Abstract
Utilizing a recently identified Sox10 distal enhancer directing Cre expression, we report S4F:Cre, a transgenic mouse line capable of inducing recombination in oligodendroglia and all examined neural crest derived tissues. Assayed using R26R:LacZ reporter mice expression was detected in neural crest derived tissues including the forming facial skeleton, dorsal root ganglia, sympathetic ganglia, enteric nervous system, aortae and melanoblasts, consistent with Sox10 expression. LacZ reporter expression was also detected in non-neural crest derived tissues including the oligodendrocytes and the ventral neural tube. This line provides appreciable differences in Cre expression pattern from other transgenic mouse lines that mark neural crest populations, including additional populations defined by the expression of other SoxE proteins. The S4F:Cre transgenic line will thus serve as a powerful tool for lineage tracing, gene function characterization and genome manipulation in these populations.
The neural crest (NC) is a transient migratory collective of cell populations that arise at the dorsal aspect of the neural tube as it closes. This emigration occurs along the length of the anterior-posterior axis and gives rise to a myriad of structures including, but not limited to, melanocytes, the sympathetic nervous system, the enteric (parasympathetic) nervous system (ENS), connective tissue of the face and neck and peripheral myelinating glia (Schwann cells) (Douarin and Kalcheim, 1999). SOX10 (SRY-box containing gene 10) encodes a critical transcription factor in neural crest development (Britsch et al., 2001; Southard-Smith et al., 1998) and is expressed in most, if not all, NC-derived lineages. Loss-of-function SOX10 mutations result in Waardenburg-Shah Syndrome in humans (WS4; OMIM Accession No. 277580) (Mollaaghababa and Pavan, 2003; Pingault et al., 1998) which is characterized by defects in multiple NC-derived populations, including epidermal hypopigmentation, absence of the pigmented stria vascularis of the inner ear, and the enteric aganglionosis characteristic of Hirschsprung disease (HSCR). Dominant-negative SOX10 mutations have been identified in patients with Peripheral demyelinating neuropathy, Central dysmyelinating leukodystrophy, Waardenburg Syndrome and HSCR (PCWH; OMIM Accession No. 609136) (Chakravarti, 2003; Inoue et al., 2004). Additionally, multiple mutant alleles with corresponding nervous system and pigmentary anomalies have been described and studied in model organisms, including mice and zebrafish, underscoring the important role played by SOX10 in neural crest development (Dutton et al., 2001; Southard-Smith et al., 1998).
Shortly after their emigration from the neural tube Sox10 expression is down-regulated in most NC derivatives, except melanocytes (mouse not zebrafish) and Schwann cells (Elworthy et al., 2003; Kim et al., 2003; Kuhlbrodt et al., 1998; Mollaaghababa and Pavan, 2003; Osawa et al., 2005; Southard-Smith et al., 1999). However, while SOX10 is also expressed in presumptive oligodendrocytes, its expression is upregulated and maintained in the mature oligodendrocytes upon myelination (Kuhlbrodt et al., 1998; Southard-Smith et al., 1999). Importantly, the NC population specific down-regulation of Sox10 expression has thus far limited its use in fate mapping of NC cells, prompting our search for sequences with broad regulatory control in many NC populations.
We have recently described the identification of a distal enhancer approximately 28.5 kb upstream of the mouse Sox10 transcriptional start site that directs reporter expression in a near pan-neural crest manner (Antonellis et al., 2008), verifying our observations in both transgenic zebrafish and transgenic mice. We termed this enhancer multi-species conserved sequence 4 (MCS4; (Antonellis et al., 2008)). Reporter expression directed by MCS4 is evident in both transgenic zebrafish and mice in early NC-derived populations, including the cranial ganglia, dorsal root ganglia, sympathetic chain ganglia, melanoblasts, and the enteric nervous system. Additionally, we demonstrated reporter expression in myelinating glial populations in the corresponding transgenic zebrafish lines. Thus we set out to use this sequence to establish a Cre-expressing mouse strain that would mark and enable analysis of most, if not all, NC-derived populations and oligodendroglia throughout the life of the animal.
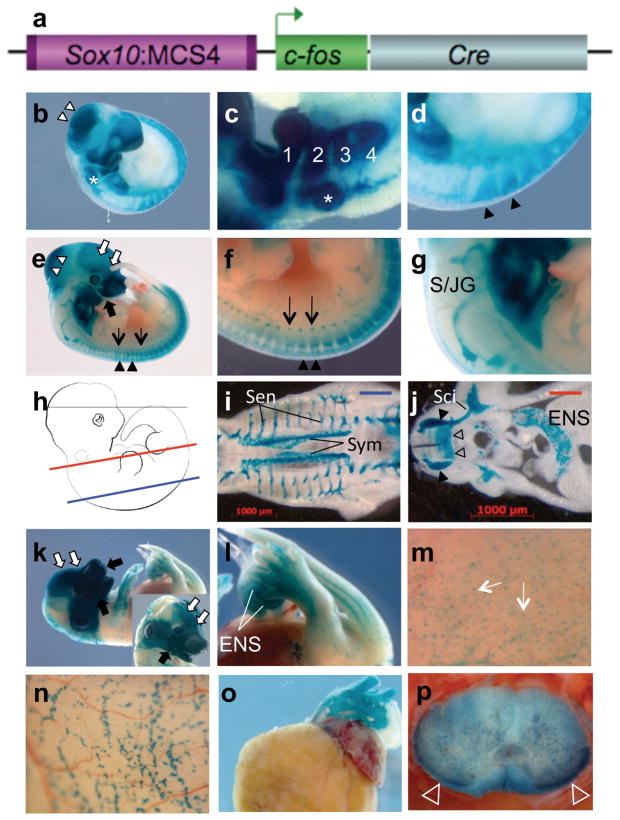
We generated a transgenic mouse line containing a construct placing MCS4 in front of the mouse c-Fos minimal promoter and the Cre recombinase open reading frame (Fig. 1A), We established timed matings between mice hemizygous for the S4F:Cre (Sox10-MCS4-c-Fos-Cre) transgene and mice heterozygous for the Cre-inducible R26R:LacZ reporter mice (Soriano, 1999). Embryos were isolated at specific time points mid-gestation (E8.75, E9-9.5, E11.5 and E13.5) and LacZ reporter expression was detected by β-galactosidase staining. All S4F:Cre;R26R:LacZ embryos older than 9.0 days displayed reporter patterns consistent with the known expression of Sox10 (Britsch et al., 2001; Southard-Smith et al., 1998) and patterns reported for Sox10_MCS4 directed LacZ reporter expression (Antonellis et al., 2008). By E9.5, S4F:Cre;R26R:LacZ embryos demonstrated LacZ reporter expression in the dorsal root ganglia pharyngeal arches (Figure 1B,C,D), and in the otic vesicle, consistent with neural crest contribution to those structures. However, there is little evidence of expression within the dorsal aspect of the neural tube, suggesting that either this enhancer sequence is not active until after presumptive neural crest have delaminated and begun their migratory journey or that there is insufficient time between activation and emigration to detect induction of reporter in the pre-migratory population. At E11.5, LacZ expression was detected in the viscero- and neurocranial crest-derived craniofacial mesenchyme, in the sympathetic and sensory ganglia, dorsal root ganglia (DRG), and the superior and jugular ganglia (Figure 1E, F, G). Sections taken through embryos at this stage similarly demonstrate LacZ signal in sympathetic and sensory ganglia, DRG, Sciatic nerve, enteric nervous system and the ventral columns of the neural tube (Figure 1H, I, J). Additionally we detect signal dorsally around the midbrain/hindbrain boundary (Figure 1E). Although Nichols (1981) reported that cranial crest migration at the level of the midbrain/hindbrain persists for an unspecified time after closure of the neural tube we hesitate to identify these as active NC émigrés. To get a clearer image of the tissues marked in this region we generated sagittal sections in E12 embryos (see supplemental figure). LacZ signal in these sections is most heavily concentrated in the roof of the midbrain and the fourth ventricle. Despite being indelibly marked, signal in this area is significantly diminished at E13.5 suggesting that a fraction of the cell population may have been migratory (Figure 1K); observed signal is superficial, restricted to cell layers consistent with presumptive connective tissue of the skull at an axial level of the boundary between midbrain and hindbrain. These facts taken in combination may be consistent with a crest contribution to the interparietal bone as proposed by Yoshida et al., (2008) but we cannot say this unequivocally. This is also consistent with the proposed existence of cephalic neural crest cells with neurogenic, melanogenic, and osteogenic differentiation capacities (Calloni et al., 2009). We cannot, however, exclude the possibility that signal reduction in neuronal populations of the midbrain and hindbrain at this stage in part reflects reduced reagent penetration in older embryos.
Figure 1.
S4F:Cre induces LacZ reporter expression in R26R:LacZ in developing transgenic mice in neural crest derived and oliogodendroglial cell populations. A) Design of S4F:Cre construct incorporating Sox10:MCS-28.5 enhancer, the c-Fos minimal promoter and Cre coding sequence. B, C and D) LacZ reporter expression is detected in tissues of whole mount embryos at embryonic day 9.5 (E9.5), including expression at the axial level of the midbrain/hindbrain boundary (B, White arrowhead), otic vesicle (asterisk, B and C), in the pharyngeal arches (1–4, C), and the dorsal root ganglia (DRG) (B and Black Arrowhead in Panel D). Panels E, F, G) show LacZ reporter expression detected in E11.5 mice, labeling structures consistent with facial mesenchyme derived from viscerocranial (E, Black filled arrow) and neurocranial (E, White filled arrow) crest, DRG (Black Arrowhead, E and F), Sympathetic chain (Black Arrows, E and F), and in the superior/jugular ganglion (S/JG, G). H) Schematic illustrating the level of vibratome section cuts viewed in panels I (Blue) and J (Red). I) LacZ reporter expression in a section at the level of the spine (Blue, panel I) of E11.5 stained mice identifying structures consistent with sensory (Sen) and sympathetic (Sym) ganglia. J) Reporter expression in a section through the viscera (Red, panel I), uncovering expression in cell populations consistent with the DRG (Black Arrowheads), Sciatic nerve (Sci) and the enteric nervous system (ENS). K, L and M) LacZ reporter expression detected in whole mount E13.5 embryos, illustrating reporter expression in structures consistent with facial mesenchyme derived from viscerocranial (Black filled arrow) and neurocranial (White filled arrow) crest (K), the ENS (L) and melanoblasts (White Arrows, M). N) LacZ staining in the myenteric plexus of the ENS of a whole mount portion of the adult small bowel. O) Reporter signal consistent with neural crest contribution to the aorta of the adult heart. P) Signal consistent with oligodendrocytes in the ventral columns of the adult cervical spine (Open white arrowheads).
At E13.5, LacZ reporter expression also clearly demarcates all of the forming craniofacial connective tissue derived from neurocranial and viscerocranial crest populations; reporter signal is clearly detected in the mid-gut loops protruding through the umbilical hernia consistent with the placement of crest-derived enteric neuroblasts; and it is also detected in the migrating presumptive melanoblasts of the forming ectoderm (Figure 1K, L and M). Confirmation of the identity of the embryonic midgut expression is found in the LacZ signal that clearly delineates the myenteric plexus of the adult small bowel (Figure 1N). At E15.5, whole mount LacZ stains the outflow tract of the developing heart (data not shown) but is perhaps more distinctly seen in the major vessels, specifically the aortae (Figure 1O) and pulmonary artery (data not shown) of the adult heart. Additionally, reporter expression is also clearly detected in the oligodendroglial populations particularly those concentrated in the ventral columns of the adult cervical spine (Figure 1P). These data are consistent with endogenous Sox10 expression and with our recent analyses of multiple independent transgenic mice expressing LacZ under direct control of Sox10 MCS 4 and GFP-expressing zebrafish lines generated using the Sox10 MCS 4 sequence (Antonellis et al., 2008).
S4F:Cre line is now one of several Cre-expressing driver lines whose spatial expression pattern includes a spectrum of neural crest-derived cell populations. While similar lines generated with regulatory sequences taken from the Wnt1 and the human tissue plasminogen activator (Ht-PA) loci also mark many NC-derived tissues, certain notable differences exist (Table 1; (Pietri et al., 2003)). In addition to NC lineages, Wnt1:Cre also directs expression the dorsal neural tube prior to NC emigration, and in cells of the midbrain/hindbrain boundary area that are identified as cephalic mesenchyme and expand subsequently to label neuronal populations throughout the midbrain (dorsal and ventral) (Brault et al., 2001; Chai et al., 2000; Danielian et al., 1998; Echelard et al., 1994; Jiang et al., 2000; Pietri et al., 2003). This pattern is different from that seen in our S4F:Cre line in two significant ways. First, in S4F:Cre the reporter expression in the region of the midbrain/hindbrain boundary remains dorsally located, predominating in what is consistent with connective tissue of the skull above the roof of the midbrain as well as in the roof of the midbrain itself. It is also unclear whether Wnt1:Cre expression in the area extends outside the CNS. Second, as described above, by E13.5 S4F:Cre-induced signal has become restricted to the vault of the forming skull. By contrast with both Wnt1:Cre and S4F:Cre mice, Ht-PA does not mark any cells at the axial position of the midbrain or hindbrain (Table 1; (Pietri et al., 2003)).
Table 1.
Comparison of the pattern of β-galactosidase activity in S4F:Cre;R26R:LacZ, Wnt1-Cre;R26R:LacZ and Ht-Pa-Cre;R26R:LacZ mice. List of tissues in which β-galactosidase activity is induced by S4F:Cre, Ht-Pa:Cre (Pietri et al., 2003)and Wnt1:Cre (Brault et al., 2001; Chai et al., 2000; Danielian et al., 1998; Echelard et al., 1994; Jiang et al., 2000; Pietri et al., 2003).
| Tissue | S4F:Cre | Wnt1:Cre | Ht-Pa:Cre |
|---|---|---|---|
| Otic Vessicle | Yes | Yes | Yes |
| Dorsal Root Ganglia* | Yes | Yes | Yes |
| Sympathetic Ganglia* | Yes | Yes | Yes |
| Craniofacial Mesenchyme* | Yes | Yes | Yes |
| Melanoblasts* | Yes | Yes | Yes |
| Enteric Nervous System* | Yes | Yes | Yes |
| Superior/Jugular Ganglion* | Yes | Yes | Yes |
| Oligodendroglia | Yes | No | No |
| Aortae* | Yes | Yes | Yes |
| Ventral Neural Tube | Yes | No | No |
| Developing Limb | Yes | No | No |
Yes= β-galactosidase detected or reported in this tissue, No= β-galactosidase not detected or reported in this tissue.
, Structures known to be colonized or established by neural crest-derived populations.
Consistent with endogenous Sox10 expression, S4F:Cre appears also to mark oligodendroglial populations of the CNS. By contrast neither Wnt1:Cre nor Ht-PA:Cre mark oligodendroglial populations. We also observed S4F:Cre marking of the ventral neural tube at E11.5 (Figure 1J), which predates the endogenous SOX10 expression in that region. While we cannot discount that the ventral neural tube expression may be caused by position effect, this expression pattern may be explained by comparison with the spatial and temporal expression of the remaining SoxE proteins (Sox8 and Sox9). In our initial analysis of MCS4 sequence in mice, we identified dimeric SoxE binding sites that are perfectly conserved from mammalian to avian species and are also directly critical for the regulatory activity of the element (Antonellis et al., 2008). Thus our observation of LacZ signal in the ventral neural tube, fated for the formation of oligodendroglia and motor neurons may reflect the activation of this element by Sox9, which is required for the genesis of oligodendrocyte precursors (Stolt et al., 2003), or Sox8 (Stolt et al., 2004). Furthermore, SoxE genes are known or predicted to be subject to transcriptional regulation by one another and to display partial functional redundancy in different tissues (Cheung and Briscoe, 2003; Cheung et al., 2005; McKeown et al., 2005; Suzuki et al., 2006). We also note two further observations that are not overtly consistent with what is established of Sox10 expression. First, we detect LacZ expression in the forming limb (Figure 1L) and second, this transgene appears to be active in the male germline (data not shown). Although the latter is consistent with the known role for Sox9 but not Sox10 in the genesis of sertoli cells (Kent et al., 1996), the former is more difficult to attribute at this stage. The expression in the limb spatially resembles expression in the developing skeletal muscle and as such may be consistent with the distribution of Sox8. However, we cannot exclude the potential that this location of reporter expression simply reflects a permissive attribute of the tested enhancer or the impact of genomic location of the inserted transgene.
In summary, S4F:Cre mice mark oligodendroglial populations as well as all evaluated ectomesenchymal and non-ectomesenchymal NC-derived populations in a manner consistent with endogenous Sox10 expression; these tissues include all craniofacial skeletal components, sympathetic and parasympathetic neuronal/glial populations as well as epidermal melanocyte precursors. Additionally, this line marks several lineages in which (or at times when) SoxE proteins other than Sox10 play critical roles. We suggest this line may thus serve as a robust in vivo reagent, not only facilitating experiments to better understand Sox10 regulatory control but also enabling genome manipulation and lineage tracing of nearly complete populations of neural crest cells and oligodendroglia, among others, in both normal and disease contexts.
Methods
Generation of Trangsgenic Line
S4F:Cre was generated using an overlapping PCR strategy (Yolov and Shabarova, 1990), peformed with Pfu Ultra Taq (Stratagene). A PCR amplicon containing a Gateway® Invitrogen targeting construct and the murine c-Fos promoter was amplified off of pTol2GW (F Primer-GGCCGGCCGATATCATCACAAGTTTGTACAAAAAAGCTGAACGAGAAACG; R Primer-GGTGTACGGTCAGTAAATTGGACATACCGGTGGATCCTCGAAGTTTGGGG) (Fisher et al., 2006). A PCR amplicon containing the Cre open reading frame and polyadenylation signal were amplified off of the pCX-Cre (Forward Primer-CCCCAAACTTCGAGGATCCACCGGTATGTCCAATTTACTGACCGTACACCAAAATTT GCC; Reverse Primer- GGCGCGCCGAGAAGAGGGACAGCTATGACTGGGAG)(Bilodeau et al., 2007). Using homologous sequences in the primers, the Gateway c-Fos promoter amplicon and the Cre-polyA sequence amplicon were merged. The Gateway c-Fos promoter Cre construct was TOPO cloned into pCR2.1 (Invitrogen). The Sox10 MCS 4 enhancer (Antonellis et al., 2008) was placed in front of the c-Fos minimal promoter directing Cre recombinase expression using Gateway recombination. The construct was injected into B6SJLF1 pronuclei. A PCR genotyping strategy was used to identify founders using primers against Cre (Cre Forward Primer- CGTACTGACGGTGGGAGAAT; Cre Reverse Primer-CCCGGCAAAACAGGTAGTTA) Four G0 transgenic mice were identified that were positive for the transgene, of which only one transmitted through the germline in crosses with R26R:LacZ heterozygous mice.
LacZ Staining
Pups used for LacZ staining were generated using timed matings, counting 12:00 pm of the day that vaginal plugs were observed as day 0.5. Whole mount LacZ staining was then performed as described previously (Jackson and Abbott, 2000). Briefly, mice E13.5 and younger were fixed for 1 hour in 4% PFA, mice older than E13.5 were fixed overnight in 4% PFA, washed in LacZ wash solution and stained at room temperature monitored under a dissecting scope. The reaction was then stopped, and mice were stored in methanol until photographing.
Supplementary Material
Acknowledgments
We would like to thank Dr Roger Reeves for his kind gift of the R26R:LacZ reporter mice and Dr David Huso for his expert assistance with analysis. We would also like to thank David McGaughey, Megana Prasad and Rachel Stine for their critical reading of the manuscript. The work was funded in part by the National Human Genome Research Institute’s (NHGRI) Intramural Research Program, by the National Institute of General Medical Sciences ((R01GM071648; NIGMS, NIH) and Institutional funds from the Johns Hopkins Department of Pediatric Urology.
References
- Antonellis A, Huynh JL, Lee-Lin S-Q, Vinton RM, Renaud G, Loftus SK, Elliot G, Wolfsberg TG, Green ED, McCallion AS, Pavan WJ. Identification of Neural Crest and Glial Enhancers at the Mouse Sox10 Locus through Transgenesis in Zebrafish. PLoS Genetics. 2008;4:e1000174–e1000174. doi: 10.1371/journal.pgen.1000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau M, Girard S, Hebert J, Sauvageau G. A retroviral strategy that efficiently creates chromosomal deletions in mammalian cells. Nat Methods. 2007;4:263–268. doi: 10.1038/nmeth1011. [DOI] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. Development. Vol. 128. Cambridge, England: 2001. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development; pp. 1253–1264. [DOI] [PubMed] [Google Scholar]
- Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave K-A, Birchmeier C, Wegner M. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes & Development. 2001;15:66?78–66?78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calloni GW, Le Douarin NM, Dupin E. High frequency of cephalic neural crest cells shows coexistence of neurogenic, melanogenic, and osteogenic differentiation capacities. Proc Natl Acad Sci U S A. 2009;106:8947–8952. doi: 10.1073/pnas.0903780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Development. Vol. 127. Cambridge, England: 2000. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis; pp. 1671–1679. [DOI] [PubMed] [Google Scholar]
- Chakravarti A, McCallion AS, Lyonnet S. Hirschsprung Disease. In: Scriver B, Valle, Sly, editors. The Metabolic and Molecular Bases of Inherited Disease. 8. Chapter 251. New York: McGraw-Hill; 2003. [Google Scholar]
- Cheung M, Briscoe J. Development. Vol. 130. Cambridge, England: 2003. Neural crest development is regulated by the transcription factor Sox9; pp. 5681–5693. [DOI] [PubMed] [Google Scholar]
- Cheung M, Chaboissier M-C, Mynett A, Hirst E, Schedl A, Briscoe J. The transcriptional control of trunk neural crest induction, survival, and delamination. Developmental Cell. 2005;8:179–192. doi: 10.1016/j.devcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Current Biology: CB. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Douarin NL, Kalcheim C. The Neural Crest. 2. Cambridge University Press; 1999. p. 445. [Google Scholar]
- Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, Rauch J, Geisler R, Haffter P, Kelsh RN. Development. Vol. 128. Cambridge, England: 2001. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates; pp. 4113–4125. [DOI] [PubMed] [Google Scholar]
- Echelard Y, Vassileva G, McMahon AP. Development. Vol. 120. Cambridge, England: 1994. Cis-acting regulatory sequences governing Wnt-1 expression in the developing mouse CNS; pp. 2213–2224. [DOI] [PubMed] [Google Scholar]
- Elworthy S, Lister JA, Carney TJ, Raible DW, Kelsh RN. Transcriptional regulation of mitfa accounts for the sox10 requirement in zebrafish melanophore development. Development. 2003;130:2809–2818. doi: 10.1242/dev.00461. [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006;312:276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- Inoue K, Khajavi M, Ohyama T, Hirabayashi S-i, Wilson J, Reggin JD, Mancias P, Butler IJ, Wilkinson MF, Wegner M, Lupski JR. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nature Genetics. 2004;36:361–369. doi: 10.1038/ng1322. [DOI] [PubMed] [Google Scholar]
- Jackson IJ, Abbott CM. A Practical Approach. Oxford University Press; USA: 2000. Mouse Genetics and Transgenics. [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Development. Vol. 127. Cambridge, England: 2000. Fate of the mammalian cardiac neural crest; pp. 1607–1616. [DOI] [PubMed] [Google Scholar]
- Kent J, Wheatley SC, Andrews JE, Sinclair AH, Koopman P. Development. Vol. 122. Cambridge, England: 1996. A male-specific role for SOX9 in vertebrate sex determination; pp. 2813–2822. [DOI] [PubMed] [Google Scholar]
- Kim J, Lo L, Dormand E, Anderson DJ. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003;38:17–31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown SJ, Lee VM, Bronner-Fraser M, Newgreen DF, Farlie PG. Sox10 overexpression induces neural crest-like cells from all dorsoventral levels of the neural tube but inhibits differentiation. Developmental Dynamics: An Official Publication of the American Association of Anatomists. 2005;233:430–444. doi: 10.1002/dvdy.20341. [DOI] [PubMed] [Google Scholar]
- Mollaaghababa R, Pavan WJ. The importance of having your SOX on: role of SOX10 in the development of neural crest-derived melanocytes and glia. Oncogene. 2003;22:3024–3034. doi: 10.1038/sj.onc.1206442. [DOI] [PubMed] [Google Scholar]
- Nichols DH. Neural crest formation in the head of the mouse embryo as observed using a new histological technique. J Embryol Exp Morphol. 1981;64:105–120. [PubMed] [Google Scholar]
- Osawa M, Egawa G, Mak SS, Moriyama M, Freter R, Yonetani S, Beermann F, Nishikawa S. Molecular characterization of melanocyte stem cells in their niche. Development. 2005;132:5589–5599. doi: 10.1242/dev.02161. [DOI] [PubMed] [Google Scholar]
- Pietri T, Eder O, Blanche M, Thiery JP, Dufour S. The human tissue plasminogen activator-Cre mouse: a new tool for targeting specifically neural crest cells and their derivatives in vivo. Developmental Biology. 2003;259:176–187. doi: 10.1016/s0012-1606(03)00175-1. [DOI] [PubMed] [Google Scholar]
- Pingault V, Bondurand N, Kuhlbrodt K, Goerich DE, Pr,hu MO, Puliti A, Herbarth B, Hermans-Borgmeyer I, Legius E, Matthijs G, Amiel J, Lyonnet S, Ceccherini I, Romeo G, Smith JC, Read AP, Wegner M, Goossens M. SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nature Genetics. 1998;18:171–173. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Southard-Smith EM, Angrist M, Ellison JS, Agarwala R, Baxevanis AD, Chakravarti A, Pavan WJ. The Sox10(Dom) mouse: modeling the genetic variation of Waardenburg-Shah (WS4) syndrome. Genome Res. 1999;9:215–225. [PubMed] [Google Scholar]
- Southard-Smith EM, Kos L, Pavan WJ. Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nature Genetics. 1998;18:60–64. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Friedrich RP, Wegner M. Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development. 2004;131:2349–2358. doi: 10.1242/dev.01114. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Sock E, Chaboissier M-C, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes & Development. 2003;17:1677–1689. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Sakai D, Osumi N, Wada H, Wakamatsu Y. Sox genes regulate type 2 collagen expression in avian neural crest cells. Development, Growth & Differentiation. 2006;48:477–486. doi: 10.1111/j.1440-169X.2006.00886.x. [DOI] [PubMed] [Google Scholar]
- Yolov AA, Shabarova ZA. Constructing DNA by polymerase recombination. Nucleic Acids Res. 1990;18:3983–3986. doi: 10.1093/nar/18.13.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Vivatbutsiri P, Morriss-Kay G, Saga Y, Iseki S. Cell lineage in mammalian craniofacial mesenchyme. Mech Dev. 2008;125:797–808. doi: 10.1016/j.mod.2008.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.