Abstract
To generate transgenic planarians we used a set of versatile vectors for animal transgenesis based on the promiscuous transposons, mariner, Hermes and piggyBac, and a universal enhanced GFP (EGFP) marker system with three Pax6 dimeric binding sites, the 3xP3-EGFP developed by Berghammer et al. [Berghammer, A. J., Klinger, M. & Wimmer, E. A. (1999) Nature 402, 370–371]. This marker is expressed specifically in the eyes of various arthropod taxa. Upon microinjection into the parenchyma of adult planarians and subsequent electroporation, these vectors transpose efficiently into the planarian genome. One of the cell types transformed are the totipotent “neoblast” stem cells present in the adults, representing 30% of total cells. The neoblast represents a unique cell type with the capacity to proliferate and to differentiate into all somatic cell types as well as into germ cells. All three transposon vectors have high transformation efficiency, but only Hermes and piggyBac show stable integration. The mariner vector is frequently lost presumably because of the presence of active mariner-type transposons in the genome of the Girardia tigrina. Transformed animals are mosaics containing both transformed and untransformed neoblasts. These differentiate to form EGFP-positive and -negative photoreceptor cells. Such mosaicism is maintained through several cycles of regeneration induced by decapitation or asexual reproduction. Transformed neoblasts also contribute to the germ line, and can give rise to pure transgenic planarian lines in which EGFP is expressed in all photoreceptor cells after sexual reproduction. The presence of the transgenes was confirmed by PCR, plasmid rescue assay, inverse PCR, and Southern blotting. Our results with the 3xP3-EGFP marker confirm the presence of Pax6 activity in the differentiated photoreceptor cells of planarian eyes. Transgenesis will be an important tool to dissect developmental molecular mechanisms in planarian regeneration, development and stem cell biology, and may also be an entry point to analyze the biology of parasitic Platyhelminthes.
Keywords: transgenesis, stem cells, Pax-6, regeneration, enhanced GFP
The name “planarian” is generically applied to the free-living Turbellaria of the Phylum Platyhelminthes (the flatworms), which are one of the classical model organisms for the study of regeneration (1–3). The most peculiar cell type in planarians are the totipotent stem cells, called neoblasts. Neoblasts are the only cells endowed with the capacity to divide and to differentiate into all cell types of the adult planarian (4, 5). They possess an extraordinary morphological plasticity as well as a large capacity for regeneration and regulation of body size in the adult organism. Elucidation of the molecular mechanisms underlying regeneration requires functional genetic analyses. To that aim, several approaches have been taken. First, it is now possible to generate loss-of-function phenotypes for particular genes by RNA interference caused by the injection of double-stranded RNA (6, 7). Another powerful method in genetic analysis is the production of gain-of-function mutants. Gene transfer experiments can be used for this purpose. In planarians, germ-line transformation depends on transgene integration into neoblasts and subsequent germ-line differentiation, and the choice of transformation markers is critical for its detection. Different methods to transform neoblasts are presently being pursued in various laboratories. Here we report successful generation of transgenic planarians using transposon vectors and an eye-specific transformation marker.
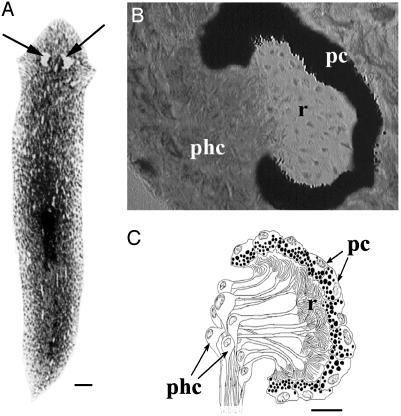
In Platyhelminthes, light is perceived by specific photoreceptor cells that form various eye types, mostly pigment-cup ocelli, located dorsally to the cephalic ganglia (reviewed in refs. 8 and 9). Such eyes are composed of two characteristic cell types, photoreceptors and pigment cells (Fig. 1). Photoreceptors are bipolar neurons with dendritic extremities forming a rhabdomeric structure, a highly ordered assembly of microvilli where opsin accumulates, and axons connecting the eyes to the cephalic ganglia (10). The pigment cells form an eyecup that surrounds the rhabdomeres and shields the eye from one side (Fig. 1). The pigment cells are also involved in the turn-over of the photoreceptor membranes (9, 11). During head regeneration, new eyes are induced after the cephalic ganglia have differentiated. Eye regeneration takes ≈5–6 days at 17°C with the initial formation of small eyespots that later grow to their normal size by gradual addition of newly differentiated eye cells (12). Some genes of the conserved eye morphogenetic pathway, such as Pax6 and sine oculis, are known to be involved in both regeneration and maintenance of eyes in planarians (13–15).
Fig. 1.
Planarian eye structure. (A) Dorsal view of a whole planarian G. tigrina; arrows indicate the location of the eyes in the dorsal head. (B) Sagittal wax section showing the pigmented eye cup produced by apposition of several pigment cells. The opening where the photoreceptors cells project their rabdomeric structures and their cell bodies is shown. (C) Drawing of B illustrating the various morphological structures. pc, pigment cells; phc, photoreceptor cell bodies; r, rhabdomeric structures. (Scale bars, 0.5 mm in A, and 0.05 mm in B and C.)
As a first step in transforming neoblast cells, we have developed a method for the introduction of exogenous DNA into neoblasts under conditions that allow stable integration of the transgene into the neoblast genome. For this purpose, we have used injection and subsequent electroporation in adult planarians of a series of the most promiscuous transposable elements that have distinct insertion specificities (mariner, Hermes, and piggyBac) and an eyespecific and potentially universal transformation marker. This marker, called 3xP3-EGFP, is based on an artificial promoter that is responsive to the transcription factor Pax6 from different animal phyla and the enhanced GFP (EGFP) reporter gene (16). P3-related sequences are found in rhodopsin and other photoreceptorspecific genes ranging from flies to humans. Here we show that this reporter gene is detectable in the eyes of transformed animals and regenerated heads as a mosaic of EGFP-positive photoreceptor cells. Transformed germ cells were also produced when electroporated animals were induced to regenerate head and gonads, in which transformed neoblasts differentiated into germ cells. The hermaphroditic sexual reproduction of such animals produced a second generation of pure transgenic lines stably expressing the EGFP in the eyes.
Materials and Methods
Species. Specimens of a sexual race of the planarian Girardia tigrina were collected near Montpellier (France) and maintained in spring water. Two-week-starved, 9- to 10-mm-long animals were used in all transgenic experiments. Planarians were cut prepharyngeally according to Saló and Baguñà (17) and allowed to regenerate in Petri dishes with spring water.
Plasmids. We used the plasmid DNA constructs 3xP3-EGFPaf with mariner, piggyBac, and Hermes as transposable elements kindly provided by Ernst Wimmer (18). The helper plasmids contain mariner and Hermes transposase sequences under the control of the Drosophila hsp82 promoter and piggyBac transposase sequence under the control of the Drosophila hsp70 promoter. Twenty micrograms of the each vector (3xP3-EGFPaf) was coprecipitated with or without 20 μg of helper plasmid (pKhsp82MOS, pKhsp82Hermes, pBacΔSac) and dissolved in 40 μl of sterilized water.
Transformation Assays. Adult planarians and posterior and anterior regenerating planarians were injected with 2 μl of plasmid DNA solution (1 μg/μl) into the intestinal cavity and parenchymal by using a nanoject injector (Drummond Scientific, Broomball, PA). They were then electroporated by a single pulse of 15 V for 30 ms with special electrodes for fish embryos (CUY701–5E and CUY701–5L Kobayashi type, Nepa Gene, Chiba, Japan) and an electro-square porator (Kobayashi T820). The phenotype was checked every week for 60 days until fluorescent photoreceptors were clearly detected. All of the images in vivo were taken on a Leica (MZ FLIII) fluorescence stereomicroscope with GFP3 filters and recorded on a Leica camera (DC 300F). Transgenic planarians were fixed with 2% HCl in Holtfreter solution (19), and mounted in a Slow Fade antifade kit (Molecular Probes) to be analyzed in a Zeiss axiophot fluorescence microscope and by Spectral TCS confocal microscopy (Serveis Científico-Tècnics, Universitat de Barcelona).
PCR Analysis on Genomic DNA. Total genomic DNA was extracted according to Garcia-Fernàndez et al. (20) from tail pieces of transformed mosaic animals and wild-type planarians as a negative control. Genomic DNA obtained from two tails of the transgenic lines (28 and 56 ng, respectively) was amplified separately by the multiple displacement amplification method (21) to get the final genomic DNA amount of 178 and 138 μg. Neither human nor Drosophila contaminations were detected in the amplified genomic DNA after Southern blot analysis with human- or Drosophila-specific probes (data not shown). DNA (1 μg) was then amplified by PCR with two specific EGFP primers: GFP3 (forward): 5′-TACGGCAAGCTGACCCTGAAGT-3′ and GFP4 (reverse): 5′-TTCAGCTCGATGCGGTTCACCA3′. Samples were subjected to a 5-min denaturation step at 94°C followed by 35 cycles of 1 min at 94°C, 30 s at 57°C, 1 min at 72°C, and a final extension at 72°C for 7 min. As an internal control of amplification efficiency, we used the homeobox gene Dth-2 (22): Dth-2 forward: 5′-CCAATGCTAGTAATGATCCGCGTAT-3′ and Dth-2 reverse: 5′-TGGGAGACCGTTCTTTATCGTCAAC-3′. Samples were subjected to a 2-min denaturation step at 94°C followed by 35 cycles of 1 min at 94°C, 30 s at 57°C, 1 min at 72°C, and a final extension at 72°C for 2 min. Amplified fragments were cloned with the TOPO TA cloning kit (Invitrogen) and sequenced with the ABI Prism kit (Perkin–Elmer).
Plasmid Rescue Assay. Part of the genomic DNA obtained from tails of the transgenic lines was introduced into Escherichia coli strain DH5α by heat shock and selected on LB agar plates containing ampicillin (0.1 mg/ml).
Inverse PCR Analysis. Genomic DNA from two adult transformed mosaic planarians, one transformed with Hermes transposon construct and the other with piggyBac, a tail of a line transgenic planarian, and a wild-type planarian were digested separately with SauIIIAI and ligated under conditions described by Exelixis (South San Francisco, CA). The amplification was performed with the following primers: for 5′ Hermes end, HLF: 5′CAGTCGCCTGCCTTATGCTTTTGGAGAGCG-3′, HLR: 5′-A ATGA ATTTTTTGTTCA AGTGGCA A AGCAC-3′, HLF2: 5′-GCCTGCCTTATGCTTTTGGAGAGCGAAAGC-3′ and HLR2: 5′-GCA AGTGGCGCATA AGTATCA A A ATA AGCC-3′; for 3′ Hermes end, HRF: 5′AAAATACTTGCACTCAAAAGGCTTGACACC, HRR: 5′GAGTATTTTTTCACAACTTAACAACAACAG-3′, HRF2: 5′-GTGCTTATCTATGTGGCTTACGTTTGCCTG-3′, and HRR2: 5′-TTTTCACAACTTAACAACAACAGTTGTTTG3′; for 5′ piggyBac end, 5F1: 5′-GACGCATGATTATCTTTTACGTGAC-3′, 5R1: 5′-TGACACTTACCGCATTGACA-3′, 5F2:5′-GCGATGACGAGCTTGTTGGTG-3′, and 5R2: 5′TCCAAGCGGCGACTGAGATG-3′; for 3′ piggyBac end, 5Forward: 5′-CAACATGACTGTTTTTAAAGTACAAA-3′, 2Reverse: 5′-GTCAGAAACAACTTTGGCACATATC-3′, 7Forward: 5′-CCTCGATATACAGACCGATA A A ACACAT-3′ and 3R1: 5′-TGCATTTGCCTTTCGCCTTAT-3′. The PCRs were performed under the following amplification conditions: 1 cycle of 95°C for 5 min, 35 cycles of 95°C for 30 s, 65°C for 30 s, 72°C for 1 min, and 1 cycle of 72°C for 7 min. Amplification products were cloned with TOPO TA Cloning kit (Invitrogen), and the DNA sequences were determined by using the inverse PCR-nested primers closer to the amplified sequence.
Southern Blot Analysis. Southern blot analysis were performed at high stringency as described (23). Ten micrograms of genomic DNA isolated from wild-type animals and amplified DNA from genomic DNA isolated from both transgenic lines were digested with EcoRI, which cuts once within the transformation construct, and with BamHI, which cuts twice within the transformation construct, prepared for Southern blot analysis and hybridized with a radiolabeled probe (see Fig. 3). Five nanograms of pHer{3xP3-EGFPaf} was digested with the same restriction enzymes and prepared for Southern as positive control. Filters were exposed for 4 days in a Phosphor Screen (Molecular Dynamics), and images were obtained by using a PhosphorImager (Molecular Dynamics).
Fig. 3.
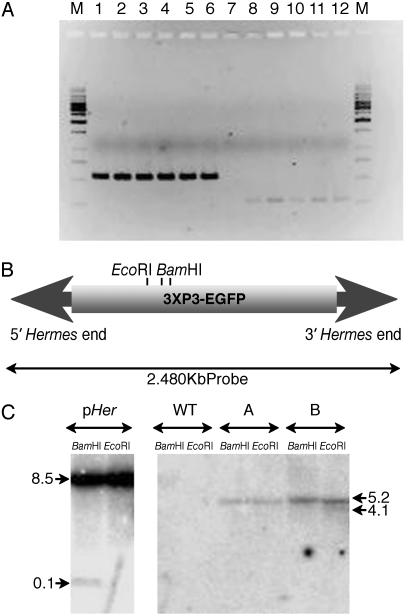
(A) PCR amplification of EGFP from transformed mosaics, transgenic lines, and wild-type (WT) control planarians. Lanes 1–6 show control amplifications with primers for the homeobox gene Dth-2 (22). Lanes 7–12 show PCR amplification with EGFP primers. Lanes 1 and 7, negative control with genomic DNA from WT planarian; lanes 2 and 8, genomic DNA from transformed pHer{3xP3-EGFPaf} mosaic planarian; lanes 3 and 9, genomic DNA from a transgenic line A carrying pHer{3xP3-EGFPaf}; lanes 4 and 10, genomic DNA from a transgenic line B carrying pHer{3xP3-EGFPaf}; lanes 5 and 11, amplified genomic DNA from the transgenic line A; lanes 6 and 12, amplified DNA from the transgenic line B. M shows the DNA molecular marker 1-kb ladder (MBI Fermentas). (B) Schematic diagram (not to scale) of the pHer{3xP3-EGFPaf} fragment used as a probe for the Southern DNA blot hybridization analysis showing the EcoRI and BamHI (with two close sites) restriction sites used to digest the genomic DNA. (C) Southern DNA blot hybridization of pHer{3xP3-EGFPaf} as a positive control, WT planarians as a negative control, line A and line B transgenic planarians carrying pHer{3xP3-EGFPaf}, all of them digested with both EcoRI and BamHI. Arrows indicate the molecular sizes in kb.
Results
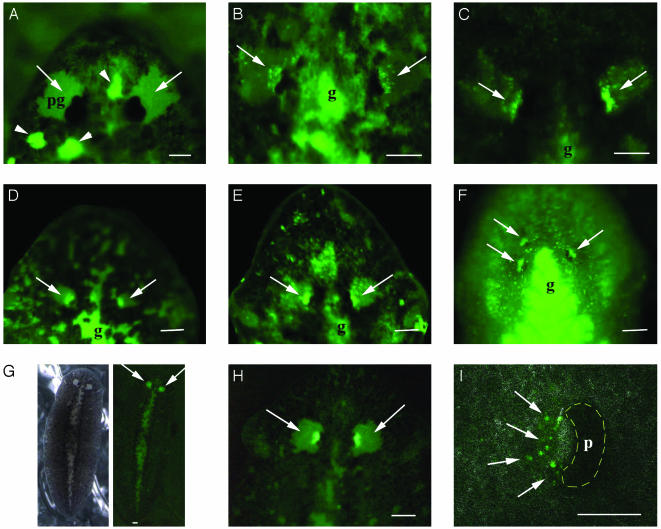
Transgenic Mosaics Expressing the GFP Marker Are Efficiently Produced with the Three Vectors Derived from mariner, Hermes, and piggyBac Transposons. We previously observed a rather variable efficiency of transfection of planarian cells when whole planarians were injected and electroporated with plasmid DNA containing a chicken β-actin promoter-driving GFP (data not shown). However, when using the 3xP3-EGFP vector containing an artificial dimeric Pax6-binding site repeated in tandem, the efficiency of transfection rose, producing planarians with photoreceptor cells expressing EGFP. A well defined phenotype appears after 30–60 days. The delay is caused by the small number (≈35) and slow turnover of differentiated photoreceptor cells, which are gradually replaced by differentiation from transformed neoblasts. The three vectors used, mariner, Hermes, and piggyBac, all show a highly efficient transformation (Fig. 2 B–D and Table 1), although the stability of transformation differs. Hermes and piggyBac transformants were stable with no significant decrease in the number of transformed animals 8 months after the initial transformation (Table 1), nor in the intense EGFP expression in a large fraction of the photoreceptor cells in a mosaic fashion (Fig. 2 C and D). By contrast, control organisms do not show any EGFP fluorescence in the unpigmented periglobular area behind the eye spots (Fig. 2A). Some autofluorescent spots, which are also present in the controls, can be observed in the epidermis and in the cephalic branch of the gut located between the eyes because of the presence of food debris (Fig. 2 B, D, and F). Integration of the mariner-derived vector is less stable presumably because of the presence of several mariner-type transposons in high copy numbers in G. tigrina (24, 25). Such mariner transposons contain an intact ORF and typical regulatory sequences, which suggests that at least some of them are functional transposons. The inverted repeats flanking the mariner-derived transposon vector MosI from Drosophila mauritania and the homologous sequences described for G. tigrina Pmar1 (25) show 36% of sequence identity and share most of the nucleotides of the consensus sequence. Another fact that supports the functionality of endogenous mariner transposons is that some transformation phenotypes were observed in control planarians transformed with mariner plasmid DNA in the absence of mariner helper plasmid DNA providing the transposase. By contrast, no transformation phenotype was observed when Hermes or piggyBac vectors were injected alone without a helper plasmid encoding the tranposase (data not shown). In addition to the lower stability of the mariner constructs, the transformed mosaics show a smaller number of EGFP-positive photoreceptor cells (Fig. 2B).
Fig. 2.
Planarians transformed by electroporation with EGFP constructs. (A) Untreated control showing no EGFP fluorescence in the eyes (arrows). This is easily seen in the periglobular nonpigmented epidermis (pg). Arrowheads indicate autofluorescence in some unpigmented patches of epidermal cells. (B–D) Transformed mosaic animals expressing the 3xP3-EGFP construct, in mariner, Hermes, and piggyBac, respectively. Differences in EGFP fluorescence between eyes in the same individual indicate mosaic expression in some but not all photoreceptor cells. The anterior branch of the gut (g) shows autofluorescence caused by food particles. (E) Regenerated head from a posterior transformed fragment of a whole animal showing mosaic EGFP expression in both regenerated eyes. The same EGFP expression can be observed in regenerated heads of planarians after being transformed and cut. (F) Ventral view of a transformed mosaic animal that regenerated multiple eyes. All three regenerated eyes show mosaic EGFP expression (arrows). Autofluorescence in the gut (g) is also detected. (G and H) Transgenic planarian line transformed with Hermes-derived vector 12 months after electroporation. Strong homogeneous fluorescence is observed in both eyes. (I) Spectral TCS confocal microscopy image of the left eye cup scanned with a transmission filter (phase contrast) and fluorescence channel. All photoreceptor cells located close to the pigment cup (p) have fluorescence (arrows). (Scale bars, 0.25 mm.)
Table 1. Transformation efficiency and stability of transfected EGFP vectors in planarian transgenic mosaics.
| No. of injected and electroporated planarians
|
No. of survivors after electroporation, %
|
Average rate of EGFP-expressing animals after, % (n)
|
|||
|---|---|---|---|---|---|
| Vectors | 2 months | 3 months* | 8 months | ||
| mariner | 69 | 100 | 70 (44/67) | 52 (38/73) | 30 (21/70) |
| piggyBac | 75 | 100 | 80 (60/75) | 71 (54/76) | 66 (44/67) |
| Hermes | 51 | 100 | 98 (49/50) | 83 (48/58) | 70 (37/53) |
Data show an increase in the total number of analyzed organisms caused by the sectioning and regeneration of some transgenic animals.
Transformation Events Are Likely to Be Dependent on Transposon Integration into Neoblasts. We investigated the stability for 8 months of the transgenic mosaics during several cycles of regeneration induced by decapitation, and during asexual reproduction by archytomy (regeneration after spontaneous fission) (Fig. 2E). A mosaic pattern of transformed neoblasts was maintained with all three vectors (Table 1), but with a reduced number of EGFP-positive cells in the mariner constructs for the reasons discussed above. The fact that regenerating animals express EGFP in newly differentiated photoreceptor cells indicates that the neoblasts transformed by electroporation are distributed throughout the parenchyma. In some animals, multiple eye induction, which normally occurs at low frequency in planarians was observed, with all regenerated eyes expressing EGFP in a mosaic pattern (Fig. 2F).
Germ-Line Transformation in Adult Planarians and Establishment of Transgenic Lines. The germ line in planarians is derived from neoblasts, which form germ cells and gonads continuously during adult life. During long periods of starvation, sexual strains shrink in size and destroy their gonads and their germ line by apoptosis (26). When feeding is resumed, planarians regrow and redifferentiate gonads and germ cells from the residual neoblast cell population. This reversible process may occur repeatedly without apparent impairment to the individual or its future ability to mature and breed (2). We took advantage of this plasticity and induced regeneration of the germ line from the mosaic population of transformed neoblasts in primary transformants. To test for germline transmission, we used a sexual strain of G. tigrina and analyzed EGFP expression in the eyes of newly hatched juveniles. From the Hermes-derived vector, three newborn planarians, A, B, and C, were obtained, which stably expressed EGFP in all photoreceptor cells, but one of them died after 1 month. Progeny produced by fission (lines A and B) all express EGFP in all photoreceptor cells. These lines were started in January 2002, and were still expressing EGFP 12 months later (Fig. 2 G–I).
Presence of the Transgene in the Planarian Genome. The presence of the transgene in planarian cells was confirmed by PCR amplification with EGFP-specific primers from genomic DNA of transformed mosaic animals and transgenic lines A and B (Fig. 3A). The amplified band was shown to be EGFP by sequencing.
The presence of the transgene in an integrated manner was indirectly confirmed by both the fact that we obtained stably transformed progeny and by plasmid rescue assays and Southern blot analysis in both lines. No plasmid rescue was obtained after transformation of E. coli strain DH5α with genomic DNA from both lines, providing no evidence for the presence of the construct as an episome. To the contrary, Southern blot analysis revealed the insertion of the transgene in the planarian genome. As illustrated in Fig. 3C, Southern blot analysis shows also no evidence for the presence of the construct as an episome, because in that case, digesting with EcoRI would give one single band of 8.5 kb in size. As mentioned earlier, the construct contains just one restriction site for EcoRI and two cleavage sites for BamHI that are very closely spaced (<30 bp). Instead, we see one band (5.2 kb) for the transgenic line A and two (5.2 and 4.1 kb) for the transgenic line B, digested with EcoRI. In the case of the digestion with BamHI, we observe one band that seems to have the same size for both lines and that is ≈5.2 kb as well. Based on these results at least two interpretations are possible; in the cases where we observe a single band of 5.2 kb, two bands with approximately the same size may be superimposed so that the sum of the two bands has a total size exceeding that of the construct. This would mean that the entire construct was integrated in the genome as a single copy. However, it is intringuing that a similar sized band is detected in both the EcoRI and the BamHI digestions. This observation suggests that not a single copy but a sort of concatemer may have been integrated into the genome leading to the superimposition of several bands in the Southern blot, which would explain why the dominant band detected looks the same size in the EcoRI and BamHI digestions. This hypothesis is also consistent with the presence of plasmid DNA being integrated along with the transposon.
On the other hand, inverse PCR experiments only worked out for the 3′ piggyBac end of a transformed mosaic planarian and for the 5′ Hermes end of the line A transgenic planarian showing that the flanking regions of the transposon ends corresponded to the flanking plasmidic regions of the transposon ends and not to genomic DNA. This supports that not only the sequence of the transposon but also the flanking plasmid sequences were integrated into the genome. Therefore, the integration followed neither the precise “cut-and-paste” mechanism nor the “replicative” transposition (27), because there are no 8.5-kb bands in the Southern in addition to the others. Therefore, another recombinational mechanism and/or a secondary rearrangement of some copies of the transgene might have occurred. However, experiments of transformation with Hermes and piggyBac constructs without the corresponding helper plasmids encoding the transposase did not show EGFP expression, indication that the gene transfer is transposase dependent. Other authors got similar results by using the Hermes transposition system (27, 28).
Discussion
We have generated transgenic planarians by injection and subsequent electroporation of transposon-derived vectors using an artificial P3 promoter driving EGFP expression in photoreceptors cells under the control of the Pax6 transcription factor. The primary cell type, which incorporates the transgene, is the neoblast, a totipotent stem cell capable of differentiating into all somatic cells as well as into the germ line. Such transformants are stably maintained through several cycles of continuous asexual reproduction by fission and regeneration. From a mosaic of transformed totipotent stem cells, homogeneous transgenic lines were produced by germ-line regeneration from transformed neoblasts. The EGFP is strongly expressed in photoreceptor cells and is easily observable in vivo in anaesthetized planarians through the periglobular area, a nonpigmented spot of epidermis located dorsal to the eyes. In contrast to higher animals whose Pax-6 gene is not expressed in differentiated photoreceptor cells (29–31), we demonstrate that Gt-Pax6 genes are also active in differentiated photoreceptors as has been suggested (13, 14). Even though the artificial 3xP3 promoter element was designed to bind three Pax-6 homodimers (32), not all tissues where Pax6 is expressed exhibit fluorescence, for example the central nervous system (14). Thus, there are additional factors required to activate the 3xP3 promoter.
When we compare the results obtained with the different vectors, they all show high transformation efficiency, but notably Hermes and piggyBac are stably integrated, whereas the mariner vector is less stable. We attribute this instability to the presence of endogenous mariner-type transposons in the planarian G. tigrina genome (25). Serial regenerative processes during asexual reproduction do not lead to a decrease of the transgenic neoblast population, even after >8 months (Table 1). Transgenic lines produced by regeneration of the germ line from transformed neoblasts are also stable. A plasmid rescue assay, a specific PCR amplification of EGFP, and Southern blot analysis confirm the presence of the transgene in the planarian genome. The fact that integration occurs through a yet undetermined mechanism does not prevent it from being a powerful laboratory tool.
In conclusion, we have developed a simple and efficient protocol that can be used routinely to generate stable transgenic planarian lines by electroporation. The use of adult animals to transform neoblasts gives both a high rate of survival and a high frequency of transformation. Our data show that Hermes- and piggyBac-derived vectors transpose at a high frequency and integrate stably into the chromosomes of G. tigrina. Stable germ-line transformation experiments are a powerful tool to analyze developmental mechanisms at the genetic and molecular levels. The gain-of-function system described in planarians can be used to explore gene expression. In addition, RNA interference can be used for gene knock out experiments, which offers a powerful tool for the study of planarian development, stem cell biology, and regeneration at a molecular and functional genetic level. Furthermore, GFP expression can now be monitored in real time in live animals to characterize the regulatory DNA sequences involved in regeneration and development. Finally, these techniques may also be extended to study other groups of platyhelminthes, such as parasites, which are of considerable medical interest.
Acknowledgments
We thank Prof. E. Wimmer for providing the vector constructs, information on primers, and helpful comments; Dr. Makiko Seimiya for accomplishing the planarian genomic DNA amplification; Dr. Dawid Ward for providing the Multiple Displacement Amplification kit; Prof. J. Baguñà and Dr. Aziz Aboobaker for useful suggestions on the manuscript; Prof. E. Neumann for advice on electroporation; and Prof. Atkinson for helpful information. This work was supported by grants from the Swiss National Science Foundation and the Kantons of Basel (to W.J.G.), Dirección General de Investigación Cientifica y Ténica (Ministerio de Educación y Ciencia, Spain) Grants PB98-1261-C02-01 and BMC2002-03992 (to E.S.), The Roche Research Foundation (T.M.), and a Ministerio de Educación, Cultura y Deporte Fellowship (to C.G.-E.).
Abbreviation: EGFP, enhanced GFP.
References
- 1.Morgan, T. H. (1901) Regeneration (Macmillan, New York).
- 2.Baguñà J. (1998) in Cellular and Molecular Basis of Regeneration: From Invertebrates to Humans, eds. Ferretti, P. & Gèraudie, J. (Wiley, Chichester, U.K.), pp. 135–165.
- 3.Saló, E. & Baguñà J. (2002) J. Exp. Zool. 292, 528–539. [DOI] [PubMed] [Google Scholar]
- 4.Baguñà J. (1981) Nature 290, 14–15. [Google Scholar]
- 5.Baguñà J. & Romero, R. (1981) Hydrobiologia 84, 181–194. [Google Scholar]
- 6.Sanchez-Alvarado, A. & Newmark, P. A. (1999) Proc. Natl. Acad. Sci. USA 96, 5049–5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pineda, D., Gonzalez, J., Callaerts, P., Ikeo, K., Gehring, W. J. & Saló, E. (2000) Proc. Natl. Acad. Sci. USA 97, 4525–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hyman, L. (1951) The Invertebrates II (McGraw–Hill, New York).
- 9.Rieger, R. M., Tyler, S., Smith, J. P. S., III, & Rieger, G. R. (1991) in Platyhelminthes and Nemertinea, Microscopic Anatomy of Invertebrates, eds. Harrison, F. W. & Bogitsch, B. J. (Wiley–Liss, New York), Vol. 3, pp. 7–140. [Google Scholar]
- 10.Orii, H., Katayama, T., Sakurai, T., Agata, K. & Watanabe, K. (1998) Hydrobiologia 383, 183–187. [Google Scholar]
- 11.Tamamaki, N. (1990) Zool. Sci. 7, 385–393. [Google Scholar]
- 12.Sakai, F., Agata, K., Orii, H. & Watanabe, K. (2000) Zool. Sci. 17, 375–381. [DOI] [PubMed] [Google Scholar]
- 13.Callaerts, P., Muñoz-Marmol, A. M., Glardon, S., Castillo, E., Sun, H., Li, W. H., Gehring, W. J. & Salo, E. (1999) Proc. Natl. Acad. Sci. USA 96, 558–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pineda, D., Rossi, L., Batistoni, R., Salvetti, A., Marsal, M., Gremigni, V., Falleni, A., Gonzalez-Linares, J., Deri, P. & Saló, E. (2002) Development (Cambridge, U.K.) 129, 1423–1434. [DOI] [PubMed] [Google Scholar]
- 15.Saló, E., Pineda, D., Marsal, M., Gonzalez, J., Gremigni, V. & Batistoni, R. (2002) Gene 287, 67–74. [DOI] [PubMed] [Google Scholar]
- 16.Berghammer, A. J., Klingler, M. & Wimmer, E. A. (1999) Nature 402, 370–371. [DOI] [PubMed] [Google Scholar]
- 17.Saló, E. & Baguñà J. (1984a) J. Embryol. Exp. Morph. 83, 63–80. [PubMed] [Google Scholar]
- 18.Horn, C. & Wimmer, E. A. (2000) Dev. Genes Evol. 210, 630–637. [DOI] [PubMed] [Google Scholar]
- 19.Betchaku, T. (1970) J. Exp. Zool. 174, 253–280. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Fernàndez, J., Baguñà J. & Saló, E. (1993) Development (Cambridge, U.K.) 118, 241–253. [DOI] [PubMed] [Google Scholar]
- 21.Dean, F. B., Hosono, S., Fang, L., Wu, X., Faruqui, A. F., Bray-Ward, P., Sun, Z., Zong, Q., Du, Y., Du, J., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 5261–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Fernàndez, J., Baguñà J. & Saló, E. (1991) Proc. Natl. Acad. Sci. USA 88, 7338–7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandbrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 24.Garcia-Fernàndez, J., Marfany, G., Baguñà J. & Saló, E. (1993) Nature 364, 109–110. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Fernàndez, J., Bayascas-Ramírez, J. R., Marfany, G., Muñoz-Mármol, A. M., Casali, A., Baguñà J. & Saló, E. (1995) Mol. Biol. Evol. 12, 421–431. [DOI] [PubMed] [Google Scholar]
- 26.Bowen, I. & Ryder, T. (1974) Cell Tissue Res. 154, 265–271. [DOI] [PubMed] [Google Scholar]
- 27.Atkinson, P. W., Pinkerton, A. C. & O'Brochta, D. A. (2001). Annu. Rev. Entomol. 46, 317–346. [DOI] [PubMed] [Google Scholar]
- 28.Jasinskiene, N., Coates, C. J. & James, A. A. (2000) Insect Mol. Biol. 9, 11–18. [DOI] [PubMed] [Google Scholar]
- 29.Hitchcock, P. F., MacDonald, R. E., VanDeRyt, J. T. & Wilson, S. W. (1996) J. Neurobiol. 29, 399–413. [DOI] [PubMed] [Google Scholar]
- 30.Belecky-Adams, T., Tomarev, S., Li, H. S., Ploder, L., McInnes, R. R., Sundin, O. & Adlere, R. (1997) Invest. Ophthalmol. Vis. Sci. 38, 1293–1303. [PubMed] [Google Scholar]
- 31.Hirsch, N. & Harris, W. A. (1997) J. Neurobiol. 32, 45–61. [PubMed] [Google Scholar]
- 32.Sheng, G., Thouvenot, E., Schmucker, D., Wilson, D. S. & Desplan, C. (1997) Genes Dev. 11, 1122–1131. [DOI] [PubMed] [Google Scholar]