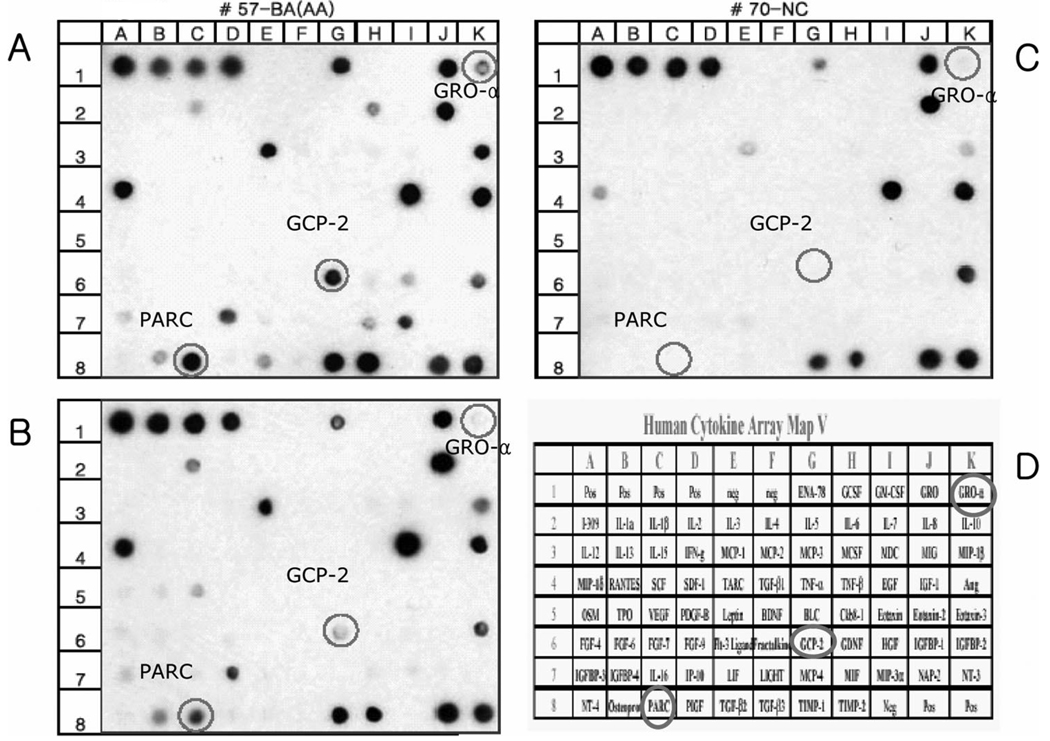
All subjects tolerated sputum induction with no adverse effects. There were no statistical differences between the three groups in age, FEV1 percentage predicted, and FEV1/FVC ratio (data not shown). Figure 1 shows the human cytokines microarray map and examples of the microarray results in each group. Notably, the microarray intensities of GRO-α, granulocyte chemotactic protein (GCP)-2, and PARC were more highly expressed in samples from BA subjects than in samples from AC and NC subjects. The protein microarray signal intensities for these chemokines are summarized in Table 1. The signal intensities of GRO-α, eotaxin-2, and PARC were significantly higher in BA subjects (p = 0.036, p = 0.042, and p = 0.033, respectively) than in AC and NC subjects. The GCP-2 signal intensity appeared higher in BA subjects than in AC and NC subjects, although the difference did not reach statistical significance (p = 0.052).
FIGURE 1.
Custom human cytokine array (RayBio Custom Human Cytokine Arrey; RayBiotech). A total of 79 antibodies toward cytokines and chemokines were placed on the array. Representative membrane protein arrays incubated with sputum supernatant of a BA subject (top left, A), an AC subject (bottom left, B), and an NC subject (top right, C). Four spots in the upper left (row 1; columns A–D) and two spots in the lower right (row 8; columns J and K) of the membranes indicate the positive controls, and two spots in the upper left (row 1; columns E and F) and one in the lower right (row 8; column I) of the membranes indicate the negative controls. The names and locations of each cytokine/chemokine custom spots for this set of experiments are listed (bottom right, D).
Table 1.
The Protein Microarray Signal Intensities for Chemokines*
| Chemokines | BA Subjects (n = 9) |
AC Subjects (n = 3) |
NC Subjects (n = 6) |
p Value |
|---|---|---|---|---|
| PARC/CCL18 | 43 ± 11.0 | 20 ± 15.0 | 11 ± 6.4 | 0.033 |
| GCP-2 | 21 ± 10.0 | 7 ± 6.4 | 2 ± 1.6 | 0.052 |
| GRO-α | 15 ± 4.3 | 12 ± 11.6 | 0 | 0.036 |
| Eotaxin-2 | 8 ± 4.3 | 0 | 0 | 0.042 |
Values are given as the mean ± SEM, unless otherwise indicated. The intensities of the dots from the protein microarray analyses were scanned and the pixel densities were analyzed using image-processing and analysis software (NIH Image; National Institutes of Health). The density values were normalized to the values of the positive controls provided by the manufacturer as 100%.
PARC Profiles
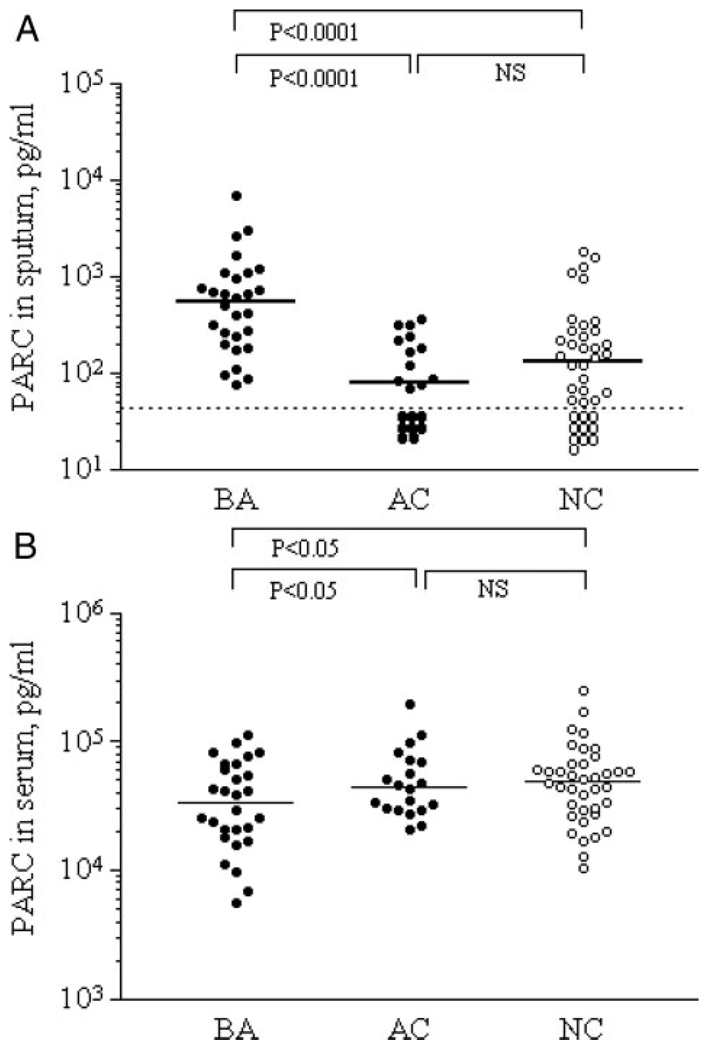
Because increases in PARC in the airway sputum specimens from patients with asthma were not reported previously, we focused on this molecule and quantitated the levels by ELISA. The PARC levels in sputum were significantly higher in the BA group than in the AC and NC groups (p < 0.0001) [Fig 2, top, A]. In contrast, in serum, the PARC levels were significantly lower in the BA group than in the AC and NC groups (p < 0.05) [Fig 2, bottom, B].
FIGURE 2.
PARC levels in sputum and serum from BA subjects, AC subjects, and NC subjects. The PARC levels in sputum specimens (top, A) and in serum specimens (bottom, B) from BA subjects, AC subjects, and NC subjects. Solid line shows medians; dashed line shows sensitivity level (top, A).
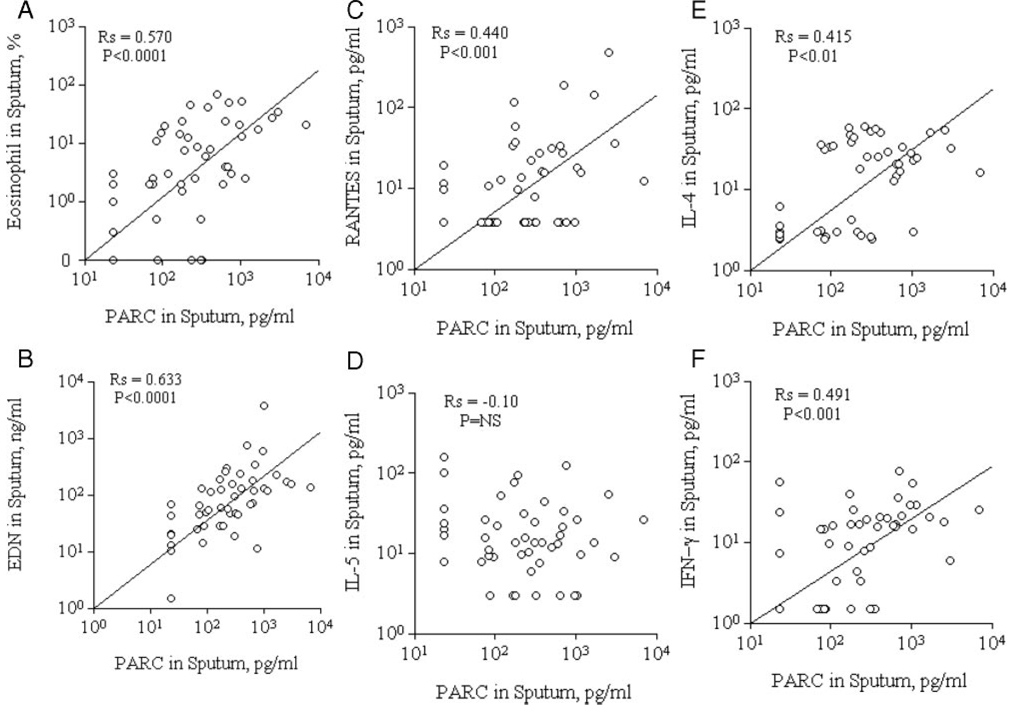
In general, the PARC levels in sputum were correlated with eosinophilic inflammatory markers. For example, the PARC levels in sputum correlated with sputum eosinophil counts (Fig 3, top left, A) and EDN levels (Fig 3, bottom left, B) [r = 0.570, p < 0.0001, and r = 0.633, p < 0.0001, respectively].
FIGURE 3.
PARC levels, eosinophil inflammatory markers, and cytokines/chemokines in sputum specimens. Correlations are shown between PARC levels and eosinophils (top left, A), EDN (bottom left, B), RANTES (top middle, C), IL-5 (bottom middle, D), IL-4 (top right, E), and IFN-γ (bottom right, F) in sputum specimens.
In addition, PARC levels correlated with RANTES levels in sputum (r = 0.440, p < 0.001) [Fig 3, top middle, C], but not with IL-5 levels (r=−0.10, p > 0.05) [Fig 3, middle bottom, D]. Furthermore, PARC levels in sputum were positively correlated with both T helper (Th) cell type 1 and Th2 inflammatory cytokines; the PARC levels correlated significantly with IL-4 (Fig 3, top right, E) and IFN-γ (Fig 3, bottom right, F) levels in sputum (r = 0.415, p < 0.01, and r = 0.491, p < 0.001, respectively).
DISCUSSION
Chemokines likely play an important role in the development of allergic responses by recruiting cells involved in inflammation, particularly Th2 cells, eosinophils, and basophils. Protein microarray technology is most accurately called a protein-detecting microarray.7 By studying many molecules simultaneously, protein microarray technology allows us to learn how proteins interact with each other to control complex processes in cells, tissues, and even whole organisms. A protein-detecting microarray comprises many different affinity reagents arrayed at high spatial density on a solid support. Each agent captures a target protein from a complex mixture, and the captured proteins are subsequently detected and quantified.4 Using this technology, we can study simultaneously many components, such as cytokines and chemokines, that have a role in asthma and learn how these interact with each other.
Allergic asthma is considered a Th2-mediated inflammatory airway disorder that is characterized by airway eosinophil and basophil infiltration, increased mucus production, and bronchial remodeling, leading to airway obstruction. High levels of specific IgE in response to common environmental allergens are characteristic as well.8 For cell recruitment to inflammatory sites, chemokines orchestrate cell trafficking, leukocyte homing, cell activation, and ultimately tissue inflammation. The chemokine receptors CCR5 and CCR3 are expressed primarily on Th1 cells,9, whereas CCR3, CCR4, and CCR8 expression predominates on Th2 cells.10 Among various chemokines, those likely important for eosinophilic inflammation are CCL5 (RANTES), CCL7 (monocyte chemoattractant protein-3), CCL11 (eotaxin-1), and CCL13 (monocyte chemoattractant protein-4), all of which exert their activities via CCR3 expressed on eosinophils. 11,12 Eotaxin is a key mediator for intrapulmonary eosinophil accumulation and development of lung injury. Eotaxin-2/CCL24 is a potent eosinophil chemoattractant in vitro and in vivo,13–15 similarly to eotaxin-1,16 and is increased in sputum cell cultures from patients with asthma.17 More recently, a range of non-CCR3-binding chemokines, which are able to influence eosinophil function, has been identified, including CCL3 (macrophage inflammatory protein-1α) and CCL22 (macrophage-derived chemokine).18 In addition, GRO-α has chemotactic activity for endothelial cells19, and GCP-2 protein is known to cause neutrophil recruitment similarly to that of IL-8.20 GRO-α and the neutrophil chemoattractant GCP-2 are released from the bronchial epithelium in asthma patients.21–23 Several other cytokines seem important for eosinophilic inflammation. New chemokines designated as PARC/CCL18,24 CCL18/dendritic cell-derived chemokine 1,25 CCL18/ alternative macrophage activation-associated CC chemokine-1,26 and CCL18/macrophage inflammatory protein 427 have been described. The PARC gene has been mapped to the long arm of chromosome 17, where most of the CC chemokine genes have been mapped, and showed 59.6% homology at the amino acid level to macrophage inflammatory protein-1α, which suggested it may act as an eosinophil chemoattractant.27 Thus, a large number of chemokines may be involved in airway eosinophilic inflammation in patients with asthma.
To sort out the complex network of the cytokines and chemokines, we took a nonbiased microarray approach to analyze sputum specimens from patients with asthma. Interestingly, we found that relatively few cytokines and chemokines appear to be elevated by this approach. Levels of GRO-α, eotaxin-2, and PARC were significantly increased in patients with asthma. We were also able to verify the increase in PARC by a specific ELISA. Therefore, PARC/CCL18 may have an important role in allergic asthma.
PARC/CCL18 is strongly expressed in the lung and is expressed to a lesser extent in the thymus and lymph nodes.24 Levels of PARC/CCL18 were also highly increased in the supernatants of peripheral blood mononuclear cells from patients with asthma (who are allergic to house dust mite) that were cultured with house dust mite.28 In addition, PARC is highly expressed in blood or tissue specimens from a number of different diseases, such as atheromatosis, 29 rheumatoid arthritis,30 cutaneous contact hypersensitivity, 31 hypersensitivity pneumonitis,32 and more recently, atopic dermatitis.33–35 Importantly, in our study, serum PARC levels were lower in patients with asthma than in NC subjects, suggesting that PARC is preferentially expressed in the lungs.
We also found that the PARC levels in sputum correlated with eosinophilic inflammatory markers, namely eosinophil counts, EDN, and RANTES, with the Th1 and Th2 inflammatory cytokines IL-4 and IFN-γ. Generally, PARC is induced by Th2 cytokines. Indeed, the Th2 cytokines IL-4, IL-13, and IL-10 up-regulate PARC expression,26,28 whereas IFN-γ inhibits its production.26 PARC was secreted by antigen-presenting cells, in particular by monocytes, alveolar macrophages, monocyte-derived dendritic cells,25,26,36 and eosinophils.37 Functionally, PARC attracts naive CD4+ and CD8+ T cells,25,27 B lymphocytes,38 and immature dendritic cells,39 and is thought to be involved in primary immune responses. Injection of synthetic PARC into the peritoneal cavity of mice led to the accumulation of both CD4+ and CD8+ T lymphocytes, but not monocytes or granulocytes.27 Furthermore, de Nadai et al28 showed that some increases in PARC levels were linked to monocyte activation by IL-4 and IL-13, which were produced by Dermatophagoides pteronyssinus-stimulated T cells. Thus, increased levels of PARC may reflect the Th2-like airway environment in patients. On the other hand, the involvement a Th1 cytokine, IFN-γ in asthma seems complex. Previously, IFN-γ was found to be absent or secreted in very low levels in asthma patients40; IFN-γ has been shown to prevent the development of antigen-induced airway eosinophilia and hyperresponsiveness. 41,42 In contrast, another report has suggested43 that production of IFN-γ, as well as IL-4 and IL-5, by sputum CD4+ and CD8+ T cells was increased in patients with asthma and was related to disease severity. In addition, murine model studies44–47 have shown that the coexistence of Th1 and Th2 cells aggravate the Th2 cell-mediated allergic responses. Therefore, in some asthma patients, IFN-γ could increase to reflect their severe airway inflammation, and IFN-γ levels may correlate with PARC levels.
In conclusion, protein microarray technology could prove useful in the study of many cytokines and chemokines simultaneously. In this study, we found that GRO-α, eotaxin-2, and PARC were increased significantly in sputum specimens from patients with asthma. In particular, PARC in the airways may play an important role in the eosinophilic inflammation of asthmatic airways.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (AI50494) and in part by the Korean Research Foundation grant funded by the Korean Government (MOEHRD) [KRF-2005-042-E00078].
Abbreviations
- AC
atopic control
- BA
bronchial asthma
- DTT
dithiothreitol
- EDN
eosinophil-derived neurotoxin
- ELISA
enzyme-linked immunosorbent assay
- GCP
granulocyte chemotactic protein
- GRO
growth-regulated oncogene
- IFN
interferon
- IL
interleukin
- NC
normal control
- PARC
pulmonary and activation-regulated chemokine
- RANTES
regulated upon activation, normal T cell expressed and secreted cytokine
- Th
T helper
Footnotes
The authors have reported to the ACCP that no significant conflicts of interest exist with any companies/organizations whose products or services may be discussed in this article.
Reproduction of this article is prohibited without written permission from the American College of Chest Physicians (www.chestjournal.org/misc/reprints.shtml).
Reprints Information about ordering reprints can be found online: http://www.chestjournal.org/site/misc/reprints.xhtml
REFERENCES
- 1.Pizzichini E, Pizzichini MM, Efthimiadis A, et al. Indices of airway inflammation in induced sputum: reproducibility and validity of cell and fluid-phase measurements. Am J Respir Crit Care Med. 1996;154:308–317. doi: 10.1164/ajrccm.154.2.8756799. [DOI] [PubMed] [Google Scholar]
- 2.Spanevello A, Migliori GB, Sharara A, et al. Induced sputum to assess airway inflammation: a study of reproducibility. Clin Exp Allergy. 1997;27:1138–1144. [PubMed] [Google Scholar]
- 3.Claman DM, Boushey HA, Liu J, et al. Analysis of induced sputum to examine the effects of prednisone on airway inflammation in asthmatic subjects. J Allergy Clin Immunol. 1994;94:861–869. doi: 10.1016/0091-6749(94)90154-6. [DOI] [PubMed] [Google Scholar]
- 4.MacBeath G. Protein microarrays and proteomics. Nat Genet. 2002;32:S526–S532. doi: 10.1038/ng1037. [DOI] [PubMed] [Google Scholar]
- 5.Fahy JV, Liu J, Wong H, et al. Cellular and biochemical analysis of induced sputum from asthmatic and from healthy subjects. Am Rev Respir Dis. 1993;147:1126–1131. doi: 10.1164/ajrccm/147.5.1126. [DOI] [PubMed] [Google Scholar]
- 6.Carlson M, Hakansson L, Peterson C, et al. Secretion of granule proteins from eosinophils and neutrophils is increased in asthma. J Allergy Clin Immunol. 1991;87:27–33. doi: 10.1016/0091-6749(91)90209-7. [DOI] [PubMed] [Google Scholar]
- 7.Kodadek T. Protein microarrays: prospects and problems. Chem Biol. 2001;8:105–115. doi: 10.1016/s1074-5521(00)90067-x. [DOI] [PubMed] [Google Scholar]
- 8.Kay AB. Allergy and allergic diseases: first of two parts. N Engl J Med. 2001;344:30–37. doi: 10.1056/NEJM200101043440106. [DOI] [PubMed] [Google Scholar]
- 9.D’Ambrosio D, Iellem A, Bonecchi R, et al. Selective up-regulation of chemokine receptors CCD4 and CCR8 upon activation of polarized human type 2 Th cells. J Immunol. 1998;161:5111–5115. [PubMed] [Google Scholar]
- 10.Sallusto F, Mackay CR, Lanzavecchia A. Selective expression of the eotaxin receptor CCR3 by human T helper 2 cells. Science. 1997;277:2005–2007. doi: 10.1126/science.277.5334.2005. [DOI] [PubMed] [Google Scholar]
- 11.Peasse JE, Weller CL, Williams TJ. Regulation of eosinophil trafficking in asthma and allergy. Ernst Schering Res Found Workshop. 2004;99:85–100. doi: 10.1007/978-3-662-05403-1_7. [DOI] [PubMed] [Google Scholar]
- 12.Bisset LR, Schmid-Grendelmeier R. Chemokines and their receptors in the pathogenesis of allergic asthma: progress and perspective. Curr Opin Pulm Med. 2004;11:35–42. doi: 10.1097/01.mcp.0000144502.50149.e0. [DOI] [PubMed] [Google Scholar]
- 13.Forssmann U, Uguccioni M, Loetscher P, et al. Eotaxin-2, a novel CC chemokine that is selective for the chemokine receptor CCR3, and acts like eotaxin on human eosinophil and basophil leukocytes. J Exp Med. 1997;185:2171–2176. doi: 10.1084/jem.185.12.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White JR, Imburgia C, Dul E, et al. Cloning and functional characterization of a novel human CC chemokine that binds to the CCR3 receptor and activates human eosinophil. J Leukoc Biol. 1997;62:667–675. doi: 10.1002/jlb.62.5.667. [DOI] [PubMed] [Google Scholar]
- 15.Ying S, Robinson DS, Meng Q, et al. C-C chemokines in allergen-induced late-phase cutaneous responses in atopic subjects: association of eotaxin with early 6-hour eosinophils, and of eotaxin-2 and monocyte chemoattractant protein-4 with the later 24-hour tissue eosinophilia, and relationship to basophils and other CC chemokines (monocyte chemoattractant protein-3 and RANTES) J Immunol. 1999;163:3976–3984. [PubMed] [Google Scholar]
- 16.Rankin SM, Conroy DM, Williams TJ. Eotaxin and eosinophil recruitment: implications for human diseases. Mol Med Today. 2000;6:20–27. doi: 10.1016/s1357-4310(99)01635-4. [DOI] [PubMed] [Google Scholar]
- 17.Scheicher EM, Teixeira MM, Cunha FQ, et al. Eotaxin-2 in sputum cell culture to evaluate asthma inflammation. Eur Respir J. 2007;29:489–495. doi: 10.1183/09031936.00060205. [DOI] [PubMed] [Google Scholar]
- 18.Yamashita U, Kuroda E. Regulation of macrophage-derived chemokine (MDC, CCL22) production. Crit Rev Immunol. 2002;22:105–114. [PubMed] [Google Scholar]
- 19.Alam R. Chemokines in cell movement and inflammation. In: Adkinson NF Jr, Yunginger JW, Busse WW, et al., editors. Middleton’s allergy principles & practice. 6th ed. St. Louis, MO: Mosby; 2003. pp. 159–176. [Google Scholar]
- 20.Baruch A, Oppenheim JJ, Proost P, et al. Characterization of synthetic human granulocyte chemotactic protein 2: usage of chemokine receptors CXCR1 and CXCR2 and in vivo inflammatory properties. Biochemistry. 1997;36:2716–2723. doi: 10.1021/bi961999z. [DOI] [PubMed] [Google Scholar]
- 21.Koch AE, Kunkel SL, Shah MR, et al. Growth-related gene product alpha: a chemotactic cytokine for neutrophils in rheumatoid arthritis. J Immunol. 1995;155:3660–3666. [PubMed] [Google Scholar]
- 22.van Damme J. Identification of a novel granulocyte chemotactic protein (GCP-2) from human tumor cells: in vitro and in vivo comparison with natural forms of GRO, IP-10, and IL-8. J Immunol. 1993;150:1000–1015. [PubMed] [Google Scholar]
- 23.Prause O, Laan M, Lotvall J, et al. Pharmacological modulation of interleukin-17-induced GCP-2-, GRO-α- and interleukin-8 release in human bronchial epithelial cells. Eur J Pharmacol. 2003;462:193–198. doi: 10.1016/s0014-2999(03)01341-4. [DOI] [PubMed] [Google Scholar]
- 24.Hieshima K, Imai T, Baba M, et al. A novel human CC chemokine PARC that is most homologous to macrophage-inflammatory protein-1 α/LD78 α and chemotactic for T lymphocytes, but not for monocytes. J Immunol. 1997;159:1140–1149. [PubMed] [Google Scholar]
- 25.Adema GJ, Hartgers F, Verstraten R, et al. A dendritic-cell-derived C-C chemokine that preferentially attracts naïve T cells. Nature. 1997;387:713–717. doi: 10.1038/42716. [DOI] [PubMed] [Google Scholar]
- 26.Kodelja V, Muller C, Politz O, et al. Alternative macrophage activation-associated CC-chemokine-1, a novel structural homologue of macrophage inflammatory protein-1-α with a Th2-associated expression pattern. J Immunol. 1998;160:1411–1418. [PubMed] [Google Scholar]
- 27.Guan P, Burghes AH, Cunningham A, et al. Genomic organization and biological characterization of the novel human CC chemokine DC-CK-1/PARC/MIP-4/SCYA18. Genomics. 1999;56:296–302. doi: 10.1006/geno.1998.5635. [DOI] [PubMed] [Google Scholar]
- 28.de Nadai P, Charbonnier A, Chenivesse C, et al. Involvement of CCL18 in allergic asthma. J Immunol. 2006;176:6286–6293. doi: 10.4049/jimmunol.176.10.6286. [DOI] [PubMed] [Google Scholar]
- 29.Reape TJ, Rayner K, Manning CD, et al. Expression and cellular localization of the CC chemokines PARC and ELC in human atherosclerotic plaques. Am J Pathol. 1999;154:365–374. doi: 10.1016/S0002-9440(10)65283-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schutyser E, Struyf S, Wuyts A, et al. Selective induction of CCL18/PARC by staphylococcal enterotoxins in mononuclear cells and enhanced levels in septic and rheumatoid arthritis. Eur J Immunol. 2001;31:3755–3762. doi: 10.1002/1521-4141(200112)31:12<3755::aid-immu3755>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 31.Goebeler M, Trautmann A, Voss A, et al. Differential and sequential expression of multiple chemokines during elicitation of allergic contact hypersensitivity. Am J Pathol. 2001;158:431–440. doi: 10.1016/s0002-9440(10)63986-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pardo A, Smith KM, Abrams J, et al. CCL18/DC-CK-1/PARC up-regulation in hypersensitivity pneumonitis. J Leukocyte Biol. 2001;70:610–616. [PubMed] [Google Scholar]
- 33.Gunther C, Bello-Fernandez C, Kopp T, et al. CCL18 is expressed in atopic dermatitis and mediates skin homing of human memory T cells. J Immunol. 2005;174:1723–1728. doi: 10.4049/jimmunol.174.3.1723. [DOI] [PubMed] [Google Scholar]
- 34.Nomura I, Gao B, Boguniewicz M, et al. Distinct patterns of gene expression in the skin lesions of atopic dermatitis and psoriasis: a gene microarray analysis. J Allergy Clin Immunol. 2003;112:1195–1202. doi: 10.1016/j.jaci.2003.08.049. [DOI] [PubMed] [Google Scholar]
- 35.Pivarcsi A, Gombert M, Dieu-Nosjean MC, et al. CC chemokine ligand 18, an atopic dermatitis-associated and dendritic cell-derived chemokine, is regulated by staphylococcal products and allergen exposure. J Immunol. 2004;173:5810–5817. doi: 10.4049/jimmunol.173.9.5810. [DOI] [PubMed] [Google Scholar]
- 36.Vissers JL, Hartgers FC, Lindhout E, et al. Quantitative analysis of chemokine expression by dendritic cell subsets in vitro and in vivo. J Leukoc Biol. 2001;69:785–793. [PubMed] [Google Scholar]
- 37.Schraufstatter I, Takamori H, Sikora L, et al. Eosinophils and monocytes produce pulmonary and activation-regulated chemokine, which activates cultured monocytes/macrophages. Am J Physiol. 2004;286:L494–L501. doi: 10.1152/ajplung.00323.2002. [DOI] [PubMed] [Google Scholar]
- 38.Lindhout E, Vissers JL, Hartgers FC, et al. The dendritic cell-specific CC-chemokine DC-CK1 is expressed by germinal center dendritic cells and attracts CD38-negative mantle zone B lymphocytes. J Immunol. 2001;166:3284–3289. doi: 10.4049/jimmunol.166.5.3284. [DOI] [PubMed] [Google Scholar]
- 39.Vulcano M, Struyf S, Scapini P, et al. Unique regulation of CCL18 production by maturing dendritic cells. J Immunol. 2003;170:3843–3849. doi: 10.4049/jimmunol.170.7.3843. [DOI] [PubMed] [Google Scholar]
- 40.Mazzarella G, Bianco A, Catena E, et al. Th1/Th2 lymphocyte polarization in asthma. Allergy. 2000;55:S6–S9. doi: 10.1034/j.1398-9995.2000.00511.x. [DOI] [PubMed] [Google Scholar]
- 41.Iwamoto I, Nakajima H, Endo H, et al. Interferon γ regulates antigen-induced eosinophil recruitment into the mouse air ways by inhibiting the infiltration of CD4+ T cells. J Exp Med. 1993;177:573–576. doi: 10.1084/jem.177.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li XM, Chopra RK, Chou TY, et al. Mucosal IFN-γ gene transfer inhibits pulmonary allergic responses in mice. J Immunol. 1996;157:3216–3219. [PubMed] [Google Scholar]
- 43.Cho SH, Stanciu LA, Holgate ST, et al. Increased interleukin- 4, interleukin-5, and interferon-γ in airway CD4+ and CD8+ cells in atopic asthma. Am J Respir Crit Care Med. 2005;171:224–230. doi: 10.1164/rccm.200310-1416OC. [DOI] [PubMed] [Google Scholar]
- 44.Randolph DA, Carruthers CJ, Szabo SJ, et al. Modulation of airway inflammation by passive transfer of allergen-specific Th1 and Th2 cells in a mouse model of asthma. J Immunol. 1999;162:2375–2383. [PubMed] [Google Scholar]
- 45.Hansen G, Berry G, DeKruyff RH, et al. Allergen-specific Th1 cells fail to counterbalance Th2 cell-induced airway hyperreactivity but cause severe airway inflammation. J Clin Invest. 1999;103:175–183. doi: 10.1172/JCI5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Randolph DA, Stephen R, Carruthers CJ, et al. Cooperation between Th1 and Th2 cells in a murine model of eosinophilic airway inflammation. J Clin Invest. 1999;104:1021–1029. doi: 10.1172/JCI7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakajima H, Takatsu K. Role of cytokines in allergic airway inflammation. Int Arch Allergy Immunol. 2007;142:265–273. doi: 10.1159/000097357. [DOI] [PubMed] [Google Scholar]