Abstract
Most organisms exhibit daily changes in physiology, behavior, and metabolism under the control of a cell-autonomous circadian clock. In the core clock mechanism, clock genes form a transcription factor network to generate circadian rhythms of gene expression. Phosphorylation of clock proteins and histone modifications also play important roles in the clock regulation. Pharmacological approaches have been making significant contributions to the clock research, for example, in characterizing the roles of protein kinases CKIδ, CKIε, CK2, and GSK-3β. Recently, high-throughput circadian functional assays of mammalian cells have been established. Chemical biology approaches utilizing high-throughput compound screening will open a new way for the circadian clock field together with RNAi-based genomic screening. Finding a set of compounds that potently affect the clock function will lead to the identification of novel clock components and form the basis for therapeutic strategies directed towards circadian disorders.
Organization of Circadian Rhythms in Mammals
The circadian clock controls daily rhythms in a variety of physiological processes such as sleep/wake, body temperature, hormone secretion, and metabolism (Hastings et al., 2003; Green et al., 2008; Takahashi et al., 2008; Eckel-Mahan and Sassone-Corsi, 2009). The identification of clock-controlled processes is expanding and includes haematopoietic stem cell release (Mendez-Ferrer et al., 2008) and blood levels of hundreds of metabolites (Minami et al., 2009). Many of the rhythms persist even under constant conditions in the absence of any external time cues. Importantly, the intrinsic period length of the rhythms is strictly regulated by the circadian clock mechanism, and perturbation of clock function results in a change in period length. To synchronize with ambient 24-h cycles, the clock has an ability to adjust its phase in response to environmental time cues primarily through light (Guler et al., 2008; Hatori et al., 2008).
The circadian clock mechanism resides at the cellular level, and single cells exhibit circadian rhythms in a cell-autonomous manner (Nagoshi et al., 2005; Welsh et al., 2005). These cellular oscillators are organized in a hierarchy, in which the suprachiasmatic nucleus (SCN), located in brain, constitutes the central circadian pacemaker controlling behavioral rhythms (Hastings et al., 2003; Liu et al., 2007a; Takahashi et al., 2008). On the other hand, peripheral clocks in other tissues control local rhythmic outputs such as retinal visual processing, hepatic glucose regulation, and vascular regulation of blood pressure and heart rate (Storch et al., 2007; Lamia et al., 2008; Wang et al., 2008). Within the SCN, the cellular clocks are synchronized to form a coherent oscillator through intercellular coupling, making the SCN clock more robust against genetic and environmental perturbations than peripheral clocks (Liu et al., 2007b).
Transcription Factor Networks of the Circadian Clock
More than a dozen transcription factors and modulators constitute transcriptional feedback loops in the mammalian circadian clock mechanism (Figure 1A) (Reppert and Weaver, 2002; Gachon et al., 2006; Liu et al., 2008; Takahashi et al., 2008). In brief, bHLH-PAS proteins CLOCK (or its homolog NPAS2) and BMAL1 activate transcription of Per and Cry genes, and PER and CRY proteins (PER1, PER2, CRY1, and CRY2) in turn inhibit their own transcription. This core loop is connected to two interlocking loops composed of bZIP proteins (DBP, TEF, HLF, and E4BP4) and nuclear hormone receptors (REV-ERBα, REV-ERBβ, RORa, RORb, and RORc). These factors act in a combinatorial manner on their three cognate cis-acting elements (E box, D box, and RORE) to form a network that generates robust rhythmic gene expression (Ukai-Tadenuma et al., 2008; Baggs et al., 2009). Importantly, many clock proteins bind to histone-modifying enzymes (Table 1), and histone acetylation and methylation show circadian rhythms on clock gene promoters (Etchegaray et al., 2003; Curtis et al., 2004; Naruse et al., 2004; Brown et al., 2005; Etchegaray et al., 2006; Ripperger and Schibler, 2006; Liu et al., 2007c; Alenghat et al., 2008), providing another essential layer of control.
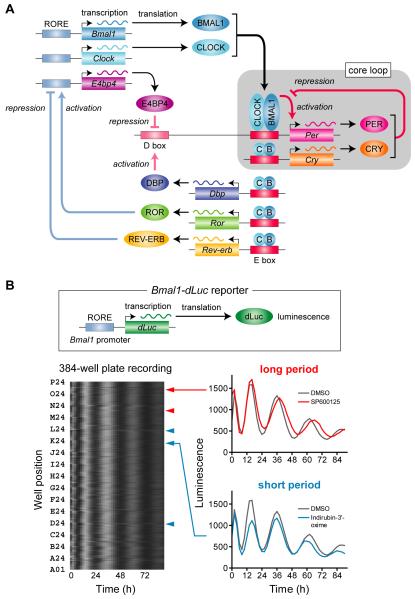
Figure 1.
Mammalian Circadian Clock Mechanism and High-throughput Circadian Assay
(A) Transcription factor feedback loops of the mammalian circadian clock. In the core loop, heterodimers of CLOCK (or NPAS2) and BMAL1 activate transcription from E box element, and PER and CRY proteins inhibit the activation. In addition, DBP (or TEF, HLF) activate and E4BP4 repress D box-mediated regulation, and ROR proteins activate and REV-ERV proteins repress RORE-mediated regulation, forming interlocking loops. These feedback loops generate the rhythmic expression of not only clock genes but also output genes to control the circadian changes in physiology and behavior.
(B) Circadian high-throughput screening of compound library. A clonal reporter cell line was established by using the circadian reporter Bmal1-dLuc (top panel). Luminescence intensity of the reporter cells showed circadian rhythm by reflecting Bmal1 promoter activity. The rhythm was monitored in the presence of compounds (final 7 μM). One screening of the compound library LOPAC contained four 384-well plates, and profiles of one 384-well plate are represented in bottom left panel. Each horizontal raster line represents a single well, with elapsed time plotted to right. Luminescence intensity data from each well are normalized for amplitude, and then indicated by gray scale: peak is white and trough is black. Red and blue arrowheads indicate the positions of long and short period compounds, respectively. Note that there are many compounds that change the phase of the rhythm without affecting the period. Bottom right panels indicate representative traces for a long period compound (SP600125) and a short period compound (Indirubin-3′- oxime).
Table 1.
Histone-modifying Enzymes and Cofactors/Ligands Binding to Clock Proteins
| Clock Proteins | Binding Partners | References |
|---|---|---|
| Histone acetylation | ||
| CLOCK/NPAS2-BMAL1 | p300/CBP and PCAF | Etchegaray et al.,2003; Curtis et al., 2004 |
| CRY1 | mSin3B and HDAC1/2 | Naruse et al., 2004 |
| REV-ERBα | NCoR and HDAC3 | Alenghat et al., 2008 |
| RORa | PGC-1α, p300, and GCN5 | Liu et al., 2007c |
| Histone methylation | ||
| CLOCK-BMAL1 | EZH2 | Etchegaray et al., 2006 |
| PER1/2 | WDR5 | Brown et al., 2005 |
| Cofactors/Ligands | ||
| CLOCK/NPAS2-BMAL1 | NAD | Rutter et al., 2001 |
| NPAS2 | Heme | Dioum et al., 2002 |
| PER2 | Heme | Kaasik and Lee, 2004 |
| CRY1/2 | FAD | Hitomi et al., 2009 |
| REB-ERBα/β | Heme | Raghuram et al., 2007; Yin et al., 2007 |
| RORa | Choresterol derivative | Kallen et al., 2002 |
In addition to transcriptional regulation, post-translational modifications of clock proteins by phosphorylation, ubiquitination, and acetylation play important roles. Most clock proteins undergo rhythmic phosphorylation (Lee et al., 2001), and many protein kinases are involved in the clock mechanism (see below). Upon phosphorylation of PER proteins, F-box proteins β-TrCP1 and β-TrCP2 lead to regulated PER degradation thorough the ubiquitin-proteasome pathway that effects period regulation (Eide et al., 2005; Shirogane et al., 2005; Reischl et al., 2007; Maier et al., 2009). Another F-box protein Fbxl3 causes proteasomal degradation of CRY proteins, and mice harboring a missense mutation of Fbxl3 gene show a long period phenotype (Busino et al., 2007; Godinho et al., 2007; Siepka et al., 2007). Acetylation of BMAL1 is regulated by CLOCK and SIRT1 and necessary for normal clock oscillation (Hirayama et al., 2007; Nakahata et al., 2008). SIRT1 also deacetylates PER2 to facilitate proteasomal degradation (Asher et al., 2008). Intriguingly, SIRT1 activity is regulated by NAD+, and the rate-limiting enzyme in NAD+ biosynthesis (NAMPT) is under circadian control via a CLOCK-BMAL1-SIRT1 circuit, forming an interlocked transcriptional-enzymatic feedback loop (Nakahata et al., 2009; Ramsey et al., 2009). cAMP signaling also constitutes a novel feedback circuit: the transcription-based loops drive rhythms of cAMP signaling, and dynamic changes in cAMP signaling regulate transcriptional output cycles (O’Neill et al., 2008).
Pharmacological Approaches for the Circadian Clock Mechanism
Application of well characterized compounds has provided important insights into the molecular mechanism of the circadian clock. Here we summarize protein kinase inhibitors which have been extensively used (Table 2). Of note, the effectiveness of a pharmacological approach is not only limited to kinases. For example, a set of pharmacological studies identified the interlocking loops of cAMP signaling and NAD+ metabolism (O’Neill et al., 2008; Nakahata et al., 2009; Ramsey et al., 2009).
Table 2.
Kinase Inhibitors Changing the Circadian Period
| Compounds | Primary targets |
Period phenotypes |
Cell types | References | Phenotypes in U2OS cells* |
|---|---|---|---|---|---|
| IC261 | CKIδ/ε | Long period | Rat-1 fibroblasts | Eide et al., 2005 | Not tested |
| CKI-7 | CKIδ/ε | Long period | NIH3T3 fibroblasts | Vanselow et al., 2006 | Not tested |
| D4476 | CKIδ/ε | Long period | NIH3T3 fibroblasts | Reischl et al., 2007 | Long period |
| PF-670462 | CKIδ/ε | Long period | Rat-1 fibroblasts | Walton et al., 2009 | Not tested |
| PF-4800567 | CKIε | No change | Rat-1 fibroblasts | Walton et al., 2009 | Not tested |
| DRB | CK2 | Long period | Aplysia eye | Raju et al., 1991 | Long period |
| DMAT | CK2 | Long period | U2OS cells NIH3T3 fibroblasts |
Maier et al., 2009 Tsuchiya et al., 2009 |
Not tested |
| Indirubin-3′-oxime | CDK&GSK-3 | Not tested | Short period | ||
| Kenpaullone | CDK&GSK-3 | Short period | Rat-1 fibroblasts | Vougogiannopoulou et al., 2008 | Short period |
| Chir99021 | GSK-3 | Not tested | Short period | ||
| 1-azakenpaullone | GSK-3 | Not tested | Short period | ||
| Indirubin derivatives | GSK-3 | Short period | Rat-1 fibroblasts | Vougogiannopoulou et al., 2008 | Not tested |
| Roscovitine | CDK | Long period | Bulla eye | Krucher et al., 1997 | Long period |
| SB203580/SB202190 | p38 | Long period | Chicken pineal gland | Hayashi et al., 2003 | Long period |
| SP600125 | JNK | Long period | Mouse SCN | Chansard et al., 2007 | Long period |
CKIδ and CKIε
Genetic and biochemical studies beautifully indicated the role of CKIδ/ε in the mammalian circadian clock. Hamster tau mutants showing a short period behavioral rhythm have a missense mutation in the CKIε gene (Lowrey et al., 2000), and human familial advanced sleep phase syndrome (FASPS) with early sleep times and early-morning awakening is attributed to missense mutations of PER2 and CKIδ genes (Toh et al., 2001; Xu et al., 2005). Mouse models harboring these mutations show a similar short period phenotype (Xu et al., 2007; Meng et al., 2008). Molecularly, CKIδ/ε phosphorylate PER proteins to cause proteasomal degradation, and tau and FASPS mutations lead higher activity of PER degradation than wild type, explaining short period phenotype (Gallego et al., 2006; Vanselow et al., 2006; Meng et al., 2008).
The functional importance of CKIδ/ε was successfully supported by known CKIδ/ε inhibitors IC261, CKI-7, and D4476, all of which cause period lengthening in cultured cells (Eide et al., 2005; Vanselow et al., 2006; Reischl et al., 2007; Hirota et al., 2008). Furthermore, the CKIδ/ε inhibitor PF-670462 and CKIε-selective inhibitor PF-4800567 have been recently developed, and their effects on the circadian period revealed minimal role of CKIε and a much more prominent role of CKIδ in the period regulation (Walton et al., 2009). This finding is consistent with a recent study using CKIδ and CKIε deficient mice (Etchegaray et al., 2009).
CK2
Our own compound screening strategy in human cells (see below) identified a CK2 inhibitor, DRB, as a long period acting compound (Hirota et al., 2008). Similarly, another CK2 inhibitor DMAT causes period lengthening (Maier et al., 2009; Tsuchiya et al., 2009). Moreover, RNAi-based screening approaches have also identified the role of CK2 in the regulation of period length (Maier et al., 2009). These observations are consistent with previous findings in Drosophila that decreased activity of CK2 causes long period behavioral rhythms (Lin et al., 2002; Akten et al., 2003). Further biochemical studies revealed that CK2 phosphorylates PER2 to regulate its stability (Maier et al., 2009; Tsuchiya et al., 2009) and also phosphorylates BMAL1 to regulate its nuclear accumulation (Tamaru et al., 2009). Importantly, PER2 and BMAL1 proteins that have missense mutations at CK2-phosphorylation sites cause abnormal oscillation of the cellular clock, indicating the importance of these modifications (Maier et al., 2009; Tamaru et al., 2009; Tsuchiya et al., 2009).
GSK-3β
Biochemical studies revealed that GSK-3β phosphorylates PER2 for nuclear localization (Iitaka et al., 2005), CRY2 for proteasomal degradation (Harada et al., 2005), and REV-ERBα for stabilization (Yin et al., 2006). Lithium has been proposed to act through GSK-3 inhibition (Quiroz et al., 2004), and it robustly lengthens the circadian period in a wide range of experimental systems (Engelmann, 1987), suggesting that GSK-3 inhibition causes period lengthening. Consistently, reduction of GSK-3 activity by genetic manipulation causes period lengthening in Drosophila (Martinek et al., 2001). However, our chemical screening strategy identified two compounds inhibiting both CDK and GSK-3 (indirubin-3′-oxime and kenpaullone) as short period compounds. Further analyses with GSK-3-selective inhibitors (Chir99021 and 1-azakenpaullone) and RNAi-mediated knock-down revealed that inhibition of GSK-3β clearly causes a short period phenotype in mammals (Hirota et al., 2008). Similar period shortening was observed with newly developed indirubin derivatives that selectively inhibit GSK-3 (Vougogiannopoulou et al., 2008). Because lithium affects inositol monophosphatase and other phosphomonoesterases as well as GSK-3 (Quiroz et al., 2004), the long period phenotype in mammals might be mediated by lithium-targeted protein(s) other than GSK-3. Identifying these additional targets is of current interest in the field.
Other kinases for period regulation
Pharmacological studies revealed that a CDK inhibitor roscovitine, a p38 MAPK inhibitor SB203580, and a JNK MAPK inhibitor SP600125 cause period lengthening in cultured Bulla eye (Krucher et al., 1997), chicken pineal gland (Hayashi et al., 2003), and mouse SCN (Chansard et al., 2007), respectively. Interestingly, all of them (or the analog) were identified as long period compounds in our screening with human cells (Hirota et al., 2008). It should be noted that these compounds possibly inhibit CKIδ/ε as well as their primary targets (Hasegawa and Cahill, 2004; Fabian et al., 2005). Further studies are necessary to clarify the role of CDK, p38, and JNK in the molecular clockwork.
Kinases for phase regulation
In addition to the period regulation, pharmacological studies identified the involvement of kinases in the phase-shifting mechanism. A MEK (ERK kinase) inhibitor U0126 attenuates light-dependent phase delays and advances in the SCN (Butcher et al., 2002; Coogan and Piggins, 2003) and serum shock-mediated rhythm induction in cultured fibroblasts (Akashi and Nishida, 2000). Similarly, a CaMKII inhibitor KN-62 attenuates phase delays and advances in the SCN (Golombek and Ralph, 1994). KN-62 also inhibits light-dependent activation of ERK, implicating CaMKII as an upstream regulator of ERK (Butcher et al., 2002). On the other hand, a PKG inhibitor KT5823 attenuates phase advances but not phase delays (Ding et al., 1998), suggesting a time-of-day specific function for PKG. Intriguingly, PKGII deficient mice show impaired phase delays but normal phase advances (Oster et al., 2003), contrary to the pharmacological study. In cultured fibroblasts, an ALK inhibitor SB431542 attenuates alkaline shock-induced phase delays, and ALK-SMAD3-Dec1 signaling was identified as a novel clock input pathway (Kon et al., 2008).
High-Throughput Screening: a New Avenue for the Circadian Assay
In addition to the individual approach using a small number of compounds, comprehensive screening of compound libraries will be effective in investigating the molecular clock mechanism. Advances in cell-based circadian assays and bioluminescence recording technology (Nagoshi et al., 2005; Welsh et al., 2005) enabled us to develop a high-throughput circadian functional assay (Hirota et al., 2008). This system consists of luminescent reporter cells, screening automation, and a data analysis pipeline. We utilized the circadian luciferase reporter Bmal1-dLuc which expresses the rapidly degradable luciferase under the control of Bmal1 gene promoter (Liu et al., 2008) to monitor circadian rhythms in cultured cells (Figure 1B, top panel). The GNF Automated Compound Profiling System (Melnick et al., 2006) was applied to record the luminescence every 2 hours over 4 days. A unique aspect of the circadian screening is that the phenotype is not only a simple intensity change but also an alternation in a repeating cycle. Because of this uniqueness, a specialized algorithm is necessary in identifying the “hits”. We developed a curve fitting program CellulaRhythm to calculate rhythm parameters such as period length from large amounts of luminescence data of 384-well plate recordings. The program can also visualize the luminescence rhythms as traces and heat maps for manual inspection of the validity of the calculated parameters. A more sophisticated software MultiCycle (Actimetrics) is recently developed, and it works similar to CellulaRhythm for the circadian parameter estimation. Between the parameters (period, phase, amplitude, and damping rate) we focused on the period, because a deficiency of the core clock mechanism can be reflected to an alternation of the period. More severe phenotype is arrhythmicity, but it is difficult to discriminate from the effect on the general health of the cells. Importantly, the period is the most robust parameter, coming from the repeating characteristics of the clock.
The success of the screening may depend on the robustness of the system. Especially, the cellular rhythmicity is a key factor for the circadian screening. Among all cell lines tested, human U2OS cells showed prominent and highly-reproducible rhythmicity even in a 384-well format. In control (untreated) condition, more than 97% of the wells are within the period range of mean ± 0.5 h. We applied this system to further dissect the circadian clock mechanism using a chemical biology approach. A structurally diverse chemical library, LOPAC (Library of Pharmacologically Active Compounds) containing 1,280 pharmacologically active compounds was initially used to analyze the effect on the circadian period length in human U2OS cells (Figure 1B. bottom panels). We identified 11 compounds causing reproducible period changes of ≥0.5 h. Among them, 7 compounds are protein kinase inhibitors/activators, including roscovitine, SP600125, and SB202190 (an analog of SB203580) previously known to change the circadian period in other organisms (Table 2). Importantly, the period effects of indirubin-3′-oxime, kenpaullone, and DRB predicted the novel roles of GSK-3β and CK2 in the mammalian circadian clock mechanism (Table 2). Together, these observations demonstrate the validity of the high-throughput circadian assay system and the effectiveness of chemical biology in exploring unidentified mechanisms of the circadian clock.
Our group and others recently developed 96 or 384-well format high-throughput circadian assays for RNAi-based genomic screening (Hirota et al., 2008; Vollmers et al., 2008; Maier et al., 2009; Zhang et al., 2009). The screening of RNAi libraries for human kinases suggested the involvement of more than 22 kinases in the cellular clock mechanism (Maier et al., 2009; Zhang et al., 2009). For example, knock-down of MAPK8 (JNK1) causes period lengthening (Zhang et al., 2009), which is consistent with the effect of a JNK inhibitor SP600125 (Chansard et al., 2007; Hirota et al., 2008). Combination of genomic approaches and further screening of kinase inhibitor libraries will likely reveal novel roles for many kinases in the mammalian clockworks.
Future Outlook of the Chemical Biology Approach for the Circadian Clock
In the screening of limited numbers of well-characterized compounds, many of the hits were related to the pathways already known to affect the circadian clock function. Therefore, it is interesting to expand the circadian screening for more comprehensive, large-scale compound libraries containing hundreds of thousands of compounds. A wide variation of chemical structures has the advantage of probing many classes of potential targets, which may include not only kinases but also other proteins such as histone modifying enzymes, metabolic enzymes, and even clock proteins. An attractive possibility of a chemical biology approach will be the identification of novel clock components that cannot be easily achieved by forward and reverse genetic screens because of lethality, pleiotropy, and functional overlapping of closely related proteins. Although our high-throughput compound screening approach is very powerful, there are several important points to keep in mind. (1) Generally, target identification for proteins specifically affected by the novel compound is technically challenging. In addition, compounds (even well-characterized ones) may have multiple targets that give rise to the observed circadian effect. After the identification of candidate protein targets, further studies are necessary by using other compounds targeting the same protein(s) (if available) and/or RNAi-mediated knock-down. The confirmation of the phenotype by multiple reagents may lead to the determination of the responsible protein(s). Simultaneous knock-down of multiple genes will be required if there is functional overlap, and this approach cannot be easily achieved in RNAi screening which targets each gene one by one. (2) The phenotype may arise not only from a direct effect on the clock but also from indirect effects such as alterations of general transcription/translation and the overall health of the cells. Although it is difficult to exclude the possibility of indirect effects, biochemical studies play important roles in exploring the mechanism of direct effects. For example, CK2 directly phosphorylates clock proteins for rhythm regulation (Maier et al., 2009; Tamaru et al., 2009; Tsuchiya et al., 2009), while it has a general effect on cell health and transcription (St-Denis and Litchfield, 2009). (3) There are potential differences in the phenotypes between different candidate cell lines that can be used for screening. Given that the majority of the genes showing circadian expression are tissue-specific (Duffield, 2003), clock modifiers could be cell type-specific. Immortalized cell lines may have aberrant signaling pathways that possibly affect the circadian phenotype. So far, however, many of the period changing compounds are effective in a variety of cell-types including primary cells and even other organisms (Table 2). On the other hand, a phase changing compound dexamethasone is effective in peripheral tissues but not in the SCN because of the absence of glucocorticoid receptor (Balsalobre et al., 2000). Testing the effect of compounds in the SCN and peripheral tissues ex vivo as well as in vivo will provide new opportunities for common and tissue-specific clock mechanisms. Together, chemical biology will play an increasingly important role along with genome-wide RNAi screening in dissecting the molecular mechanism of the circadian clock.
Furthermore, compound screening will generate novel proof-of-concept probes for manipulating clock functions in a dose-dependent and inducible manner. Such proof-of-concept probes will provide the chemical starting points for the identification of small molecule therapeutics designed for circadian disorders. For example, a novel CKIδ/ε inhibitor might be useful in treating the observed short period phenotype caused by FASPS and tau mutations. Interestingly, many clock proteins bind to cofactors or ligands (Table 1) (Rutter et al., 2001; Dioum et al., 2002; Kallen et al., 2002; Kaasik and Lee, 2004; Raghuram et al., 2007; Yin et al., 2007; Hitomi et al., 2009), and CLOCK protein has an acetyltransferase activity (Doi et al., 2006). Compounds affecting these activities may alter circadian clock function and can be used as specific modulators of circadian oscillations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akashi M, Nishida E. Involvement of the MAP kinase cascade in resetting of the mammalian circadian clock. Genes Dev. 2000;14:645–649. [PMC free article] [PubMed] [Google Scholar]
- Akten B, Jauch E, Genova GK, Kim EY, Edery I, Raabe T, Jackson FR. A role for CK2 in the Drosophila circadian oscillator. Nat Neurosci. 2003;6:251–257. doi: 10.1038/nn1007. [DOI] [PubMed] [Google Scholar]
- Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, Bucan M, Ahima RS, Kaestner KH, Lazar MA. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Baggs JE, Price TS, DiTacchio L, Panda S, Fitzgerald GA, Hogenesch JB. Network features of the mammalian circadian clock. PLoS Biol. 2009;7:e52. doi: 10.1371/journal.pbio.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- Brown SA, Ripperger J, Kadener S, Fleury-Olela F, Vilbois F, Rosbash M, Schibler U. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science. 2005;308:693–696. doi: 10.1126/science.1107373. [DOI] [PubMed] [Google Scholar]
- Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF, Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- Butcher GQ, Doner J, Dziema H, Collamore M, Burgoon PW, Obrietan K. The p42/44 mitogen-activated protein kinase pathway couples photic input to circadian clock entrainment. J Biol Chem. 2002;277:29519–29525. doi: 10.1074/jbc.M203301200. [DOI] [PubMed] [Google Scholar]
- Chansard M, Molyneux P, Nomura K, Harrington ME, Fukuhara C. c-Jun N-terminal kinase inhibitor SP600125 modulates the period of mammalian circadian rhythms. Neuroscience. 2007;145:812–823. doi: 10.1016/j.neuroscience.2006.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coogan AN, Piggins HD. Circadian and photic regulation of phosphorylation of ERK1/2 and Elk-1 in the suprachiasmatic nuclei of the Syrian hamster. J Neurosci. 2003;23:3085–3093. doi: 10.1523/JNEUROSCI.23-07-03085.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis AM, Seo SB, Westgate EJ, Rudic RD, Smyth EM, Chakravarti D, FitzGerald GA, McNamara P. Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J Biol Chem. 2004;279:7091–7097. doi: 10.1074/jbc.M311973200. [DOI] [PubMed] [Google Scholar]
- Ding JM, Buchanan GF, Tischkau SA, Chen D, Kuriashkina L, Faiman LE, Alster JM, McPherson PS, Campbell KP, Gillette MU. A neuronal ryanodine receptor mediates light-induced phase delays of the circadian clock. Nature. 1998;394:381–384. doi: 10.1038/28639. [DOI] [PubMed] [Google Scholar]
- Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA, McKnight SL. NPAS2: a gas-responsive transcription factor. Science. 2002;298:2385–2387. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Duffield GE. DNA microarray analyses of circadian timing: the genomic basis of biological time. J Neuroendocrinol. 2003;15:991–1002. doi: 10.1046/j.1365-2826.2003.01082.x. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan K, Sassone-Corsi P. Metabolism control by the circadian clock and vice versa. Nat Struct Mol Biol. 2009;16:462–467. doi: 10.1038/nsmb.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide EJ, Woolf MF, Kang H, Woolf P, Hurst W, Camacho F, Vielhaber EL, Giovanni A, Virshup DM. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann W. Effects of lithium salts on circadian rhythms. In: Halaris A, editor. Chronobiology and Psychiatric Disorders. Elsevier Science; Amsterdam: 1987. pp. 263–289. [Google Scholar]
- Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- Etchegaray JP, Machida KK, Noton E, Constance CM, Dallmann R, Di Napoli MN, DeBruyne JP, Lambert CM, Yu EA, Reppert SM, Weaver DR. Casein kinase 1 delta regulates the pace of the mammalian circadian clock. Mol Cell Biol. 2009;29:3853–3866. doi: 10.1128/MCB.00338-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray JP, Yang X, DeBruyne JP, Peters AH, Weaver DR, Jenuwein T, Reppert SM. The polycomb group protein EZH2 is required for mammalian circadian clock function. J Biol Chem. 2006;281:21209–21215. doi: 10.1074/jbc.M603722200. [DOI] [PubMed] [Google Scholar]
- Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Gallego M, Eide EJ, Woolf MF, Virshup DM, Forger DB. An opposite role for tau in circadian rhythms revealed by mathematical modeling. Proc Natl Acad Sci U S A. 2006;103:10618–10623. doi: 10.1073/pnas.0604511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho SI, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwailid MM, Romero MR, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- Golombek DA, Ralph MR. KN-62, an inhibitor of Ca2+/calmodulin kinase II, attenuates circadian responses to light. Neuroreport. 1994;5:1638–1640. doi: 10.1097/00001756-199408150-00024. [DOI] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guler AD, Ecker JL, Lall GS, Haq S, Altimus CM, Liao HW, Barnard AR, Cahill H, Badea TC, Zhao H, et al. Melanopsin cells are the principal conduits for rod-cone input to non-image-forming vision. Nature. 2008;453:102–105. doi: 10.1038/nature06829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y, Sakai M, Kurabayashi N, Hirota T, Fukada Y. Ser-557-phosphorylated mCRY2 is degraded upon synergistic phosphorylation by glycogen synthase kinase-3 beta. J Biol Chem. 2005;280:31714–31721. doi: 10.1074/jbc.M506225200. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Cahill GM. Regulation of the circadian oscillator in Xenopus retinal photoreceptors by protein kinases sensitive to the stress-activated protein kinase inhibitor, SB 203580. J Biol Chem. 2004;279:22738–22746. doi: 10.1074/jbc.M401389200. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- Hatori M, Le H, Vollmers C, Keding SR, Tanaka N, Buch T, Waisman A, Schmedt C, Jegla T, Panda S. Inducible ablation of melanopsin-expressing retinal ganglion cells reveals their central role in non-image forming visual responses. PLoS ONE. 2008;3:e2451. doi: 10.1371/journal.pone.0002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Sanada K, Hirota T, Shimizu F, Fukada Y. p38 mitogen-activated protein kinase regulates oscillation of chick pineal circadian clock. J Biol Chem. 2003;278:25166–25171. doi: 10.1074/jbc.M212726200. [DOI] [PubMed] [Google Scholar]
- Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- Hirota T, Lewis WG, Liu AC, Lee JW, Schultz PG, Kay SA. A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3beta. Proc Natl Acad Sci U S A. 2008;105:20746–20751. doi: 10.1073/pnas.0811410106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi K, DiTacchio L, Arvai AS, Yamamoto J, Kim ST, Todo T, Tainer JA, Iwai S, Panda S, Getzoff ED. Functional motifs in the (6-4) photolyase crystal structure make a comparative framework for DNA repair photolyases and clock cryptochromes. Proc Natl Acad Sci U S A. 2009;106:6962–6967. doi: 10.1073/pnas.0809180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iitaka C, Miyazaki K, Akaike T, Ishida N. A role for glycogen synthase kinase-3beta in the mammalian circadian clock. J Biol Chem. 2005;280:29397–29402. doi: 10.1074/jbc.M503526200. [DOI] [PubMed] [Google Scholar]
- Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- Kallen JA, Schlaeppi JM, Bitsch F, Geisse S, Geiser M, Delhon I, Fournier B. X-ray structure of the hRORalpha LBD at 1.63 A: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORalpha. Structure. 2002;10:1697–1707. doi: 10.1016/s0969-2126(02)00912-7. [DOI] [PubMed] [Google Scholar]
- Kon N, Hirota T, Kawamoto T, Kato Y, Tsubota T, Fukada Y. Activation of TGF-beta/activin signalling resets the circadian clock through rapid induction of Dec1 transcripts. Nat Cell Biol. 2008;10:1463–1469. doi: 10.1038/ncb1806. [DOI] [PubMed] [Google Scholar]
- Krucher NA, Meijer L, Roberts MH. The cyclin-dependent kinase (cdk) inhibitors, olomoucine and roscovitine, alter the expression of a molluscan circadian pacemaker. Cell Mol Neurobiol. 1997;17:495–507. doi: 10.1023/A:1026358821640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R. A role for casein kinase 2alpha in the Drosophila circadian clock. Nature. 2002;420:816–820. doi: 10.1038/nature01235. [DOI] [PubMed] [Google Scholar]
- Liu AC, Lewis WG, Kay SA. Mammalian circadian signaling networks and therapeutic targets. Nat Chem Biol. 2007a;3:630–639. doi: 10.1038/nchembio.2007.37. [DOI] [PubMed] [Google Scholar]
- Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007b;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007c;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B, Wendt S, Vanselow JT, Wallach T, Reischl S, Oehmke S, Schlosser A, Kramer A. A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 2009;23:708–718. doi: 10.1101/gad.512209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek S, Inonog S, Manoukian AS, Young MW. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell. 2001;105:769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- Melnick JS, Janes J, Kim S, Chang JY, Sipes DG, Gunderson D, Jarnes L, Matzen JT, Garcia ME, Hood TL, et al. An efficient rapid system for profiling the cellular activities of molecular libraries. Proc Natl Acad Sci U S A. 2006;103:3153–3158. doi: 10.1073/pnas.0511292103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Ferrer S, Lucas D, Battista M, Frenette PS. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008;452:442–447. doi: 10.1038/nature06685. [DOI] [PubMed] [Google Scholar]
- Meng QJ, Logunova L, Maywood ES, Gallego M, Lebiecki J, Brown TM, Sladek M, Semikhodskii AS, Glossop NR, Piggins HD, et al. Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58:78–88. doi: 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami Y, Kasukawa T, Kakazu Y, Iigo M, Sugimoto M, Ikeda S, Yasui A, van der Horst GT, Soga T, Ueda HR. Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci U S A. 2009;106:9890–9895. doi: 10.1073/pnas.0900617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi E, Brown SA, Dibner C, Kornmann B, Schibler U. Circadian gene expression in cultured cells. Methods Enzymol. 2005;393:543–557. doi: 10.1016/S0076-6879(05)93028-0. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse Y, Oh-hashi K, Iijima N, Naruse M, Yoshioka H, Tanaka M. Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol Cell Biol. 2004;24:6278–6287. doi: 10.1128/MCB.24.14.6278-6287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster H, Werner C, Magnone MC, Mayser H, Feil R, Seeliger MW, Hofmann F, Albrecht U. cGMP-dependent protein kinase II modulates mPer1 and mPer2 gene induction and influences phase shifts of the circadian clock. Curr Biol. 2003;13:725–733. doi: 10.1016/s0960-9822(03)00252-5. [DOI] [PubMed] [Google Scholar]
- Quiroz JA, Gould TD, Manji HK. Molecular effects of lithium. Mol Interv. 2004;4:259–272. doi: 10.1124/mi.4.5.6. [DOI] [PubMed] [Google Scholar]
- Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju U, Koumenis C, Nunez-Regueiro M, Eskin A. Alteration of the phase and period of a circadian oscillator by a reversible transcription inhibitor. Science. 1991;253:673–675. doi: 10.1126/science.1871602. [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischl S, Vanselow K, Westermark PO, Thierfelder N, Maier B, Herzel H, Kramer A. Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythms. 2007;22:375–386. doi: 10.1177/0748730407303926. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- Shirogane T, Jin J, Ang XL, Harper JW. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem. 2005;280:26863–26872. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Denis NA, Litchfield DW. Protein kinase CK2 in health and disease: From birth to death: the role of protein kinase CK2 in the regulation of cell proliferation and survival. Cell Mol Life Sci. 2009;66:1817–1829. doi: 10.1007/s00018-009-9150-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch KF, Paz C, Signorovitch J, Raviola E, Pawlyk B, Li T, Weitz CJ. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 2007;130:730–741. doi: 10.1016/j.cell.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru T, Hirayama J, Isojima Y, Nagai K, Norioka S, Takamatsu K, Sassone-Corsi P. CK2alpha phosphorylates BMAL1 to regulate the mammalian clock. Nat Struct Mol Biol. 2009;16:446–448. doi: 10.1038/nsmb.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptacek LJ, Fu YH. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Akashi M, Matsuda M, Goto K, Miyata Y, Node K, Nishida E. Involvement of the protein kinase CK2 in the regulation of mammalian circadian rhythms. Sci Signal. 2009;2:ra26. doi: 10.1126/scisignal.2000305. [DOI] [PubMed] [Google Scholar]
- Ukai-Tadenuma M, Kasukawa T, Ueda HR. Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat Cell Biol. 2008;10:1154–1163. doi: 10.1038/ncb1775. [DOI] [PubMed] [Google Scholar]
- Vanselow K, Vanselow JT, Westermark PO, Reischl S, Maier B, Korte T, Herrmann A, Herzel H, Schlosser A, Kramer A. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers C, Panda S, DiTacchio L. A high-throughput assay for siRNA-based circadian screens in human U2OS cells. PLoS ONE. 2008;3:e3457. doi: 10.1371/journal.pone.0003457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vougogiannopoulou K, Ferandin Y, Bettayeb K, Myrianthopoulos V, Lozach O, Fan Y, Johnson CH, Magiatis P, Skaltsounis AL, Mikros E, Meijer L. Soluble 3′,6-substituted indirubins with enhanced selectivity toward glycogen synthase kinase -3 alter circadian period. J Med Chem. 2008;51:6421–6431. doi: 10.1021/jm800648y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton KM, Fisher K, Rubitski D, Marconi M, Meng QJ, Sladek M, Adams J, Bass M, Chandrasekaran R, Butler T, et al. Selective inhibition of casein kinase 1epsilon minimally alters circadian clock period. J Pharmacol Exp Ther. 2009;330:430–439. doi: 10.1124/jpet.109.151415. [DOI] [PubMed] [Google Scholar]
- Wang N, Yang G, Jia Z, Zhang H, Aoyagi T, Soodvilai S, Symons JD, Schnermann JB, Gonzalez FJ, Litwin SE, Yang T. Vascular PPARgamma controls circadian variation in blood pressure and heart rate through Bmal1. Cell Metab. 2008;8:482–491. doi: 10.1016/j.cmet.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Imaizumi T, Kay SA. Real-time reporting of circadian-regulated gene expression by luciferase imaging in plants and mammalian cells. Methods Enzymol. 2005;393:269–288. doi: 10.1016/S0076-6879(05)93011-5. [DOI] [PubMed] [Google Scholar]
- Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptacek LJ, Fu YH. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- Xu Y, Toh KL, Jones CR, Shin JY, Fu YH, Ptacek LJ. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wang J, Klein PS, Lazar MA. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science. 2006;311:1002–1005. doi: 10.1126/science.1121613. [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, Atwood A, Huss JW, 3rd, Janes J, et al. A genome-wide siRNA screen for modifiers of the circadian clock in human cells. Cell. 2009 doi: 10.1016/j.cell.2009.08.031. in press please include DOI number. [DOI] [PMC free article] [PubMed] [Google Scholar]