Abstract
3′-Phosphoinositide-dependent protein kinase 1 (PDK-1) phosphorylates and activates members of the AGC protein kinase family and plays an important role in the regulation of cell survival, differentiation, and proliferation. However, how PDK-1 is regulated in cells remains elusive. In this study, we demonstrated that PDK-1 can shuttle between the cytoplasm and nucleus. Treatment of cells with leptomycin B, a nuclear export inhibitor, results in a nuclear accumulation of PDK-1. PDK-1 nuclear localization is increased by insulin, and this process is inhibited by pretreatment of cells with phosphatidylinositol 3-kinase (PI3-kinase) inhibitors. Consistent with the idea that PDK-1 nuclear translocation is regulated by the PI3-kinase signaling pathway, PDK-1 nuclear localization is increased in cells deficient of PTEN (phosphatase and tensin homologue deleted on chromosome 10). Deletion mapping and mutagenesis studies unveiled that presence of a functional nuclear export signal (NES) in mouse PDK-1 located at amino acid residues 382 to 391. Overexpression of constitutively nuclear PDK-1, which retained autophosphorylation at Ser-244 in the activation loop in cells and its kinase activity in vitro, led to increased phosphorylation of the predominantly nuclear PDK-1 substrate p70 S6KβI. However, the ability of constitutively nuclear PDK-1 to induce anchorage-independent growth and to protect against UV-induced apoptosis is greatly diminished compared with the wild-type enzyme. Taken together, these findings suggest that nuclear translocation may be a mechanism to sequestrate PDK-1 from activation of the cytosolic signaling pathways and that this process may play an important role in regulating PDK-1-mediated cell signaling and function.
The 3′-phosphoinositide-dependent protein kinase-1 (PDK-1) is a 64-kDa protein comprised of a Ser/Thr kinase domain near the N terminus and a C-terminal pleckstrin homology (PH) domain (1). This pivotal kinase plays a crucial role in mediating signal transduction downstream of phosphatidylinositol 3-kinase (PI3-kinase) in response to mitogen stimulation. Growth factor stimulation activates PI3-kinase, which converts phosphatidylinositol (4,5)-bisphosphate [PtdIns(4,5)P2] to phosphatidylinositol (3,4,5)-trisphosphate [PtdIns(3,4,5)P3]. This lipid product is bound by the PH domain of PDK-1, resulting in the recruitment of PDK-1 to the plasma membrane. Protein kinase B (PKB), the best characterized substrate of PDK-1, is recruited to the plasma membrane when its own PH domain binds to PtdIns(3,4,5)P3, leading to colocalization of these two proteins and phosphorylation of PKB by PDK-1 on Thr-308 (1), resulting in PKB activation. Downstream substrates of PKB include the antiapoptotic protein BAD and Forkhead (FH) transcription factors. In addition to PKB, PDK-1 has also been reported to phosphorylate many other kinases, including ribosomal p70 protein kinases, that are involved in the regulation of protein translation (2–5).
At the cellular level, PDK-1 regulates key insulin effects such as glycogen synthesis, glucose transporter (GLUT4) membrane translocation, protein synthesis, and cell survival (reviewed in refs. 6 and 7). Forced expression of PDK-1 induced anchorage-independent growth in vitro, a hallmark of cellular transformation, and isografts of PDK-1 transforming cell lines into syngeneic mice induced the formation of poorly differentiated mammary carcinomas, indicating that dysregulation of PDK-1 may be involved in tumorigenesis (8).
Establishing the in vivo role of PDK-1 in animal models has proven difficult because complete loss of PDK-1 results in embryonic lethality in fruit flies and mice (9, 10). Murine PDK-1–/– embryos die at embryonic day 9.5, displaying gross abnormalities such as lack of somites, forebrain, and neural crest-derived tissues (9). Hypomorphic mice with reduced PDK-1 expression are smaller than their wild-type littermates due to a reduction in cell volume, therefore implicating PDK-1's involvement in regulating cell size (9).
Many components of the PI3-kinase pathway such as the insulin receptor, insulin receptor substrates (IRS-1 and -2), PI3-kinase, and PKB are capable of nuclear shuttling (11–14). Synthesis of PtdIns(3,4,5)P3 from PtdIns(4,5)P2 by nuclear PI3-kinase have been reported (15). These observations suggest that an intact PI3-kinase pathway may be reconstituted in the nucleus to regulate nuclear events such as gene transcription. Sequence analysis of Drosophila PDK-1 Dstpk61 revealed the presence of a putative bipartite nuclear localization signal (16). In this study, we demonstrate that PDK-1 is a cytoplasmic-nuclear-shuttling protein. This discovery is further verified by the identification of a functional nuclear export signal (NES) in murine PDK-1 (mPDK-1). Constitutive nuclear localization of PDK-1 does not dampen its kinase activity; however, the ability of constitutively nuclear PDK-1 to promote anchorage-independent growth and protect against UV-induced apoptosis is impaired. These results imply that nuclear localization may be a novel regulatory mechanism of PDK-1 function.
Materials and Methods
Cell Culture. CHO/IR (Chinese hamster ovary cells overexpressing the insulin receptor) cells (17) and murine hepatocyte cells transformed with the SV40 antigen (18) were maintained as described. PTEN+/+, PTEN–/– (19), NMuMg, and HeLa cells were maintained in DMEM supplemented with 10% FCS and 1% penicillin/streptomycin. Transfections of all cell lines except murine hepatocytes (transfected with Lipofectamine 2000) were performed with Lipofectamine (GIBCO/BRL). Comma-1D cells were maintained in DMEM:F12 (10 mM Hepes, pH 7.6) containing 5 μg/ml gentamycin, 10 μg/ml insulin, 5 ng/ml epidermal growth factor, and 2% FCS.
Plasmid Construction. The mammalian expression vector-pCDNA3.1A encoding mPDK-1 tagged with N-terminal eYFP and C-terminal Myc epitope was used to generate C-terminal deletion constructs and site-directed mutagenesis. All site-directed mutagenesis products were verified by restriction mapping and DNA sequencing. The cDNA encoding hGrb10ζ has been described (20).
PDK-1 in Vitro Kinase Assays. CHO/IR cells transiently expressing wild-type Myc-tagged PDK-1, kinase-inactive PDK-1 (K114G), constitutively nuclear PDK-1 (Δ382–391), or kinase-inactive and constitutively nuclear PDK-1 (K114G/Δ382–391) were lysed, and the Myc-tagged proteins were immunoprecipitated by using an anti-Myc monoclonal antibody. In vitro kinase assays were carried out by using a synthetic peptide derived from the activation loop of PKB (KTFCGTPEYLAPEVRR) as described (17).
Phosphorylation of p70 S6KβI in Cells. HeLa cells transiently expressing Myc-tagged mPDK-1 proteins with FLAG-p70 S6KβI-GFP (4) were lysed, and the proteins were immunoprecipitated by using a monoclonal anti-FLAG (Sigma) antibody. p70 S6KβI phosphorylation was detected by blotting with an anti-phospho-(Thr) PDK-1 substrate antibody (Cell Signaling Technology, Beverly, MA). The relative phosphorylation level of p70 S6KβI was calculated by normalizing the phosphorylation level on the phospho-blot by the p70 S6KβI and PDK-1 loading levels [Western blots were quantified by using Scion (Frederick, MD) image]. The basal phosphorylation of p70 S6KβI in the presence of PDK-1 was arbitrarily set to 100%.
Cellular Fractionation. Subconfluent cultures growing on 100-mm plates were harvested in ice-cold PBS and pelleted by centrifugation at 2,500 × g for 1 min at 4°C. Cell pellets were resuspended in cytoplasmic lysis buffer (see Supporting Materials and Methods and Figs. 5 and 6, which are published as supporting information on the PNAS web site) and incubated on ice for 15 min. Lysates were passaged 12 times through a 26-gauge syringe followed by centrifugation at 7,200 × g for 1 min at 4°C. The supernatant was collected as the cytoplasmic fraction whereas the nuclear pellet was lysed in nuclear lysis buffer. The nuclear extracts were incubated at 4°C for 90 min; the last 60 min was performed with gentle rotation. This step was followed by vortexing for 3 min and incubation on ice for 5 min, and this step was repeated twice. The nuclear extracts were clarified by centrifugation at 7,200 × g for 5 min, and the supernatant was collected as nuclear fraction. Nuclear fractions were dialyzed in dialysis buffer at 4°C for 2 h to remove excess salt. Equal amounts of protein (5–10 μg) were loaded onto 10% SDS/PAGE.
Immunofluorescence Studies. Cells growing on glass coverslips were fixed with 4% paraformaldehyde for 20 min and permeabilized with 0.2% Triton X-100/PBS for 3 min. Fixed cells were blocked in 5% normal goat serum for 1 h before incubation with primary antibodies for another hour. Overexpressed PDK-1 was detected by using either monoclonal anti-Myc (generated in our laboratory) or anti-HA (Babco, Richmond, CA). Endogenous PDK-1 was detected by staining with a monoclonal anti-PDK-1 antibody (Santa Cruz Biotechnology). The anti-phospho-PDK-1 (Ser-241) antibody was purchased from Cell Signaling Technology. After a few washes with PBS, cells were incubated with goat anti-mouse Alexa Fluor 488 (Molecular Probes) for another hour. For double staining experiments, the secondary antibodies used were anti-mouse Alexa Fluor 568 and anti-rabbit Alexa Fluor 488. Nuclear staining was performed with 4′,6-diamidino-2-phenylindole (DAPI, Sigma) before mounting on glass slides by using Pro-Long Antifade mounting reagent (Molecular Probes).
Soft Agar Overlay Assay. Comma-1D mouse mammary epithelial cells were transfected with pMSCV-puro/eYFP-mPDK-1 or pMSCV-puro/eYFP-mPDK-1Δ382–391. After selection in 2 μg/ml puromycin for 10 passages, the population of enhanced yellow fluorescent protein (eYFP)-expressing cells was enriched by flow cytometry. The assay was performed as described in ref. 8, with the exception that the cells were not fed each week. Colonies were visually counted at the end of 3 weeks after staining with 1 mg/ml p-iodonitrotetrazolium (Sigma).
DNA Fragmentation Assay. NMuMg mouse epithelial stable cell lines were generated by cotransfection of pSV2-puro with either pcDNA/eYFP, pcDNA/eYFP-mPDK-1, or pcDNA/eYFP-mPDK-1Δ382–391. Twenty-four hours after 2 × 106 cells growing on 100-mm dishes were exposed to 70 J/m2 UV radiation, the floating and adherent cells were collected and lysed in buffer 1(10 mM Tris·HCl,pH7.4/0.15 M Tris·HCl/2 mM MgCl2/1mMDTT/0.5% Nonidet P-40) for 15 min. Crude nuclei were pelleted by centrifugation (10,000 × g, 10 min) and resuspended in ice-cold buffer 2 (10 mM Tris·HCl, pH7.4/0.35 M NaCl/1 mM MgCl2/1mMDTT) for 30 min. The nuclear lysates were extracted once with phenol/chloroform/isoamyl alcohol (15 min at room temperature with gentle rocking). Total DNA was precipitated with ethanol after the addition of 0.1 M MgCl2. Approximately 10 μg of DNA was loaded onto a 1.8% agarose gel.
Results
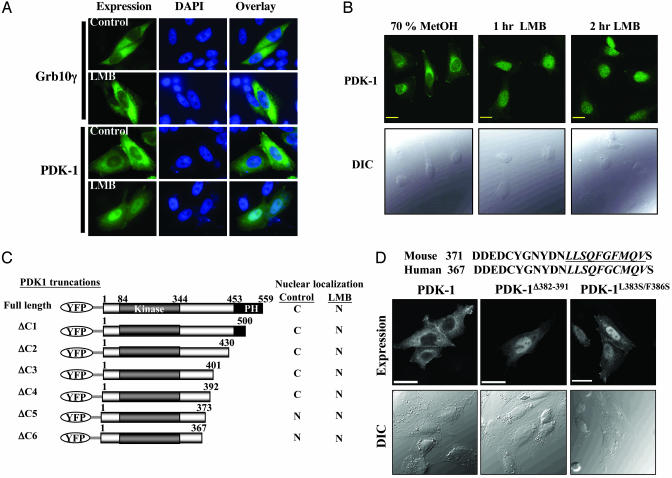
PDK-1 Is a Cytoplasmic-Nuclear-Shuttling Protein and Contains a Nuclear Export Sequence. We found that cell staining for mPDK-1 transiently expressed in several different cell lines revealed faint, and occasionally strong, nuclear staining that is excluded from the nucleoli (data not shown). This finding suggests that PDK-1 may be a cytoplasmic-nuclear-shuttling protein. Consistent with this hypothesis, it has been reported that Drosophila PDK-1 possesses a putative bipartite nuclear localization signal (16) that is also conserved in the mouse and human protein. To examine whether PDK-1 undergoes nuclear translocation, we treated CHO/IR cells expressing Myc-mPDK-1 with leptomycin B (LMB), a specific inhibitor of CRM1-mediated nuclear export (21, 22). LMB treatment induced mPDK-1 nuclear accumulation (Fig. 1A) although it had no effect on the localization of hGrb10ζ (Fig. 1A Top Left and Center), an unrelated cytosolic protein. To exclude the possibility that LMB-induced mPDK-1 nuclear localization is an artifact of overexpression, we investigated the effect of LMB treatment on endogenous PDK-1 localization in HeLa cells by cell staining. HeLa cells were used because these cells stained strongly for endogenous PDK-1 as opposed to other cell lines such as mouse hepatocytes or mouse embryonic fibroblast cells (data not shown). In untreated cells, endogenous PDK-1 can be found in the perinuclear region, cytoplasm and at the plasma membrane (Fig. 1B). LMB treatment resulted in loss of perinuclear staining and nuclear accumulation of endogenous PDK-1 (Fig. 1B). Taken together, the data strongly suggest that PDK-1 is a novel cytoplasmic-nuclear-shuttling protein.
Fig. 1.
PDK-1 is a cytoplasmic nuclear-shuttling protein. (A) Localization of HA-Grb10γ or Myc-mPDK-1 in CHO/IR cells treated with vehicle (70% methanol) or 5 nM LMB for 2.5 h. Fixed cells were stained with an antibody to the tag, followed by staining with anti-mouse Alexa Fluor 488 and 4′,6-diamidino-2-phenylindole (DAPI). Images were captured with an inverted fluorescent microscope (IX70). (B) Confocal images depicting the localization of endogenous PDK-1 in HeLa cells treated with vehicle or 20 nM LMB for the indicated time points. (Upper) Endogenous PDK-1 was visualized by staining with a monoclonal anti-PDK-1 antibody. (Lower) Confocal differential image contrast. (Bars = 20 μm.) (C) Schematic representation of the C-terminal truncations of mPDK-1 and summary of their localization in CHO/IR cells treated with either vehicle or 5 nM LMB. (D Top) Amino acid alignment of mouse and human PDK-1. (Middle and Bottom) Confocal images showing the localization of Myc-tagged mPDK-1, mPDK-1Δ382–391, or mPDK-1L383S/F386S in CHO/IR cells. DIC, Differential image contrast.
Because LMB acts by inhibiting CRM1 binding to NES-containing proteins (21–23), the observation that LMB induces PDK-1 nuclear retention raises the possibility that PDK-1 contains a bona fide NES. We used deletion mapping analysis using mPDK-1 fused to eYFP to identify the putative NES. PDK-1 was fused to eYFP to prevent passive diffusion of the smaller truncated PDK-1 proteins into the nucleus. C-terminal truncations of eYFP-mPDK-1 were created as shown in Fig. 1C, and the expression of these proteins was verified by Western blotting (see Supporting Materials and Methods and Figs. 5 and 6). The localization of these proteins was tested in CHO/IR cells in the presence or absence of LMB. mPDK-1(ΔC5) but not mPDK-1(ΔC4) localized to the nucleus in the absence of LMB (Figs. 1C, 5, and 6 and Supporting Materials and Methods), indicating that the region between amino acids 373 and 392 contains a putative NES. Closer examination of this sequence revealed the presence of a highly conserved stretch of hydrophobicrich sequence that could function as a NES (Fig. 1D Top). Indeed, deletion of amino acids 382–391 (Fig. 1D Top, underlined) causes nuclear retention of Myc-tagged mPDK-1 (Fig. 1D). We further confirmed that these ten amino acids contain a functional NES by examining the localization of a mutant in which two hydrophobic residues were mutated to serines (mPDK-1L383S/F386S). As demonstrated in Fig. 1D, Myc-mPDK-1L383S/F386S is retained in the nucleus of CHO/IR in the absence of LMB. The predominant nuclear localization of mPDK-1Δ382–391 and L383S/F386S has been verified in other cell lines (murine hepatocytes, C2C12 myoblasts, and NMuMG mammary epithelial cells), suggesting that it is not a cell line-specific event (data not shown).
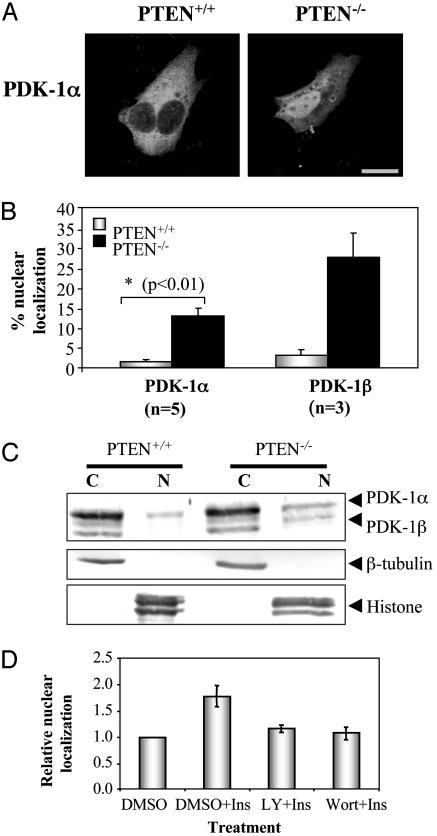
PDK-1 Nuclear Translocation Is Regulated by the PI3-Kinase Pathway. Since its discovery as the kinase that phosphorylates PKB (1), PDK-1 has emerged as a central player in the PI3-kinase-signaling pathway. Therefore, we investigated whether PDK-1 nuclear translocation depends on activation of this pathway. Our system of choice is a cell line deficient in PTEN (phosphatase and tensin homologue deleted on chromosome 10) (19), a tumor suppressor protein found deleted in a wide variety of human cancers (24). PTEN functions as a negative regulator of the PI3-kinase pathway by dephosphorylating PtdIns(3,4,5)P3; therefore, loss of this protein results in activation of this pathway (25, 26). Cell staining of mouse embryonic fibroblast cells lacking PTEN (PTEN–/– cells) and wild-type cells (PTEN+/+ cells) revealed a higher incidence of Myc-tagged mPDK-1 in the nucleus of PTEN-deficient cells (Fig. 2 A and B). We also examined the nuclear localization of mPDK-1β, a splice variant of PDK-1 that lacks 27 aa at its N terminus (27). The nuclear localization frequency of this PDK-1 isoform is even higher compared with that of mPDK-1α (Fig. 2B). How the loss of the first 27 aa resulted in increased mPDK-1β nuclear localization remains to be established.
Fig. 2.
PDK-1 nuclear localization depends on the PI3-kinase pathway. (A) Confocal images showing localization of Myc-mPDK-1α in PTEN+/+ and PTEN–/– cells. (Bar = 20 μm.) (B) Graphical representation of PDK-1α and β localization in PTEN+/+ and PTEN–/– cells; bars represent mean ± SEM from at least three independent experiments. P value was determined by using Student's t test. (C) Subconfluent PTEN+/+ and Pten–/– cells were lysed and fractionated into cytoplasmic (C) and nuclear (N) fractions. (Top) Anti-PDK-1 blot. (Middle) Anti-β-tubulin blot. (Bottom) Anti-histone blot. (D) CHO/IR transiently expressing Myc-mPDK-1 were serum-starved in the presence/absence of DMSO, 50 nM wortmannin (Wort), or 50 μM LY 294002 (LY) for 1 h before stimulation with 20 nM insulin (Ins) for 20 min. Localization of PDK-1 was visualized by staining with an antibody to the tag. Data are mean ± SEM from three independent experiments.
To further confirm that PDK-1 nuclear localization is increased in PTEN–/– cells, lysates from PTEN+/+ and PTEN–/– cells were fractionated into cytoplasmic and nuclear fractions, and the localization of endogenous mPDK-1 was detected by using a rabbit polyclonal anti-PDK-1 antibody (27). As shown in Fig. 2C, elevated levels of mPDK-1α and mPDK-1β were observed in the nuclear fraction of PTEN-deficient cells. Overexpression results (Fig. 2B) showed a more impressive increase in PDK-1 nuclear localization in PTEN–/– cells compared with endogenous results (Fig. 2C), which may indicate that overexpression increases nuclear localization of this shuttling protein. However, both overexpression and endogenous results indicate that nuclear localization of PDK-1 is increased in PTEN–/– cells, thereby supporting our hypothesis that nuclear localization of PDK-1 is regulated by the PI3-kinase pathway.
Because PDK-1 is a central player in the insulin signal transduction pathway, we examined whether insulin stimulation can induce PDK-1 nuclear accumulation, and whether this translocation depends on the PI3-kinase pathway. As shown in Fig. 2D, insulin treatment of CHO/IR cells produced a modest but reproducible increase in the nuclear accumulation of exogenous mPDK-1. This insulin-induced PDK-1 nuclear retention is inhibited by pretreatment with either wortmannin or LY 294002, indicating that this process is PI3-kinase dependent.
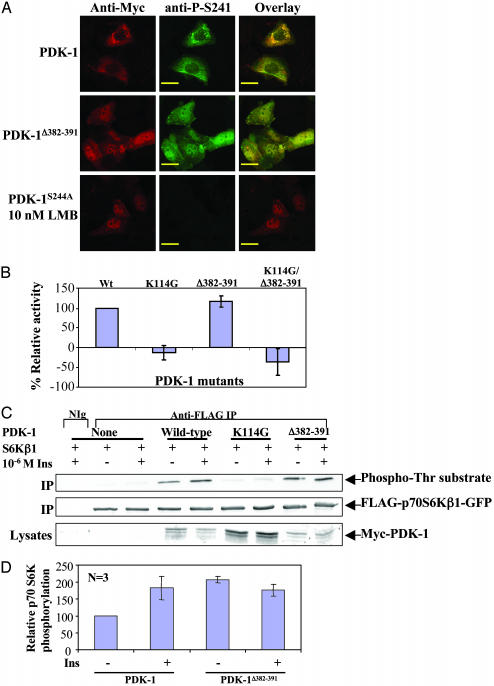
Nuclear Localization of PDK-1 Does Not Inhibit Its Kinase Activity. The negative regulation of PDK-1 has not been characterized. Thus, PDK-1 nuclear translocation may be a mechanism for down-regulation of PDK-1 function, either by sequestration in the nucleus and/or dephosphorylation resulting in down-regulation of its kinase activity. Such a scenario has been reported for p42/p44 MAP kinases, where chronic stimulation resulted in nuclear sequestration, dephosphorylation, and loss of kinase activity (28). We therefore tested whether phosphorylation at S244 is retained in constitutively nuclear mPDK-1 because phosphorylation at this serine residue has been demonstrated to be essential for its kinase activity. Murine hepatocytes transiently expressing Myc-mPDK-1, Myc-mPDK-1S244A, and Myc-mPDK-1Δ382–391 were treated with vehicle or LMB for 2.5 h, followed by fixation and staining with anti-Myc and anti-phospho-hPDK-1 (S241) antibodies (S241 in hPDK-1 corresponds to S244 of mPDK-1). The phospho-S244 staining is specific because Myc-mPDK-1S244A was not recognized by this antibody (Fig. 3A). Phosphorylation of endogenous mPDK-1 at S244 was not detected in these hepatocytes, possibly due to low expression of endogenous PDK-1 and/or low S244 autophosphorylation levels. No loss of phosphorylation at S244 was observed in constitutively nuclear PDK-1 (Myc-mPDK-1Δ382–391) (Fig. 3A Middle). Similar results were obtained in CHO/IR cells (data not shown).
Fig. 3.
Nuclear PDK-1 is phosphorylated in its activation loop and is kinaseactive. (A) Confocal images showing localization of Myc-mPDK-1, mPDK-1Δ382–391, or mPDK-1S244A in murine hepatocytes treated with vehicle or 10 nM LMB for 2.5 h. (Bars = 20 μm.) (B) In vitro kinase activities of immunopurified Myc-tagged mPDK-1, mPDK-1K114G, mPDK-1Δ382–391, or mPDK-1K114G/Δ382–391 against a PKB-based peptide substrate. (C) Phosphorylation of immunopurified FLAG-p70 S6KβI by PDK-1 proteins from HeLa cells treated with 10–6 M insulin for 10 mins. p70 S6KβI phosphorylation (Top), p70 S6KβI expression (Middle), and PDK-1 expression (Bottom) levels were detected by blotting with anti-phospho-Thr PDK-1 substrate, anti-FLAG, and anti-Myc antibodies, respectively. (D) Graphical representation of phosphorylation of p70 S6KβI by PDK-1. Bars represent the mean ± SEM from three independent experiments.
To directly measure whether nuclear PDK-1 is kinase-active, we performed in vitro kinase assays of immunopurified Myc-tagged mPDK-1, Myc-mPDK-1K114G, Myc-mPDK-1Δ382–391, or Myc-mPDK-1K114G/Δ382–391 from CHO/IR cells by using a PKB-based peptide as substrate. As demonstrated in Fig. 3B, no significant difference was observed between the kinase activities of wild type and the constitutively nuclear form of mPDK-1.
We investigated whether constitutively nuclear PDK-1 can phosphorylate a predominantly nuclear substrate by examining the ability of mPDK-1Δ382–391 to phosphorylate p70 S6KβI (4). HeLa cells transiently expressing p70 S6KβI alone or with mPDK-1 proteins were stimulated with insulin and lysed, and the phosphorylation of the immunoprecipitated p70 S6KβI was detected by blotting with a phospho-Thr PDK-1 substrate antibody (Fig. 3C). An approximately 2-fold increase in the basal phosphorylation level of p70 S6KβI was observed with mPDK-1Δ382–391 compared with wild-type mPDK-1 (Fig. 3 C and D). Insulin treatment did not further increase the phosphorylation of p70 S6KβI by mPDK-1Δ382–391, presumably because the basal phosphorylation level was already high. These results suggest that increased nuclear colocalization of PDK-1 with p70 S6KβI resulted in elevated basal phosphorylation of the latter, further supporting the hypothesis that nuclear PDK-1 retains its kinase activity.
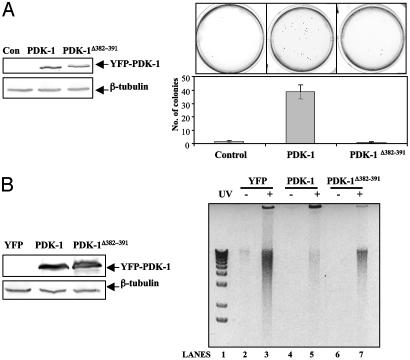
Nuclear Localization of PDK-1 Negatively Regulates Its Function. Overexpression of hPDK-1 in the Comma-1D mouse mammary epithelial cell lines was reported to promote anchorage-independent growth, a hallmark of cellular transformation (8). We generated Comma-1D cell lines stably expressing eYFP-mPDK-1 or eYFP-mPDK-1Δ382–391 (Fig. 4A Left) to investigate the efficiency of nuclear PDK-1 at inducing cellular transformation. Consistent with a previous report by using human PDK-1 (8), we found that overexpression of eYFP-mPDK-1 also induced anchorage-independent growth (Fig. 4A). In contrast, the Comma-1D cell line expressing eYFP-mPDK-1Δ382–391 did not produce any colonies above background, indicating that nuclear localization disrupted the transformation ability of PDK-1.
Fig. 4.
Effect of nuclear PDK-1 localization on cellular transformation and UV-induced apoptosis. (A Left) Western blot of whole cell lysates from Comma-1D cell lines showing expression levels of eYFP-mPDK-1 and eYFP-mPDK-+Δ382–391. (Upper Left) Anti-PDK-1 blot. (Lower Left) β-Tubulin blot (loading level control). (Right) Pictures of colony formation on soft agar (Upper Right) and the graphical representation of colony formation assay (Lower Right). Bars represent the mean ± SEM from three independent experiments. (B Left) Western blot of whole cell lysates from NMuMg cell lines expressing eYFP, eYFP-mPDK-1, and eYFP-mPDK-1Δ382–391 showing expression of mPDK-1. (Upper Left) Anti-PDK-1 blot. (Lower Left) β-Tubulin blot. (Right) Inverted contrast image of an ethidium bromide-stained gel showing DNA fragmentation of nuclear DNA purified from NMuMg cells exposed to 70 J/m2 UV light. Data are representative of three independent experiments.
PDK-1 promotes cell survival by phosphorylating PKB (10, 29–31), whose antiapoptotic roles are well characterized (32). We sought to verify that nuclear localization negatively regulates PDK-1 function by investigating the ability of mPDK-1 and mPDK-+Δ382–391 to protect against UV-induced cell death. A different cell line (NMuMg mouse mammary epithelial cells) was used for this experiment to avoid cell line-specific responses. After exposure to 70 J/m2 UV, NMuMg cell lines stably expressing eYFP, eYFP-mPDK-1, or eYFP-mPDK-1Δ382–391 (Fig. 4B) were harvested, and the DNA fragmentation of nuclear DNA (indicator of apoptosis) was examined. Consistent with the established role of PDK-1 in promoting cell survival (6), NMuMg cells stably expressing eYFP-PDK-1 survived better compared with cells expressing YFP (Fig. 4B Right, lane 5 vs. lane 3). Although overexpression of eYFP-mPDK-1Δ382–391-protected cells against UV-induced apoptosis compared with control cells expressing eYFP alone (Fig. 4B Right, lane 7 vs. lane 3), it was not as efficient as PDK-1 at protecting cells against nuclear DNA fragmentation (lane 7 vs. lane 5). Taken together, the results indicated that nuclear localization of PDK-1 may be a mechanism to negatively regulate PDK-1 function.
Discussion
We demonstrate that PDK-1, previously thought to shuttle only between the cytoplasm and plasma membrane, also undergoes cytoplasmic-nuclear shuttling. LMB treatment induced nuclear accumulation of overexpressed and endogenous PDK-1 in different cell lines, indicating that it is not a cell line-specific event (Fig. 1 and data not shown). We further confirmed this result by identifying and mutating two residues within the NES of PDK-1, which resulted in constitutive PDK-1 nuclear localization. These results suggest that PDK-1 cytoplasmic localization may be maintained by the presence of a strong NES.
Although PDK-1 has been shown to be a cytoplasmic-nuclear-shuttling protein, many questions remains unanswered. For example, the physiological stimulus that induces PDK-1 nuclear translocation remains to be clarified. We obtained a modest but reproducible increase of exogenous mPDK-1 nuclear localization (1.5- to 2-fold) in CHO/IR (Fig. 2D) and hepatocytes treated with insulin (data not shown). The insulin-induced PDK-1 nuclear retention observed in CHO/IR cells can be inhibited by pretreatment with the PI3-kinase inhibitors wortmannin and LY 294002 (Fig. 2D). These data suggest that PDK-1 nuclear translocation is regulated by the PI3-kinase pathway. However, the physiological significance of insulin-stimulated PDK-1 nuclear translocation remains unaddressed.
A major question that our discovery raises pertains to the physiological function of nuclear PDK-1. Because constitutively nuclear PDK-1 retains its kinase activity (Fig. 3B), localization in the nucleus does not seem to down-regulate the activation state of this enzyme. This hypothesis is supported by the observation that PDK-1Δ382–391 seems to be more efficient at phosphorylating its predominantly nuclear substrate, p70 S6KβI (Fig. 3 D and E), than the wild-type PDK-1. However, we cannot rule out the possibility that nuclear localization acts to sequester PDK-1 away from its known substrates in the cytoplasm and at the plasma membrane. The observation that PDK-1Δ382–391 is less efficient at promoting anchorage-independent cell growth and protecting against UV-induced apoptosis (Fig. 4) lends credibility to this hypothesis. Taken altogether, our data imply that nuclear localization of PDK-1 may be a form of negative regulation of its function, rather than its kinase activity. It is tempting to speculate that prolonged growth factor stimulation may trigger the nuclear translocation of a fraction of PDK-1, thereby dampening the activation of the PI3-kinase pathway.
Interestingly, both PDK-1-dependent cellular transformation and cell survival have been shown to be dependent on PKB activation although, in the case of cellular transformation, the activation of PKCα is also required (8). Although nuclear localization of PDK-1 may sequester PDK-1 from endogenous PKB, thereby leading to inhibition of Akt-mediating signaling, it may also position PDK-1 next to a nuclear substrate and consequently activate another signaling pathway. The observation that exogenous nuclear expression of PDK-1 resulted in increased basal phosphorylation of p70 S6KβI would support this idea. More work is needed to determine whether PDK-1 has a nuclear substrate and the consequences, if any, of phosphorylation of this putative substrate.
Supplementary Material
Acknowledgments
We thank Dr. Domenico Accili, Dr. LuZhe Sun (University of Texas Health Science Center, San Antonio), Dr. Daniel Medina (M. D. Anderson Cancer Center, University of Texas), and Dr. Hong Wu for their generous gifts of murine hepatocyte, NMuMg, Comma-1D, and PTEN cell lines, respectively. We thank Kazuyoshi Yonegawa for providing the p70 S6KβI construct, Derong Hu for excellent technical assistance, and Dr. Feng Liu for insightful comments and reagents. This work was supported in part by National Institutes of Health Grant DK56166 and by an award from the Howard Hughes Medical Institute Research Resource Program.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PDK-1, 3′-phosphoinositide-dependent protein kinase 1; mPDK-1, murine PDK-1; CHO/IR, Chinese hamster ovary cells overexpressing the insulin receptor; LMB, leptomycin B; PI3-kinase, phosphatidylinositol 3-kinase; NES, nuclear export signal; eYFP, enhanced yellow fluorescent protein; PTEN, phosphatase and tensin homologue deleted on chromosome 10; PtdIns(4,5)P2, phosphatidylinositol (4,5)-bisphosphate; PKB, protein kinase B.
References
- 1.Alessi, D. R., James, S. R., Downes, C. P., Holmes, A. B., Gaffney, P. R., Reese, C. B. & Cohen, P. (1997) Curr. Biol. 7, 261–269. [DOI] [PubMed] [Google Scholar]
- 2.Alessi, D. R., Kozlowski, M. T., Weng, Q. P., Morrice, N. & Avruch, J. (1998) Curr. Biol. 8, 69–81. [DOI] [PubMed] [Google Scholar]
- 3.Pullen, N., Dennis, P. B., Andjelkovic, M., Dufner, A., Kozma, S. C., Hemmings, B. A. & Thomas, G. (1998) Science 279, 707–710. [DOI] [PubMed] [Google Scholar]
- 4.Minami, T., Hara, K., Oshiro, N., Ueoku, S., Yoshino, K., Tokunaga, C., Shirai, Y., Saito, N., Gout, I. & Yonezawa, K. (2001) Genes Cells 6, 1003–1015. [DOI] [PubMed] [Google Scholar]
- 5.Lee-Fruman, K. K., Kuo, C. J., Lippincott, J., Terada, N. & Blenis, J. (1999) Oncogene 18, 5108–5114. [DOI] [PubMed] [Google Scholar]
- 6.Vanhaesebroeck, B. & Alessi, D. R. (2000) Biochem. J. 346, 561–576. [PMC free article] [PubMed] [Google Scholar]
- 7.Wick, K. L. & Liu, F. (2001) Curr. Drug Targets Immune Endocr. Metabol. Disord. 1, 209–221. [DOI] [PubMed] [Google Scholar]
- 8.Zeng, X., Xu, H. & Glazer, R. I. (2002) Cancer Res. 62, 3538–3543. [PubMed] [Google Scholar]
- 9.Lawlor, M. A., Mora, A., Ashby, P. R., Williams, M. R., Murray-Tait, V., Malone, L., Prescott, A. R., Lucocq, J. M. & Alessi, D. R. (2002) EMBO J. 21, 3728–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho, K. S., Lee, J. H., Kim, S., Kim, D., Koh, H., Lee, J., Kim, C., Kim, J. & Chung, J. (2001) Proc. Natl. Acad. Sci. USA 98, 6144–6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu, A., Sciacca, L. & Baserga, R. (2003) J. Cell. Physiol. 195, 453–460. [DOI] [PubMed] [Google Scholar]
- 12.Kim, S. J. (1998) Biochem. Mol. Biol. Int. 46, 187–196. [DOI] [PubMed] [Google Scholar]
- 13.Podlecki, D. A., Smith, R. M., Kao, M., Tsai, P., Huecksteadt, T., Brandenburg, D., Lasher, R. S., Jarett, L. & Olefsky, J. M. (1987) J. Biol. Chem. 262, 3362–3368. [PubMed] [Google Scholar]
- 14.Meier, R., Alessi, D. R., Cron, P., Andjelkovic, M. & Hemmings, B. A. (1997) J. Biol. Chem. 272, 30491–30497. [DOI] [PubMed] [Google Scholar]
- 15.Lu, P. J., Hsu, A. L., Wang, D. S., Yan, H. Y., Yin, H. L. & Chen, C. S. (1998) Biochemistry 37, 5738–5745. [DOI] [PubMed] [Google Scholar]
- 16.Clyde, D. & Bownes, M. (2000) J. Endocrinol. 167, 391–401. [DOI] [PubMed] [Google Scholar]
- 17.Dong, L. Q., Zhang, R. B., Langlais, P., He, H., Clark, M., Zhu, L. & Liu, F. (1999) J. Biol. Chem. 274, 8117–8122. [DOI] [PubMed] [Google Scholar]
- 18.Rother, K. I., Imai, Y., Caruso, M., Beguinot, F., Formisano, P. & Accili, D. (1998) J. Biol. Chem. 273, 17491–17497. [DOI] [PubMed] [Google Scholar]
- 19.Liliental, J., Moon, S. Y., Lesche, R., Mamillapalli, R., Li, D., Zheng, Y., Sun, H. & Wu, H. (2000) Curr. Biol. 10, 401–404. [DOI] [PubMed] [Google Scholar]
- 20.Dong, L. Q., Du, H., Porter, S. G., Kolakowski, L. F., Jr., Lee, A. V., Mandarino, L. J., Fan, J., Yee, D. & Liu, F. (1997) J. Biol. Chem. 272, 29104–29112. [DOI] [PubMed] [Google Scholar]
- 21.Fornerod, M., Ohno, M., Yoshida, M. & Mattaj, I. W. (1997) Cell 90, 1051–1060. [DOI] [PubMed] [Google Scholar]
- 22.Fukuda, M., Asano, S., Nakamura, T., Adachi, M., Yoshida, M., Yanagida, M. & Nishida, E. (1997) Nature 390, 308–311. [DOI] [PubMed] [Google Scholar]
- 23.Kudo, N., Wolff, B., Sekimoto, T., Schreiner, E. P., Yoneda, Y., Yanagida, M., Horinouchi, S. & Yoshida, M. (1998) Exp. Cell Res. 242, 540–547. [DOI] [PubMed] [Google Scholar]
- 24.Li, J., Yen, C., Liaw, D., Podsypanina, K., Bose, S., Wang, S. I., Puc, J., Miliaresis, C., Rodgers, L., McCombie, R., et al. (1997) Science 275, 1943–1947. [DOI] [PubMed] [Google Scholar]
- 25.Wu, X., Senechal, K., Neshat, M. S., Whang, Y. E. & Sawyers, C. L. (1998) Proc. Natl. Acad. Sci. USA 95, 15587–15591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stambolic, V., Suzuki, A., de la Pompa, J. L., Brothers, G. M., Mirtsos, C., Sasaki, T., Ruland, J., Penninger, J. M., Siderovski, D. P. & Mak, T. W. (1998) Cell 95, 29–39. [DOI] [PubMed] [Google Scholar]
- 27.Dong, L. Q., Ramos, F. J., Wick, M. J., Ann Lim, M., Guo, Z., Strong, R., Richardson, A. & Liu, F. (2002) Biochem. Biophys. Res. Commun. 294, 136–144. [DOI] [PubMed] [Google Scholar]
- 28.Volmat, V., Camps, M., Arkinstall, S., Pouyssegur, J. & Lenormand, P. (2001) J. Cell Sci. 114, 3433–3443. [DOI] [PubMed] [Google Scholar]
- 29.Arico, S., Pattingre, S., Bauvy, C., Gane, P., Barbat, A., Codogno, P. & Ogier-Denis, E. (2002) J. Biol. Chem. 277, 27613–27621. [DOI] [PubMed] [Google Scholar]
- 30.Flynn, P., Wongdagger, M., Zavar, M., Dean, N. M. & Stokoe, D. (2000) Curr. Biol. 10, 1439–1442. [DOI] [PubMed] [Google Scholar]
- 31.Sato, S., Fujita, N. & Tsuruo, T. (2002) Oncogene 21, 1727–1738. [DOI] [PubMed] [Google Scholar]
- 32.Hill, M. M. & Hemmings, B. A. (2002) Pharmacol. Ther. 93, 243–251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.