Abstract
Background
The contribution of low frequency drug-resistant HIV-1 variants to failure of antiretroviral therapy is not well-defined in treatment-experienced patients.
Objective
We sought to detect minor non-nucleoside reverse transcriptase inhibitor (NNRTI)-resistant variants at the onset of multidrug efavirenz-containing therapy in both NNRTI-naïve and NNRTI-experienced patients and to determine their association with virologic response.
Methods
Plasma samples at entry and virologic failure from patients enrolled in the AIDS Clinical Trials Group study 398 were analyzed by standard genotype, single-genome sequencing and allele-specific PCR (K103N and Y181C) to detect and quantify minor NNRTI-resistant variants.
Results
Minor populations of NNRTI-resistant variants that were missed by standard genotype were detected more often at study entry in NNRTI-experienced patients than NNRTI-naïve patients by both single-genome sequencing (8 of 12 vs. 3 of 15; P=0.022) and allele-specific PCR (>1% Y181C: 5 of 22 vs. 3 of 72, respectively, P = 0.016). K103N variants at frequencies >1% were associated with inferior HIV-1 RNA response to efavirenz-containing therapy between entry and week 24 (+0.5 vs −1.1 log10 copies/ml; P <0.001).
Conclusions
Minor NNRTI-resistant variants were more prevalent in NNRTI-experienced patients and were associated with reduced virologic response to efavirenz-containing multidrug regimens.
Keywords: HIV-1 Drug-resistance, Minority Variants, Virologic Response
INTRODUCTION
HIV-1 diversity within infected individuals arises from continuous, high-level virus turnover (~1011 virions and 108 infected cells per day), from nucleotide misincorporations by error-prone reverse transcriptase (RT) during viral DNA synthesis, and possibly from misincorporations by host cell RNA polymerase II during viral RNA synthesis [1–3]. Many of these mutations do not have a large negative effect on viral fitness and thus accumulate during successive rounds of viral replication, generating a diverse population of variants termed a “quasispecies”. This diversity supports the hypothesis that drug-resistant HIV-1 variants exist within an infected individual before antiretroviral drug therapy is started [3, 4]. The frequency of such pre-existing drug-resistant variants can increase rapidly with partially suppressive antiretroviral therapy and become dominant in the virus population. Following removal of drug selective pressure by cessation of therapy, drug-resistant variants often decline such that they become a minor fraction of the virus population, but at higher frequencies than in drug naïve individuals [5]. Such variants can rapidly re-emerge after restarting antiretroviral therapy with drugs to which the variants are resistant or cross-resistant [5].
HIV-1 drug resistance testing using genotype analysis of PCR-amplified sequences from the virus population in plasma is recommended for management of antiretroviral therapy [6]. However, this standard method identifies only the dominant viruses present and fails to detect variants comprising less than 10–20% of the virus population [7]. More sensitive techniques have been developed that can detect minor populations of drug-resistant variants that are missed by standard genotype, but their clinical utility has yet to be established [5, 7, 8, 9, 10]. Minor drug-resistant variants have been detected in treatment naïve individuals and are associated with higher risk of failure of initial antiretroviral therapy [9, 10]. In treatment-experienced patients, minor drug-resistant variants may persist for months and even years during and after treatment [5, 11, 12]. It is not known, however, whether such drug-resistant variants influence response to subsequent treatment regimens. We therefore assessed the influence of minor NNRTI-resistant variants on response to efavirenz-containing multidrug regimens.
AIDS Clinical Trials Group (ACTG) study 398 was a randomized, multi-center trial that tested the efficacy of one or two protease inhibitors in combination with abacavir, adefovir, and efavirenz, in patients on a failing regimen containing a protease inhibitor. All patients had prior exposure to NRTI and 44% had prior exposure to the NNRTI delavirdine or nevirapine [13]. Treatment response was strongly associated with prior NNRTI exposure: NNRTI-experienced patients were more than twice as likely to reach a protocol-defined virologic failure endpoint compared to NNRTI-naive patients. NNRTI experience predicted poor virologic outcome independent of detecting NNRTI resistance at entry by standard population genotype [14]. This finding lead to the hypothesis that minor NNRTI-resistant variants, selected by prior NNRTI exposure, but below the limit of detection by population genotype, were contributing to failure of the efavirenz-containing regimens. To test this, we compared the frequency of minor NNRTI-resistant variants between NNRTI-naive and NNRTI-experienced patients, determined if their detection was predictive of virologic response, and analyzed the genetic relatedness of NNRTI-resistant variants detected at study entry and virologic failure.
METHODS
Subject selection and samples
Plasma samples were obtained at study entry and at protocol-defined virologic failure from subjects in ACTG study 398 [13]. For single-genome sequencing analyses, a total of 27 subjects (15 NNRTI-naïve and 12 NNRTI-experienced) were randomly selected from enrollees meeting the following criteria: i) entry sample negative for NNRTI-resistance mutations by standard genotype analysis (ViroSeq platform; Celera, Alameda, CA); ii) reached a protocol-defined virologic failure endpoint by study week 24 [13], and iii) the virologic failure sample had one or more major NNRTI-resistance mutations (L100I, K101E, K103N, V106A or M, V108I, Y181C or I, Y188C, H, or L, G190A or S, P225H, or P236L [15]) detected by standard genotype (ViroSeq).
NNRTI resistance at study entry by allele-specific real-time PCR was determined for 103 participants, consisting of 27 NNRTI-experienced and 76 randomly selected NNRTI-naive patients. Both groups had available entry plasma samples and a negative standard genotype for NNRTI resistance.
ACTG 398 was approved by the institutional review boards of the participating centers, and all patients provided written consent for participation in the study and testing of samples.
Single-genome sequencing
Single-genome sequencing was performed as reported [8] with the following modifications: 500ul of plasma (diluted to contain 5,000 copies of HIV-1 RNA) was extracted, cDNA was prepared using the Superscript 1st Strand Synthesis System (InVitrogen, Carlsbad, CA), and was serially diluted 1:2 in 5mM Tris-HCl (pH 8.0) to a maximum dilution of 1:16. Ten independent PCR reactions were setup in a screening plate for each cDNA dilution (1:2, 1:4, 1:8, and 1:16, respectively). Amplification products from cDNA dilutions yielding approximately 30% positive PCR reactions were sequenced using the Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). All sequences were assembled using Sequencher software (Gene Codes, Ann Arbor, MI) or by the automated Phrap program in conjunction with Phred and CLUSTAL W [16–19]. The Polyphred program was used to identify potential mixtures, which were excluded unless they were at a single nucleotide position [20]. Drug-resistance mutations in single sequences were identified using the Stanford database HIValg web-based program [21].
Phylogenetic analyses
Phylogenetic analyses were performed as follows: sequences were trimmed using Sequencher, aligned by CLUSTAL W (DAMBE), and phenograms were generated with HIV-1LAI as the outgroup using the Neighbor-Joining method (MEGA v2.1) with the p-distance model analysis preference [22, 23]. Distance was determined as the proportion (p) of nucleotide sites at which two sequences being compared differ and calculated by dividing the number of differences by total number of nucleotides compared. Bootstrap values were calculated using 1000 replicates and values ≥60 are shown in the phenograms.
Allele-specific PCR for NNRTI-resistant variants
Allele-specific PCR was performed as published [5, 24] with the following modifications: 250–1800µl of plasma containing ≥10,000 HIV-1 RNA copies were analyzed at codons 103 and 181 using 3 discriminatory primers for 3 different mutant alleles (AAC and AAT for K103N and TGT for Y181C). A first round of real-time RT-PCR generated a 637-bp pol amplification product, which was diluted to 107 copies and re-amplified by a second round of real-time PCR utilizing both non-discriminatory and discriminatory primers specific for codons 103 and 181 in RT. Amplicon generation was monitored by Sybr Green fluorescence and specificity determined by thermal denaturation analysis. Results were reported as % mutant or wild-type.
Statistical analysis
All analyses were preformed using the R statistical software package [25]. The association between presence of NNRTI resistance and virologic failure by week 24 was assessed using Fisher’s exact test. The association between a history of NNRTI experience and virologic failure by week 24, controlling for baseline NNRTI resistance mutations, was determined using a logistic regression model (Wald test, odds ratios and % confidence interval). The association between NNRTI experience and the detection of NNRTI-resistant variants at study entry by single-genome sequencing was determined using a Fisher’s exact test. The association between NNRTI experience and the proportion of sequences containing NNRTI resistance mutations at entry or virologic failure were determined by a logistic regression. A Gehan-Wilcoxon rank test was used to compare changes in HIV-1 RNA between subgroups with and without NNRTI resistant variants detected by single-genome sequencing, stratified by baseline NNRTI experience and accounting for HIV-1 RNA censored below the limit of detection. Seven of 27 subjects experienced virologic failure before week 24, thus their available HIV-1 RNA data were imputed (week 16 for 5 cases and week 8 for 2). HIV-1 RNA values below the limit of detection (200 copies/mL) were replaced by 200 copies/mL. Statistical analyses of data from allele-specific PCR were performed using mutant frequency cutoffs of >0.5% and >1.0%. The association between NNRTI experience and the detection of K103N or Y181C was determined using a Fisher’s exact test. The association between mutant frequency and change in HIV-1 RNA from entry to week 24 was determined using a non-parametric log-rank test. Fifteen of 103 subjects experienced virologic failure prior to week 24; therefore, their available data were imputed (week 16 for 7 cases, week 8 for 4 cases, week 4 for 3, and week 2 for 1 case). The association between mutant frequency and virologic failure (HIV-1 RNA >200 copies/ml at week 24) was determined by a logistic regression.
RESULTS
Influence of prior NNRTI experience on virologic response to therapy
As previously reported [13], virologic failure in ACTG study 398 was strongly associated with prior NNRTI exposure either to nevirapine or delavirdine (efavirenz exposure was not permitted). By study week 24, protocol-defined virologic failure had occurred in 83% (176 of 212) of NNRTI experienced patients versus 58% (156 of 269) of NNRTI naive participants (ITT analysis, P <0.001) [13]. Standard genotype analysis was performed on baseline samples from 452 of the 481 study participants. NNRTI resistance mutations were detected in 165 participants (9 of 246 NNRTI-naïve and 156 of 206 NNRTI-experienced patients) and the presence of resistance was associated with virologic failure by week 24 (P <0.001). In the current study, we found that a history of NNRTI experience was significantly associated with virologic failure at week 24 after controlling for baseline NNRTI resistance (OR 2.07 95% CI 1.1–3.88; P = 0.024, Wald test of logistic regression). To explore this observation further, we examined the change in plasma HIV-1 RNA from baseline to week 24 among three subgroups: 1) NNRTI-naïve with baseline standard genotype negative for NNRTI resistance (N = 235); 2) NNRTI-experienced with NNRTI resistance detected at baseline by standard genotype (N = 156); and 3) NNRTI-experienced with baseline standard genotype negative for NNRTI resistance (N = 48). The fourth group of NNRTI-naïve subjects with NNRTI resistance at baseline (N = 9) was too small to include in this analysis.
Figure 1 shows that the change in HIV-1 RNA from entry to week 24 in the NNRTI-experienced subgroup without baseline NNRTI resistance by standard genotype resembled that of the genotype-positive NNRTI-experienced subgroup, which was inferior to that of the NNRTI-naïve subgroup. This observation led to the hypothesis that minor populations of NNRTI-resistant variants compromised the response to efavirenz-containing multidrug therapy in NNRTI-experienced patients with negative standard genotypes for NNRTI resistance.
Figure 1. Median change in HIV-1 RNA (log10 copies/ml) by NNRTI experience and baseline NNRTI resistance.
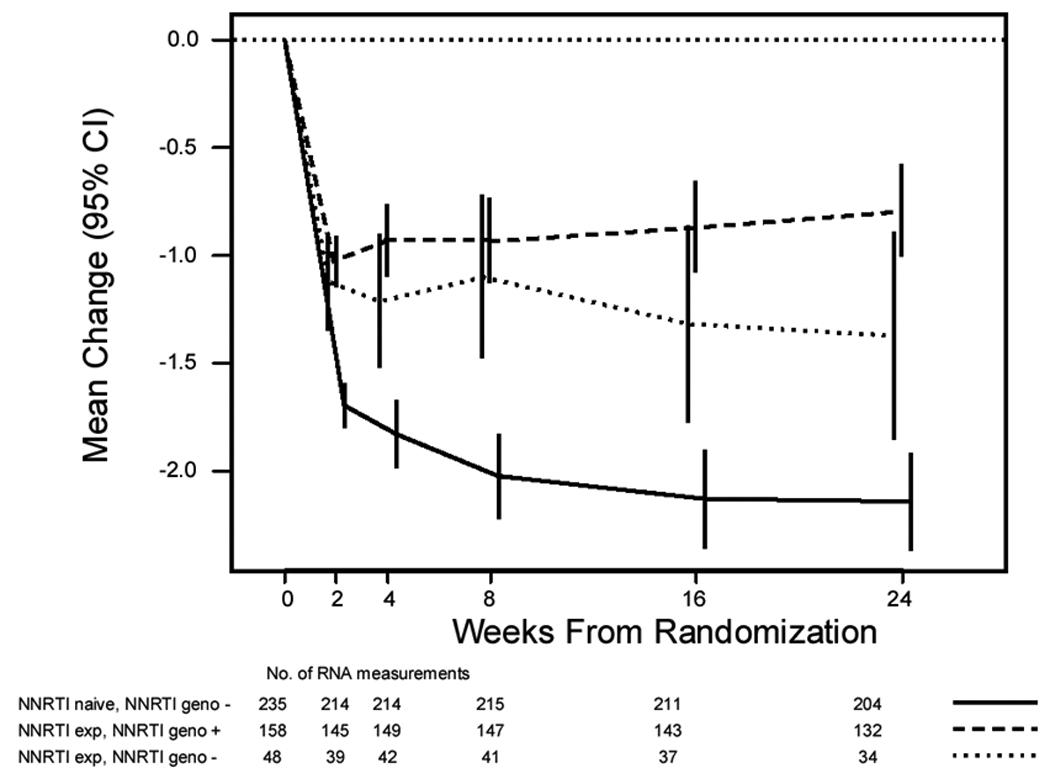
NNRTI resistance was assessed by standard genotype (ViroSeq v2.0). Subgroups shown are 1) NNRTI naïve, standard genotype negative for NNRTI resistance; 2) NNRTI experienced, standard genotype positive for NNRTI resistance; 3) NNRTI experienced, standard genotype negative for NNRTI resistance. Vertical bars are 95% confidence intervals.
To test this hypothesis, we used two more sensitive methods, single-genome sequencing and allele-specific PCR, to detect minor NNRTI-resistant variants in plasma samples from study entry and to assess their impact on virologic response.
NNRTI-resistance mutations detected by single-genome sequencing
For single-genome sequencing analyses, 27 baseline and virologic failure sample pairs were randomly selected as described in Methods. For NNRTI-experienced patients, the median duration after discontinuation of prior NNRTI therapy was 366 days (range 0–555 days). Sequences were generated and analyzed at entry and failure time points except for participant 11E for whom no sample was available at virologic failure.
Single-genome sequencing results are summarized in Table 1. A total of 1566 sequences from study entry were analyzed (an average of 46 sequences/sample), allowing the detection of variants present at a frequency of ≥ 5% with 90% confidence. Only ~12 sequences/sample were analyzed at virologic failure because NNRTI-resistant variants were dominant as determined by standard genotype analysis.
Table 1. NNRTI resistance detected by single-genome sequencing.
| A. NNRTI-naive subjects. | ||||||
|---|---|---|---|---|---|---|
| NNRTI Resistance at Study Entrya | NNRTI Resistance at Virologic Failure | |||||
| Patient | Standard Genotype |
Mutant Sequencesc |
NNRTI Resistance Mutations |
Standard Genotype |
Mutant Sequences |
NNRTI Resistance Mutations |
| 1N | None | 0 of 52 | None | K103N, V108I | 15 of 15 | K103N, V108I, M230L |
| 2N | None | 0 of 52 | None | K103N | 13 of 13 | L100I, K103N, V108I |
| 3N | None | 0 of 49 | None | K103N, V108I | 13 of 13 | L100I, K103N, V108I |
| 4N | None | 0 of 55 | None | K103N | 14 of 14 | K103N |
| 5N | None | 1 of 53 | P225H | K103N | 12 of 12 | K103N |
| 6N | None | 0 of 51 | None | K103N | 6 of 7 | K103N |
| 7N | None | 1 of 40 | K103N | K103N, M230L | 16 of 16 | K103N, Y188C, M230L |
| 8N | None | 0 of 50 | None | K103N, G190A | 11 of 13 | K103N, V106M |
| 9N | None | 0 of 63 | None | K103N | 11 of 11 | K103N |
| 10N | None | 0 of 46 | None | K103N | 14 of 14 | K103N |
| 11N | None | 0 of 51 | None | L100I, K103N | 12 of 12 | L100I, K103N, V108I |
| 12N | None | 1 of 48 | L100I | G190S | 15 of 15 | G190S |
| 13N | None | 0 of 48 | None | K103N, Y181C | 5 of 16 | K103N, Y181C |
| 14N | None | 0 of 45 | None | K103N, V108I | 14 of 14 | K103N, V108I |
| 15N | None | 0 of 70 | None | K103N, G190A | 8 of 8 | K103N, G190A |
| B. NNRTI-experienced subjects. | ||||||
|---|---|---|---|---|---|---|
| NNRTI Resistance at Entry | NNRTI Resistance at Virologic Failure | |||||
| Patient | Standard Genotype |
Mutant Sequences |
NNRTI Resistance Mutations |
Standard Genotype |
Mutant Sequences |
NNRTI Resistance Mutations |
| 1E | None | 2 of 32 | V108I | K103N, V108I | 12 of 12 | K103N, V108I |
| 2E | None | 0 of 48 | None | L100I, K103N | 10 of 10 | L100I, K103N |
| 3E | None | 8 of 41 | K101E | L100I, K101E, Y188H/L |
5 of 11 | L100I, K101E, Y188L, G190A |
| 4E | None | 0 of 45 | None | K103N, P225H | 10 of 10 | L100I, K103N, P225H |
| 5E | None | 10 of 30 | K101E, Y181C, G190A |
K101E, V108I, Y181C, G190A/S |
15 of 15 | K101E, V108I, Y181C, G190A, G190S |
| 6E | None | 3 of 19 | Y181C | K103N, Y181C | 14 of 14 | K103N, V106M, Y181C, G190A, G190S |
| 7E | None | 1 of 33 | K103N | K103N, V108I | 11 of 11 | K103N, V108I |
| 8E | None | 1 of 34 | K103N | K103N, V108I | 9 of 9 | K103N, V108I |
| 9E | None | 0 of 46 | None | K103N | 13 of 13 | K103N |
| 10E | None | 0 of 48 | None | L100I, K103N | 9 of 9 | L100I, K103N |
| 11E | None | 1 of 45 | G190E | K103N | NA | NAd |
| 12E | None | 5 of 47 | Y181C, G190A | K101E, Y181C, G190A |
18 of 18 | K101E, V108I, Y181C, G190A |
Major NNRTI resistance mutations designated by the International AIDS Society-USA [6].
Determined by ViroSeq version 2.0.
Number of sequences containing NNRTI resistance mutations/total sequences by single-genome sequencing.
Sample not available.
The number and types of NNRTI-resistance mutations detected at study entry were significantly different between NNRTI-naïve and -experienced groups (table 1A and B). Specifically, one or more sequences encoding an NNRTI resistance mutation were detected in 8 of 12 NNRTI-experienced patients compared with 3 of 15 NNRTI-naïve patients (P=0.022). Furthermore, the fraction of sequences encoding NNRTI-resistance mutations was higher in the NNRTI-experienced group (31 of 468 sequences) than in the NNRTI naïve group (3 of 773 sequences; P<0.001).
In this small subset of 27 participants analyzed by single-genome sequencing, the HIV-1 RNA response from entry to virologic failure was inferior in the NNRTI-experienced group (+0.13 ± 0.19 [SE] log10 copies/ml) compared with the NNRTI-naïve group (−0.35 ± 0.13 [SE] log10 copies/ml; P=0.032).
Phylogenetic analyses
Phylogenetic analyses were performed to assess the relatedness between sequences with NNRTI resistance mutations at entry and virologic failure. Close clustering between baseline and failure sequences containing NNRTI resistance mutations, as evidenced by high bootstrap values, was observed in 1 of 3 NNRTI-naïve subjects and 1 of 8 NNRTI-experienced subjects, with the suggestion of clustering in a second NNRTI-experienced subject. Fig. 2 illustrates these phylogenetic relationships. In subject 7N from the NNRTI-naive group (Fig 2A), a baseline sequence containing K103N clustered closely (bootstrap value 96) with sequences at failure containing K103N linked to Y188C or M230L. By contrast, baseline sequences from NNRTI-naïve subject 5N and 12N containing P225H and L100I, respectively, did not cluster with the predominant resistant variants at failure (Fig 2B–C).
Figure 2. Phylogenetic analysis comparing relatedness between single genome sequences at baseline and virologic failure.
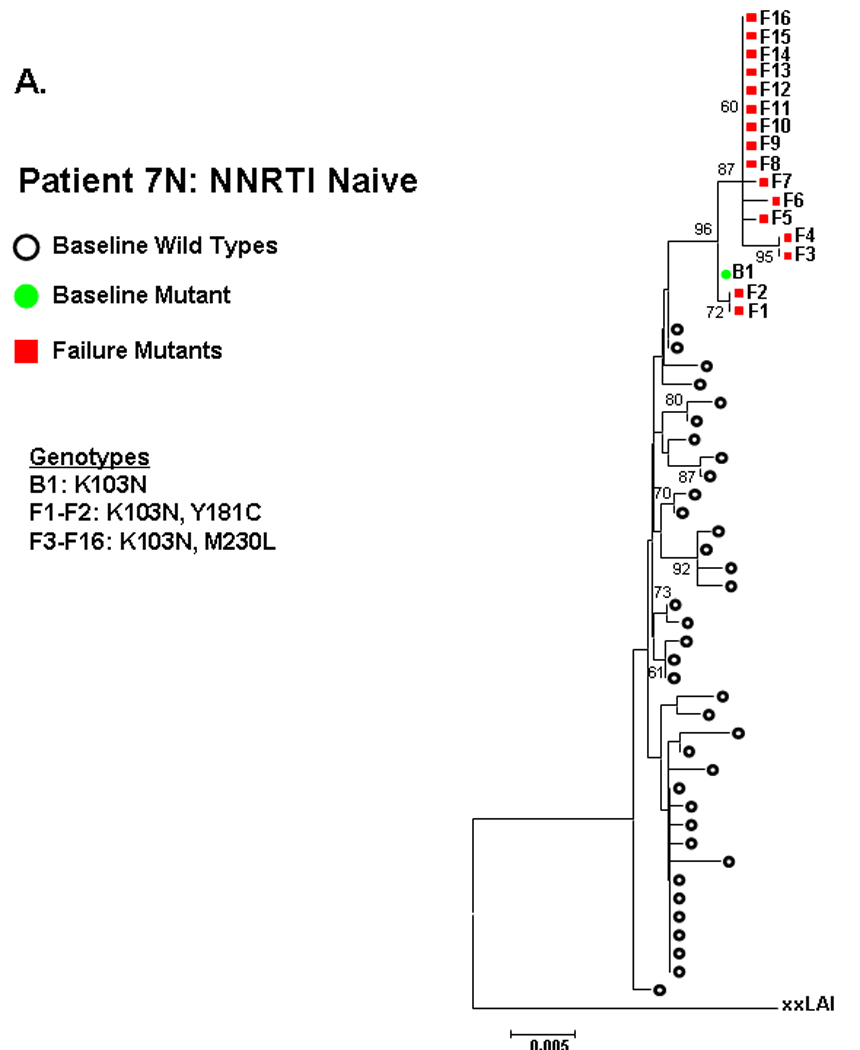
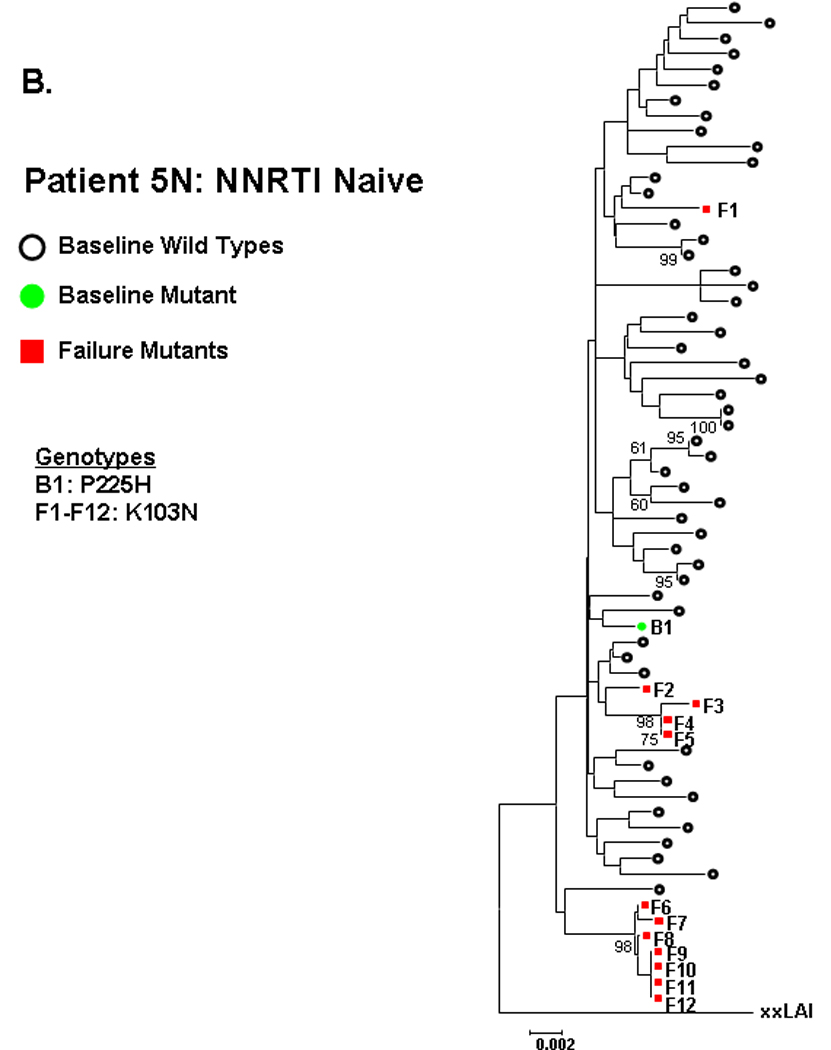
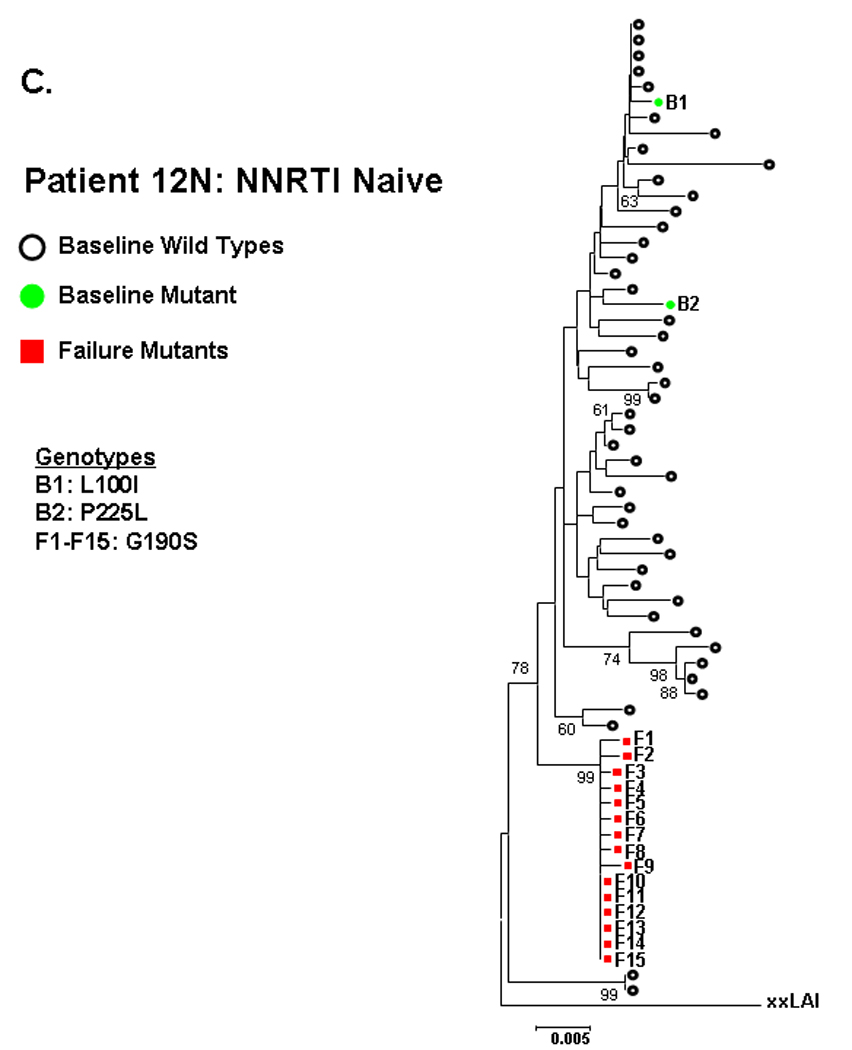
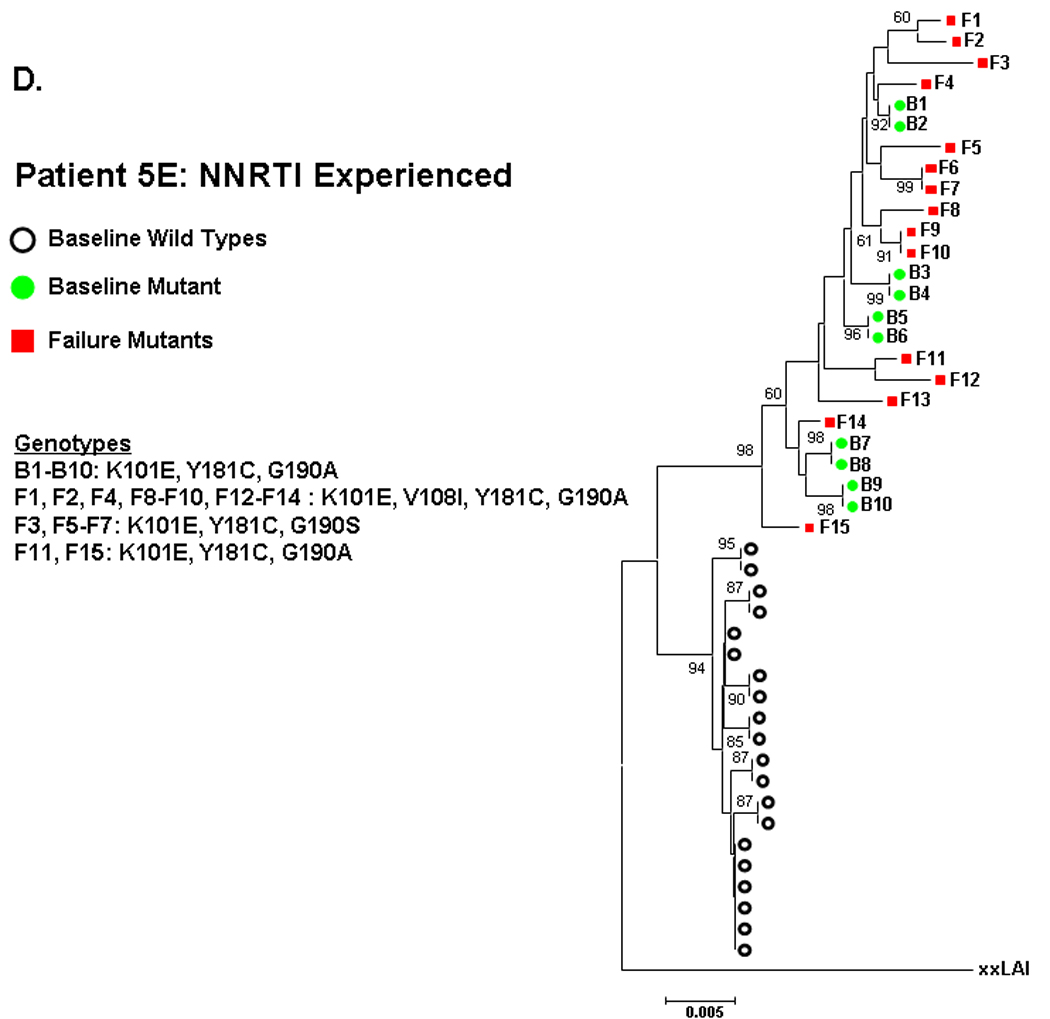
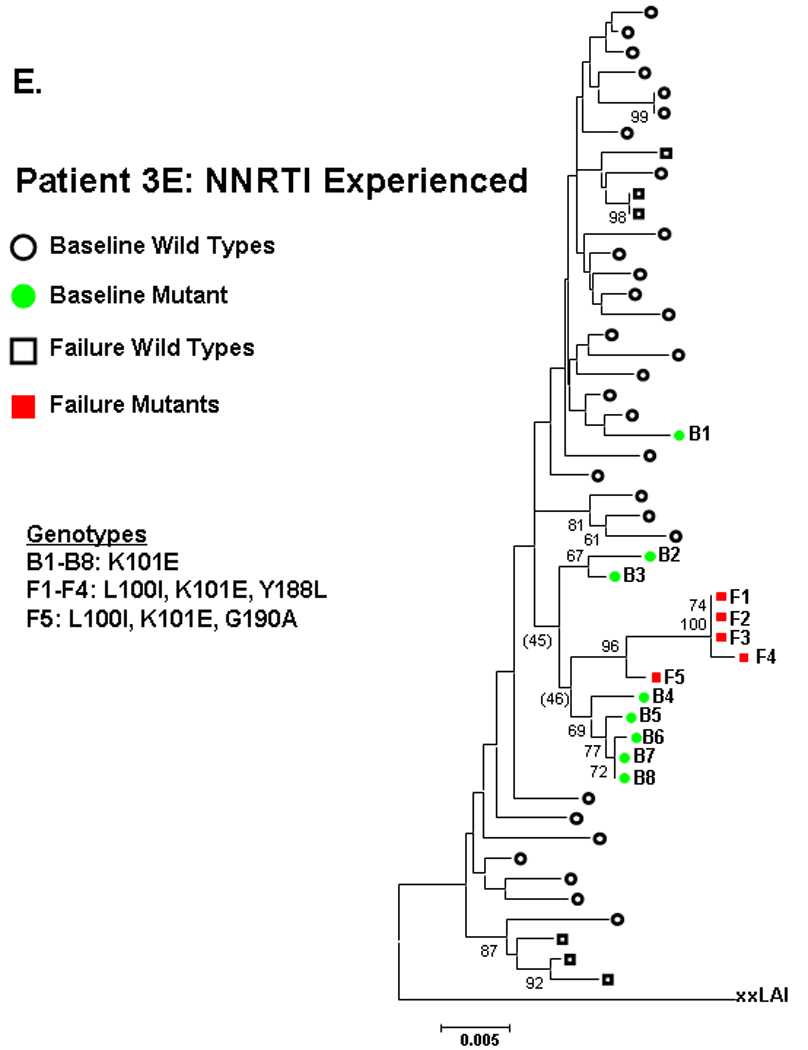
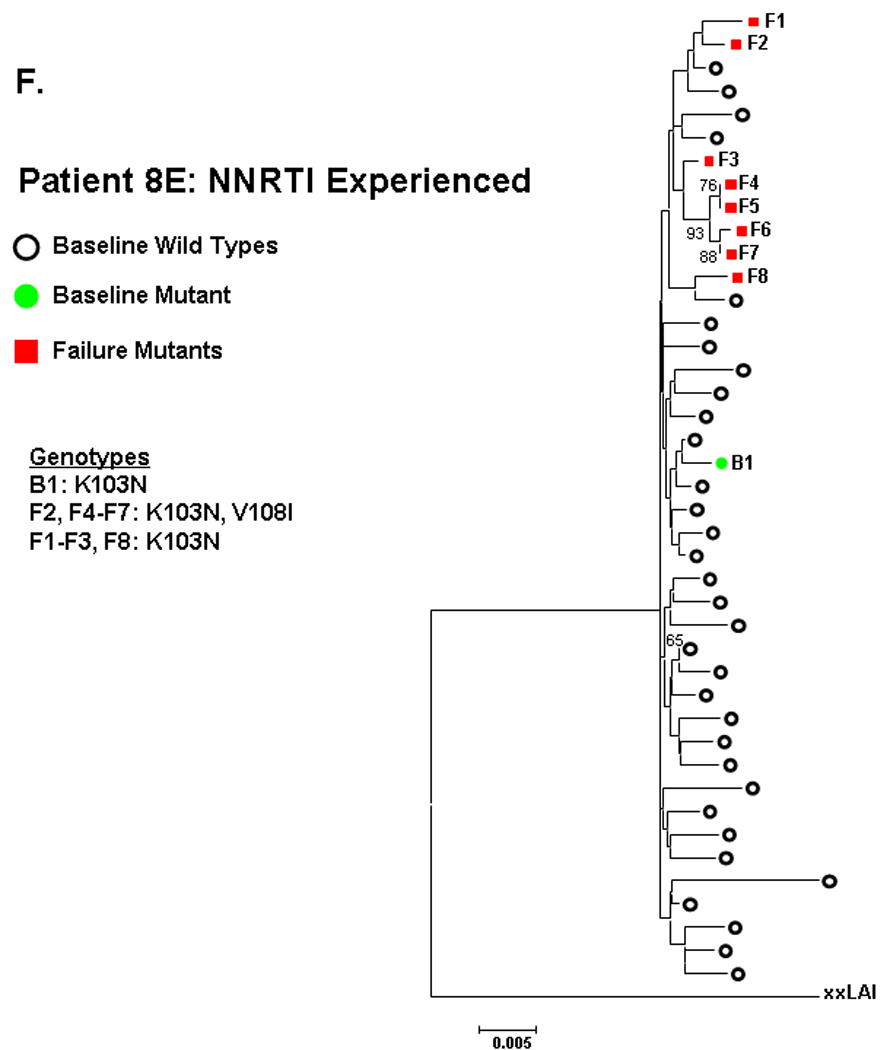
NNRTI naive subjects are shown in panels A–C and NNRTI experienced in panels D–F. Solid circles are baseline sequences with NNRTI resistance mutations. Open circles are baseline sequences without NNRTI resistance mutations. Solid squares are sequences at virologic failure having NNRTI resistance mutations. Open squares are failure sequences without NNRTI resistance mutations. Bootstrap values >60 are shown.
NNRTI-experienced subject 5E had two distinct populations of sequences at entry, one wildtype and one containing several linked NNRTI-resistance mutations (K101E/Y181C/G190A). The mutant sequences at entry intermingled in multiple clusters (bootstrap values >60) with the mutant sequences at failure (Fig 2D). For NNRTI-experienced subject 3E, entry sequences containing K101E showed a trend toward clustering (bootstrap value 46) with failure sequences containing L100I/K101E/Y188L or L100I/K101E/G190A (figure 2E). The observed clustering of entry and failure mutant sequences for these subjects was not altered by changing drug-resistance mutations to wildtype (not shown).
The NNRTI-resistant mutants detected at entry for the 5 other NNRTI-experienced subject (table 1B) did not cluster with the mutant sequences at failure. To 13 illustrate, patient 8E had sequences at entry and failure containing K103N, but these were not closely related (figure 2F).
NNRTI-resistance mutations detected by allele-specific PCR
To further assess if minor populations of NNRTI-resistant variants were more frequent in the NNRTI-experienced patients and associated with virologic failure, baseline samples from 103 subjects (76 NNRTI-naive and 27 NNRTI-experienced) with negative standard genotype for NNRTI resistance were tested using allele-specific PCR for mutants K103N and Y181C. Of the 103 subjects selected, 5 subjects (2 NNRTI-naïve and 3 NNRTI-experienced) were excluded since follow-up HIV-1 RNA measurements were not available. In addition, allele-specific PCR could not be obtained in 4 subjects for Y181C and 3 for K103N, leaving 95 subjects evaluated for K103N and 94 for Y181C (table 2). A significant association was found between NNRTI-experience and Y181C variants at mutant cutoff frequencies >0.5% (5/22 versus 3/72, P=0.016) and >1% (5/22 versus 3/72, P=0.016). A trend for association was found between NNRTI-experience and K103N at frequencies >0.5% (4/24 versus 4/71, P = 0.11) and >1% (3/24 versus 2/7, P = 0.10).
Table 2. Impact of K103N and Y181C Variants on HIV-1 RNA Response.
Change in HIV-1 RNA from Baseline to Week 24 (log10 copies/ml)a
| % Mutant | K103N (Nb) | Y181C (Nb) |
|---|---|---|
| >1.0% | +0.50 (5) | −0.40 (8) |
| ≤1.0% | −1.10 (90) | −1.10 (86) |
| P-value | 0.001 | 0.38 |
Intent-to-treat analysis
Number of subjects
K103N at frequencies >1.0% was strongly associated with inferior HIV-1 RNA response from baseline to week 24 by intent-to-treat (ITT) analysis (P <0.001; table 2). Specifically, K103N >1.0% was associated with a rise in HIV-1 RNA of 0.5 log10 copies/ml, whereas K103N ≤1.0% was associated with a decrease in HIV-1 RNA of 1.1 log10 copies/ml. K103N at >0.5% frequency was also significantly associated with inferior HIV-1 RNA response, although the strength of association was lower (P = 0.006). K103N at frequencies >1% was also associated with protocol-defined virologic failure in both on-treatment (P = 0.036) and ITT analyses (P = 0.053). By contrast, Y181C variants were not significantly associated with either protocol-defined virologic failure or change in HIV-1 RNA from entry to week 24 (table 2), although the average reduction in HIV-1 was lower for those with >1% Y181C (−0.4 log10 copies/ml) versus those with ≤1.0% mutant (−1.1 log10 copies/ml).
DISCUSSION
This work provides several lines of evidence that minor populations of NNRTI-resistant variants not detected by currently recommended resistance assays can contribute to failure of antiretroviral regimens containing efavirenz, especially among patients with prior exposure to nevirapine or delavirdine. This evidence includes i) more frequent detection of minor NNRTI-resistant variants in NNRTI-experienced than NNRTI–naïve patients by two different methods (single-genome sequencing and allele-specific PCR), ii) strong phylogenetic relatedness in several subjects between single-genome sequences containing NNRTI resistance mutations detected before the initiation of efavirenz-containing therapy and those present at the time of virologic failure, and iii) a significant association between K103N variants at frequencies >0.5–1% and reduced HIV-1 RNA response to efavirenz-containing regimens. These findings contribute to the growing body of evidence that the current generation of resistance assays used in clinical practice may miss minor but clinically important populations of drug-resistant variants [9, 10].
Although the regimens compared in ACTG 398 included adefovir and single or dual unboosted protease inhibitors, which are no longer relevant for management of HIV-1 infection, the study provided an important opportunity to examine the potential role of minor drug-resistant variants on treatment response to multidrug regimens. We focused on NNRTI-resistant variants because 44% of study subjects had a history of prior exposure to nevirapine or delavirdine, and all subjects were randomized to a new regimen containing efavirenz. The limited potency of the other regimen components - adefovir, abacavir, and single or dual unboosted protease inhibitors - in NRTI- and PI-experienced patients, efavirenz activity is a key driver of treatment response [13]. In addition, it is known that single point mutations in HIV-1 RT (e.g. K103N) can confer high-level cross-resistance to other NNRTI and that such mutations can be detected by several methods, including single-genome sequencing and allele-specific PCR [7]. As such, ACTG 398 provided key opportunities to investigate the influence of major and minor populations of NNRTI-resistant variants on response to efavirenz-containing regimens. The results of these investigations provide strong evidence that both major (detected by standard genotype) and minor populations of NNRTI-resistant variants can compromise virologic response to efavirenz-containing regimens [13, 14].
Allele-specific PCR revealed that prior exposure to delavirdine or nevirapine was more strongly associated with the presence of Y181C variants at frequencies >0.5 – 1.0% in entry samples than with K103N variants at similar frequencies. This difference is likely explained by the preferential selection of Y181C over K103N by nevirapine and to a lesser extent by delavirdine [26, 27, 28]. By contrast, K103N at frequencies >0.5 – 1% was strongly associated with decreased virologic response to efavirenz-containing regimens, whereas Y181C variants were not significantly associated with response. This finding is consistent with greater fitness and higher level resistance of K103N variants to efavirenz than Y181C variants [29–31].
Phylogenetic analyses were performed on single-genome sequences to assess the genetic relatedness between NNRTI-resistant variants at entry and virologic failure. These analyses revealed several important findings. In one of 15 NNRTI-naïve subjects (7N; figure 2A), an entry sequence containing K103N clustered closely with all the mutant sequences at failure containing K103N with M230L or Y181C. This observation suggests that the virus population at failure evolved from a K103N mutant population detected at entry. Although anecdotal, this example is consistent with other reports of major and minor NNRTI-resistant mutants in treatment naïve individuals that arise spontaneously or are acquired through transmission of resistant virus and can compromise response to NNRTI-containing regimens [9, 10, 32]. In two other NNRTI-naïve patients (figure 2B–C), sequences at entry had NNRTI resistance mutations, but completely different NNRTI-resistant mutant sequences were present at virologic failure. These examples illustrate the complexity of assigning significance to a specific variant that is identified using more sensitive methods of detection. In 1 of 8 NNRTI-experienced patients, entry sequences containing NNRTI-resistance mutations clustered closely with mutant sequences at failure, again suggesting that the virus population at failure evolved from the mutant population at entry. Identifying such associations is noteworthy given the small number of sequences (N = 45) examined at entry relative to the total population of virus. Indeed, in the 5 other NNRTI-experienced patients, there was no obvious clustering of mutant sequences from entry and failure time points (patient 8E, figure 2F as an example). The lack of clustering may be due recombination and the large potential impact of few nucleotide substitutions on phylogenetic relationships between closely related sequences from the same individual. In addition, the phylogenetic analyses highlight the limited sampling possible by single genome sequencing and the advantage of allele-specific PCR for detecting specific variants at low frequency in virus populations as well as relating their frequency to treatment response as describe above.
In summary, our findings support the general concept that minor populations of NNRTI-resistant variants that are missed by current clinical testing can persist after NNRTI exposure and contribute to failure of multidrug regimens containing an NNRTI. This concept is particularly relevant for women in developing countries who have received single dose nevirapine for prevention of mother-to-child transmission of HIV-1 and who begin therapy with an NNRTI-containing regimen [33]. Although one report suggests that NNRTI-resistant variants decline to clinically insignificant levels in most women 6 months after single dose nevirapine [34], others have reported long-term persistence of variants and poor response to NNRTI-containing regimens [24, 35, 36]. Additional studies are thus needed to define the clinical significance and optimal management of minor variants that are resistant to NNRTI or other drug classes.
Acknowledgements
We will like to acknowledge the ACTG 398 Protocol Team, the pharmaceutical co-sponsors (Agouron, DuPont-Merck, Gilead Sciences, GlaxoSmithKline, Merck, Roche, and ViroLogic), volunteers, and research staff. We would like to thank Bob Stephen and Beena Neelam of the Advanced Biomedical Computing Center, SAIC-Frederick, National Cancer Institute, National Institutes of Health, Frederick, Maryland, for the help in assembling single genome sequences.
Financial support: The University of Pittsburgh Virology Support Laboratory (204VC009) subcontracted under U01 AI-68636, sponsored by Social and Scientific Systems, Inc; NIH/NIAID/ACTG and ACTU grant AI-68636. National Cancer Institute HIV Drug-resistance Program (25XS119) through SAIC-Frederick, Inc; The Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, under contract N01-CO-12400.
Footnotes
Potential conflicts of interest: JWM is a consultant to Gilead Sciences, Merck, and Chimerix. JWM has received grant support from Merck. SMH is a scientific advisor to Merck and Progenics. EKH, AW, VFB, MK, DN, MW, SP, FV, and JMC report no relevant conflict of interests.
Presented in part: 11th Conference on Retroviruses and Opportunistic Infections. San Francisco, CA: February 2004.
References
- 1.Ho DD, Neumann AV, Perelson AS, Chen W, Leonard JM, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 2.Mansky L, Temin H. Lower in vivo mutation rate of human immunodeficiency Virus Type 1 than that predicted from the fidelity of purified reverse transcriptase. Journal of Virology. 1995;69(8):5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coffin JM. HIV population dynamics in Vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 4.Kearney M, Palmer S, Maldarelli F, et al. Frequent polymorphism at drug resistance sites in HIV-1 Protease and reverse transcriptase. AIDS. 2008;22:497–501. doi: 10.1097/QAD.0b013e3282f29478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmer S, Boltz V, Maldarelli F, et al. Selection and persistence of non-nucleoside reverse transcriptase inhibitor-resistant HIV-1 in patients starting and stopping non-nucleoside therapy. AIDS. 2006;20(5):701–710. doi: 10.1097/01.aids.0000216370.69066.7f. [DOI] [PubMed] [Google Scholar]
- 6.Hirsch MS, Gunthard HF, Schapiro JM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an international AIDS Society-USA Panel. Clin Infect Dis. 2008;47(2):266–285. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 7.Halvas EK, Aldrovandi G, Balfe P, et al. Blinded, multicenter comparison of methods to detect a drug-resistant mutant of Human Immunodeficiency Virus Type 1 at low frequency. Journal of Clinical Microbiology. 2006;44(7):2612–2614. doi: 10.1128/JCM.00449-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer S, Kearney M, Maldarelli F, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. Journal of Clinical Microbiology. 2005;43(1):406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simen BB, Simons JF, Hullsiek KH, et al. Low abundance drug-resistant viral variants in chronically infected HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis. 2009;199:693–701. doi: 10.1086/596736. [DOI] [PubMed] [Google Scholar]
- 10.Johnson JA, Li JF, Wei X, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naïve populations and associate with reduced treatment efficacy. PLoS Med. 2008;5(7):e158. doi: 10.1371/journal.pmed.0050158. PMID: 18666824 [PubMed - in process] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hance AJ, Lemiale V, Izopet J, et al. Changes in Human Immunodeficiency Virus Type 1 populations after treatment interruption in patients failing antiretroviral therapy. Journal of Virology. 2001;75(14):6410–6417. doi: 10.1128/JVI.75.14.6410-6417.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lecossier D, Shulman N, Morand-Joubert L, et al. Detection of minority populations of HIV-1 expressing the K103N resistance mutation in patients failing Nevirapine. Journal of Acquired Immune Deficiency Syndrome. 2005;38:37–42. doi: 10.1097/00126334-200501010-00007. [DOI] [PubMed] [Google Scholar]
- 13.Hammer SM, Vaida F, Bennett KK, et al. For the AIDS Clinical Trials Group 398 Study Team. Dual vs single protease inhibitor therapy following antiretroviral treatment failure: A randomized trial. JAMA. 2002;288:169–180. doi: 10.1001/jama.288.2.169. [DOI] [PubMed] [Google Scholar]
- 14.Mellors JW, Palmer S, Nissley D, et al. For the ACTG 398 Study Team. Low frequency NNRTI-resistant variants contribute to failure of efavirenz-containing regimens. [Abstract #39]. 11th Conference on Retroviruses and Opportunistic Infections; February 2004; San Francisco, CA. [Google Scholar]
- 15.Johnson VA, Brun-Vézinet F, Clotet B, Kuritzkes DR, Pillay D, Schapiro JM, Richman DD. The International AIDS Society-USA (IAS-USA) Drug Resistance Mutations Group-Update of the Drug Resistance Mutations in HIV-1: Fall 2006. Topics in HIV Medicine. 2006;14(3):125–130. [PubMed] [Google Scholar]
- 16.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Research. 1998;8:186–194. [PubMed] [Google Scholar]
- 18.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Research. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 19.Chenna R, Sugawara H, Koike T, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Research. 2003;31(13):3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nickerson DA, Tobe V, Taylor SL. PolyPhred: Automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Research. 1997;25(14):2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu TF, Shafer RW. Web Resources for HIV type 1 Genotypic-Resistance Test Interpretation. Clin Infect Dis. 2006;42(11):1608–1618. doi: 10.1086/503914. Epub 2006 Apr 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia X, Xie Z. DAMBE: Software package for data analysis in molecular biology and evolution. Boston: Kluwer Academic Publishers; 2000. p. 276. [DOI] [PubMed] [Google Scholar]
- 23.Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics. 2001;17(12):1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 24.Palmer S, Boltz V, Martinson N, et al. Persistence of nevirapine-resistant HIV-1 in women after single dose nevirapine for prevention of maternal to fetal transmission. Proceed Natl Acad Sci USA. 2006;103(18):7094–7099. doi: 10.1073/pnas.0602033103. PMID: 16641095 [PubMed - indexed for MEDLINE] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ihaka R, Gentleman RR. A language for data analysis and graphics. Journal of Computational and Graphical Statistics. 1996;5(3):299–314. [Google Scholar]
- 26.Mellors JW, Dutschman G, Guang-Jin IM, Tramontano E, Winkler SR, Cheng Y-C. In vitro selection and molecular characterization of Human Immunodeficiency Virus-1 resistant to non-nucleoside inhibitors of reverse transcriptase. Molecular Pharmacology. 1992;41:446–451. [PubMed] [Google Scholar]
- 27.Halvir DV, Eastman S, Gamst A, Richman DD. Nevirapine-resistant Human Immunodeficiency Virus: Kinetics of replication and estimated prevalence in untreated patients. Journal of Virology. 1996;70(11):7894–7899. doi: 10.1128/jvi.70.11.7894-7899.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Demeter LM, Shafer RW, Meehan PM, et al. Delavirdine susceptibilities and associated reverse transcriptase mutation in human immunodeficiency virus type 1 isolates from patients in a phase I/II trial of delavirdine monotherapy (ACTG 260) Antimicrobial Agents and Chemotherapy. 2000;44(3):794–797. doi: 10.1128/aac.44.3.794-797.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wainberg MA. HIV resistance to Nevirapine and other non-nucleoside reverse transcriptase inhibitors. Journal of Acquired Immune Deficiency Syndromes. 2003;34 Supplement 1:S2–S7. doi: 10.1097/00126334-200309011-00002. [DOI] [PubMed] [Google Scholar]
- 30.Collins JA, Gregory Thompson M, Paintsil E, Ricketts M, Gedzior J, Alexander L. Competitive fitness of nevirapine-resistant human immunodeficiency virus type 1 mutants. Journal of Virology. 2004;78(2):603–611. doi: 10.1128/JVI.78.2.603-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bacheler L, Jeffrey S, Hanna G, et al. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. Journal of Virology. 2001;75(11):4999–5008. doi: 10.1128/JVI.75.11.4999-5008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Little SJ, Holte S, Routy J-P, et al. Antiretroviral-drug resistance among patients recently infected with HIV. The New England Journal of Medicine. 2002;347(6):385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 33.Mellors JW, Chow J. Single-Dose Nevirapine to Prevent Maternal to Child Transmission of HIV-1: Balancing the Benefits and Risks. Clinical Infectious Diseases. 2009;48:473–475. doi: 10.1086/596489. [DOI] [PubMed] [Google Scholar]
- 34.Lockman S, Shapiro RL, Smeaton LM, et al. Response to antiretroviral therapy after a single, peripartum dose of nevirapine. New England Journal of Medicine. 2007;356(2):135–147. doi: 10.1056/NEJMoa062876. PMID: 17215531 [PubMed - indexed for MEDLINE] [DOI] [PubMed] [Google Scholar]
- 35.Lockman S A5208/OCTANE Study Team. Lopinavir/ritonavir +Tenofovir/Emtricitabine is superior to Nevirapine + Tenofovir/emtricitabine for women with prior exposure to single-dose nevirapine: A5208 (“OCTANE”)[Abstract #94LB]. 16th Conference on Retroviruses and Opportunistic Infections; February 2009; San Francisco, CA. [Google Scholar]
- 36.Coovadia A, Hunt G, Abrams EJ, et al. Persistent minority K103N mutations among women exposed to single-dose nevirapine and virologic response to nonnucleoside reverse-transcriptase inhibitor-based therapy. Clinical Infectious Diseases. 2009;48:462–472. doi: 10.1086/596486. [DOI] [PMC free article] [PubMed] [Google Scholar]


