Abstract
GATA transcription factors, together with Friend of GATA (FOG) cofactors, are required for the differentiation of diverse cell types. Multiple aspects of hematopoiesis are controlled by the interaction of FOG-1 with the GATA-1/2/3 subfamily. Likewise, FOG-2 is coexpressed with the GATA-4/5/6 subfamily at other sites, including the heart and gonads. FOG-2 and GATA-4 are required for cardiac development. Through transgenic rescue of hematopoietic defects of FOG-1–/– embryos we define an unsuspected role for FOG-1 in heart development. In particular, rescued FOG-1–/– mice die at embryonic day (E) 14.5 with cardiac defects that include double outlet right ventricle and a common atrioventricular valve. Using conditional inactivation of Fog-1 we assign the cell of origin in which FOG-1 function is required. Neural crest cells migrate properly into FOG-1–/– hearts and mice with FOG-1 conditionally excised from neural crest derivatives fail to develop cardiac abnormalities. In contrast, conditional inactivation of FOG-1 in endothelial-derived tissues by means of Tie-2-expressed Cre recapitulates the rescue-knockout defects. These findings establish a nonredundant requirement for FOG-1 in the outlet tract and atrioventricular valves of the heart that depend on expression in endothelial-derived tissue and presumably reflect cooperation with the GATA-4/5/6 subfamily.
Keywords: GATA-4, neural crest, endothelium, double outlet right ventricle, cardiogenesis
GATA transcription factors play essential roles throughout development in diverse organ systems and species (1). The six mammalian GATA factors each contain two highly homologous zinc fingers (2). The carboxy-finger is principally responsible for recognition of the WGATAR DNA motif, although the amino finger contributes to binding at a subset of palindromic GATA sites (3). In addition, the amino finger mediates interaction with transcriptional cofactors known as FOGs (for Friend of GATA) (4). There are two mammalian FOG factors, FOG-1 and FOG-2 (4, 5).
Of the six GATA factors, GATA-1/2/3 constitute a subfamily based on sequence and expression similarities (1). GATA-1, the founding member of the family, is highly restricted in its expression to the hematopoietic system save for expression in Sertoli cells (6, 7). Likewise, GATA-2 and GATA-3 are expressed in hematopoietic stem cells or progenitors of specific lineages (8, 9). Gene targeting has revealed functions for GATA-1 in four hematopoietic lineages (erythroid, megakaryocytic, mast, and eosinophil) (10–13), GATA-2 in immature hematopoietic progenitors and mast cells (9, 14), and GATA-3 in T lymphocytes (8).
FOG-1 is expressed within hematopoietic lineages in an overlapping fashion with respect to GATA-1, -2, and -3 (5). During embryogenesis, FOG-1 and GATA-1 are coexpressed in yolk sac and fetal liver. Knockout of FOG-1, or a specific knock-in mutation of GATA-1 that disrupts its physical interaction with FOG-1, results in arrest in erythroid cell maturation mimicking loss of GATA-1 (10, 15, 16). Within the megakaryocyte lineage, FOG-1 acts with either GATA-1 or -2 to direct differentiation of the earliest precursors (17). Within naïve T cells FOG-1 is coexpressed with GATA-3, where it may repress GATA-3-dependent activation and promotion of T helper 2 cell development (18). Hence, multiple aspects of hematopoiesis are controlled by the interaction of FOG-1 with the GATA-1/2/3 subfamily.
In contrast to GATA-1/2/3, GATA-4/5/6 are expressed within various mesoderm- and endoderm-derived tissues, including the heart, liver, lung, gut, and urogenital system (reviewed in ref. 19). Embryos lacking GATA-4 die between embryonic days (E) 7.0 and 9.5 with cardia bifida because of a defect of ventral morphogenesis. Although GATA-4–/– embryonic stem cells generate cardiac myocytes, formation of visceral endoderm and definitive endoderm of the foregut is defective. Mice with a targeted mutation of GATA-5 are reported to be normal except for defects in the female genitourinary tract. GATA-6 loss results in early embryonic lethality at E6.5–E7.5 because of impaired extraembryonic endoderm development. A block in the differentiation of heart precursors on forced expression suggests additional roles for GATA-6 in heart development (20).
Similar to FOG-1's coexpression and function with GATA-1/2/3 in hematopoiesis, a second FOG member, FOG-2, is coexpressed with GATA-4/5/6 in the developing heart (4). FOG-2, like FOG-1, interacts with the amino finger of all GATA proteins (4, 21, 22). Loss of FOG-2 results in a constellation of cardiac defects that resembles the tetralogy of Fallot malformation, including an overriding aorta, subpulmonic stenosis, and a subaortic ventricular septal defect (23, 24). In addition, FOG-2–/– hearts lack coronary vasculature, despite the presence of an intact epicardium. This defect is rescued by transgenic expression of FOG-2 in the myocardium (24). These phenotypes are remarkably similar to those seen in mice harboring a GATA-4 knock-in mutation (called GATA-4KI/KI) that disrupts interaction with FOG proteins (25). However, GATA-4KI/KI mice demonstrate a subtle variation of phenotype, including a double outlet right ventricle (DORV) as opposed to an overriding aorta seen in FOG-2–/– mice (25).
To explore nonhematopoietic roles of FOG-1, possibly within the heart, we have rescued FOG-1–/– mice with a FOG-1 cDNA–transgene that is specifically expressed in the hematopoietic compartment. On hematopoietic rescue, the survival of FOG-1–/– embryos is extended from E10.5 to E14.5. Transgene rescued mice succumb because of heart defects, including DORV and a common atrioventricular (AV) valve. Significantly, in contrast to FOG-2–/– mice, transgene-rescued FOG-1–/– mice have normal coronary vasculature. Expression analysis reveals that FOG-1 is expressed within the outflow tract of developing hearts. Cell lineage-specific deletion of FOG-1 assigns its role in cardiac development to the endothelial lineage rather than neural crest cells. Thus, FOG-1 plays an important role in the development of the heart that is not redundant with that of the other FOG factor, FOG-2, and presumably reflects function with the GATA-4/5/6 subfamily.
Methods
FOG-1 Rescue Vector and Generation of Transgenic Mice. A 3.0-kb murine FOG-1 cDNA fragment was inserted into a GATA-1 gene expression cassette (26). Insert DNA, released by SalI digestion, was introduced by pronuclear injection into fertilized CD1 strain (Charles River Breeding Laboratories) oocytes. An expressing transgenic line was interbred with FOG-1+/– mice (129 × C57BL/6N) to obtain transgene-rescued heterozygotes that were further intermated to produce transgene-rescued FOG-1–/– embryos.
In Vitro Hematopoietic Progenitor Colony Assay. Fetal liver cells of E12.5 embryos were disaggregated and 1 × 105 cells were plated in α-MEM supplemented with 1% methylcellulose, 20% FCS, 2 units/ml erythropoietin, and 20 ng/ml c-kit ligand (Amgen Biologicals). Colonies of erythroid colony-forming units were counted on day 2 and colonies of erythroid burst-forming units were counted on day 6.
Expression Assays. In situ hybridization with 5′-[α-[35S]thio]UTP-labeled riboprobes (27), whole-mount hybridization with riboprobes labeled with digoxigenin-UTP (Roche Molecular Biochemicals), and whole-mount immunohistochemistry on isolated hearts were performed as described (24). A multiple-tissue Northern blot (Clontech) was hybridized to radiolabeled FOG-1 cDNA probe by using standard methods. The Wnt-1 Cre (28), floxed ROSA26 locus (29), and FOG-1 rescue transgene alleles were bred into the FOG-1–/– background. Hearts were isolated from E14.5 embryos and stained for β-galactosidase (LacZ) expression as described (29).
Generation of a Floxed FOG-1 Allele. A 10-kb KpnI–NotI fragment of Fog-1 genomic DNA was subcloned into a polylinker-modified Litmus 28 plasmid (New England Biolabs). We then inserted a loxP site in the intron preceding zinc finger 1, a floxed neomycin expression cassette ≈500 bp 3′ of the terminal exon, and a herpes simplex virus thymidine kinase cassette. Construct DNA was linearized with SspI and electroporated into TL1 embryonic stem cells. A correctly targeted clone was injected into C57BL/6 blastocysts after in vitro excision of the neomycin cassette. Genotyping was performed by PCR and Southern analysis.
Results
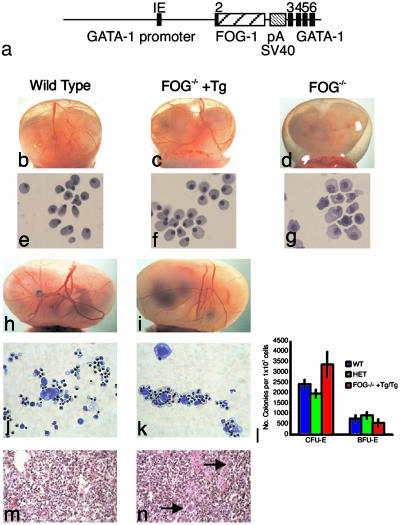
Transgenic Rescue of Hematopoiesis in FOG-1–/– Mice. To explore nonhematopoietic requirements for FOG-1 in development, we first rescued the hematopoietic defects of FOG-1–/– mice with FOG-1 cDNA expressed from regulatory sequences of the GATA-1 gene (Fig. 1a) (5). Transgene-derived RNA was expressed at levels closely approximating that of endogenous FOG-1 mRNA in spleen (data not shown). FOG-1–/– embryos die between E10.5 and E12.5 because of anemia, as reflected by a pale yolk sac (Fig. 1 b–d). Transgene-rescued FOG-1–/– embryos survive to E14.5–E15.5 with normal-appearing primitive blood cells in the yolk sac and fetal liver (Fig. 1 e–i). Definitive erythropoiesis was also normal in the rescued mice based on morphology and assay of erythroid colony-forming and burst-forming units from E14.5 livers (Fig. 1 j–l). Megakaryopoiesis was rescued in the transgene-rescued FOG-1–/– embryos, as shown by the presence of megakaryocytes in liver sections (Fig. 1 m and n). Death of transgene-bearing FOG-1–/– embryos despite normal hematopoiesis indicates that FOG-1 is essential for development of a nonhematopoietic tissue.
Fig. 1.
Transgenic rescue of hematopoiesis in FOG-1–/– embryos. (a) Structure of the GATA-1/FOG-1 transgenic construct. Seven kilobases of the GATA-1 upstream sequence and ≈1.5 kb of the downstream sequence of exon 3 flank the FOG-1 cDNA. The GATA-1 exon sequence is shown in black, FOG-1 cDNA is shown in wide stripes, and the rabbit globin poly(A) addition region and simian virus 40 sequence for polyadenylation signals and transgene detection are shown in narrow stripes. Phenotypes of wild-type (b), transgene-rescued FOG-1–/– (c), and FOG-1–/– (d) embryos with intact visceral yolk sacs (E11.5) are shown. Blood-filled vessels are visible in wild-type and rescued embryos. May–Grünwald–Giemsa-stained blood cells from E12.5 wild-type (e), rescued (f), and FOG-1–/– (g) yolk sacs are shown. (Original magnification: ×1,000.) Phenotypes of wild-type (h) and rescued (i) embryos with intact visceral yolk sacs (E14.5) and May–Grünwald–Giemsa staining of mixed colonies at day 7 of differentiation from wild-type (j) and rescued (k) embryonic livers on E14.5 are shown. Mixed colonies were harvested and cytospun before staining. (Original magnification: ×400.) (l) Number of colonies of erythroid colony-forming units (CFU-E) and burst-forming units (BFU-E) observed after 2 and 6 days of culture, respectively. Eight homozygous, 10 heterozygous, and 2 rescued embryos were analyzed. Error bars represent SEM. Cross section of livers stained with H+E from E14.5 wild-type (m) and rescued (n) embryos are shown. Arrows mark megakaryocytes in the rescued liver section. (Original magnification: ×200.)
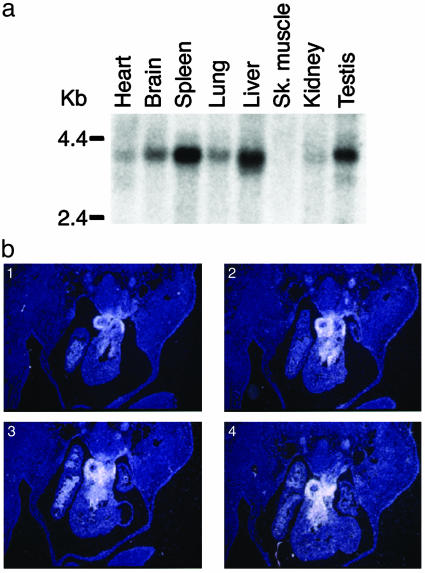
Cardiac Expression of FOG-1. FOG-1 expression was assessed by Northern analysis to determine where FOG-1 might also be required. A major ≈3.5-kb RNA transcript was detected strongly in spleen, testis, and liver. Lower level expression was also noted in the heart, brain, lung, and kidney (Fig. 2a). Examination of FOG-1 expression by 35S in situ hybridization of transverse sections of E12.5 embryos demonstrated localized expression in the developing heart. Specifically, FOG-1 message is found in the outflow tract region (Fig. 2b). Thus, FOG-1 is coexpressed in the heart along with members of the GATA-4/5/6 subfamily.
Fig. 2.
Expression analysis of FOG-1 mRNA. (a) Clontech multiple-tissue Northern blot membrane was hybridized to a FOG-1 cDNA probe. (b) Serial sections of wild-type E12.5 hearts were probed with antisense FOG-1 riboprobe.
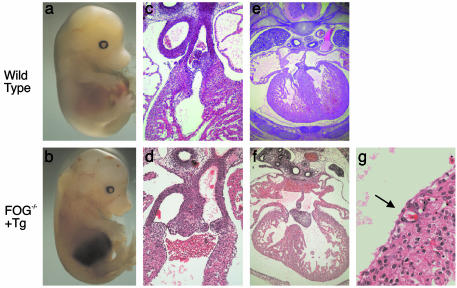
Defects in Cardiac Development in FOG-1–/– Transgene-Rescued Embryos. Timed matings demonstrated that transgene-rescued FOG-1–/– embryos died between E14.5 and E15.5, accompanied by pallor, edema, and hemorrhages (Fig. 3 a and b). These findings are consistent with cardiac failure and similar to the appearance of FOG-2–/– and GATA-4KI/KI embryos (24, 25). Heterozygous transgene-rescued FOG-1–/+ mice are born at the expected frequency and develop normally.
Fig. 3.
Cardiac defects in transgene-rescued FOG-1–/– embryos. Wild-type (a) and mutant (b) embryos at E14.5 show edema and hemorrhage in the mutant. Transverse sections of wild-type (c) and mutant (d) hearts at E14.5 show aorta and pulmonary trunk arising from the right ventricle in the mutant. (Original magnification: ×100.) Transverse sections of wild-type (e) and mutant (f) hearts at E14.5 show a common AV valve with a ventricular septal defect in the mutant. (Original magnification: ×100.) (g) Section of the ventricular wall of a mutant embryo at E14.5. The arrow identifies a coronary vessel. (Original magnification: ×400.)
The histology of wild-type and transgene-rescued FOG-1–/– embryos at E14.5 was examined after serial sectioning in the transverse plane. All mutant hearts exhibited DORV, where both the aorta and the pulmonary trunk arise from the right ventricle (Fig. 3 c and d). This pathology resembles that observed in the GATA-4KI/KI embryos (25). In addition, transgene-rescued FOG-1–/– embryos exhibited atrial and ventricular septal defects along with an AV canal defect that included a single common AV valve (as opposed to separate tricuspid and mitral valves) (Fig. 3 e and f). These findings are similar to observations made in FOG-2–/– and GATA-4KI/KI embryos (24, 25). However, in contrast to those embryos, transgene-rescued FOG-1–/– embryos have coronary vessels (Fig. 3g). In addition, the transgene-rescued FOG-1–/– myocardium is variably thinner than wild type but not as thin as that seen in the FOG-2–/– embryos.
Proper Neural Crest Cell Migration in FOG-1–/– Transgene-Rescued Embryos. The defects seen in the FOG-1 rescue mice could be attributed to the loss of FOG-1 action in one of several cardiac cell lineages. Both neural crest cells and endothelially derived mesenchymal cells of the endocardial cushions are essential cell lineages in the development of the cardiac outflow tract and the aorticopulmonary septum (28, 30). We sought to determine whether FOG-1 was required in either endothelium or neural crest.
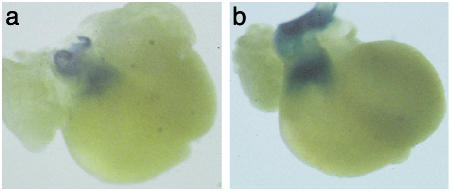
We first used a Cre-LoxP fate-mapping strategy to determine whether FOG-1 might be important for cardiac neural-crest migration. We bred a Wnt-1-Cre transgene and a stopped LacZ reporter allele into FOG-1-rescue mice to mark neural crest cells and their progeny by LacZ expression on Cre-mediated removal of a stopper fragment (29). Although the gross morphology of the hearts from the rescued embryos was abnormal, the abundance and location of neural-crest cells were similar to those seen in wild-type hearts (Fig. 4). This result suggests that a defect in neural crest cell migration is unlikely to account for cardiac defects in transgene-rescued FOG-1–/– embryos.
Fig. 4.
Cardiac neural crest cells populate the outflow tract of wild-type and transgene-rescued FOG-1–/– hearts. Hearts from E14.5 embryos that contained a Wnt-1 Cre, ROSA26 LacZ reporter of either wild-type (a) or rescued (b) genotype were dissected and stained for β-galactosidase. Positive cells populate the aorticopulmonary septum of wild-type and mutant hearts.
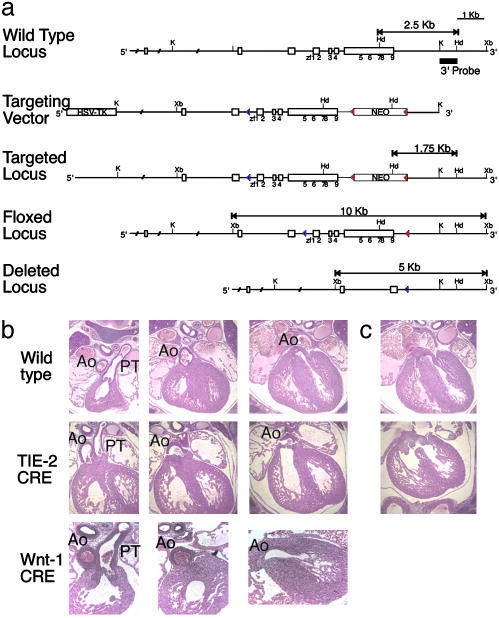
Conditional Inactivation of FOG-1 from Neural Crest Cells and Endothelial Cells. Although neural crest cells migrate into the heart of mutant embryos, we could not attest to their functionality. To address this concern we created a conditional mutant FOG-1 strain (FOG-1fl/fl; Fig. 5a) in which FOG-1 function could be ablated within specific lineages. The floxed Fog-1 allele is functional, as FOG-1fl/– and FOG-1fl/fl animals are viable. On deletion of the floxed allele in the germ line, the original FOG-1–/– phenotype is recapitulated (data not shown).
Fig. 5.
Neural crest-specific and endothelial-specific excision of a floxed Fog-1 locus. (a) Partial map of the murine Fog-1 locus (Top), the Fog-1 conditional knockout targeting vector (Top Middle), targeted homologous recombination before Cre-mediated excision of the selection cassette (Middle), after removal of the neomycin resistance (neoR) cassette (Bottom Middle), and complete deletion of the locus (Bottom). The targeting construct contains the herpes simplex virus thymidine kinase and neoR genes under the control of a phosphoglycerate kinase (PGK) promoter. Recombination results in replacement of wild-type FOG-1 with genomic DNA harboring a LoxP site in an intronic SphI site 5′ to zinc finger 1, as shown as empty boxes with positions of zinc fingers underneath. Solid triangles represent LoxP sites. The probe used in Southern blotting is shown as a black line. K, KpnI; Xb, XbaI; Hd, HindIII; S, SpeI. (b) Comparable transverse serial sections (Left to Right) of hearts from E14.5 wild-type (Top), Tie-2-Cre-mediated excision (Middle), and Wnt-1-Cre-mediated excision (Bottom) embryos. Note in the middle row, last column, that the aorta of the Tie-2-Cre-mediated FOG-1-excised heart arises from the right ventricle. (Original magnification: ×100.) Ao, aorta; PT, pulmonary trunk. (c) Hearts at E14.5 show a common AV valve with a ventricular septal defect in the Tie-2-Cre-mediated FOG-1-excised heart (Lower) and not in the wild-type heart (Upper).
Wnt-1 and Tie-2-Cre transgenes were used to inactivate FOG-1 specifically within neural crest and endothelial-derived cells, respectively (28, 30). Although Wnt-1-Cre is specific to neural crest lineages, the Tie-2-Cre exhibits partial excision within immature hematopoietic cells (unpublished data). To mitigate any consequences of Tie-2-Cre expression in hematopoietic cells, we crossed our FOG-1 rescue allele into the Tie-2-Cre and FOG-1 floxed allele matings. Mice with FOG-1 deleted from the neural crest are viable and fail to display DORV or other cardiac defects at E14.5 (Fig. 5b). On the other hand, endothelial-specific inactivation of Fog-1 recapitulates the cardiovascular phenotype observed in the FOG-1 rescue embryos (Fig. 5b). All embryos with FOG-1 deleted from the endothelium by Tie-2-Cre display DORV, a common AV valve, and an atrial and a ventricular septal defect. These results indicate that the role of FOG-1 in cardiac development is in endothelial-derived structures.
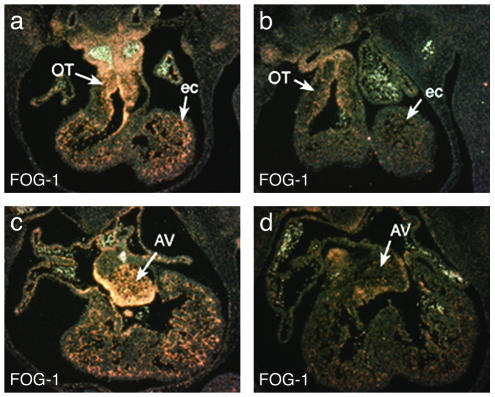
One difference between the rescue mice and the endothelial-specific FOG-1 knockout mice remained, namely in the relationship of the aorta to the pulmonary trunk. In the rescue embryos, both main arteries arise from the right ventricle at nearly the same level. However, in the endothelial-specific FOG-1 knockout, a small degree of conal rotation is present, resulting in a slightly more rostral origin of the aorta from the right ventricle compared with that of the pulmonary artery. To explore whether FOG-1 was completely ablated in the Tie-2-Cre conditional knockout, we performed FOG-1 35S in situ analysis (Fig. 6). From this analysis, we suggest that the subtle differences in phenotypes of the Tie-2-Cre-mediated deletion of FOG-1fl/fl and transgene-rescued FOG-1–/– embryos may result from residual expression of Fog-1 in endothelial cells, although we cannot rule out a contribution from an alternative cardiac lineage.
Fig. 6.
Expression of Fog-1 in wild-type (a and c) and Tie-2-Cre-mediated Fog-1-excised (b and d) hearts in E12.5 embryos in the conotruncal region (a and b) and AV valves (c and d). Note the reduced expression of Fog-1 in the heart in b and d but continued expression in the blood. OT, outflow tract; ec, endocardial cells; AV, atrioventricular valve.
Discussion
Cardiac morphogenesis is complex and reflects the interplay of different cell types, structures, multiple regulatory proteins, and signaling pathways. Congenital heart disease, the largest class of birth defects, is often caused by mutations in developmental control genes (reviewed in ref. 31). Mutations in cardiomyocyte-specific transcription factors such as GATA-4, Nkx2–5, and Tbx5 have already been implicated in congenital human cardiac disease. Whereas it has been established that the GATA cofactor FOG-2 is required for normal heart development, the work described here demonstrates that FOG-1, a cofactor not shown previously to function with the GATA-4/5/6 subfamily, is also essential for proper cardiac development.
The defects seen in the transgene-rescued FOG-1–/– embryos and the Tie-2-Cre knockout embryos are most similar to those in FOG-2–/– and GATA-4KI/KI mice. One phenotype shared by all mutants is the presence of atrial and ventricular septal defects. Recently, mutations in GATA-4 have been associated with isolated cardiac septal defects in a large human pedigree (32).
A second defect shared by the GATA-4KI/KI FOG-2–/– and transgene-rescued FOG-1–/– embryos is a common AV valve. Multiple transcription factors have been implicated in AV valve formation, including Smad6 and NF-ATc1 (reviewed in ref. 31). Of particular note, NF-ATc1 is known to cooperate with GATA-5 in endocardial development (33). In addition, NF-ATc1 binds to GATA-4 and promotes transcription of several cardiac promoters (34). Perhaps a GATA–FOG–NF-AT complex may participate in formation of the AV valves.
Two differences in the cardiac phenotypes of the FOG-2–/– and transgene-rescued FOG-1–/– mice are notable. First, transgene-rescued FOG-1–/– mice develop coronary vessels in distinction to the absence of coronary vasculature seen in FOG-2––/– and GATA-4KI/KI mice (24, 25). Second, both the transgene-rescued FOG-1–/– and GATA-4KI/KI embryos display DORV, as opposed to the overriding aorta seen in the FOG-2–/– mice. This observation suggests that GATA-4 might perform distinct roles with FOG-1 and FOG-2 in different aspects of cardiogenesis.
It is surprising perhaps that the closely related FOG family proteins do not compensate functionally for one another, but rather, loss of either results in similar cardiac defects. One possibility is that they may coordinate a common morphogenetic pathway for the great vessels within distinct cell populations. Formation of the outflow tract and AV valves requires the interaction of diverse cell types. Two cell lineages critical for the formation of the outflow tract of the heart are the endothelially derived mesenchyme of the endocardial cushions and cardiac neural crest cells which migrate into the cardiac outflow tract and form the aorticopulmonary septum (28, 30). Endothelial cells that undergo endocardial–mesenchymal transformation are also essential for the development of the AV valves (30). Disruption of several different genes expressed within either lineage [e.g., NF-1 (35) in endothelium and Pax3 (27) and platelet-derived growth factor α (36) in neural crest] results in cardiac outflow tract abnormalities. From myocardial-specific transgenic rescue, we previously concluded that FOG-2 is required within the myocardium for the initiation of coronary vasculature (24). In this work we have shown that FOG-1 acts specifically within the endothelium for its role in heart development.
It is likely that FOG-1 cooperates with one (or multiple) of the GATA-4/5/6 subfamily within the endothelium of the developing heart. All three factors are coexpressed in the endocardium (2). Mutations of the GATA-5 gene in the zebrafish faust mutant lead to defects of endocardial differentiation (37). In vitro cell assays also point to a pivotal role for GATA-5 in the differentiation of committed cardiogenic precursors to endothelial endocardial cells (33). However, GATA-4 is also present in endocardial cells and the GATA-4KI/KI mice display a remarkably similar phenotype of DORV and a common AV valve. In either scenario, our findings suggest that FOG-1 functions with the GATA-4/5/6 subfamily in the endothelium of the heart, similar to its ability to serve as a transcriptional cofactor to GATA-1/2/3 in the blood. The strong conservation of critical amino acid residues that mediate association of GATA and FOG proteins is consistent with mixing and matching of factors in different developmental contexts (38).
It is notable that endothelium and blood are postulated to share a common precursor, the hemangioblast (39), within the early embryo. Recently, mutations in the gene encoding the phosphatase Shp2 have been associated with Noonan syndrome, a condition characterized by endocardial cushion defects (40). Noonan patients also have an increased incidence of leukemia, and mutations in the Shp2 gene are found in additional patients with juvenile myelomonocytic leukemia (41). Likewise, the type 1 neurofibromatosis gene Nf1 plays an essential role in endothelial cells, and Tie-2-Cre-mediated inactivation of Nf1 leads to both cardiac defects and hematological disorders (ref. 35 and unpublished observations of J.A.E.). Thus, FOG-1 may be a component of a conserved regulatory pathway that functions in derivatives of a precursor to the blood and vascular lineages.
Acknowledgments
We thank S. Tevosian for advice and discussions, S. Litovsky for his review of cardiac pathology, and M. Hamblen, C. Browne, and M. Sasaluxanon for technical assistance. S.G.K. is supported by the Howard Hughes Medical Institute Predoctoral Fellowship Program and the National Institutes of Health Medical Scientist Training Program. S.H.O. is an Investigator of the Howard Hughes Medical Institute. Partial support for these studies was derived from a National Institutes of Health grant to S.H.O.
Abbreviations: AV, atrioventricular; DORV, double outlet right ventricle; En, embryonic day n; FOG, Friend of GATA.
References
- 1.Orkin, S. H. (1992) Blood 80, 575–581. [PubMed] [Google Scholar]
- 2.Laverriere, A. C., MacNeill, C., Mueller, C., Poelmann, R. E., Burch, J. B. & Evans, T. (1994) J. Biol. Chem. 269, 23177–23184. [PubMed] [Google Scholar]
- 3.Trainor, C. D., Omichinski, J. G., Vandergon, T. L., Gronenborn, A. M., Clore, G. M. & Felsenfeld, G. (1996) Mol. Cell. Biol. 16, 2238–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tevosian, S. G., Deconinck, A. E., Cantor, A. B., Rieff, H. I., Fujiwara, Y., Corfas, G. & Orkin, S. H. (1999) Proc. Natl. Acad. Sci. USA 96, 950–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsang, A. P., Visvader, J. E., Turner, C. A., Fujiwara, Y., Yu, C., Weiss, M. J., Crossley, M. & Orkin, S. H. (1997) Cell 90, 109–119. [DOI] [PubMed] [Google Scholar]
- 6.Tsai, S. F., Martin, D. I., Zon, L. I., D'Andrea, A. D., Wong, G. G. & Orkin, S. H. (1989) Nature 339, 446–451. [DOI] [PubMed] [Google Scholar]
- 7.Yomogida, K., Ohtani, H., Harigae, H., Ito, E., Nishimune, Y., Engel, J. D. & Yamamoto, M. (1994) Development (Cambridge, U.K.) 120, 1759–1766. [DOI] [PubMed] [Google Scholar]
- 8.Ting, C. N., Olson, M. C., Barton, K. P. & Leiden, J. M. (1996) Nature 384, 474–478. [DOI] [PubMed] [Google Scholar]
- 9.Tsai, F. Y., Keller, G., Kuo, F. C., Weiss, M., Chen, J., Rosenblatt, M., Alt, F. W. & Orkin, S. H. (1994) Nature 371, 221–226. [DOI] [PubMed] [Google Scholar]
- 10.Fujiwara, Y., Browne, C. P., Cunniff, K., Goff, S. C. & Orkin, S. H. (1996) Proc. Natl. Acad. Sci. USA 93, 12355–12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shivdasani, R. A., Fujiwara, Y., McDevitt, M. A. & Orkin, S. H. (1997) EMBO J. 16, 3965–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Migliaccio, A. R., Rana, R. A., Sanchez, M., Lorenzini, R., Centurione, L., Bianchi, L., Vannucchi, A. M., Migliaccio, G. & Orkin, S. H. (2003) J. Exp. Med. 197, 281–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu, C., Cantor, A. B., Yang, H., Browne, C., Wells, R. A., Fujiwara, Y. & Orkin, S. H. (2002) J. Exp. Med. 195, 1387–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai, F. Y. & Orkin, S. H. (1997) Blood 89, 3636–3643. [PubMed] [Google Scholar]
- 15.Tsang, A. P., Fujiwara, Y., Hom, D. B. & Orkin, S. H. (1998) Genes Dev. 12, 1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nichols, K. E., Crispino, J. D., Poncz, M., White, J. G., Orkin, S. H., Maris, J. M. & Weiss, M. J. (2000) Nat. Genet. 24, 266–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang, A. N., Cantor, A. B., Fujiwara, Y., Lodish, M. B., Droho, S., Crispino, J. D. & Orkin, S. H. (2002) Proc. Natl. Acad. Sci. USA 99, 9237–9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou, M., Ouyang, W., Gong, Q., Katz, S. G., White, J. M., Orkin, S. H. & Murphy, K. M. (2001) J. Exp. Med. 194, 1461–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molkentin, J. D. (2000) J. Biol. Chem. 275, 38949–38952. [DOI] [PubMed] [Google Scholar]
- 20.Gove, C., Walmsley, M., Nijjar, S., Bertwistle, D., Guille, M., Partington, G., Bomford, A. & Patient, R. (1997) EMBO J. 16, 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu, J. R., McKinsey, T. A., Xu, H., Wang, D. Z., Richardson, J. A. & Olson, E. N. (1999) Mol. Cell. Biol. 19, 4495–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svensson, E. C., Tufts, R. L., Polk, C. E. & Leiden, J. M. (1999) Proc. Natl. Acad. Sci. USA 96, 956–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Svensson, E. C., Huggins, G. S., Lin, H., Clendenin, C., Jiang, F., Tufts, R., Dardik, F. B. & Leiden, J. M. (2000) Nat. Genet. 25, 353–356. [DOI] [PubMed] [Google Scholar]
- 24.Tevosian, S. G., Deconinck, A. E., Tanaka, M., Schinke, M., Litovsky, S. H., Izumo, S., Fujiwara, Y. & Orkin, S. H. (2000) Cell 101, 729–739. [DOI] [PubMed] [Google Scholar]
- 25.Crispino, J. D., Lodish, M. B., Thurberg, B. L., Litovsky, S. H., Collins, T., Molkentin, J. D. & Orkin, S. H. (2001) Genes Dev. 15, 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Visvader, J. E., Fujiwara, Y. & Orkin, S. H. (1998) Genes Dev. 12, 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Epstein, J. A., Li, J., Lang, D., Chen, F., Brown, C. B., Jin, F., Lu, M. M., Thomas, M., Liu, E., Wessels, A. & Lo, C. W. (2000) Development (Cambridge, U.K.) 127, 1869–1878. [DOI] [PubMed] [Google Scholar]
- 28.Jiang, X., Rowitch, D. H., Soriano, P., McMahon, A. P. & Sucov, H. M. (2000) Development (Cambridge, U.K.) 127, 1607–1616. [DOI] [PubMed] [Google Scholar]
- 29.Mao, X., Fujiwara, Y. & Orkin, S. H. (1999) Proc. Natl. Acad. Sci. USA 96, 5037–5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kisanuki, Y. Y., Hammer, R. E., Miyazaki, J., Williams, S. C., Richardson, J. A. & Yanagisawa, M. (2001) Dev. Biol. 230, 230–242. [DOI] [PubMed] [Google Scholar]
- 31.Srivastava, D. & Olson, E. N. (2000) Nature 407, 221–226. [DOI] [PubMed] [Google Scholar]
- 32.Garg, V., Kathiriya, I. S., Barnes, R., Schluterman, M. K., King, I. N., Butler, C. A., Rothrock, C. R., Eapen, R. S., Hirayama-Yamada, K., Joo, K., et al. (2003) Nature 424, 443–447. [DOI] [PubMed] [Google Scholar]
- 33.Nemer, G. & Nemer, M. (2001) Ann. Med. 33, 604–610. [DOI] [PubMed] [Google Scholar]
- 34.Molkentin, J. D., Lu, J. R., Antos, C. L., Markham, B., Richardson, J., Robbins, J., Grant, S. R. & Olson, E. N. (1998) Cell 93, 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gitler, A. D., Zhu, Y., Ismat, F. A., Lu, M. M., Yamauchi, Y., Parada, L. F. & Epstein, J. A. (2003) Nat. Genet. 33, 75–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orr-Urtreger, A., Bedford, M. T., Do, M. S., Eisenbach, L. & Lonai, P. (1992) Development (Cambridge, U.K.) 115, 289–303. [DOI] [PubMed] [Google Scholar]
- 37.Reiter, J. F., Alexander, J., Rodaway, A., Yelon, D., Patient, R., Holder, N. & Stainier, D. Y. (1999) Genes Dev. 13, 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crispino, J. D., Lodish, M. B., MacKay, J. P. & Orkin, S. H. (1999) Mol. Cell 3, 219–228. [DOI] [PubMed] [Google Scholar]
- 39.Choi, K. (2002) J. Hematother. Stem Cell Res. 11, 91–101. [DOI] [PubMed] [Google Scholar]
- 40.Masheshwari, M., Belmont, J., Fernbach, S., Ho, T., Molinari, L., Yakub, I., Yu, F., Combes, A., Towbin, J., Craigen, W. J. & Gibbs, R. (2002) Hum. Mutat. 20, 298–304. [DOI] [PubMed] [Google Scholar]
- 41.Neel, B. G., Gu, H. & Pao, L. (2003) Trends Biochem. Sci. 28, 284–293. [DOI] [PubMed] [Google Scholar]