Abstract
The composition of the lower genital tract microbiota in women is believed to affect the risk of sexually acquiring HIV. Since macaque genital microbiota could similarly impact vaginal infection with SIV we identified microbiota in 11 rhesus macaques using multitag pyrosequencing of the 16S rRNA gene. The microbiota was polymicrobial with a median of nine distinct bacterial taxa per macaque (range 3–16 taxa, each constituting 1% or more of the sequences). Taxa frequently found included Peptoniphilus, Sneathia, Porphyromonas, Mobiluncus, Atopobacter, Dialister, Thioreductor, Prevotella, and Streptococcus, many of which are also frequently found in women with bacterial vaginosis. Lactobacillus sequences (mostly L. johnsonii) were found in only four macaques but were not predominant in any (median of 0% of sequences, range 0–39%). All macaques were resampled 6 months after the first time point to determine the stability of the microbiota. The microbiota remained polymicrobial with a median of 10 taxa (range 6–18). Microbial patterns remained similar for six of the macaques, changed substantially in two, and had a mixed pattern in three. Significant sialidase enzyme activity, a marker of bacteria vaginosis in women, was detected in genital fluid from 9/11 and 8/11 macaques from the first and second time points, respectively. These results show that the macaque lower genital microbiota resembled a bacteria vaginosis-type microbiota in women and suggest that the microbiota of macaques in captivity promote rather than protect against vaginal infection with SIV. These results also suggest macaques could be used as an animal model to study some aspects of bacterial vaginosis.
Introduction
The microbiota of the lower genital tract in many women consists predominantly of Lactobacillus species. Common lactobacilli in women are L. crispatus, L. jensenii, L. iners, and L. gasseri.1 Epidemiologic studies show that women who have a genital microbiota that is predominantly lactobacilli have a decreased risk of acquiring several sexually transmitted diseases (STDs), including HIV and HSV-2, when compared to women with bacterial vaginosis.2–4 Bacterial vaginosis is a condition in which the predominant genital microbiota is not lactobacilli, but is instead a polymicrobial mixture of bacteria including Gardnerella vaginalis, Mobiluncus species, Prevotella species, and other anaerobic bacteria.5
Vaginal infection of macaques with SIV or SHIV is used frequently as a nonhuman primate model for HIV infection of women. Vaginal infection of macaques with SIV or SHIV is also used in vaccine protection studies and in studies of microbicide safety and efficacy. It is possible that the genital microbiota of macaques could have an impact on these types of studies. Although the types of bacteria that make up macaque genital microbiota have been investigated, all previous studies have used culture to identify bacteria. Culture studies of genital bacteria in women have been recently recognized to have the drawback of missing organisms that are uncultivable or difficult to cultivate.6,7 Thus, it has become apparent, using molecular techniques that identify genital bacteria by their 16S rRNA sequence, that many of the types of bacteria that are present in the genital tract of women are not cultivatable, that these noncultivatable bacteria can make up a substantial proportion the microbiota in some women, and that some of these bacteria are associated with pathogenesis.6–8 These studies in humans suggest the importance of extending those same types of analyses to identify genital microbiota in macaques.
Whereas lactobacilli are recognized as important for genital health in women, previous studies of macaque genital microbiota show conflicting data on the relative frequency and predominance of lactobacilli. For example, in two studies of microbicides, Patton et al.9,10 reported that the baseline vaginal pH in pigtailed macaques was approximately 6–7, suggesting that lactobacilli are not dominant in the microbiota of these macaques. In contrast, several groups cultured lactobacilli from both rhesus and pigtailed macaque vaginal samples and reported little or no change in total lactobacilli or in H2O2-producing lactobacilli during treatment with microbicides.9–13 On the other hand, hydrogen peroxide-producing lactobacilli were cultured in only half of pigtailed macaques at study baseline, but the levels of these lactobacilli were one to two logs lower than several other types of cultivable bacteria.9 Doyle et al.14 assessed the microbiota in rhesus macaques by culture and reported that Streptococcus, Staphylococcus, Mobiluncus, Peptostreptococcus, and a Corynebacterium-like bacterium were all isolated frequently (41–65% of macaques). However, colony counts were not provided in that study and isolation of lactobacilli was not mentioned.
The goal of this study was to determine the composition of genital microbiota in rhesus macaques using multitag pyrosequencing of the 16S rRNA gene, which directly identifies organisms without culture. The stability of the macaque genital microbiota was also assessed by pyrosequencing of samples collected 6 months later. Sialidase activity, a marker of bacterial vaginosis in humans, was also measured in the genital fluid of the macaques.
Materials and Methods
Sample collection
Genital tract samples were collected from macaques by cervicovaginal lavage (CVL) that was performed by irrigation of the vaginal vault with 4 ml of sterile nonbacteriostatic saline, followed by aspiration from the posterior fornix. CVL was held on ice until processing, which occurred within 6 h of collection. CVL was gently vortexed to evenly distribute cells before storage at −70°C.
Animals
All animals used for the microbiota studies were rhesus macaques (Macaca mulatta) of Indian origin housed at the Tulane National Primate Research Center (TNPRC). Macaques were purchased and imported from the Caribbean Primate Research Center 7 months prior to first sample collection. Prior to importation, the animals were born and raised in group breeding colonies, so their parentage and prior sexual activity were unknown. Following arrival at the TNPRC, macaques were given complete physical examinations, blood work (CBC, chemistries), and routine stool examinations for parasites and all were deemed to be normal, healthy, cycling mature females 4–14 years of age at sample collection. The 10 macaques whose vaginal pH was monitored were normal, cycling rhesus macaques of Chinese (n = 6) or Indian (n = 4) origin between 8 and 13 years of age who were born and raised at the TNPRC. All animals were housed in accordance with the American Association for Accreditation of Laboratory Animal Care, and the Guide for Care and Use of Laboratory Animals, National Academy Press. All animal studies were reviewed and approved by the Tulane University Animal Care and Use Committee.
DNA isolation
DNA was isolated as described elsewhere.11 Briefly, bacteria were pelleted by centrifugation and lysed with buffers containing lysosyme, SDS, and proteinase K. DNA was purified using phenol/chloroform and precipitated by incubation with absolute ethanol, with glycogen added as a carrier.
Multitag pyrosequencing and phylogenetic analysis
To perform multiplexed pyrosequencing, a set of 12 primer sets (tags) was generated that each contained either the 27F or 355R 16S primers that were tagged on the 5 prime end with a 4 base “barcode.” PCR was performed on individual macaque samples using the unique barcoded primers as follows: extracted DNA (10 ng) was amplified by PCR using primers L27F (5′-AGAGTTTGATCCTGGCTCA G-3′) and 355R′ (5′-GCTGCCTCCCGTAGGAGT-3′), which are broad range primers for bacteria.15 The reactions were performed using 20 μl (final volume) mixtures containing 1 × PCR buffer, 0.01% bovine serum albumin, 2.5 mM MgCl2, 0.5 mM of each deoxynucleoside triphosphate, 0.2 μM of each primer, and 2 U of TaqGold DNA polymerase (Applied Biosystems, Foster City, CA). The initial denaturation step was 95°C for 11 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 48°C for 30 s, and extension at 72°C for 2 min, followed by a final extension step that consisted of 72°C for 35 min. Pyrosequencing of the amplified, tagged DNA was performed by 454 Life Sciences (Branford, CT) with 10 to 12 separately tagged samples included in a single slot. Custom PERL scripts were used to sort the sequences based on the barcodes, search the rRNA database, identify the reads, and exclude sequences that had multiple tags, which indicated chimeric sequences. The Bayesian Classifier provided by the Ribosomal Database II Project (RDP 10) was used to identify bacteria.16 For each CVL sample at the first collection time point, duplicates of PCR and pyrosequencing were performed. Because only minor differences were observed between the duplicates (see Fig. 1), data from only one of the duplicates are shown in the tables and figures.
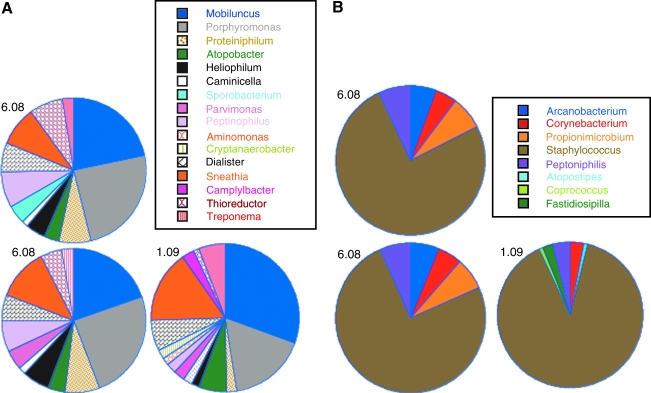
FIG. 1.
Frequencies of Bacterial Taxa Identified in 11 Macaques
Quantification of total bacteria numbers
Bacteria were quantified in DNA isolated from CVL using real time PCR. Primers F340 (ACTCCTACGGGAGGCAGCAGT) and R514 (ATTACCGCGGCTGCTGGC),17 which bind to an area of the 16S rRNA gene conserved in all bacteria, were used along with Syber Green (Applied Biosystems) in an ABI 7900HT Fast Real Time PCR System (Applied Biosystems). DNA from Gardnerella vaginalis (American Type Culture Collection, Manassas, VA) was used to produce a standard curve.
Sialidase activity
The sialidase assay was adapted from a previous study.18 Enzyme reactions were run at a total volume of 150 μl in wells of a 96-well plate. Equal volumes (50 μl) of macaque CVL (cleared by centrifugation), sodium acetate buffer (0.1 M, pH 4.6), and 2′-(4-methylumbellifery)-α-d-N-acetylneuraminic acid (0.4 mM, Sigma Chemical Co., St. Louis, MO) were mixed and incubated at 37°C. Fluorescence was read every 5 min at 460 nm with an excitation of 360 nm and compared with a standard curve made with 4-methylumbelliferone (Sigma). The protein concentration in macaque CVL samples was measured using the BCA protein assay (Thermo Scientific, Rockford, IL) and final sialidase activity was adjusted to activity per microgram of protein.
Results
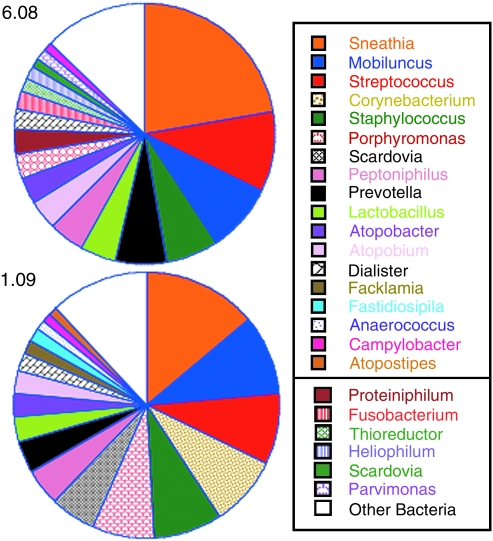
Multitag pyrosequencing of the 16S rRNA gene was performed on DNA isolated from lower genital tract samples collected from 11 healthy rhesus macaques. In all, 9672 16S sequences were identified from the 11 macaques with an average length of 244 base pairs. Analysis was focused on taxa found at ≥1% of the total community in each macaque with the premise that the most abundant taxa contribute most significantly to the functionality of the microbial community. Thirty-eight separate taxa of bacteria were found in at least one of the 11 macaques at ≥1% of the sequences from each macaque at this sampling time point (Table 1). The number of taxa found in each macaque was relatively large, with a median of nine different types found (range 3–16). Replicate PCRs were performed and pyrosequenced for each macaque, which resulted in an additional 8500 sequences. Only minor differences between the replicates of PCR and pyrosequencing were observed (Fig. 1 and not shown).
Table 1.
Frequencies of Bacterial Taxa Identified in 11 Macaques
| Taxaa | June 2008 | January 2009 |
|---|---|---|
| Peptoniphilus | 8 | 8 |
| Sneathia | 8 | 6 |
| Porphyromonas | 5 | 6 |
| Mobiluncus | 5 | 5 |
| Atopobacter | 5 | 4 |
| Dialister | 5 | 3 |
| Thioreductor | 5 | 2 |
| Prevotella | 4 | 4 |
| Streptococcus | 4 | 3 |
| Parvimonas | 4 | 3 |
| Heliophilum | 4 | 2 |
| Campylobacter | 3 | 5 |
| Atopobium | 3 | 2 |
| Proteiniphilum | 3 | 2 |
| Corynebacterium | 2 | 7 |
| Anaerococcus | 2 | 4 |
Listed are bacteria that comprised at least 1% of the sequences found in three or more macaques at one of the two sampling times. Taxa found in only one or two macaques include (listed in decreasing frequency) Lactobacillus, Fusobacterium, Centipeda, Bacteroides, Marinilabilia, Helocococcus, Fastidiosipila, Actinobaculum, Gardnerella, Parascardovia, Soehngenia, Treponema, Arcanobacterium, Propionimicrobium, Alkaliflexus, Abiotrophia, Staphylococcus, Catonella, Aminomonas, Dendrosporobacter, Mycoplasma, Caminicella, Atopostipes, Propionibacterium, Scardovia, Facklamia, Peptococcus, Achromobacter, Tropheryma, Fluviicola, Aerococcus, Eremococcus, Ignavigranum, Dolosigranulum, Sinococcus, Gemella, Bryantella, Coprococcus, Lachnobacterium, Sporobacterium, Cryptanaerobacter, Veillonella, Delftia, and Aggregatibacter.
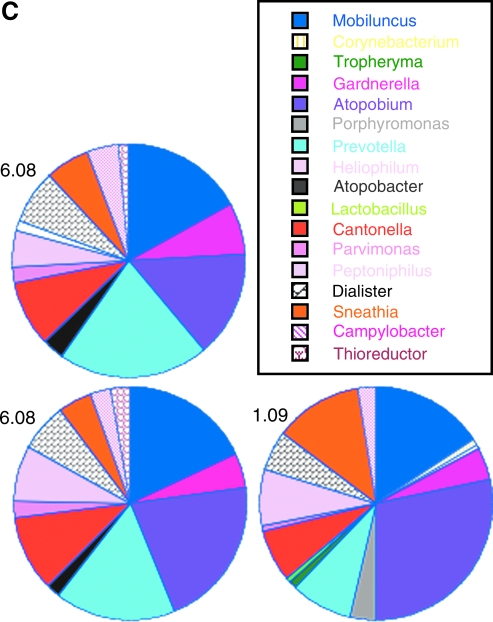
Two genera, Sneathia and Peptoniphilis, were found in 8/11 of the macaques at ≥ 1% of the microbiota (Table 1). Sneathia also constituted a large fraction of the microbiota in macaques on average (Fig. 2). Other taxa found frequently were Porphyromonas, Mobiluncus, Atopobacter, Dialister, Thioreductor, Prevotella, and Streptococcus (Table 1). Four macaques had one single taxa as the predominant bacteria type and they were Sneathia in two macaques (77% and 75% of sequences), Staphylococcus in one (72%), and Streptococcus in one (70%, Table 2). The other seven macaques did not have microbiota dominated by one bacteria type, but instead consisted of mixtures of bacteria (Table 2).
FIG. 2.
Predominant Bacterial Sequences in Macaques
Table 2.
Predominant Bacterial Sequences in Macaques
|
Macaque |
976 |
978 |
979 |
981 |
982 |
|---|---|---|---|---|---|
| Age (years) | 4 | 6 | 6 | 6 | 6 |
| Reads | 661 | 1129 | 1128 | 851 | 970 |
| 6-08 | Mobiluncus 24a | Sneathia 77 | Prevotella 24 | Porphyromonas 22 | Sneathia 75 |
| Sneathia 23 | Peptoniphilus 11 | Sneathia 21 | Mobiluncus 20 | ||
| Thioreductor 10 | Atopobium 18 | Sneathia 8 | |||
| Proteiniphilum 9 | Lactobacillus 11 | Peptoniphilus 7 | |||
| Peptoniphilus 7 | Atopobacter 8 | Thioreductor 7 | |||
| Heliophilum 6 | Proteiniphilum 7 | ||||
| Dialister 6 | |||||
| Reads | 481 | 1853 | 699 | 1011 | 1541 |
| 1-09 | Sneathia 38 | Scardovia 50 | Corynebacterium 51 | Mobiluncus 29 | Sneathia 66 |
| Atopobacter 20 | Streptococcus 23 | Facklamia 18 | Porphyromonas 16 | Mobiluncus 14 | |
| Mobiluncus 16 | Sneathia 19 | Fastidiosipila 11 | Sneathia 15 | Peptoniphilus 12 | |
| Atopostipes 9 | Dialister 6 | ||||
| Atopobacter 6 | |||||
| Treponema 6 | |||||
| 975 | 973 | 974 | 972 | 980 | 977 |
|
9 |
10 |
10 |
13 |
13 |
14 |
| 716 | 991 | 1003 | 984 | 810 | 429 |
| Staphylococcus 72 | Lactobacillus 39 | Sneathia 18 | Mobiluncus 20 | Atopobium 20 | Streptococcus 70 |
| Propionimicrobium 7 | Streptococcus 36 | Proteiniphilum 16 | Prevotella 20 | Mobiluncus 18 | Bacterioides 8 |
| Peptoniphilus 7 | Fusobacterium 24 | Atopobacter 16 | Sneathia 15 | Prevotella 16 | Scardovia 8 |
| Mobiluncus 13 | Dialister 8 | Catonella 10 | |||
| Peptoniphilus 7 | Atopobium 8 | Peptoniphilus 8 | |||
| Alkaliflexus 7 | Dialister 6 | ||||
| 680 | 819 | 1198 | 1072 | 3425 | 322 |
| Staphylococcus 89 | Porphyromonas 36 | Corynebacterium 33 | Mobiluncus 30 | Atopobium 27 | Streptococcus 74 |
| Lactobacillus 33 | Porphyromonas 20 | Prevotella 26 | Mobiluncus 15 | Scardovia 9 | |
| Propionimicrobium 10 | Peptoniphilus 9 | Dialister 11 | Sneathia 12 | Prevotella 7 | |
| Peptoniphilus 7 | Anaerococcus 7 | Porphyromonas 7 | Prevotella 8 | Corynebacterium 6 | |
| Arcanobacterium 5 | Peptoniphilus 6 | Peptoniphilus 8 |
Numbers to the right of each taxa are percentage of sequences. Only taxa that comprised >5% of sequences are listed.
All 11 macaques were resampled 6 months after the first time point to determine the stability of the vaginal microbiota and pyrosequencing of these samples resulted in 13,101 sequences. The microbiota in the macaques remained relatively polymicrobial with a median of 10 different types found (range 6–18). The numbers of taxa found in macaques at the second time point were not significantly different than the numbers of taxa found at the first time point (p > 0.05, Mann–Whitney test). The major types of microbiota in six of the macaques were relatively stable between the two time points (macaques 972, 975, 977, 980, 981, and 982; Table 2 and Fig. 1). For example, microbiota of macaque 972 consisted of mixtures of Mobiluncus, Prevotella, Dialister, Porphyromonas, Peptoniphilius, and Atopobacter at both sampling times while animals 975, 977, and 982 at the second sampling time remained predominately Staphylococcus, Streptococcus, or Sneathia, respectively (Table 2). The macaques ranged in age from 4 to 14 years and there was no apparent relationship between age and types or diversity of the microbiota (Table 2).
In contrast, two macaques had substantially different microbiota at the second time point (macaques 974 and 979). Macaque 974 had only one minor constituent (Peptoniphilus) that was found at the two sampling times whereas 979 had no taxa that matched between the two times. The microbiota of the other three macaques (973, 976, and 978) had some similarity between the two samplings, but also some differences. For example, microbiota in macaque 973 was 39% and 33% Lactobacillus at the two time points, but none of the other taxa matched.
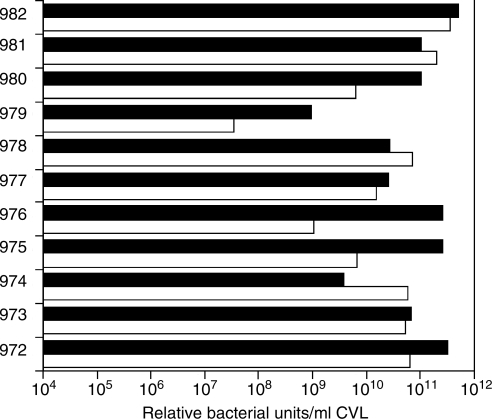
The relative bacterial numbers in the macaque samples were estimated using real time PCR with universal bacterial 16S rRNA gene primers. The number of bacteria genomes in CVL ranged from 2 × 107/ml to 4 × 1011/ml with a median of 8 × 1010/ml at the first time point and 1 × 1011/ml at the second time point (Fig. 3). There was a trend toward stability in the bacteria counts in the samples (linear regression, p = 0.057). Thus, macaque 879 had the lowest counts at both time points whereas macaque 982 had the highest counts at both time points. However, the counts for several of the macaques differed by one to two logs between the two times.
FIG. 3.
Macaque Genital Fluid pH over 14 Months
Lactobacillus sequences were found in four of the macaques, but in only two of the macaques at ≥1% of the microbiota. Lactobacillus constituted 39% and 11% of the sequences at the first sampling time in macaques 973 and 979, respectively, but was a major constituent of microbiota in only macaque 973 (33%) at the second time point (Table 2). The presence of lactobacilli in samples from 973 and 979 taken at the first time point was confirmed by culture, whereas the absence of lactobacilli was confirmed for macaques 976 and 972, macaques that did not have Lactobacillus sequences. Essentially all of the Lactobacillus 16S sequences corresponded to L. johnsonii, although a few sequences corresponding to L. gasseri and L. salivarius were also found.
The above results showed that the levels of lactobacilli in these macaques were relatively low compared to women with normal healthy Lactobacillus-dominated microbiota. Several recent studies that used 16S gene sequencing to identify genital microbiota6,19 showed that in most women with healthy genital microbiota, >95% of 16S sequences corresponded to Lactobacillus. In humans, microbiota that is predominantly Lactobacillus causes a genital fluid pH <4.5 whereas women with bacterial vaginosis generally have a genital fluid pH >4.5. Undiluted genital fluid was not available from the macaques analyzed by pyrosequencing. However, genital fluid pH was measured for a similarly housed group of 10 macaques over a 14-month period (Table 3). At each time, the median pH for the macaques was 7.0 and only one macaque at one time point had a vaginal pH <4.5. The results suggest that relatively low levels of Lactobacillus were present in the microbiota of these macaques, similar to the macaques whose microbiota was sequenced. Furthermore, the fact that these 10 animals were a mixture of Indian and Chinese origin all born and raised at the TNPRC, whereas the other 11 were all Indian origin and born at the Caribbean Primate Research Center, suggested that the results may be similar in rhesus macaques, regardless of their genetic or geographic origin. However, additional studies of macaques from different locales will be necessary to confirm this hypothesis.
Table 3.
Macaque Genital Fluid pH over 14 Months
| Macaque | 12/30/02 | 1/14/03 | 2/25/03 |
|---|---|---|---|
| AK46 | 7 | 7 | 7 |
| AL01 | 7 | 7 | 9 |
| AL37 | 6.5 | 7 | NDa |
| BA93 | 7 | 7 | 8 |
| L384 | 7 | 8 | 9 |
| N400 | 7 | 8 | 7 |
| N967 | 7 | 6 | 5 |
| R323 | 7 | 7 | 8 |
| T314 | 7 | 7 | 5 |
| T619 | 6.5 | 8 | 4 |
Unable to read due to menses.
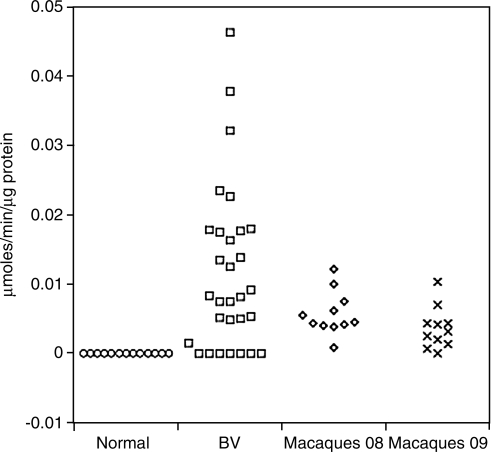
Genital fluid from most women with bacterial vaginosis contains sialidase activity since several of the BV-associated bacteria produce this enzyme. To determine if the macaque microbiota also expressed sialidase, the fluid from the 11 macaques was assayed for sialidase activity (Fig. 4). Detectable sialidase activity was found in 10/11 samples from the first time point and 9/11 samples from the second time point. In contrast, genital fluid from women with Lactobacillus-predominant microbiota had essentially no sialidase activity whereas fluid from women with BV had a range of sialidase activity that substantially overlapped the activity seen in the macaques (Fig. 4). Interestingly, samples that had lower numbers of taxa (975, 977, 978, and 982) had significantly lower sialidase activity (p = 0.045, Mann–Whitney test) than all other samples. Samples with high Sneathia levels, however, were not significantly different than other samples. There was no significant relationship (linear regression) between bacterial numbers (Fig. 3) and sialidase activity (Fig. 4).
FIG. 4.
Sialidase activity in macaque CVL samples. Genital fluid from 11 macaques, sampled at two different times, was tested for sialidase activity. Sialidase activity in CVL obtained from 13 women with no bacterial vaginosis (Normal) and 30 women with bacterial vaginosis (BV) is also shown for comparison. CVL was collected from women using 10 ml of saline. BV in the women was determined using Nugent gram stain values 7–10 while normal was 0–3.
Discussion
This study is the first to identify lower genital tract microbiota in rhesus macaques using molecular techniques and several potentially important findings resulted from this analysis. Foremost, the data suggest that the genital microbiota of rhesus macaques has similarities to the condition called bacterial vaginosis in women. There are five features of the macaque microbiota observed here that are similar to bacterial vaginosis. First, lactobacilli were relatively infrequent in the macaques. Lactobacillus sequences were found in only two of the macaques at appreciable levels but were not the dominant sequences in either. In women with a Lactobacillus-dominant microbiota, lactobacilli frequently make up >95% of the sequences.6,7,19
Second, the macaque genital microbiota was relatively polymicrobial, which is a hallmark of bacterial vaginosis.6,7,19,20 Thus, macaque microbiota consisted of between three and 18 taxa and seven of the macaques had microbiota that was not dominated by any one bacterium. The other four macaques had one predominant bacterium, but each of the dominant types constituted only up to 62–89% of the sequences.
Third, many of the bacterial taxa found in the macaques were analogous to those found in women with bacterial vaginosis. Mobiluncus, Sneathia, Peptoniphilis, Porphyromonas, Atopobium, Dialister, and Prevotella were all found in the rhesus macaques frequently and are also often found in women with bacterial vaginosis.6,21,22 Some of these had been previously found by culture in rhesus macaques including Mobiluncus, Porphyromonas, and Peptostreptococcus (closely related to Peptoniphilis).14 Sneathia, Atopobium, and Dialister had not been reported previously in macaques. A difference between the macaque microbiota and bacterial vaginosis, however, is that Gardnerella vaginalis is found frequently in women with bacteria vaginosis and at fairly high levels, but was found in only one of the macaques at the first time point and two at the second time point at relatively low levels.
The fourth feature of macaque genital microbiota that is similar to bacterial vaginosis is that sialidase activity is present in both. Briseldin et al.18 reported that cultures of Prevotella, Bacterioides, and Gardnerella all produced sialidase whereas Mobiluncus, Peptostreptococcus, and Porphyromonas did not. Prevotella was found in four of the macaques and Gardnerella in only one. Uncultivable bacteria such as Sneathia cannot be directly tested for production of sialidase.
A fifth feature of the macaque genital microbiota that has similarities to bacterial vaginosis is that the genital fluid of macaques consistently has a pH that is >4.5 (Fig. 4). Production of acid by lactobacilli is highly associated with vaginal acidification in women23 and, therefore, the paucity of lactobacilli in the macaques is a likely explanation for the pH of 7.
The finding that the genital microbiota of rhesus macaques is relatively low in lactobacilli, but high in bacterial vaginosis-like bacteria could have implications for the use of rhesus macaques as a model for HIV infection of women. In a number of cross-sectional and longitudinal studies, bacterial vaginosis has been consistently associated with an increased risk of HIV infection.2 Although macaques are susceptible to vaginal infection with SIV and SHIV, there are no studies of the relationship of the macaque microbiota on susceptibility to infection. Importantly, the macaque may represent a model that could be used to address many of the questions about the contribution of microbiota to transmission susceptibility. Macaques could be used to investigate the relative contribution of the many factors that have been hypothesized to affect the susceptibility of women with bacteria vaginosis to HIV infection such as increased vaginal pH, decreased H2O2, degradation of mucous by sialidases, toll-like receptor activation of immune cells, and increased levels of proinflammatory cytokines.24,25
This study also suggests the possibility that rhesus macaques could be used as an animal model to study some aspects of bacterial vaginosis. There is no current animal model of bacterial vaginosis, but there are many important unresolved questions about bacterial vaginosis that an animal model could be used to explore. As mentioned above, susceptibility to immunodeficiency virus infection could be studied, but bacterial vaginosis also appears to affect susceptibility to other genital viruses such as HSV-1, HSV-2, and HPV.26 Also, bacterial vaginosis is recurrent in many women and microbiologic factors are currently being explored as a cause for this,27 which could also be tested in macaques. Bacterial vaginosis is also a well-established risk factor for preterm delivery and spontaneous abortion in women, which could be investigated in macaques,28 although the reproductive biology and immunology may be quite different between rhesus macaques and humans and prevent the use of macaques for these types of studies.
Staphylococcus and Streptococcus were found in macaques previously by culture14 and in the current study Streptococcus was found in five different macaques during at least one of the sampling times and was found in one macaque as the predominant bacterium. Staphylococcus was also observed to be predominant in one of the macaques in this study. Both of these bacteria can be found by culture in the genital microbiota of women with or without bacterial vaginosis. Streptococcus has been reported to be the predominant bacterium in some women by molecular identification.7,29 Staphylococcus is also commonly isolated by culture from the genital microbiota of women, but has not yet been reported to be the predominant bacterium in women using molecular identification.6,7,19
We observed that the microbiota in several of the macaques appeared to be stable over a 6-month period. Lactobacillus has been suggested to be a stable colonizer, but there are few reports of other specific microbiota being found consistently over time. Stable Lactobacillus colonization is believed to be stable due to production of lactic acid and H2O2, which prevents overgrowth by other bacteria.5
This study has several limitations. Here only 11 rhesus macaques were analyzed from one primate center, and these animals had been recently imported from another primate center, so it is not certain this flora would be representative of all colony-housed rhesus macaques. However, the “neutral” pH of the samples from the 10 animals from the TNPRC suggested a similar microbiota may exist in animals from both primate facilities. Furthermore, both TNPRC and the Caribbean primate centers are in a tropical climate, so it is possible that genital microbiota could vary in different climates or in different housing conditions (single housing, etc.). Finally, the microbiota of animals in the wild could be very different, although wild animals are not used in laboratory animal model studies. In addition, it is possible that rhesus macaques differ from pigtailed, cynomolgus, or other macaque species that have been used in animal models of HIV infection. Comparative studies between species might be informative in explaining the susceptibility of different macaques to vaginal SIV and SHIV transmission.
Acknowledgments
This work was supported by NIH grant AI076981 and the James B. Pendleton Charitable Trust.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Antonio MA. Hawes SE. Hillier SL. The identification of vaginal Lactobacillus species and the demographic and microbiologic characteristics of women colonized by these species. J Infect Dis. 1999;180:1950–1956. doi: 10.1086/315109. [DOI] [PubMed] [Google Scholar]
- 2.Atashili J. Poole C. Ndumbe PM. Adimora AA. Smith JS. Bacterial vaginosis and HIV acquisition: A meta-analysis of published studies. AIDS. 2008;22:1493–1501. doi: 10.1097/QAD.0b013e3283021a37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaul R. Nagelkerke NJ. Kimani J, et al. Prevalent herpes simplex virus type 2 infection is associated with altered vaginal flora and an increased susceptibility to multiple sexually transmitted infections. J Infect Dis. 2007;196:1692–1697. doi: 10.1086/522006. [DOI] [PubMed] [Google Scholar]
- 4.Cherpes TL. Meyn LA. Krohn MA. Lurie JG. Hillier SL. Association between acquisition of herpes simplex virus type 2 in women and bacterial vaginosis. Clin Infect Dis. 2003;37:319–325. doi: 10.1086/375819. [DOI] [PubMed] [Google Scholar]
- 5.Cherpes TL. Hillier SL. Meyn LA. Busch JL. Krohn MA. A delicate balance: Risk factors for acquisition of bacterial vaginosis include sexual activity, absence of hydrogen peroxide-producing lactobacilli, black race, and positive herpes simplex virus type 2 serology. Sex Transm Dis. 2008;35:78–83. doi: 10.1097/OLQ.0b013e318156a5d0. [DOI] [PubMed] [Google Scholar]
- 6.Fredricks DN. Fiedler TL. Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353:1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 7.Hyman RW. Fukushima M. Diamond L. Kumm J. Giudice LC. Davis RW. Microbes on the human vaginal epithelium. Proc Natl Acad Sci USA. 2005;102:7952–7957. doi: 10.1073/pnas.0503236102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han YW. Shen T. Chung P. Buhimschi IA. Buhimschi CS. Uncultivated bacteria as etiologic agents of intra-amniotic inflammation leading to preterm birth. J Clin Microbiol. 2009;47:38–47. doi: 10.1128/JCM.01206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patton DL. Sweeney YC. Cummings PK. Meyn L. Rabe LK. Hillier SL. Safety and efficacy evaluations for vaginal and rectal use of BufferGel in the macaque model. Sex Transm Dis. 2004;31:290–296. doi: 10.1097/01.olq.0000124614.91448.d4. [DOI] [PubMed] [Google Scholar]
- 10.Patton DL. Sweeney YT. Balkus JE, et al. Preclinical safety assessments of UC781 anti-human immunodeficiency virus topical microbicide formulations. Antimicrob Agents Chemother. 2007;51:1608–1615. doi: 10.1128/AAC.00984-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patton DL. Sweeney YT. Balkus JE. Hillier SL. Vaginal and rectal topical microbicide development: Safety and efficacy of 1.0% Savvy (C31G) in the pigtailed macaque. Sex Transm Dis. 2006;33:691–695. doi: 10.1097/01.olq.0000216022.18321.d3. [DOI] [PubMed] [Google Scholar]
- 12.Ratterree M. Gettie A. Williams V, et al. Safety and distribution of cellulose acetate 1,2-benzenedicarboxylate (CAP), a candidate anti-HIV microbicide in rhesus macaques. AIDS. 2005;19:1595–1599. doi: 10.1097/01.aids.0000185990.16477.47. [DOI] [PubMed] [Google Scholar]
- 13.Schlievert PM. Strandberg KL. Brosnahan AJ, et al. Glycerol monolaurate does not alter rhesus macaque (Macaca mulatta) vaginal lactobacilli and is safe for chronic use. Antimicrob Agents Chemother. 2008;52:4448–4454. doi: 10.1128/AAC.00989-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doyle L. Young CL. Jang SS. Hillier SL. Normal vaginal aerobic and anaerobic bacterial flora of the rhesus macaque (Macaca mulatta) J Med Primatol. 1991;20:409–413. [PubMed] [Google Scholar]
- 15.Lane DJ. 16S/23S rRNA sequencing. In: Goodfellow ESaM., editor. Nucleic Acid Techniques in Bacterial Systematics (pp. 115–175) West Sussex, England: John Wiley & Sons Ltd; 1991. [Google Scholar]
- 16.Cole JR. Chai B. Farris RJ, et al. The Ribosomal Database Project (RDP-II): Sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:294–296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barman M. Unold D. Shifley K, et al. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Briselden AM. Moncla BJ. Stevens CE. Hillier SL. Sialidases (neuraminidases) in bacterial vaginosis and bacterial vaginosis-associated microflora. J Clin Microbiol. 1992;30:663–666. doi: 10.1128/jcm.30.3.663-666.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spear GT. Sikaroodi M. Zariffard MR. Landay AL. French AL. Gillevet PM. Comparison of the diversity of the vaginal microbiota in HIV-infected and HIV-uninfected women with or without bacterial vaginosis. J Infect Dis. 2008;198:1131–1140. doi: 10.1086/591942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spiegel CA. Davick P. Totten PA, et al. Gardnerella vaginalis and anaerobic bacteria in the etiology of bacterial (nonspecific) vaginosis. Scand J Infect Dis Suppl. 1983;40:41–46. [PubMed] [Google Scholar]
- 21.Hill GB. The microbiology of bacterial vaginosis. Am J Obstet Gynecol. 1993;169:450–454. doi: 10.1016/0002-9378(93)90339-k. [DOI] [PubMed] [Google Scholar]
- 22.Fredricks DN. Fiedler TL. Thomas KK. Oakley BB. Marrazzo JM. Targeted PCR for detection of vaginal bacteria associated with bacterial vaginosis. J Clin Microbiol. 2007;45:3270–3276. doi: 10.1128/JCM.01272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boskey ER. Telsch KM. Whaley KJ. Moench TR. Cone RA. Acid production by vaginal flora in vitro is consistent with the rate and extent of vaginal acidification. Infect Immun. 1999;67:5170–5175. doi: 10.1128/iai.67.10.5170-5175.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hillier SL. The vaginal microbial ecosystem and resistance to HIV. AIDS Res Hum Retroviruses. 1998;14(Suppl 1):S17–21. [PubMed] [Google Scholar]
- 25.Spear GT. St John E. Zariffard MR. Bacterial vaginosis and human immunodeficiency virus infection. AIDS Res Ther. 2007;4:25. doi: 10.1186/1742-6405-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allsworth JE. Lewis VA. Peipert JF. Viral sexually transmitted infections and bacterial vaginosis: 2001–2004. National Health and Nutrition Examination Survey data. Sex Transm Dis. 2008;35:791–796. doi: 10.1097/OLQ.0b013e3181788301. [DOI] [PubMed] [Google Scholar]
- 27.Marrazzo JM. Thomas KK. Fiedler TL. Ringwood K. Fredricks DN. Relationship of specific vaginal bacteria and bacterial vaginosis treatment failure in women who have sex with women. Ann Intern Med. 2008;149:20–28. doi: 10.7326/0003-4819-149-1-200807010-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leitich H. Bodner-Adler B. Brunbauer M. Kaider A. Egarter C. Husslein P. Bacterial vaginosis as a risk factor for preterm delivery: A meta-analysis. Am J Obstet Gynecol. 2003;189:139–147. doi: 10.1067/mob.2003.339. [DOI] [PubMed] [Google Scholar]
- 29.Cherpes TL. Melan MA. Kant JA. Cosentino LA. Meyn LA. Hillier SL. Genital tract shedding of herpes simplex virus type 2 in women: Effects of hormonal contraception, bacterial vaginosis, and vaginal group B Streptococcus colonization. Clin Infect Dis. 2005;40:1422–1428. doi: 10.1086/429622. [DOI] [PubMed] [Google Scholar]