Abstract
The retinoic acid (RA) signal, produced locally from vitamin A by retinaldehyde dehydrogenase (Raldh) and transduced by the nuclear receptors for retinoids (RA receptor and 9-cis-RA receptor), is indispensable for ontogenesis and homeostasis of numerous tissues. We demonstrate that Raldh3 knockout in mouse suppresses RA synthesis and causes malformations restricted to ocular and nasal regions, which are similar to those observed in vitamin A-deficient fetuses and/or in retinoid receptor mutants. Raldh3 knockout notably causes choanal atresia (CA), which is responsible for respiratory distress and death of Raldh3-null mutants at birth. CA is due to persistence of nasal fins, whose rupture normally allows the communication between nasal and oral cavities. This malformation, which is similar to isolated congenital CA in humans and may result from impaired RA-controlled down-regulation of Fgf8 expression in nasal fins, can be prevented by a simple maternal treatment with RA.
Keywords: Harderian gland, nasolacrimal duct, development, nuclear receptor, llama
It has been known for more than half of a century that vitamin A deficiency during gestation induces numerous malformations (1). During the last 10 years, it has been demonstrated that the ability of vitamin A (retinol) to influence development is made possible by a collection of enzymes controlling a two-step metabolic pathway (2, 3), in which retinol is first oxidized into retinaldehyde, which is subsequently converted locally into retinoic acid (RA). At least four enzymes [retinaldehyde dehydrogenases 1–4 (Raldh1–4)], which belong to the aldehyde dehydrogenases family and display tissue-specific expression patterns during development (4, 5), oxidize retinaldehyde into RA (2, 3, 6). Then, RA acts as a hormonal signal by binding to retinoid nuclear receptors [RA receptor (RAR)α, β, and γ and 9-cis-RA receptor (RXR)α, β, and γ], which control, in heterodimers, the transcription of target genes (7). Analysis of mice carrying loss-of-function mutations of retinoid receptors has indicated that RAR/RXRα heterodimers are essential for most of the developmental processes (8, 9). However, some defects displayed by these mutants have not been described during gestational vitamin A deficiency, suggesting that unliganded retinoid receptors could also be involved during development (8–10). To identify the processes that require RA-liganded receptors, genetic approaches have been used, in which local synthesis of RA is abrogated by targeted disruption of Raldh genes in mice. Raldh2 knockout results in lethality around embryonic day 9.5 (E9.5) because of severe trunk, hindbrain, and heart defects (11–13) that are reminiscent of those of vitamin A-deficient embryos (14–16) and of mutants lacking both RARα and RARγ (17). By using a RA-responsive reporter transgene (18), Raldh2-null embryos were shown to lack any detectable RA-dependent activity (11, 19, 20), except in the eye field, where Raldh1 and Raldh3 are expressed (5, 21–24). Taken together, these results indicate that RA synthesized by Raldh2 is required for early embryonic development and that Raldh1 and Raldh3 may be involved in eye formation. However, genetic ablation of Raldh1 has no apparent effects, even though Raldh1-dependent RA synthesis is abrogated in dorsal retina (25). Here we report the effects of Raldh3 (aldh1a3) inactivation on mouse development. We show that eye formation is mildly affected, whereas nasal development critically requires Raldh3-synthesized RA.
Methods
Mice. All mice were of a mixed (50%) C57BL/6–(50%)129/Sv genetic background and housed in an animal facility licensed by the French Ministry of Agriculture (agreement no. B67-218-5, 1999-02-09). Heterozygous mice (aldh1a3+/L–) were mated overnight, and animals having a vaginal plug at noon the next day were considered as E0.5. All-trans-RA (Sigma) was suspended in ethanol (5 mg/ml) and then either diluted in sunflower oil (125 μg/ml) and administered by oral gavage to pregnant females (2 mg/kg of body weight) every 12 h from E9 to E11 or diluted in 50 ml of water and mixed with 50 g of powdered food (R03 breeding diet from UAR, Villemoisson, France) at a final concentration of 100 mg/kg of food (26). RA-containing food paste, wrapped in a black plastic bag, was provided to the pregnant mice ad libitum and renewed each day from E8.5 to E14.5. Animal experiments were supervised by M.M., who is qualified for experimenting with mice (agreement no. 67-62, 2003-05-30), in compliance with the European legislation on care and use of laboratory animals.
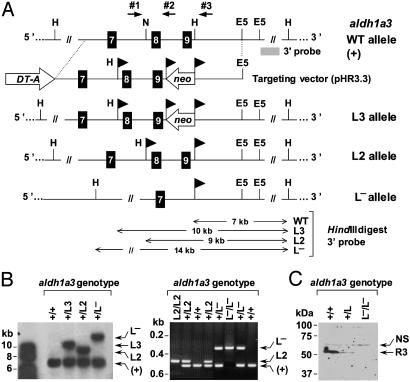
Generation and Genotyping of the Mice. To construct the targeting vector (see Fig. 1 A), a 6-kb long EcoRI–SphI fragment, isolated from a 129/Sv mouse genomic DNA library and containing exons E7–E9, was inserted into pBluescript II SK+ (Stratagene). A loxP-flanked neomycin-resistance cassette (neo) was cloned, in the antisense orientation, between the two HindIII sites located downstream of exon 9. Oligonucleotides 5′-GAGCTCATAACTTCGTATAGCATACATTATACGAAGTTATAAGCTTTGCA-3′ and 5′-AAGCTTATAACTTCGTATAATGTATGCTATACGAAGTTATGAGCTCTGCA-3′, containing SacI, loxP, and HindIII sites, were then inserted into the NsiI site located upstream of exon 8. Exons 8 and 9 encode the catalytic site of Raldh3 (amino acid residues 261–356). A diphtheria toxin A (DT-A) expression cassette was inserted at the 5′-side of the genomic DNA, yielding the pHR3.3 vector, which was linearized with KpnI and electroporated into embryonic stem (ES) cells. One clone (of 20), targeted as expected (HF9, L3 allele), was identified by Southern blot analysis (aldh1a3+/L3). Transient transfection of HF9 ES cells with a Cre-expressing plasmid allowed excision of the neo gene, yielding cells bearing the conditional loxP-flanked L2 allele. Clone HF9.24 (aldh1a3+/L2) was injected into C57BL/6 blastocysts, and two chimeras transmitted this conditional allele to their germ line. Homozygous mice (aldh1a3L2/L2) were indistinguishable from WT littermates. They were crossed with CMV–Cre transgenic mice (27), and the resulting mice bearing the excised allele (aldh1a3+/L–) were identified by Southern blot and PCR analysis. Tail DNA was genotyped by PCR using primers no. 1 (5′-AAACCAGCACCACCTCCATA-3′) and no. 2 (5′-GACCAGCTTTCCAACCTTCA-3′) (see Fig. 1 A) to amplify the WT (252 bp long) and the loxP-flanked L2 allele (302 bp long), or primers nos. 1 and 3 (5′-AAACAACACAGAACCTCCTT-3′) to amplify the excised, null L–allele (541 bp long). Conditions were 30 cycles with denaturation for 30 sec at 92°C, annealing for 30 sec at 61°C, and elongation for 30 sec at 72°C.
Fig. 1.
Targeted disruption of the aldh1a3 (Raldh3) gene. (A) Structure of the targeting vector (pHR3.3) and partial restriction map (not to scale) of the aldh1a3 locus before (WT allele, +) and after homologous recombination (L3 allele) and Cre-mediated recombination (L2 and L–alleles). Black boxes (7–9) stand for exons. The location of restriction sites (E5, EcoRV; H, HindIII; N, NsiI) and of the 3′ external probe is indicated. On the bottom part of the diagram, the sizes of the restriction fragments obtained for each allele are in kilobases. Arrowhead flags represent loxP sites. Arrows indicate the location of primers 1–3 used for PCR genotyping. (B Left) Southern blot analysis of HindIII-digested genomic DNA from ES cell clones with the indicated aldh1a3 genotype, using the 3′ probe. (Right) PCR analysis of tail genomic DNA from mice with the indicated aldh1a3 genotype. The identities of the different alleles are indicated on the right. (C) Western blot analysis of head proteins (100 μg) from E13.5 fetuses with the indicated aldh1a3 genotype, using an anti-Raldh3 polyclonal Ab. Excision of exons 8 and 9 resulted in a null allele, as confirmed by the absence of Raldh3 (R3) in E13.5 aldh1a3L–/L– fetuses. The upper band is a nonspecific signal (NS), showing similar amounts of protein on each lane.
Western Blot Analysis. Cytosolic extracts were resolved by electrophoresis on SDS/12% polyacrylamide gels and blotted onto nitrocellulose membranes (Schleicher & Schüll). Raldh3 was detected by using rabbit polyclonal antisera (a gift from U. Dräger, Eunice K. Shriver Center, Waltham, MA; crude serum at a dilution of 1:250) and purified IgG (a gift from J. Napoli, University of California, Berkeley; at a final concentration of 0.1 μg/ml). Immunoreactions were visualized by using staphylococcal protein A coupled to horseradish peroxidase (dilution 1:10,000), followed by chemiluminescence according to the protocol of the supplier of the kit (Amersham Biosciences).
Histology, Stainings, and in Situ RNA Analysis. Samples were fixed in Bouin's fluid for 5 days, embedded in paraffin, serially sectioned, and stained with Groat's hematoxylin or Mallory's trichrome (28). Before paraffin embedding, adult heads were fixed for 7 days in Bouin's fluid and then treated for 30 h in DC3 decalcifier (Labonord, Templemars, France). Staining for β-galactosidase activity was as described (18), and embryos were post-fixed in 4% buffered paraformaldehyde for 16 h at 4°C and processed for histology. Whole-mount in situ RNA hybridization was as described (17). For in situ hybridization on paraffin sections, embryos were fixed by 4% paraformaldehyde in PBS (1 h; 4°C). Digoxigenin-labeled antisense riboprobes were synthesized by T7 RNA polymerase using cDNA as templates (17).
Results and Discussion
To establish the role of Raldh3 (aldh1a3), we have generated an aldh1a3-null allele in the mouse through homologous recombination in ES cells (Fig. 1). To demonstrate that aldh1a3-null homozygotes (aldh1a3L–/L–, hereafter designated aldh1a3KO) actually lacked RA in ventral retina, nasal epithelium, and nasolacrimal groove, all of which normally contain high levels of aldh1a3 transcripts (5, 21–24) (Fig. 2 A and D), we used a RA-responsive transgenic line (18). No reporter activity could be detected in these structures of E11.5 aldh1a3KO embryos (Fig. 2 B, C, E, and F), establishing that Raldh3 is responsible for the generation of RA in these tissues. Strikingly, no aldh1a3KO mutants could be found at postnatal day 8 (among 328 mice born from aldh1a3+/L– intercrosses), whereas they were recovered at Mendelian frequency at birth (26 aldh1a3KO from 99 newborns). Their external appearance and weight (1.25 ± 0.27 g, n = 10) were similar to those of WT littermates (1.27 ± 0.23 g, n = 7). However, all null mutants became cyanotic and died from respiratory distress within 10 h. Thus, aldh1a3 inactivation is lethal shortly after birth.
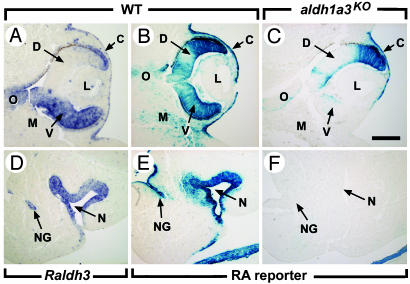
Fig. 2.
Effect of Raldh3 inactivation on RA synthesis at E11.5. (A and D) In situ hybridization with a digoxigenin-labeled Raldh3 antisense probe. (B, C, E, and F) Distribution of β-galactosidase activity driven by the RARE-hsp68-lacZ transgene (18). Raldh3 inactivation suppresses RA synthesis in the ventral retina, ventral cornea, nasal epithelium, and nasolacrimal groove. The disappearance of the RA responsiveness in the mesenchyme adjacent to the ventral retina and to the nasal epithelium may reflect a low level of Raldh3 expression, not detected by in situ hybridization. Note that the areas responsive to RA in the mutant dorsal retina and cornea express Raldh1 at this developmental stage (21, 36). C, cornea; D, dorsal retina; L, lens; M, mesenchyme; N, nasal cavity; NG, nasolacrimal groove; O, optic nerve; V, ventral retina. [Scale bar = 150 μm (A–C) and 100 μm (D–F).]
The E14.5 aldh1a3KO fetuses (n = 5) exhibited fully penetrant bilateral defects restricted to ocular and nasal regions. It is noteworthy that neither ears nor kidneys, both of which express high levels of Raldh3 (5, 21–24), display histological abnormalities (data not shown). Shortening of the ventral retina associated with lens rotation and persistence of primary vitreous body (retrolenticular membrane) (Fig. 3C; see also Fig. 4F) have been observed in vitamin A-deficient embryos (1), as well as in Rxra–/– or Rarb–/– mutants, Rarb–/–/Rarg–/– mutants, and mutants lacking either the RXRα A/B domain or its activation function 2 (AF-2) together with RARβ or RARγ (28–33). Thus, Raldh3 expression in ventral retina yields RA that activates RXRα/RARβ and RXRα/RARγ heterodimers controlling ventral retina growth, lens position, and involution of the primary vitreous body. Interestingly, Rxra–/–/Rarg–/– mutants (29, 31), but not aldh1a3KO mutants, may totally lack ventral retina. Thus, although only Raldh3 is expressed in the ventral retina (5, 22), other Raldh, notably Raldh2 (4, 26), may also provide the RA required for its development. In this respect, note that ventral retina is apparently normal in E18.5 aldh1a3KO mutants (data not shown), whereas the retrolenticular membrane has not further developed (Fig. 3D). The expression of Vax2 (34) and Tbx5 (35) in ventral and dorsal retina, respectively, were not altered in aldh1a3KO at E13.5 (data not shown), and ablation of the dorsally expressed Raldh1 does not alter Tbx5 expression (25). Thus, in contrast to a previous suggestion (36), we conclude that RA segregation into separate territories across the eye anlage is not mandatory for dorsoventral patterning of the retina.
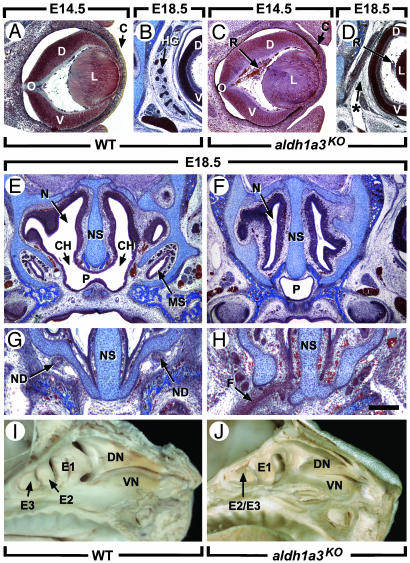
Fig. 3.
Raldh3 inactivation results in ocular and nasal defects. (A–H) Frontal histological sections through WT and aldh1a3KO heads at E14.5 (A and C) and E18.5 (B and D–H). (I and J) Views of the internal surface of the nasal cavities at E18.5. C, cornea; CH, choana; D, dorsal retina; DN, dorsal nasal concha; E1–E3, ethmoturbinates; E2/E3, fused second and third ethmoturbinates; F, persistent nasal fin; L, lens; MS, maxillary sinus; N, nasal cavity; ND, nasolacrimal duct (which drains tears from the conjunctiva of the eye to the nasal cavity); NS, nasal septum; O, optic nerve; P, nasopharynx; R, retrolenticular membrane (persistent primary vitreous body); V, ventral retina; VN, ventral nasal concha. The asterisk in D denotes the absence of the HG. [Scale bar = 150 μm (A and C), 300 μm (B, D, G, and H), and 500 μm (E and F).]
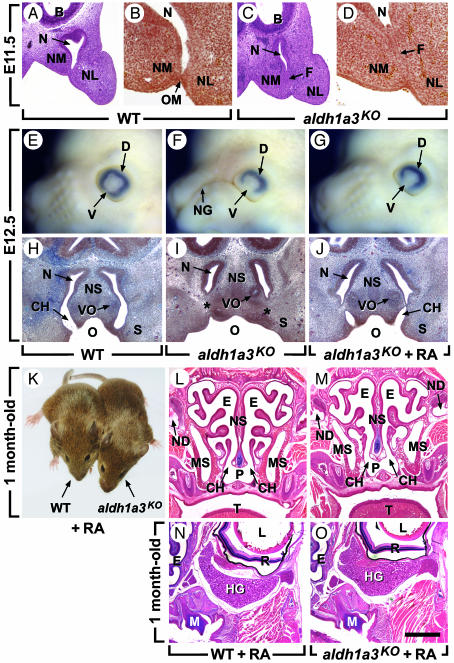
Fig. 4.
CA, which is caused by the persistence of the nasal fin, is prevented by maternal administration of RA. (A–D) Frontal histological sections of WT and aldh1a3KO embryos through the posterior portion of the nasal cavities at E11.5. In the WT embryo, the nasal cavity is large and the nasal fin has undergone a cleavage process, giving rise to the oropharyngeal membrane. In the aldh1a3KO mutant, the cavity is narrow and the fin persists. (E–G) External lateral views of E12.5 heads. (H–J) Frontal histological sections at the level of the vomeronasal organ at E12.5. The embryos in G and J were from a mother treated with RA by oral gavage between E9 and E11. This RA treatment permits opening of the choana into the oral cavity and allows invagination of the nasolacrimal groove ectoderm but at this stage does not fully restore the growth of the ventral retina. (K) WT and aldh1a3KO mice (1 mo old) born from a mother given dietary nonteratogenic doses of RA from E8.5 to E14.5. (L–O) Frontal histological sections through heads of mice shown in K. The nasal defects and HG agenesis caused by aldh1a3 knockout are prevented. Note, however, that a normal shape of the ethmoturbinates is not totally restored. Additionally, examination of serial sections shows that the opening of the nasal cavity into the nasopharynx is reduced in length (0.2 mm instead of 1 mm in WT). B, brain; CH, choana; D, dorsal retina; E, ethmoturbinate; F, nasal fin; L, lens; M, molar; MS, maxillary sinus; N, nasal cavity; ND, nasolacrimal duct; NG, nasolacrimal groove; NL, nasolateral process; NM, nasomedial process; NS, nasal septum; O, oropharyngeal cavity; OM, oropharyngeal membrane; P, nasopharynx; R, retina; S, secondary palate; T, tongue; V, ventral retina; VO, vomeronasal organ; *, blind choanal pit. [Scale bar = 150 μm (A and C), 50 μm (B and D), 300 μm (H–J), and 1.2 mm (L–O).]
The formation of Harderian glands (HGs) requires RXRα/RARγ heterodimers (30, 32, 37), whereas HG agenesis was not reported in vitamin A-deficient fetuses (1). Because corepressor-associated unliganded RAR can repress gene transcription (10, 38), HG agenesis in RA-receptor mutants might reflect artefactual activation of genes normally repressed by unliganded RARγ. However HG agenesis is constant in E18.5 aldh1a3KO fetuses (n = 4/4) (Fig. 3D), demonstrating that RA-liganded RARγ is actually required to activate target genes involved in HG ontogenesis. Moreover, this requirement is a cell-autonomous phenomenon, because the glandular epithelium, when it first appears at E16.5, expresses Raldh3 strongly and Raldh1 weakly, whereas Raldh2 is present in the surrounding mesenchyme (unpublished data).
The E18.5 aldh1a3KO mutants displayed multiple defects of the nasal region, including choanal atresia (CA, lack of communication between nasal and oral cavities; Fig. 3F), absence of maxillary sinuses (Fig. 3F) and nasolacrimal ducts (Fig. 3H), and hypoplasia of the ethmoturbinates (Fig. 3J). Because newborn mammals cannot breathe through their mouths, CA leads to a respiratory distress (39) accounting for aldh1a3KO mutants' death. All of these defects can be observed in Rarb–/–/Rarg–/– and Rara–/–/Rarg–/–mutants (refs. 28 and 30 and unpublished data). Thus, Raldh3 appears essential to generate the receptor-transduced RA signal that controls the formation of ethmoturbinates, nasolacrimal ducts, and the opening between the nasal and oropharyngeal cavities.
Nasal cavity development starts at E9.5, when nasal placodes invaginate to form nasal pits. At E10.5, epithelia lining medial and lateral nasal processes merge in flat plates, called nasal fins. Mesenchyme then grows through and replaces the anterior portion of the fins. At E11.5, interstitial gaps appear in fin posterior portions and coalesce to form oronasal membranes (Fig. 4 A and B) that break down at E12.5 (Fig. 4H), permitting the nasal cavities to communicate with the oropharynx (40). From E9.5 to E12.5, Raldh3 is the only RA-synthesizing enzyme expressed in the nasal region (5, 22). Nasal pit formation and mesenchymal invasion of nasal fins appeared, however, normal in E9.5, E10.5, and E11 aldh1a3KO mutants (Fig. 5 and data not shown). Strikingly, at E11.5, the mutant nasal cavities were narrower than their WT counterparts, and the fins failed to undergo cleavage (Fig. 4 C and D), thus leading to CA (Fig. 4I). We also analyzed the development of the nasolacrimal ducts arising in E11.5 WT embryos through invagination of the nasolacrimal grooves, which are no longer externally visible at E12.5 (Fig. 4E). In aldh1a3KO embryos, the persistence of these grooves (Fig. 4F) resulted in agenesis of the corresponding ducts (Fig. 3H).
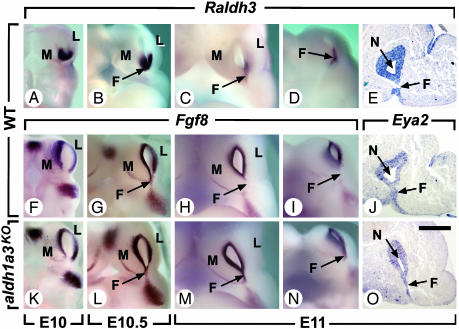
Fig. 5.
Persistence of Fgf8 expression in the nasal fin of aldh1a3KO mutants. (A–D, F–I, and K–N) External views of whole-mount in situ hybridizations of the nasal region of WT and aldh1a3KO embryos from E10 to E11 seen from the front with digoxigenin-labeled Raldh3 and Fgf8 antisense probes. (E, J, and O) In situ hybridization with digoxigenin-labeled Raldh3 or Eya2 antisense probes on frontal histological sections through the posterior portion of the nasal cavities at E11. Embryos in C, H, and M are identical to D, I, and N, respectively, but their heads were slightly tilted to better visualize the ventral portion of the nasal fin. Note that expression of Fgf8 in E10 and E10.5 aldh1a3KO embryos was indistinguishable from that in WT. At E11, the nasal fin no longer expresses Fgf8 in WT embryos, even though it is still present as demonstrated by Eya2 expression. F, nasal fin; L, nasolateral process; M, nasomedial process; N, nasal cavity. [Scale bar = 80 μm (E, J, and O).]
Importantly, oral gavage of pregnant mothers with RA (2 mg/kg) from E9 to E11 mostly prevented CA and nasolacrimal groove persistence (Fig. 4 G and J). Moreover, supplementing maternal food with nonteratogenic doses of RA (100 mg/kg of food) between E8.5 and E14.5 resulted in viable aldh1a3KO adult mice, in which choanae were opened, ethmoturbinates were developed, and bilateral nasolacrimal ducts, maxillary sinuses, and HGs were formed (Fig. 4 K–O). Thus, all of the malformations induced by aldh1a3 knockout are causally related to a RA synthesis deficiency, which can be largely prevented by maternal administration of RA.
During WT early development of the nasal cavities (E9.5–E11), Raldh3 was expressed in the posterior-central ectoderm of nasal pits and in nasal fins (Fig. 5 A–E). During the same period, Fgf8, which may act downstream of RA (41, 42), was expressed in the anterior-peripheral ectoderm of nasal pits at E10 (Fig. 5F), and then ventrally in nascent fins at E10.5 (Fig. 5G), where it disappeared at E11 (Fig. 5 H and I; the presence of fins at E11 was checked by expression of the marker Eya2, Fig. 5 J and O). In contrast in aldh1a3KO embryos, expression of Fgf8 persisted in nasal fins at E11 (Fig. 5 M and N), where its receptor Fgfr2 was also expressed (data not shown). The observations that (i) Raldh3 and Fgf8 are transiently and selectively coexpressed in the nascent nasal fins just a few hours before the disappearance of Fgf8, and (ii) Fgf8 expression persists in this structure in the absence of Raldh3, together strongly suggest that RA signaling is required at E11 to inhibit Fgf8 expression in nasal fins. Along these lines, it is noteworthy that RA signaling also down-regulates Fgf8 expression in tail bud mesoderm (43) and that mutant mice bearing a hypomorphic Fgf8 allele display malformations resembling those induced by excess RA (44–46). Interestingly, choanal stenosis or atresia is observed in human craniosynostosis syndromes resulting from constitutive activation of Fgfr (47). Thus, persistence of local activation of Fgf-signaling pathways in E11 aldh1a3KO nasal fins may account for the occurrence of CA.
The present study reveals that aldh1a3-null mutants display severe malformations of the nasal cavities, including CA, which results in a lethal respiratory distress syndrome at birth. On the other hand, the associated ocular defects are much milder and may remain functionally silent. Importantly, the occurrence of CA can be prevented by maternal administration of nonteratogenic doses of RA, thus providing the demonstration that a lethal genetic defect can be prevented by a simple treatment with a vitamin derivative. Isolated CA is a rare, heritable, developmental defect in humans (39). Our present data suggest that it could be due to mutation(s) at the aldh1a3 locus. Such mutation(s) could also be at the origin of the isolated cases of CA that have devastating consequences on breeding programs of llamas (48). Whether maternal RA administration could prevent CA in this species remains to be investigated.
Acknowledgments
We thank B. Féret, G. Kimmich, B. Weber, and the staff of Institut de Génétique et de Biologie Moléculaire et Cellulaire and Institut Clinique de la Souris common services, ES cell culture, and animal facility for their technical assistance. We also thank U. Dräger and J. Napoli for Raldh3 Abs and Dr. R. Maas for the Eya2 probe. This work was supported by funds from the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, the Hôpital Universitaire de Strasbourg, the Collège de France, the Institut Universitaire de France, and the European Economic Community (EU-QLK4-CT-02-02528). N.M. was supported by a Ministère de l'Education Nationale et de la Recherche fellowship.
Abbreviations: CA, choanal atresia; En, embryonic day n; HG, Harderian gland; RA, retinoic acid; Raldh, retinaldehyde dehydrogenase; RAR, RA receptor; RXR, 9-cis-RA receptor; ES, embryonic stem.
References
- 1.Wilson, J. G., Roth, C. B. & Warkany, J. (1953) Am. J. Anat. 92, 189–217. [DOI] [PubMed] [Google Scholar]
- 2.Duester, G. (2000) Eur. J. Biochem. 267, 4315–4324. [DOI] [PubMed] [Google Scholar]
- 3.Napoli, J. L. (1999) Prog. Nucleic Acid Res. Mol. Biol. 63, 139–188. [DOI] [PubMed] [Google Scholar]
- 4.Niederreither, K., McCaffery, P., Dräger, U. C., Chambon, P. & Dollé, P. (1997) Mech. Dev. 62, 67–78. [DOI] [PubMed] [Google Scholar]
- 5.Niederreither, K., Fraulob, V., Garnier, J. M., Chambon, P. & Dollé, P. (2002) Mech. Dev. 110, 165–171. [DOI] [PubMed] [Google Scholar]
- 6.Lin, M., Zhang, M., Abraham, M., Smith, S. M. & Napoli, J. L. (2003) J. Biol. Chem. 278, 9856–9861. [DOI] [PubMed] [Google Scholar]
- 7.Chambon, P. (1996) FASEB J. 10, 940–954. [PubMed] [Google Scholar]
- 8.Kastner, P., Mark, M. & Chambon, P. (1995) Cell 83, 859–869. [DOI] [PubMed] [Google Scholar]
- 9.Mark, M. & Chambon, P. (2003) Pure Appl. Chem. 75, 1709–1732. [Google Scholar]
- 10.Weston, A. D., Blumberg, B. & Underhill, T. M. (2003) J. Cell Biol. 161, 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niederreither, K., Subbarayan, V., Dollé, P. & Chambon, P. (1999) Nat. Genet. 21, 444–448. [DOI] [PubMed] [Google Scholar]
- 12.Niederreither, K., Vermot, J., Schuhbaur, B., Chambon, P. & Dollé, P. (2000) Development (Cambridge, U.K.) 127, 75–85. [DOI] [PubMed] [Google Scholar]
- 13.Niederreither, K., Vermot, J., Messaddeq, N., Schuhbaur, B., Chambon, P. & Dollé, P. (2001) Development (Cambridge, U.K.) 128, 1019–1031. [DOI] [PubMed] [Google Scholar]
- 14.Dickman, E. D., Thaller, C. & Smith, S. M. (1997) Development (Cambridge, U.K.) 124, 3111–3121. [DOI] [PubMed] [Google Scholar]
- 15.White, J. C., Highland, M., Kaiser, M. & Clagett-Dame, M. (2000) Dev. Biol. 220, 263–284. [DOI] [PubMed] [Google Scholar]
- 16.Zile, M. H., Kostetskii, I., Yuan, S., Kostetskaia, E., St. Amand, T. R., Chen, Y. & Jiang, W. (2000) Dev. Biol. 223, 323–338. [DOI] [PubMed] [Google Scholar]
- 17.Wendling, O., Ghyselinck, N. B., Chambon, P. & Mark, M. (2001) Development (Cambridge, U.K.) 128, 2031–2038. [DOI] [PubMed] [Google Scholar]
- 18.Rossant, J., Zirngibl, R., Cado, D., Shago, M. & Giguère, V. (1991) Genes Dev. 5, 1333–1344. [DOI] [PubMed] [Google Scholar]
- 19.Mic, F. A., Haselbeck, R. J., Cuenca, A. E. & Duester, G. (2002) Development (Cambridge, U.K.) 129, 2271–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niederreither, K., Vermot, J., Fraulob, V., Chambon, P. & Dollé, P. (2002) Proc. Natl. Acad. Sci. USA 99, 16111–16116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, H., Wagner, E., McCaffery, P., Smith, D., Andreadis, A. & Dräger, U. C. (2000) Mech. Dev. 95, 283–289. [DOI] [PubMed] [Google Scholar]
- 22.Mic, F. A., Molotkov, A., Fan, X., Cuenca, A. E. & Duester, G. (2000) Mech. Dev. 97, 227–230. [DOI] [PubMed] [Google Scholar]
- 23.Grün, F., Hirose, Y., Kawauchi, S., Ogura, T. & Umesono, K. (2000) J. Biol. Chem. 275, 41210–41218. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki, R., Shintani, T., Sakuta, H., Kato, A., Ohkawara, T., Osumi, N. & Noda, M. (2000) Mech. Dev. 98, 37–50. [DOI] [PubMed] [Google Scholar]
- 25.Fan, X., Molotkov, A., Manabe, S., Donmoyer, C. M., Deltour, L., Foglio, M. H., Cuenca, A. E., Blaner, W. S., Lipton, S. A. & Duester, G. (2003) Mol. Cell. Biol. 23, 4637–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niederreither, K., Vermot, J., Schuhbaur, B., Chambon, P. & Dollé, P. (2002) Development (Cambridge, U.K.) 129, 3563–3574. [DOI] [PubMed] [Google Scholar]
- 27.Dupé, V., Davenne, M., Brocard, J., Dollé, P., Mark, M., Dierich, A., Chambon, P. & Rijli, F. M. (1997) Development (Cambridge, U.K.) 124, 399–410. [DOI] [PubMed] [Google Scholar]
- 28.Ghyselinck, N. B., Dupé, V., Dierich, A., Messaddeq, N., Garnier, J. M., Rochette-Egly, C., Chambon, P. & Mark, M. (1997) Int. J. Dev. Biol. 41, 425–447. [PubMed] [Google Scholar]
- 29.Kastner, P., Grondona, J. M., Mark, M., Gansmuller, A., LeMeur, M., Décimo, D., Vonesch, J. L., Dollé, P. & Chambon, P. (1994) Cell 78, 987–1003. [DOI] [PubMed] [Google Scholar]
- 30.Lohnes, D., Mark, M., Mendelsohn, C., Dollé, P., Dierich, A., Gorry, P., Gansmuller, A. & Chambon, P. (1994) Development (Cambridge, U.K.) 120, 2723–2748. [DOI] [PubMed] [Google Scholar]
- 31.Kastner, P., Mark, M., Ghyselinck, N., Krezel, W., Dupé, V., Grondona, J. M. & Chambon, P. (1997) Development (Cambridge, U.K.) 124, 313–326. [DOI] [PubMed] [Google Scholar]
- 32.Mascrez, B., Mark, M., Dierich, A., Ghyselinck, N. B., Kastner, P. & Chambon, P. (1998) Development (Cambridge, U.K.) 125, 4691–4707. [DOI] [PubMed] [Google Scholar]
- 33.Mascrez, B., Mark, M., Krezel, W., Dupé, V., LeMeur, M., Ghyselinck, N. B. & Chambon, P. (2001) Development (Cambridge, U.K.) 128, 2049–2062. [DOI] [PubMed] [Google Scholar]
- 34.Barbieri, A. M., Broccoli, V., Bovolenta, P., Alfano, G., Marchitiello, A., Mocchetti, C., Crippa, L., Bulfone, A., Marigo, V., Ballabio, A., et al. (2002) Development (Cambridge, U.K.) 129, 805–813. [DOI] [PubMed] [Google Scholar]
- 35.Uemonsa, T., Sakagami, K., Yasuda, K. & Araki, M. (2002) Dev. Biol. 248, 319–330. [DOI] [PubMed] [Google Scholar]
- 36.McCaffery, P., Wagner, E., O'Neil, J., Petkovich, M. & Dräger, U. C. (1999) Mech. Dev. 85, 203–214. [DOI] [PubMed] [Google Scholar]
- 37.Lohnes, D., Kastner, P., Dierich, A., Mark, M., LeMeur, M. & Chambon, P. (1993) Cell 73, 643–658. [DOI] [PubMed] [Google Scholar]
- 38.Chen, J. D. & Evans, R. M. (1995) Nature 377, 454–457. [DOI] [PubMed] [Google Scholar]
- 39.Menasse-Palmer, L., Bogdanow, A. & Marion, R. W. (1995) Pediatr. Rev. 16, 475–476. [PubMed] [Google Scholar]
- 40.Diewert, V. M. & Wang, K. Y. (1992) Crit. Rev. Oral Biol. Med. 4, 111–130. [DOI] [PubMed] [Google Scholar]
- 41.LaMantia, A. S., Bhasin, N., Rhodes, K. & Heemskerk, J. (2000) Neuron 28, 411–425. [DOI] [PubMed] [Google Scholar]
- 42.Schneider, R. A., Hu, D., Rubenstein, J. L., Maden, M. & Helms, J. A. (2001) Development (Cambridge, U.K.) 128, 2755–2767. [DOI] [PubMed] [Google Scholar]
- 43.Abu–Abed, S., Dollé, P., Metzger, D., Wood, C., MacLean, G., Chambon, P. & Petkovich, M. (2003) Development (Cambridge, U.K.) 130, 1449–1159. [DOI] [PubMed] [Google Scholar]
- 44.Mulder, G. B., Manley, N., Grant, J., Schmidt, K., Zeng, W., Eckhoff, C. & Maggio-Price, L. (2000) Teratology 62, 214–226. [DOI] [PubMed] [Google Scholar]
- 45.Abu-Issa, R., Smyth, G., Smoak, I., Yamamura, K. & Meyers, E. N. (2002) Development (Cambridge, U.K.) 129, 4613–4625. [DOI] [PubMed] [Google Scholar]
- 46.Matt, N., Ghyselinck, N. B., Wendling, O., Chambon, P. & Mark, M. (2003) Development (Cambridge, U.K.) 130, 2083–2093. [DOI] [PubMed] [Google Scholar]
- 47.Hehr, U. & Muenke, M. (1999) Mol. Genet. Metab. 68, 139–151. [DOI] [PubMed] [Google Scholar]
- 48.Smith, B. B., Timm, K. I., Huber, M. Watrous, B. J. & Heidel, J. (2000) Alpaca Registry Journal, Vol. 5, No. 1 Available at www.alpacaregistry.net/journal/spr2000_08.html.