Abstract
The extrahepatic UDP-glucuronosyltransferase 1A10 (UGT1A10) is a phase II metabolizing enzyme that is active against a number of potent carcinogens. In the present study, UGT1A10 was examined for activity against 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL), the major procarcinogenic metabolite of the potent tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, and the promoter region of UGT1A10 was examined for variants that could lead to altered UGT1A10 expression. UGT1A10-overexpressing cell homogenates exhibited high O-glucuronidation activity against NNAL (KM = 5.95 mM). A 2000-base pair (bp) product corresponding to the UGT1A10 proximal promoter region was polymerase chain reaction (PCR)-amplified using genomic DNA from 97 white subjects, and 42 of these were sequenced. In addition to a previously reported C/G single-nucleotide polymorphism at −1271 bp (rs2741032), a novel 1664-bp deletion located between nucleotides −190 to −1856 relative to the UGT1A10 translation start site was identified. Using real-time multiplex PCR, this deletion exhibited a prevalence of 0.022 in whites (n = 156) and 0.056 in blacks (n = 133). To determine whether either polymorphism altered gene expression, in vitro assays were performed using luciferase constructs containing up to 2000 bp of the proximal UGT1A10 promoter. Constructs containing the 1664-bp deletion exhibited a significant (p = 0.009) 3-fold increase in luciferase activity compared with constructs containing the wild-type UGT1A10 promoter. No effect on luciferase activity was observed for the UGT1A10−1271G promoter variant. These data are consistent with previous studies that indicate the presence of a transcriptional repressor element within the newly identified deletion and that this deletion polymorphism may contribute to altered UGT1A10 expression and altered carcinogen detoxification between individuals.
The UDP-glucuronosyltransferase (UGT) superfamily of enzymes catalyze the glucuronidation of a variety of endogenous compounds such as bilirubin and steroid hormones, as well as xenobiotics such as drugs and environmental carcinogens (Tephly and Burchell, 1990; Owens and Ritter, 1995; Guéraud and Paris, 1998; Ren et al., 2000). Several UGTs have been implicated in the detoxification of a number of carcinogens, including metabolites of the tobacco-specific nitrosamine, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) (Ren et al., 2000; Wiener et al., 2004a), metabolites of polycyclic aromatic hydrocarbons (PAHs) such as benzo[a]pyrene (B[a]P) (Ciotti et al., 1997; Lévesque et al., 1997; Beaulieu et al., 1998; Bélanger et al., 1998; Carrier et al., 2000), and heterocyclic amines such as 2-amino-1-methyl-6-phenylimidazo[4,5-f]pyridine (PhIP) and its major metabolite, N-hydroxy-PhIP (Dellinger et al., 2007). UGTs are classified into several families and subfamilies based on their structural and sequence homology (Jin et al., 1993). The UGT1A family is derived from a single gene locus on chromosome 2q37 that spans more than 200 kilobases (Ritter et al., 1992). This locus codes for nine functional proteins that differ only in their amino terminus as a result of alternate splicing of independent exon 1 regions to a shared carboxyl terminus encoded by exons 2 through 5 (Beaulieu et al., 1997). Despite the high homology of the exon 1 regions, UGT1A enzymes display a wide range of substrate specificity and affinity (Nagar and Remmel, 2006; Kubota et al., 2007). Expression also differs among the UGT1As, with the majority expressed in liver (Ciotti et al., 1997; Lévesque et al., 1997; Burchell and Hume, 1999; Guillemette et al., 2000) and several expressed extrahepatically (Tukey and Strassburg, 2000; Oguri et al., 2004).
UGT1A10 is an extrahepatic enzyme that has been detected in a variety of tissues, including those within the gastrointestinal tract (Strassburg et al., 1997; Nakamura et al., 2008), the aerodigestive tract (Zheng et al., 2002), and in lung (Dellinger et al., 2006). Two single-nucleotide polymorphisms (SNPs) in the coding region of the UGT1A10 enzyme were previously identified, with the codon 139 Glu>Lys SNP shown to result in altered UGT1A10 activity in vitro (Dellinger et al., 2006, 2007) and was associated with altered risk for orolaryngeal carcinoma (Elahi et al., 2003). Although most UGTs are localized within the endoplasmic reticulum of the cell, it was recently shown that UGT1A10 was localized to an extramicrosomal fraction independent of the endoplasmic reticulum (Dellinger et al., 2007). This differential localization of UGT1A10 compared with other UGTs could explain the differences in glucuronidation activity observed for UGT1A10-overexpressing cell homogenates versus microsomes against PhIP and N-hydroxy-PhIP (Dellinger et al., 2007). The goals of the present study were 1) to examine whether UGT1A10-overexpressing cell homogenates exhibit activity against 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL), the major procarcinogenic metabolite of NNK, and 2) to screen the UGT1A10 promoter region for polymorphisms and determine their potential effect on UGT1A10 expression.
Materials and Methods
Chemicals and Materials.
14C-UDP-glucuronic acid (UDPGA) (200 mCi/mmol) was purchased from American Radiolabeled Chemicals (St. Louis, MO). The high-pressure liquid chromatography (HPLC) scintillation solution, Ecoscint Flow, was purchased from National Diagnostics (Atlanta, GA). UDPGA, alamethicin, and β-glucuronidase were purchased from Sigma-Aldrich (St. Louis, MO). NNAL was obtained from Toronto Research Chemicals Inc. (North York, ON, Canada). Minimal essential medium (MEM), Dulbecco's phosphate-buffered saline (minus calcium chloride and magnesium chloride), fetal bovine serum, MEM nonessential amino acids, sodium pyruvate, penicillin/streptomycin, T4 DNA ligase, Lipofectamine 2000, and One Shot competent cells were all obtained from Invitrogen (Carlsbad, CA). Taq DNA polymerase was purchased from Bio-Rad Laboratories (Hercules, CA), and PfuTurbo DNA polymerase and Quick Change site-directed mutagenesis kits were purchased from Stratagene (La Jolla, CA). QuantiTect Multiplex polymerase chain reaction (PCR) Master Mix, the Hi-Speed plasmid maxi kit, and the QIAEX II gel extraction kit were all purchased from QIAGEN (Valencia, CA). The pGL3-Basic and pRL-TK vectors were purchased from Promega (Madison, WI). The UGT1A10 probes for real-time PCR were purchased from Applied Biosystems (Foster City, CA), and PCR primers were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). All the restriction enzymes were purchased from New England Biolabs (Ipswich, MA). The Caco-2 cell line was purchased from the American Type Culture Collection (Manassas, VA). All the other chemicals were purchased from Thermo Fisher Scientific (Waltham, MA) unless specified otherwise.
Subjects.
To screen the UGT1A10 promoter region for polymorphisms, genomic DNA purified from 97 normal human liver specimens excised during surgery for removal of hepatocellular carcinoma was used as described previously (Wiener et al., 2004a). UGT1A10 promoter polymorphic prevalences were examined in healthy whites (n = 156) and blacks (n = 133) as described previously (Richie et al., 1997; Park et al., 1999; Elahi et al., 2002).
Glucuronidation Assays.
The rate of NNAL glucuronidation by UGT1A10 cell homogenates was determined essentially as described previously (Ren et al., 2000; Dellinger et al., 2006; Sun et al., 2006). After an initial incubation of cell homogenates (250 μg of protein) with alamethicin (50 μg/mg protein) for 15 min in an ice bath, glucuronidation reactions were performed in a final reaction volume of 50 μl at 37°C for 2 h in 50 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 4 mM UDPGA, 1 μCi of 14C-UDPGA, and 5 mM NNAL. Reactions were terminated by the addition of 50 μl of cold acetonitrile. After 10 min on ice, the mixtures were centrifuged for 10 min at 4°C at 16,000g; the supernatants were collected; and the solvent in the mixture was evaporated.
Samples (50 μl) were analyzed for NNAL-glucuronide formation by HPLC using a Beckman Coulter (Fullerton, CA) System Gold 126 Solvent Module HPLC system equipped with an automatic injector (model 508), a UV detector operated at 254 nm (model 166), and a radioactive flow detector with 1000-μl flow cell (IN/US Systems, Tampa, FL). HPLC was performed using an Aquasil 4-μm C18 analytical column (4.6 × 250 mm; Thermo Fisher Scientific). The gradient elution conditions were as follows: starting with 100% buffer A (50 mM KH2PO4, pH 6.0) for 10 min, a subsequent linear gradient to 78% B (90% acetonitrile) over 20 min was performed and then maintained at 78% B for 10 min. The elution flow rate was 1 ml/min, and the scintillation solution flow rate was 3 ml/min. The amount of glucuronide formed was calculated based on the ratio of the radioactivity of the glucuronide versus total radioactivity. The NNAL-glucuronide was confirmed by sensitivity to β-glucuronidase as described previously (Wiener et al., 2004a). As controls, glucuronidation assays were regularly performed using human liver microsomes (as a positive control for glucuronidation activity) and untransfected human embryonic kidney 293 cell homogenate protein (as a negative control for glucuronidation activity) as described previously (Fang et al., 2002; Wiener et al., 2004b). Experiments were always performed in triplicate in independent assays. Kinetic constants were determined using GraphPad Prism4 software (GraphPad Software Inc., San Diego, CA).
Identification and Prevalence of UGT1A10 Polymorphisms.
The 2000 base pair (bp) located immediately upstream of the UGT1A10 ATG translational start site were amplified by PCR using a sense primer (5′-TAGCTCTTCTTGTTACATTGATCCC-3′, corresponding to nucleotides −2006 to −1982 relative to the UGT1A10 translation start site) and an antisense primer (5′-CGGTTCTGATCTTCCAGAGTG-3′, corresponding to nucleotides +243 to +263 relative to the UGT1A10 translation start site). PCR amplifications were performed in a 50-μl reaction volume containing 50 to 100 ng of purified genomic DNA in 1× Taq buffer with 0.4 mM of each deoxynucleotide triphosphate, 20 pmol of both sense and antisense, and 1.25 U of Taq DNA polymerase. The reaction mixtures underwent the following incubations in a MyCycler (Bio-Rad Laboratories): one cycle of 95°C for 2 min, 41 cycles of 95°C for 30 s, 64°C for 30 s, and 72°C for 3 min, and a final extension cycle of 72°C for 10 min. PCR amplicons were run on 0.8% agarose gels that were subsequently stained with ethidium bromide and examined over UV light using a computerized photo imaging system (AlphaImager 2000; Alpha Innotech, San Leandro, CA). The expected band size was 2269 bp. Samples were gel-purified using QIAquick Spin (QIAGEN) and sequenced on an ABI 377 DNA Sequencer (Applied Biosystems). Sequences were analyzed for polymorphisms using Basic Local Alignment Search Tool (BLAST; National Center for Biotechnology Information, Bethesda, MD), with 100% of all the nucleotides covered for all the amplicons.
A newly designed multiplex allelic discrimination assays was performed to assess the prevalence of the UGT1A10 promoter polymorphism. Identification of the wild-type UGT1A10 promoter included sense (5′-TCCTGAATAATATCCTGAAGAGG-3′) and antisense (5′-CATGCTTTGCCTTTGACG-3′) primers (corresponding to −1153 to −1131 and −899 to −882, respectively, relative to the wild-type UGT1A10 translation start site) and a probe (5′-FAM-AGATCTGGGATACATGTA-MGB-3′, corresponding to −1096 to −1079 relative to the wild-type UGT1A10 translation start site). Identification of the UGT1A10 promoter deletion variant included sense (5′-TTGATCTCTGTTGATTTAAAGTCTG-3′) and antisense (5′-GCAGTAGACACACACATAAA-3′) primers (corresponding to −281 to −257 and +33 to +52, respectively, relative to the UGT1A10 deletion variant translation start site) and a probe (5′-VIC-TTGGTAAATATTCCTCAGCA-MGB-3′, corresponding to −203 to −185 relative to the UGT1A10 deletion variant translation start site). Reactions included all the wild-type and deletion primers and probes. Reactions (25 μl) were performed in 96-well plates using the ABI 7900 HT Sequence Detection System (Applied Biosystems), with reaction conditions performed with an initial cycle of 50°C for 2 min and 95°C for 15 min, and 50 cycles of 94°C for 30 s, 55 for 30 s, and 60°C for 1 min. Reactions included QuantiTect Multiplex PCR Master Mix (1× final concentration; QIAGEN), 0.4 μM of each primer, 0.2 μM of each probe, and 20 ng of genomic DNA. Negative controls (no DNA template) were run on every plate, and genotypes were assigned by the automatic calling feature of the allelic discrimination option in SDS 2.2.2 software (Applied Biosystems).
UGT1A10 Promoter Cloning and Luciferase Assays.
UGT1A10 promoter regions were cloned into the pGL3-Basic vector, which expresses the firefly luciferase reporter enzyme. PCR amplifications of the UGT1A10 promoter were performed in 50-μl reactions containing 100 ng of purified genomic DNA in 1× Pfu DNA polymerase reaction buffer with 0.4 mM of each deoxynucleotide triphosphate, 20 pmol of both sense and antisense primers, and 2.5 U of PfuTurbo DNA polymerase (Stratagene). All the regions were amplified in 50-μl reactions with the following cycling conditions: 1 cycle of 95°C for 2 min, 41 cycles of 95°C for 30 s, 65 for 30 s, and 72°C for 3 min, and a final extension cycle of 72°C for 10 min. Genomic DNA from an individual who exhibited a homozygous wild-type UGT1A10 promoter was used to amplify 186, 441, and 2204 bp of wild-type UGT1A10 5′ flanking region beginning at −5 bp upstream of the UGT1A10 translation start site. Genomic DNA from an individual who exhibited a homozygous UGT1A10 deletion variant promoter was used to amplify 441 and 2204 bp of UGT1A10 5′ deletion promoter flanking region beginning at −5 bp upstream of the UGT1A10 translation start site. The primers used for PCR amplifications of UGT1A10 wild-type and deletion promoter regions were designed with the addition of restriction enzyme sites at their 5′ proximal ends for vector insertion in the correct orientation and are listed in Table 1. The 186- and 441-bp PCR-amplified UGT1A10 promoter fragments (wild-type and/or variant promoters) were inserted into the KpnI and MluI restriction enzyme sites of the pGL3-Basic vector. Because of the presence of a KpnI site within the 2204-bp PCR fragment, the 2204-bp PCR fragments for both the wild-type and variant UGT1A10 promoters were inserted into the MluI and XmaI restriction enzyme sites of the pGL3-Basic vector. All the PCR fragments were inserted into their corresponding restriction enzyme-digested pGL3-Basic vectors by ligation with T4 DNA ligase. UGT1A10 plasmid-containing One-Shot bacterial colonies (Invitrogen) were selected by ampicillin resistance after transformation and were analyzed by restriction fragment length polymorphism analysis to ensure the presence of an insert. All the vectors containing inserts were purified using the Hi-Speed plasmid maxi kit (QIAGEN) and sequenced in full to confirm the construct.
TABLE 1.
List of primers used for PCR amplifications of UGT1A10 wild-type and deletion promoter regions were designed with the addition of restriction enzyme sites at their 5′ proximal ends for vector insertion in the correct orientation
| Primer Namea | Restriction Site | Primer sequence (5′–3′) | Primer Location |
|---|---|---|---|
| WT186S | KpnI | 5′-AGTAGGTACCTCAGCAAATGATACT-3′ | −190 to −176b |
| WT186AS | MluI | 5′-AGCCACGCGTGAACTGCAGCCCGAGCC-3′ | −21 to −5b |
| WT441S | KpnI | 5′-CTGTGGTACCCCTGGAACATGAGATGCC-3′ | −445 to −428b |
| WT441AS | MluI | 5′-AGCCACGCGTGAACTGCAGCCCGAGCC-3′ | −21 to −5b |
| WT2204S | MluI | 5′-AGCCACGCGTGTATTAGGTTTGCTTGGT-3′ | −2208 to −2187b |
| WT2204AS | XmaI | 5′-TATACCCGGGGAACTGCAGCCCGAGCC-3′ | −21 to −5b |
| Del441S | KpnI | 5′-GCGGGCGGTACCTAAAGTCTCCCACTA-3′ | −445 to −431c |
| Del441AS | MluI | 5′-AGCCACGCGTGAACTGCAGCCCGAGCC-3′ | −21 to −5c |
| DEL 2204S | MluI | 5′-CGGCCACGCGTGAATGCTTGTG-3′ | −2208 to −2198c |
| DEL 2204AS | XmaI | 5′-TATACCCGGGGAACTGCAGCCCGAGCC-3′ | −21 to −5c |
Primer names with S or AS refer to sense and antisense primers, respectively.
Relative to the UGT1A10 wild-type translation start site.
Relative to the UGT1A10 deletion variant translation start site.
The −1271C>G variant was generated with the Quick Change site-directed mutagenesis kit using the 2204-bp wild-type UGT1A10 promoter-containing vector as template. Primers used were 5′-TTCCTTCATTTCAACGTTGGTGAATCTGATG-3′ (sense) and 5′-CATCAGATTCACCAACGTTGAAATGAAGGAA-3′ (antisense), and reactions were carried out as described previously (Dellinger et al., 2006). The underlined bases indicate the bp change.
Caco-2 cells were cultured in MEM with 1 mM MEM nonessential amino acids, 0.1 mM sodium pyruvate, 10% fetal bovine serum, and 100 U/ml penicillin/streptomycin. For luciferase assays, cells were plated in six-well plates and grown in 5% CO2 to 80 to 90% confluence. Cotransfection of 4 μg of the UGT1A10 promoter-containing pGL3-Basic constructs and 0.05 μg of the pRL-TK plasmid was performed using Lipofectamine 2000 as recommended by the manufacturer (Invitrogen). The Dual-Luciferase reporter assay system (Promega) was used for all the luciferase assays. Luciferase and Renilla luminescence, corresponding to the pGL3-Basic and pRL-TK plasmids, respectively, was determined using the Synergy HT multidetection microplate reader (BioTek Instruments, Winooski, VT). All the assays were performed in triplicate in three independent experiments.
Results
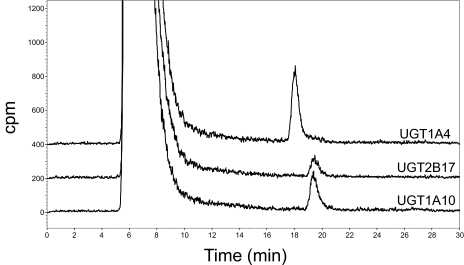
Previous studies have shown that UGT1A10 resides primarily in the extramicrosomal fraction of the cell, suggesting that studies excluding this fraction could underestimate the role of UGT1A10 against substrates (Dellinger et al., 2007). Because previous studies examining UGT1A10 glucuronidation activity against NNAL used the microsomal fraction of UGT1A10-overexpressing cells, homogenates of UGT1A10-overexpressing human embryonic kidney 293 cells were used in the in vitro glucuronidation assays against NNAL. As shown in Fig. 1, homogenates of UGT1A10-overexpressing cells exhibited glucuronidating activity against NNAL. The retention time of the UGT1A10-induced peak (19 min; Fig. 1, bottom peak) was identical to that observed for assays using UGT2B17-overexpressing cell homogenates (Fig. 1, middle peak), which was previously shown to specifically exhibit O-glucuronide formation activity against NNAL (Lazarus et al., 2005), but not UGT1A4 (Fig. 1, top peak), which was previously shown to specifically form NNAL-N-glucuronide (Wiener et al., 2004a). Similar to that observed previously for NNAL-O-glucuronide (Ren et al., 2000; Wiener et al., 2004a,b), the UGT1A10-induced peak was sensitive to treatment with β-glucuronidase but not NaOH (results not shown). Kinetic analysis showed that UGT1A10 exhibited a KM of 5.95 mM against NNAL.
Fig. 1.
Representative HPLC traces of UGT1A10 O-glucuronidation activity against NNAL. HPLC traces of glucuronidation assays using homogenates of UGT1A4- and UGT2B17-overexpressing cells are also shown. All the assays were performed using 250 μg of total protein and incubated at 37°C for 2 h with 5 mM NNAL and 1 μCi of 14C-UDPGA before analysis by HPLC.
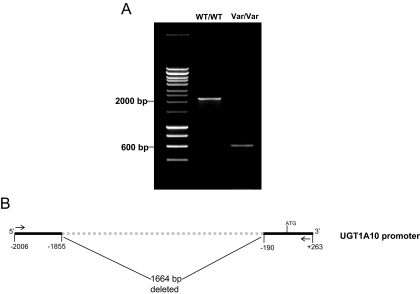
Previous studies have identified several SNPs within the UGT1A10 coding region, all of which were of low prevalence (<5%) in both whites and blacks (Elahi et al., 2002). To identify polymorphisms in the promoter and 5′-untranslated region (UTR) of the UGT1A10 gene, PCR was used to amplify a 2269-bp product corresponding to −2006 to +263 of the UGT1A10 gene relative to the translational start site using high-quality genomic DNA purified from normal liver specimens from 97 independent white subjects. Two amplicons were observed in several of the amplifications after electrophoresis on 0.8% agarose gels—the expected 2269-bp fragment and a smaller fragment of approximately 600 bp (Fig. 2A). Sequencing analysis of each purified amplicon (performed for PCR amplifications of genomic DNA from two independent subjects) indicated that the larger 2269-bp amplicon corresponded to the wild-type UGT1A10 promoter and that the smaller fragment corresponded to a UGT1A10 promoter containing a deletion spanning the region −191 to −1854 relative to the UGT1A10 translational start site (Fig. 2B). Of the 97 samples amplified, three were heterozygous and one was homozygous for this deletion polymorphism (allelic prevalence = 2.6%).
Fig. 2.
UGT1A10 promoter deletion identification and schematic. A, agarose gel (0.8%) containing PCR-amplified products using genomic DNA from subjects exhibiting a homozygous wild-type (WT/WT) or homozygous variant (Var/Var) genotypes for the UGT1A10 promoter deletion polymorphism. B, schematic representation of the UGT1A10 promoter deletion polymorphism. Arrows indicate sense (UGT1A10S) and antisense (UGT1A10AS) primers used for PCR amplification of the UGT1A10 promoter.
To better assess the prevalence of the deletion among different racial groups, a multiplex real-time assay was designed, and genotypes were assigned by the automatic calling feature of the allelic discrimination option in SDS 2.2.2 software (Applied Biosystems). In addition to the 97 subjects analyzed in the initial screen, an additional 156 white and 133 black healthy control subjects were analyzed for the deletion genotype using oral buccal cell genomic DNA. Of the additional 156 whites examined, seven were heterozygous for the UGT1A10 deletion (no additional white subjects homozygous for the deletion were identified). Of the 133 blacks examined, 13 were heterozygous and 1 was homozygous for the deletion. The resulting allelic frequencies of the deletion polymorphism in these control populations were 2.2% in whites and 5.6% in blacks. The allelic frequencies followed Hardy-Weinberg equilibrium for both populations.
In addition to the promoter deletion polymorphism, sequencing analysis of the 2269-bp UGT1A10 promoter amplicon from 47 of the 97 UGT1A10 promoter-amplified specimens from individual subjects resulted in the identification of a single polymorphism—a C>G SNP corresponding to rs2741032 in the National Center for Biotechnology Information's Entrez SNP database located −1271 upstream of the UGT1A10 ATG translational start site. No additional SNPs were identified within this region for any of the specimens sequenced in this analysis.
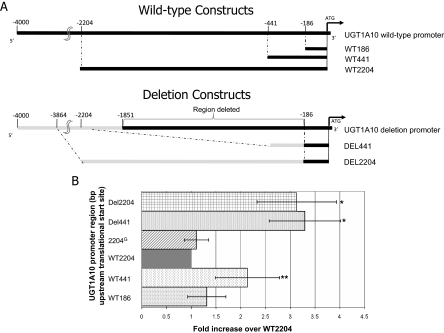
To determine whether either the 1664-bp polymorphic deletion or the C>G SNP located in the UGT1A10 promoter could affect gene expression, UGT1A10 promoter-containing luciferase constructs were designed to evaluate various regions of the ∼2000 bp of upstream UGT1A10 promoter sequences. As shown in Fig. 3A, up to 2204 bp of UGT1A10 promoter immediately upstream of the UGT1A10 translational start site were analyzed from subjects wild-type or deleted for the UGT1A10 promoter. UGT1A10 promoter constructs consisted of 186 bp (termed WT186), 441 bp (termed WT441), and 2204 bp (termed WT2204) of 5′ flanking UGT1A10 5′ UTR and promoter sequences. In addition, a 2204-bp UGT1A10 5′ UTR/promoter construct containing the C>G transversion at bp −1271 (termed 2204G) was constructed. In addition, two constructs were made for the deletion promoter containing 441 and 2204 bp of UGT1A10 5′ promoter sequences derived from an individual homozygous for the UGT1A10 promoter deletion (termed DEL441 and DEL2204, respectively). Each construct was cotransfected with pRL-TK as an internal control for transfection and assayed for luciferase expression in three independent experiments. For transfection, the Caco-2 cell line was chosen because it was previously shown to exhibit high levels of endogenous UGT1A10 expression (Gregory et al., 2004b) and therefore contains the necessary transcriptional machinery for in vitro analysis of the UGT1A10 promoter. As shown in Fig. 3B, cells transfected with either deletion construct (DEL441 or DEL2204) exhibited a significant (p = 0.009) 3-fold increase in luciferase activity in Caco-2 cells compared with cells transfected with the wild-type UGT1A10 promoter. Cells transfected with the 2204G variant construct did not exhibit any change in luciferase activity. Cells transfected with the WT441 construct also exhibited a significant (p = 0.05) 2-fold increase in luciferase activity compared with cells transfected with the wild-type UGT1A10 promoter.
Fig. 3.
Role of the UGT1A10 promoter deletion in regulating heterologous gene expression. A, schematic representation of the PCR amplification strategy used for the UGT1A10 promoter luciferase assays. Top constructs illustrate the PCR-amplified fragments containing 186, 441, or 2204 bp of wild-type (WT) UGT1A10 promoter. The bottom constructs illustrate the PCR-amplified fragments containing either 441 or 2204 bp of deletion (DEL) variant UGT1A10 promoter. The gray lines indicate the promoter region located directly upstream of the UGT1A10 deletion; UGT1A10 deletion-containing luciferase vectors were constructed so that the UGT1A10 deletion and wild-type promoters were the same size. B, UGT1A10 wild-type, UGT1A10–2204G (containing 2204 bp of adjacent wild-type UGT1A10 promoter sequences except with a C>G SNP located at bp −1271 upstream of the UGT1A10 translation start site), and UGT1A10 deletion promoter regulation of luciferase activity in Caco-2 cells. Data are presented relative to the WT2204-overexpressing cell line. Bars indicate S.D. *, p = 0.009; **, p = 0.048, a significant increase in luciferase activity was observed compared with the WT2204-overexpressing cell line.
Discussion
Previous studies have shown that UGT1A10 exhibits high O-glucuronidation activity against a variety of substrates, including several hydroxylated metabolites of polycyclic aromatic hydrocarbons (Dellinger et al., 2006) and hormones such as estradiol (Basu et al., 2004; Starlard-Davenport et al., 2008). Although UGT1A10 was also shown to exhibit high N-glucuronidation activity against PhIP and its major metabolite, N-hydroxy-PhIP (Dellinger et al., 2007), no N-glucuronidation activity was reported against NNAL (Chen et al., 2008). In the present study, we show that UGT1A10 exhibits O-glucuronidation activity against NNAL. The KM exhibited by UGT1A10 against NNAL (5.95 mM) was close to that observed for other active UGTs, including UGT2B17 (1.76 mM; Lazarus et al., 2005), the most active NNAL glucuronidation enzyme. These data suggest that UGT1A10 may be an important NNAL detoxification enzyme in UGT1A10-expressing tissues where tobacco smoke exposure may be an important risk factor for cancer and potentially other diseases. Although UGT1A10 is expressed in lung, its levels of expression are low (Dellinger et al., 2006). UGT1A10 is highly expressed in aerodigestive tract and gastrointestinal tissues (Strassburg et al., 1997; Zheng et al., 2002); therefore, it could be an important detoxifier of NNAL and potentially other carcinogens in these tissues.
Two low-prevalence SNPs in the coding region of the UGT1A10 enzyme were previously identified, with the codon 139 Glu>Lys SNP shown to result in altered UGT1A10 activity in vitro (Dellinger et al., 2006, 2007) and was associated with altered risk for orolaryngeal carcinoma (Elahi et al., 2003). However, previous studies have not characterized the UGT1A10 promoter and variants in this region. Data from the present study indicate that the proximal promoter region of the UGT1A10 gene is relatively nonvariable, with only a 1664-bp deletion polymorphism and a single C>G SNP at nucleotide −1271 of the UGT1A10 promoter identified after direct sequencing of more than 2000 bp of UGT1A10 proximal promoter for a total of 84 UGT1A10 alleles from a white population. Whereas the UGT1A10−1271G variant was previously identified by the International HapMap Project, the identified UGT1A10 promoter deletion polymorphism has not been previously reported and was not identified by HapMap. This UGT1A10 promoter deletion allele exhibited a prevalence of 2.2% in whites but was higher in blacks, exhibiting an allelic prevalence of 5.6% in this group. Although no functional effect was observed in vitro for the UGT1A10−1271G variant, functional experiments suggest that the deletion may result in increased UGT1A10 gene expression because there was an approximately 3-fold increase in heterologous gene expression in cells containing UGT1A10 promoter sequences derived from genomic DNA containing the UGT1A10 promoter deletion allele. Increased heterologous gene expression was also observed for cells containing only 441 bp of wild-type UGT1A10 proximal promoter, suggesting that sequences located upstream of bp −441 within the deleted portion of the variant UGT1A10 promoter potentially contain transcriptional repressor elements that could be important in the regulation of UGT1A10 gene expression. These data are highly consistent with that observed in previous studies, where transcriptional repressor elements upstream of UGT1A10 bp −441 were proposed (Gregory et al., 2004a,b).
Together, these and previous studies suggest that UGT1A10 is a metabolizing enzyme important in the detoxification of NNAL as well as other potent carcinogens, and that polymorphism-induced variation in gene expression may be important in individual susceptibility to the carcinogenic activities of these agents. Genotype-phenotype and large case-control studies will be required to better establish the role of the UGT1A10 promoter deletion polymorphism in cancer susceptibility.
Acknowledgments.
We thank the Functional Genomics Core and the Molecular Genetics/DNA Sequencing Core at the Penn State University College of Medicine for real-time PCR and sequencing services, respectively.
This work was supported in part by the National Institutes of Health National Institute of Dental and Craniofacial Research [Grant R01-DE13158]; the National Institutes of Health National Cancer Institute [Grant P01-CA68384]; and Pennsylvania Department of Health's Health Research Formula Funding Program [Grants 4100038714, 4100038715].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.109.030569.
- UGT
- UDP-glucuronosyltransferase
- NNK
- 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- PAH
- polycyclic aromatic hydrocarbons
- B[a]P
- benzo[a]pyrene
- PhIP
- 2-amino-1-methyl-6-phenylimidazo[4,5-f]pyridine
- SNP
- single-nucleotide polymorphism
- NNAL
- 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol
- UDPGA
- UDP-glucuronic acid
- HPLC
- high-pressure liquid chromatography
- MEM
- minimal essential medium
- bp
- base pair
- PCR
- polymerase chain reaction
- UTR
- untranslated region.
References
- Basu NK, Kubota S, Meselhy MR, Ciotti M, Chowdhury B, Hartori M, Owens IS. (2004) Gastrointestinally distributed UDP-glucuronosyltransferase 1A10, which metabolizes estrogens and nonsteroidal anti-inflammatory drugs, depends upon phosphorylation. J Biol Chem 279:28320–28329 [DOI] [PubMed] [Google Scholar]
- Beaulieu M, Lévesque E, Hum DW, Bélanger A. (1998) Isolation and characterization of a human orphan UDP-glucuronosyltransferase, UGT2B11. Biochem Biophys Res Commun 248:44–50 [DOI] [PubMed] [Google Scholar]
- Beaulieu M, Lévesque E, Tchernof A, Beatty BG, Bélanger A, Hum DW. (1997) Chromosomal localization, structure, and regulation of the UGT2B17 gene, encoding a C19 steroid metabolizing enzyme. DNA Cell Biol 16:1143–1154 [DOI] [PubMed] [Google Scholar]
- Bélanger A, Hum DW, Beaulieu M, Lévesque E, Guillemette C, Tchernof A, Bélanger G, Turgeon D, Dubois S. (1998) Characterization and regulation of UDP-glucuronosyltransferases in steroid target tissues. J Steroid Biochem Mol Biol 65:301–310 [DOI] [PubMed] [Google Scholar]
- Burchell B, Hume R. (1999) Molecular genetic basis of Gilbert's syndrome. J Gastroenterol Hepatol 14:960–966 [DOI] [PubMed] [Google Scholar]
- Carrier JS, Turgeon D, Journault K, Hum DW, Bélanger A. (2000) Isolation and characterization of the human UGT2B7 gene. Biochem Biophys Res Commun 272:616–621 [DOI] [PubMed] [Google Scholar]
- Chen G, Dellinger RW, Sun D, Spratt TE, Lazarus P. (2008) Glucuronidation of tobacco-specific nitrosamines by UGT2B10. Drug Metab Dispos 36:824–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciotti M, Obaray R, Martín MG, Owens IS. (1997) Genetic defects at the UGT1 locus associated with Crigler-Najjar type I disease, including a prenatal diagnosis. Am J Med Genet 68:173–178 [PubMed] [Google Scholar]
- Dellinger RW, Chen G, Blevins-Primeau AS, Krzeminski J, Amin S, Lazarus P. (2007) Glucuronidation of PhIP and N-OH-PhIP by UDP-glucuronosyltransferase 1A10. Carcinogenesis 28:2412–2418 [DOI] [PubMed] [Google Scholar]
- Dellinger RW, Fang JL, Chen G, Weinberg R, Lazarus P. (2006) Importance of UDP-glucuronosyltransferase 1A10 (UGT1A10) in the detoxification of polycyclic aromatic hydrocarbons: decreased glucuronidative activity of the UGT1A10139Lys isoform. Drug Metab Dispos 34:943–949 [DOI] [PubMed] [Google Scholar]
- Elahi A, Bendaly J, Zheng Z, Muscat JE, Richie JP, Jr, Schantz SP, Lazarus P. (2003) Detection of UGT1A10 polymorphisms and their association with orolaryngeal carcinoma risk. Cancer 98:872–880 [DOI] [PubMed] [Google Scholar]
- Elahi A, Zheng Z, Park J, Eyring K, McCaffrey T, Lazarus P. (2002) The human OGG1 DNA repair enzyme and its association with orolaryngeal cancer risk. Carcinogenesis 23:1229–1234 [DOI] [PubMed] [Google Scholar]
- Fang JL, Beland FA, Doerge DR, Wiener D, Guillemette C, Marques MM, Lazarus P. (2002) Characterization of benzo(a)pyrene-trans-7,8-dihydrodiol glucuronidation by human tissue microsomes and overexpressed UDP-glucuronosyltransferase enzymes. Cancer Res 62:1978–1986 [PubMed] [Google Scholar]
- Gregory PA, Lewinsky RH, Gardner-Stephen DA, Mackenzie PI. (2004a) Coordinate regulation of the human UDP-glucuronosyltransferase 1A8, 1A9, and 1A10 genes by hepatocyte nuclear factor 1alpha and the caudal-related homeodomain protein 2. Mol Pharmacol 65:953–963 [DOI] [PubMed] [Google Scholar]
- Gregory PA, Lewinsky RH, Gardner-Stephen DA, Mackenzie PI. (2004b) Regulation of UDP glucuronosyltransferases in the gastrointestinal tract. Toxicol Appl Pharmacol 199:354–363 [DOI] [PubMed] [Google Scholar]
- Guéraud F, Paris A. (1998) Glucuronidation: a dual control. Gen Pharmacol 31:683–688 [DOI] [PubMed] [Google Scholar]
- Guillemette C, Ritter JK, Auyeung DJ, Kessler FK, Housman DE. (2000) Structural heterogeneity at the UDP-glucuronosyltransferase 1 locus: functional consequences of three novel missense mutations in the human UGT1A7 gene. Pharmacogenetics 10:629–644 [DOI] [PubMed] [Google Scholar]
- Jin C, Miners JO, Lillywhite KJ, Mackenzie PI. (1993) Complementary deoxyribonucleic acid cloning and expression of a human liver uridine diphosphate-glucuronosyltransferase glucuronidating carboxylic acid-containing drugs. J Pharmacol Exp Ther 264:475–479 [PubMed] [Google Scholar]
- Kubota T, Lewis BC, Elliot DJ, Mackenzie PI, Miners JO. (2007) Critical roles of residues 36 and 40 in the phenol and tertiary amine aglycone substrate selectivities of UDP-glucuronosyltransferases 1A3 and 1A4. Mol Pharmacol 72:1054–1062 [DOI] [PubMed] [Google Scholar]
- Lazarus P, Zheng Y, Aaron Runkle E, Muscat JE, Wiener D. (2005) Genotype-phenotype correlation between the polymorphic UGT2B17 gene deletion and NNAL glucuronidation activities in human liver microsomes. Pharmacogenet Genomics 15:769–778 [DOI] [PubMed] [Google Scholar]
- Lévesque E, Beaulieu M, Green MD, Tephly TR, Bélanger A, Hum DW. (1997) Isolation and characterization of UGT2B15(Y85): a UDP-glucuronosyltransferase encoded by a polymorphic gene. Pharmacogenetics 7:317–325 [DOI] [PubMed] [Google Scholar]
- Nagar S, Remmel RP. (2006) Uridine diphosphoglucuronosyltransferase pharmacogenetics and cancer. Oncogene 25:1659–1672 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Nakajima M, Yamanaka H, Fujiwara R, Yokoi T. (2008) Expression of UGT1A and UGT2B mRNA in human normal tissues and various cell lines. Drug Metab Dispos 36:1461–1464 [DOI] [PubMed] [Google Scholar]
- Oguri T, Takahashi T, Miyazaki M, Isobe T, Kohno N, Mackenzie PI, Fujiwara Y. (2004) UGT1A10 is responsible for SN-38 glucuronidation and its expression in human lung cancers. Anticancer Res 24:2893–2896 [PubMed] [Google Scholar]
- Owens IS, Ritter JK. (1995) Gene structure at the human UGT1 locus creates diversity in isozyme structure, substrate specificity, and regulation. Prog Nucleic Acid Res Mol Biol 51:305–338 [DOI] [PubMed] [Google Scholar]
- Park JY, Schantz SP, Stern JC, Kaur T, Lazarus P. (1999) Association between glutathione S-transferase pi genetic polymorphisms and oral cancer risk. Pharmacogenetics 9:497–504 [PubMed] [Google Scholar]
- Ren Q, Murphy SE, Zheng Z, Lazarus P. (2000) O-Glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) by human UDP-glucuronosyltransferases 2B7 and 1A9. Drug Metab Dispos 28:1352–1360 [PubMed] [Google Scholar]
- Richie JP, Jr, Carmella SG, Muscat JE, Scott DG, Akerkar SA, Hecht SS. (1997) Differences in the urinary metabolites of the tobacco-specific lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in black and white smokers. Cancer Epidemiol Biomarkers Prev 6:783–790 [PubMed] [Google Scholar]
- Ritter JK, Chen F, Sheen YY, Tran HM, Kimura S, Yeatman MT, Owens IS. (1992) A novel complex locus UGT1 encodes human bilirubin, phenol, and other UDP-glucuronosyltransferase isozymes with identical carboxyl termini. J Biol Chem 267:3257–3261 [PubMed] [Google Scholar]
- Starlard-Davenport A, Lyn-Cook B, Radominska-Pandya A. (2008) Novel identification of UDP-glucuronosyltransferase 1A10 as an estrogen-regulated target gene. Steroids 73:139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassburg CP, Oldhafer K, Manns MP, Tukey RH. (1997) Differential expression of the UGT1A locus in human liver, biliary, and gastric tissue: identification of UGT1A7 and UGT1A10 transcripts in extrahepatic tissue. Mol Pharmacol 52:212–220 [DOI] [PubMed] [Google Scholar]
- Sun D, Chen G, Dellinger RW, Duncan K, Fang JL, Lazarus P. (2006) Characterization of tamoxifen and 4-hydroxytamoxifen glucuronidation by human UGT1A4 variants. Breast Cancer Res 8:R50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tephly TR, Burchell B. (1990) UDP-glucuronosyltransferases: a family of detoxifying enzymes. Trends Pharmacol Sci 11:276–279 [DOI] [PubMed] [Google Scholar]
- Tukey RH, Strassburg CP. (2000) Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol 40:581–616 [DOI] [PubMed] [Google Scholar]
- Wiener D, Doerge DR, Fang JL, Upadhyaya P, Lazarus P. (2004a) Characterization of N-glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) in human liver: importance of UDP-glucuronosyltransferase 1A4. Drug Metab Dispos 32:72–79 [DOI] [PubMed] [Google Scholar]
- Wiener D, Fang JL, Dossett N, Lazarus P. (2004b) Correlation between UDP-glucuronosyltransferase genotypes and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone glucuronidation phenotype in human liver microsomes. Cancer Res 64:1190–1196 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Fang JL, Lazarus P. (2002) Glucuronidation: an important mechanism for detoxification of benzo[a]pyrene metabolites in aerodigestive tract tissues. Drug Metab Dispos 30:397–403 [DOI] [PubMed] [Google Scholar]