Abstract
Variability in hepatic CYP3A4 cannot be explained by common CYP3A4 coding variants. We previously identified polymorphisms in pregnane X receptor (PXR) and ATP-binding cassette subfamily B member 1 (ABCB1) associated with CYP3A4 mRNA levels in small cohorts of human livers. However, the relative contributions of these genetic variations or of polymorphisms in other CYP3A4 regulators to variable CYP3A4 expression were not known. We phenotyped livers from white donors (n = 128) by quantitative real-time polymerase chain reaction for expression of CYP3A4, CYP3A5, and CYP3A7 and nine transcriptional regulators, coactivators, and corepressors. We resequenced hepatic nuclear factor-3-β (HNF3β, FoxA2), HNF4α, HNF3γ (FoxA3), nuclear receptor corepressor 2 (NCoR2), and regions of the CYP3A4 promoter and genotyped informative single-nucleotide polymorphisms in PXR and ABCB1 in the same livers. CYP3A4 mRNA was positively correlated with PXR and FoxA2 and negatively correlated with NCoR2 mRNA. A common silent polymorphism and a polymorphic trinucleotide (CCT) repeat in FoxA2 were associated with CYP3A4 expression. The transcriptional activity of the FoxA2 polymorphic CCT repeat alleles (wild-type, n = 14 and variant, n = 13, 15, and 19) when assayed by luciferase reporter transactivation assays was greatest for the wild-type repeat, with deviations from this number having decreased transcriptional activity. This corresponded with higher expression of FoxA2 mRNA and its targets PXR and CYP3A4 in human livers with (CCT) n = 14 genotypes. Multiple linear regression analysis was used to quantify the contributions of selected genetic polymorphisms to variable CYP3A4 expression. This approach identified sex and polymorphisms in FoxA2, HNF4α, FoxA3, PXR, ABCB1, and the CYP3A4 promoter that together explained as much as 24.6% of the variation in hepatic CYP3A4 expression.
CYP3A4 metabolizes 45 to 60% of currently prescribed drugs, metabolizes endogenous steroid hormones, and bioactivates environmental carcinogens such as aflatoxin B1 (Shimada et al., 1989). CYP3A4 expression and activity are major determinants of drug efficacy, toxicity, and hence, therapeutic outcome. It is noteworthy that CYP3A4 expression and activity demonstrate high interindividual variation. Because it was reported that 90% of the variation in CYP3A4 activity variation is attributable to genetic factors (Ozdemir et al., 2000), there have been extensive efforts to identify genetic variation within the CYP3A4 gene associated with altered enzyme activity. However, extensive resequencing of CYP3A4 exons, introns, and the proximal promoter by multiple groups has failed to identify common CYP3A4 genotypes that can explain variable CYP3A4 expression (Lamba et al., 2002).
We previously hypothesized that CYP3A4 expression may be related to genetic variation in cis-regulatory sequences and/or in transcription factors regulating CYP3A4 (Schuetz, 2004) transcription, because CYP3A4 activity and protein expression are significantly correlated with its mRNA expression (Lin et al., 2002; Watanabe et al., 2004; Wortham et al., 2007). CYP3A4 transcription is known to be regulated by the dynamic interaction of numerous transcription factors (Martínez-Jiménez et al., 2007), including liver-enriched transcription factors (LETFs): HNF1, HNF3β (FoxA2), HNF3γ (FoxA3), HNF4α, and CCAAT/enhancer binding protein (C/EBP-α and -β) (Ourlin et al., 1997; Jover et al., 2002; Rodríguez-Antona et al., 2003; Tirona et al., 2003; Martínez-Jiménez et al., 2007). At least three C/EBPα-responsive elements have been identified in the proximal CYP3A4 promoter, and C/EBPα and HNF3γ act synergistically in transactivation of the CYP3A4 promoter (Rodríguez-Antona et al., 2003). CYP3A4 is also regulated by the nuclear hormone receptor PXR and constitutive androstane receptor and their associated coactivators such as PGC1α and corepressors such as NCoRs and SHP (Lim and Huang, 2008). Recent studies by us and others have also shown that there is significant correlation between CYP3A4 and PXR mRNA expression (Lamba et al., 2004).
The transcription factor FoxA2 [a LETF that helps to regulate energy metabolism (Wolfrum et al., 2004), bile duct development (Li et al., 2009), and bile acid homeostasis] has not previously been evaluated for its association with human CYP3A4 activity despite the fact that hepatic deletion of FoxA2 in mice resulted in a significant decrease in a number of genes important to bile acid homeostasis, including PXR and Cyp3a11 (Bochkis et al., 2008). This finding in mouse liver is intriguing because we found that single-nucleotide polymorphisms (SNPs) within putative FoxA2 binding sites in the human PXR promoter and intron 1 are associated with hepatic PXR and CYP3A4 expression (Lamba et al., 2008). Further, FoxA2 interacts with the mouse PXR promoter during liver development (Kyrmizi et al., 2006), and cross-talk between PXR and FoxA2 has been implicated in drug-induced activation of lipid metabolism in fasting mice (Nakamura et al., 2007).
In this study, we tested the hypothesis that genetic variation in these transcription factors, in particular, FoxA2 (or their cognate binding sites in the CYP3A4 promoter), contributes to the observed variation in CYP3A4 constitutive expression. We resequenced FoxA2 and screened for polymorphisms in other CYP3A4 transcriptional regulators by resequencing or genotyping known polymorphisms in DNA from human livers phenotyped for CYP3A4 mRNA expression. Because genetic variants in multiple genes probably contribute toward the observed variation in CYP3A4 expression, we performed multiple linear regression analysis and calculated the extent of variation in CYP3A4 that is explained by the genetic variation in these multiple regulators of CYP3A4.
Materials and Methods
Human Liver Tissue.
One hundred twenty eight human livers from white donors (males, 76; females, 52) were provided by the Liver Tissue Procurement and Distribution System (National Institutes of Health Contract number N01-DK-9-2310) and by the Cooperative Human Tissue Network. Of the livers, 35 and 26 were previously analyzed for the association of CYP3A4 mRNA with the PXR and MDR1 genotypes (Lamba et al., 2006, 2008), respectively, but only 12 samples were overlapping between all three studies. The institutional review boards at St. Jude Children's Research Hospital and the University of Pittsburgh approved the use of tissue samples from organ donors. Donor demographics are given in Supplemental Table 1.
Relative mRNA Quantitation of Target Genes by the Standard Curve Method.
Total RNA was isolated from the liver tissue by use of TRIzol (Invitrogen, Carlsbad, CA). First-strand cDNA was prepared from 3 μg of total RNA by use of oligo(dT) primers and the Invitrogen Superscript II kit. Before real-time PCR, 20 μl of cDNA was diluted to 50 μl with diethyl pyrocarbonate-treated water. Two microliters of cDNA from each sample was analyzed in duplicate by real-time PCR on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). ABI gene expression assays were used for quantitative real-time PCR of target genes (Table 1). Human PPIA (cyclophilin A, VIC/MGB probe, primer limited; product 4326316E) was used as the endogenous control to normalize the relative mRNA expression of the target genes according to the manufacturer's instructions. The reaction was run by use of 1 μl of the 20× Gene Expression Assay mix along with 7 μl of diethyl pyrocarbonate-treated water and 10 μl of 2× Taqman Universal PCR Master mix (with Amperase UNG; TaqMan Universal PCR Master Mix; part 4304437) in a 20-μl final volume. Standard amplification conditions consisted of 2 min (UNG activation) at 50°C, 10 min at 95°C followed by 40 cycles of 15 s (denaturing) at 95°C and 1 min (annealing/extension) at 60°C. Standard curves (in triplicate) were prepared for each gene with use of cDNA from a high-expression sample serially diluted over a 3125-fold range, and the amounts of target genes and cyclophilin mRNA were determined by interpolation. The average target gene amount was divided by the average cyclophilin amount to obtain a normalized value. One of the experimental samples was then chosen as the calibrator, and each of the normalized values was divided by the normalized calibrator value to get the relative target gene mRNA expression levels for the study samples.
TABLE 1.
FoxA2 SNPs identified by resequencing the genomic DNA from 127 whites
| Position in Ref Seq a | Position wrt ATG b Position in cDNA GenBank Accession Number NM_153675 | Position wrt TSS0.2 c (22514102) | Position wrt TSS0.1 c (22513101) GenBank Accession Number NM_021784 | rs Number | Site in Gene | Str d | Change in Amino Acid | SNP and Flanking Sequence | Minor Allele | MAF e |
|---|---|---|---|---|---|---|---|---|---|---|
| 22515614 | −2716 | −1512 | −2513 | Prom. | (−) | ccgagtcccc(C>G)cgaaggcgtt | G | 0.004 | ||
| 22515608 | −2710 | −1506 | −2507 | rs1337918 | Prom. | (−) | ccccccgaag(G>T)cgttttcgga | T | 0.054 | |
| 22515585 | −2687 | −1483 | −2484 | Prom. | (−) | gactgggag▵G>A)cgggggagaa | A | 0.008 | ||
| 22514586 | −1688 | −484 | −1485 | Prom. | (+) | aaattggggg(C>A)gatggtggca | A | 0.004 | ||
| 22514124 | −1226 | −22 | −1023 | rs2277764 | Prom. | (−) | aatatcagag(A>G)caaatctcag | G | 0.048 | |
| 22513313 | −415 | 789 | −212 | Intron 1 | (−) | gcggtccac▵CCT>−/−)cctcctcctc | (CCT)13 | 0.083 | ||
| 22513313 | −415 | 789 | −212 | Intron 1 | (−) | gcggtccac▵CCT>−/−)cctcctcctc | (CCT)15 | 0.038 | ||
| 22513313 | −415 | 789 | −212 | Intron 1 | (−) | gcggtccac▵CCT>−/−)cctcctcctc | (CCT)17 | 0.033 | ||
| 22513313 | −415 | 789 | −212 | Intron 1 | (−) | gcggtccac▵CCT>−/−)cctcctcctc | (CCT)19 | 0.008 | ||
| 22513312 | −414 | 790 | −211 | Intron 1 | (+) | ggtggaggag(G>A)aggaggagga | A | 0.038 | ||
| 22513307 | −409 | 795 | −206 | Intron 1 | (+) | gtggtggtgg(A>T)ggaggaggag | T | 0.169 | ||
| 22512479 | 420 | Intron 2 | (−) | gaggagaag(T>C)gggtaggagt | C | 0.012 | ||||
| 22511603 | 1296 (450) | Exon3 | (−) | M87V | cgcgggcgcc(A>G)tggcgggcat | G | 0.004 | |||
| 22511571 | 1328 (482) | rs1800847 | Exon3 | (−) | A97A | ccggggcggc(C>T)ggcgtggcgg | T | 0.044 | ||
| 22511501 | 1398 (552) | Exon3 | (−) | M121V | ggccggggcc(A>G)tgggcggcct | G | 0.004 | |||
| 22511052 | 1848 (1001) | Exon3 | (+) | A270A | tgccgctgcc(G>A)gcggcgcctg | A | 0.004 | |||
| 22511025 | 1874 (1028) | rs1203910 | Exon3 | (+) | G279G | aggcctgggc(T>C)ccggcggccg | C | 0.056 | ||
| 22510674 | 2225 (1379) | rs1212275 | Exon3 | (+) | Q396Q | ttttgtgggg(C>T)tggtggtggt | T | 0.160 | ||
| 22510438 | 2461 | rs1055080 | UTR | (−) | ggatcgagg(C>T)aagtgagaga | A | 0.070 | |||
| 22510311 | 2588 | rs1974 | UTR | (−) | ccgctgcagc(C>T)gttccgtccc | G | 0.054 | |||
| 22509981 | 2918 | UTR | (+) | cttttttttt(T>C)ctttttcttg | C | 0.045 | ||||
| 22509960 | 2939 | UTR | (+) | accgtgtca(G>T)attgggaatg | T | 0.008 | ||||
| 22509953 | 2946 | UTR | (+) | ggatttcacc(G>A)tgtcaagatt | A | 0.020 | ||||
| 22509865 | 3034 | UTR | (+) | tggaactctg(G>A)cccttgcagc | A | 0.004 |
wrt, with respect to; prom., promoter.
NC_000020 (FoxA2: 22509823 … 0.22514102) from the human genome assembly build 36.2.
Complement; ATG = 22512898-96; with the nucleotide 22512898 being +1, the base immediately 5′ is −1.
Position of the FoxA2 transcription start site (TSS) in NC_000020 (build 36.2): for isoform 1 (TSS0.1) = 22513101; for isoform 2 (TSS0.2) = 22514102; with the base immediately 5′ to the TSS being −1.
Strand orientation with respect to GenBank sequence.
Minor allele frequency; polymorphisms significantly associated with FoxA2 or target gene expression are in bold.
In Silico Analysis of CYP3A4 Promoter Region to Select Regions for Resequencing.
Based on the following criteria, specific regions in the 45-kb CYP3A4 promoter-CYP3A43 intergenic region were selected for resequencing: 1) not previously resequenced in whites; 2) demonstrated high multispecies evolutionary conservation deduced by using the programs 28 Way Conservation track in University of California Santa Cruz genome browser (http://genome.ucsc.edu and http://genome.ucsc.edu/cgi-bin/hgTrackUi? hgsid = 104205615&c = chr7&g = multiz28way), Evolutionary conserved region browser (http://ecrbrowser.dcode.org/), and rVISTA (http://genome.lbl.gov/vista/rvista/submit.shtml); 3) demonstrated clustering of liver-enriched transcription factor binding sites determined by Cister plot (http://zlab.bu.edu/∼mfrith/cister.shtml) as we described previously (Lamba et al., 2008) [the matrices in the analysis included LETF matrices derived from Transfac professional (version 11.1) and ubiquitously present and essential transcription factors]; and 4) demonstrated high regulatory potential (University of California Santa Cruz genome browser; http://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid = 104205615&c = chr7&g = regPotential7X).
PCR Amplification and DNA Sequencing of the Selected Genomic Regions.
Based on the criteria described, three regions were selected for resequencing analysis in the CYP3A4 promoter: −6, −12, and −19 kb upstream of the CYP3A4 gene. The entire FoxA2 gene and 2 kb of its proximal promoter region were resequenced. We resequenced four selected regions in the HNF4α promoter and 3′ region. These selections were based on intersections of these regions with multiple regulatory criteria and with locations of known high-frequency SNPs previously shown to have regulatory significance in multiple association studies. The proximal promoter of SMRT was resequenced because this region had multiple CpG islands and DNase-hypersensitive sites, and based on Transfac and Cister, analyses were predicted to be highly expressed in liver. The coding region of HNF3γ was resequenced to identify any SNPs of potential significance. In addition, regulatory and coding SNPs in selected regions of LETFs that occurred, with a minimum allele frequency of >0.01 in whites, were present in a multispecies conserved region and intersected with at least one of several regulatory parameters, such as DNase hypersensitive sites, CpG Islands, Transcription factor binding sites, high regulatory potential, microRNA sites or splice sites, that was genotyped. In addition, selected SNPs in PXR and MDR1/ABCB1 genes were genotyped. All genes were genotyped or resequenced in DNA extracted from the phenotyped human livers using PCR primers indicated (Supplemental Table 2).
CYP3A5*3 was genotyped by use of a restriction fragment length polymorphism method described earlier (Fukuen et al., 2002). Other SNPs were identified by direct sequencing. PCR amplification was carried out in a 1× PCR buffer using 50 ng of DNA, 10 pmol each of forward and reverse primers, 0.2 mM dNTPs, and 1.5 units of Taq polymerase (Expand High Fidelity PCR System; Roche Diagnostics, Indianapolis, IN). The PCR conditions included initial denaturation at 95°C for 3 min followed by 32 to 34 cycles of denaturation at 95°C, annealing at appropriate temperatures, and synthesis at 72°C, with final synthesis at 72°C for 10 min. PCR products were checked for the correct size by agarose gel electrophoresis. Before sequencing, unincorporated nucleotides and primers were removed by incubation with shrimp alkaline phosphatase (USB, U.S. Biochemical Corp., Cleveland, OH) and exonuclease I (USB) for 30 min at 37°C, followed by enzyme inactivation at 80°C for 15 min. Sequencing was performed on an ABI Prism 3700 Automated Sequencer using the PCR primers or internal sequencing primers (Supplemental Table 1). Sequences were assembled by use of the Phred-Phrap-Consed package (University of Washington, Seattle, WA; http://droog.mbt.washington.edu/PolyPhred.html), which automatically detects the presence of heterozygous single-nucleotide substitutions by fluorescence-based sequencing of PCR products (Nickerson et al., 1997).
Statistical Analysis.
Because of the skewed distribution of the mRNA levels for the target genes, they were log-transformed (to base 2) before analysis. Of the 128 samples with gene expression data, one sample had to be excluded from genotype-phenotype association analysis because of missing genotype data.
Group differences relative to SNPs were analyzed parametrically by use of the t test with unequal variances and nonparametrically using the Wilcoxon rank-sum test to compare binary groups (e.g., GG + GT versus TT), and the Kruskal-Wallis test was used to compare three groups of genotype for each polymorphism (e.g., GG versus GT versus TT) with the phenotype.
All the statistical calculations were performed by use of the statistical program R [R Development Core Team (2007) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org]. No adjustments have been made for multiple testing.
Both the Pearson's test and Spearman's nonparametric test were used to compare correlations between the mRNA expression levels. Multiple linear regression analysis was used to determine which SNPs or mRNA expressions significantly explained CYP3A4 mRNA expression variability. For the SNPs, we either considered all 62 SNPs with a minor allele frequency of >0.01 (referred to as “less filtered”) or we filtered them and only considered SNPs with p value <0.1 in univariate analysis and with more than three samples per genotype category (referred to as “more filtered”). For mRNA expression analysis, we considered a subset of six transcription factors for which mRNA expression was available from 88 livers. Because of the high correlation between the mRNA expressions, we used factor analysis, via the function “factanal” from the package “stats” in R, to reduce the dimension of the problem (i.e., reduce the number of independent variables) and generate a set of independent factors. The following “factanal” options were used: the varimax rotation, Thompson's scores, and three factors of a possible six (which explained 84.7% of the variation). The factors generated by this method are linear combinations of the six mRNA expressions and are orthogonal to each other (i.e., linearly independent). Multiple linear regression was then used to relate the factors to CYP3A4 mRNA expression. Because there was the possibility for the interaction between sex and either the SNPs or mRNA expression, we allowed for this interaction in the multiple regression models. Stepwise regression, using the function “stepAIC” from the package “MASS” in R and the Bayesian Information Criteria, was used to build the models (i.e., terms were either added to or removed from the model based on the change in this criteria).
Transient Transactivation Assays.
We amplified and cloned −1945/+1204 (relative to TSS.2) and −3149/−1 (relative to TSS.1) of the FoxA2 promoter between the KpnI/XhoI sites in PGL3 basic to create the FoxA2-Luc reporter vector. The FoxA2 luciferase reporter vectors representing different haplotypes for the polymorphic −415 (CCT)n = 14 repeat (wild type), or the variants n = 13, 15, and 19 were generated by direct amplification from DNA with these sequences. The plasmids were sequenced to confirm the presence of appropriate numbers of CCT repeats. Transient transfection assays were conducted to determine the transactivation potential of each FoxA2 promoter haplotype. In brief, HepG2 cells (1.5 × 105 cells/well, 24-well plate) were maintained in minimum Eagle's medium α-medium supplemented with 10% fetal bovine serum. Twenty-four hours after plating, cells were transfected by calcium phosphate precipitation overnight with FoxA2-LUC reporter plasmids representing different promoter haplotypes (100 ng/well) and an internal control plasmid for transfection efficiency (CMX-β-galactosidase). The media were changed, and 24 h later the cells were harvested, lysed by reporter lysis buffer (Promega, Madison, WI), and centrifuged at 2000g; β-galactosidase and luciferase assays were performed according to the manufacturer's instructions. Luciferase activities were determined on an aliquot of supernatant according to the manufacturer's instructions (Luciferase Assay System; Promega) using an automated luminometer (model OPTOCOMP 1; MGM Instruments, Hamden, CT) and were normalized to the β-galactosidase transfection values. Within each experiment, the transfections were done in triplicates and experiments were replicated independently. The P values for the difference between the transcriptional activity of the wild-type and variant FoxA2 reporters were calculated using a two-tailed t test, assuming equal variance.
Results
FoxA2 mRNA Expression Is Positively Correlated with CYP3A4 and PXR Hepatic mRNAs.
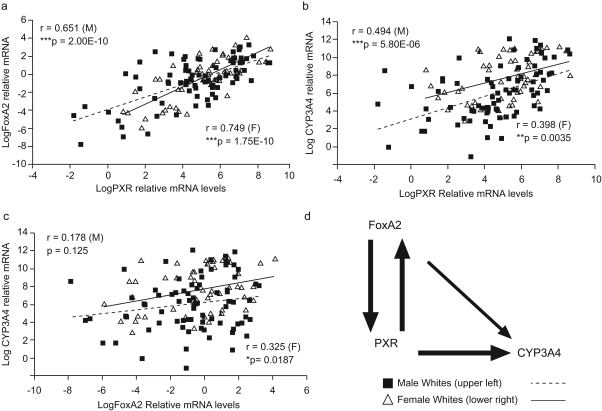
We quantitated the relative mRNA expression of CYP3A4, FoxA2, and PXR in hepatic samples from livers of white donors. A strong correlation was seen between PXR and CYP3A4 mRNA levels (Fig. 1) as described previously in smaller cohorts (Wolbold et al., 2003; Lamba et al., 2004; Schirmer et al., 2007). A significant correlation was found between FoxA2 and PXR mRNA and CYP3A4 levels, with the relationship stronger in females than in males.
Fig. 1.
Correlation plots for mRNA expression of hepatic PXR, FoxA2, and CYP3A4. PXR, FoxA2, and CYP3A4 mRNA levels were determined by use of Taqman-based gene expression assays. Scatter plots indicate correlations between the relative mRNA levels for each gene in males (■) and females (▵). a, FoxA2 and PXR. b, CYP3A4 and PXR. c, CYP3A4 and FoxA2. The p values and correlation coefficient are indicated. d, regulatory relationship between FoxA2, PXR, and CYP3A4 suggested by the correlation values. The strength of correlation between the mRNA expression values is represented by the thickness of the arrows.
Identification of FoxA2 Sequence Variations.
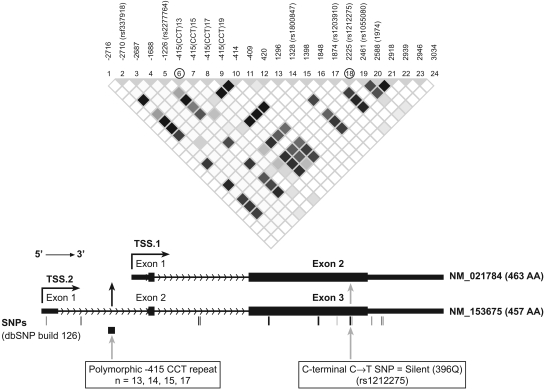
FoxA2 consists of three exons. The two major FoxA2 mRNA transcripts differ by inclusion or exclusion of exon 1 (Fig. 2). Transcript 1 is transcribed from a promoter downstream of exon 1 (from TSS.1, GenBank accession number NM_021784) and has two exons encoding a 463-aa FoxA2 protein (Uetzmann et al., 2008). CISTER analysis identified a LETF cluster in this promoter region (not shown). Transcript 2 (from TSS.2, GenBank accession number NM_153675) encodes a 457-aa FoxA2 protein. This variant has a different splice pattern at the 5′ end compared with isoform 1 that results in translation initiation from a downstream ATG in exon 2 and a shorter isoform missing 6 aa from the N terminus compared with isoform 1. Both isoforms seem to be expressed in liver (Uetzmann et al., 2008).
Fig. 2.
Schematic showing the location of the two most informative SNPs in the FoxA2 gene. Top, haploview (http://www.broadinstitute.org/haploview) generated LD plot for the 24 SNPs in the FoxA2 gene showing LD (measured as r2). The darker the color, the higher the LD, with white indicating no LD between the SNPs. Bottom, structure of FoxA2 mRNAs and the location in the FoxA2 gene of the silent exon 3 SNP and the polymorphic trinucleotide repeat at −415 bp (for simplicity, numbering is relative to the translation start site ATG designated as +1 GenBank accession number NM_153675).
We sequenced the entire FoxA2 gene and 2 kb of its proximal promoter in DNA from 128 livers of white donors and identified 25 SNPs: six exonic (all in exon 3), eight intronic, five promoter, and six 3′-UTR (Fig. 2 and Table 1). Two of the exonic SNPs were nonsynonymous, resulting in 87M>V and 121M>V amino acid changes. Both changes were predicted to be damaging by the program “Polyphen” (http://genetics.bwh.harvard.edu/pph/). The other four exonic SNPs were synonymous (A97A, rs1800847; A270A; G279G, rs1203910; and Q396Q, rs1212275). The intronic SNPs included a polymorphic (CCT)n repeat at −415 in intron 1. We identified five alleles with the polymorphic CCT repeat, ranging from n = 13 to 19 repeats with 14 repeats as the major allele.
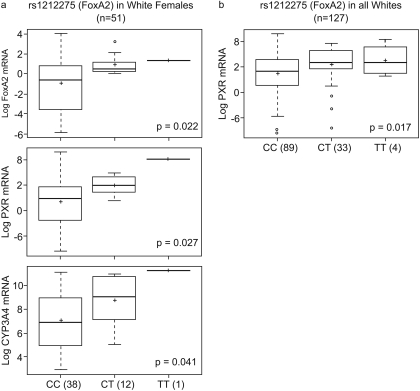
The FoxA2 2225C>T (rs1212275) Silent Polymorphism Is Associated with Hepatic FoxA2, PXR, and CYP3A4 mRNA Expression.
A silent coding change at 2225C>T (Q396Q; rs1212275) located in the C terminus of FoxA2 demonstrated significant association with FoxA2, PXR, and CYP3A4 mRNA (Fig. 3) levels. The 2225T allele was associated with higher FoxA2 (CC versus CT versus TT; −0.93 ± 2.5 versus 0.88 ± 0.96 versus 1.33; p = 0.02), PXR (CC versus CT versus TT; 4.65 ± 1.95 versus 5.95 ± 0.74 versus 8.0; p = 0.02), and CYP3A4 mRNA expression (CC versus CT versus TT; 7.1 ± 2.36 versus 8.7 ± 2.24 versus 11.25; p = 0.04) in white females (n = 51) and with PXR mRNA expression in all whites (n = 127, CC versus CT versus TT; 4.42 ± 2.17 versus 5.42 ± 1.93 versus 5.85 ± 1.76; p = 0.02).
Fig. 3.
Relationship of FoxA2 rs1212275 with FoxA2, PXR, and CYP3A4 mRNA expression in female (a) and PXR in all livers (b). Box plots indicate first and third quartiles of mRNA expression for each FoxA2 genotype, with the bold line within the box representing the median value; the whiskers representing the range after excluding the outliers. The outliers are defined by the R package as data points that fall outside of the first and third quartiles by more than 1.5 times the interquartile range, and circles falling outside the whiskers represent outliers; the + sign represents the mean. The p value from the Kruskal-Wallis nonparametric test comparing the significance of the difference between the three genotype groups is shown.
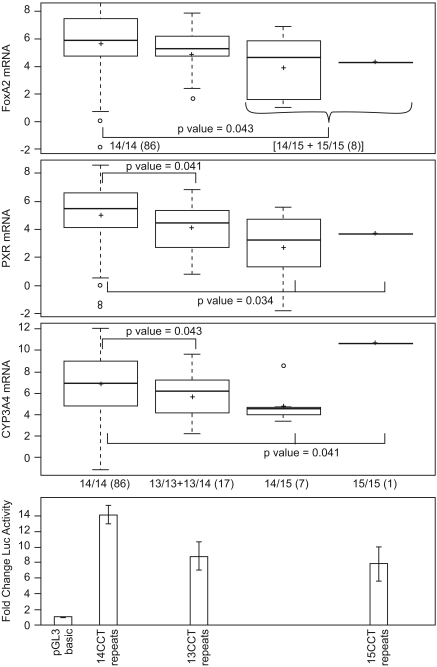
The FoxA2 Trinucleotide Repeat Polymorphism Is Associated with Hepatic FoxA2, PXR, and CYP3A4 mRNAs.
A FoxA2 polymorphic −415(CCT)n repeat in intron 1 was also significantly associated with FoxA2, PXR, and CYP3A4 mRNAs (Fig. 4). Neither the −415 trinucleotide polymorphic repeat nor the 2225C>T silent polymorphism was in linkage disequlibrium (LD) with other FoxA2 SNPs (Fig. 2). Among females (n = 51), subjects homozygous for 14 CCT repeats had 1.3- and 1.9-fold higher PXR mRNA expression compared with subjects with one or two n = 13 repeat alleles, respectively (p = 0.036) (not shown). In the combined population (Fig. 4), any deviation from (CCT) n = 14, even an increase, resulted in decreased expression of FoxA2 and the target mRNAs. FoxA2, PXR, and CYP3A4 mRNAs were decreased 1.7- (p = 0.043), 1.9- (p = 0.034), and 1.5-fold (p = 0.041), respectively, in livers with variant FoxA2 alleles. Some of the associations were even stronger if the subjects were segregated by sex (not shown). The exception was a single liver with a 15/15 polymorphic repeat that had greater CYP3A4 expression compared with livers with 14/14 and 14/15 genotypes.
Fig. 4.
Relationship of the FoxA2 −415(CCT)n polymorphic repeat with FoxA2, PXR, and CYP3A4 mRNA expression and FoxA2 transcriptional activity. Top three box plots, mRNA expression associated with the FoxA2 wild-type −415(CCT)n = 14 genotype and variant n = 13 and n = 15 genotypes. Group differences were analyzed by use of the Kruskal-Wallis test as in the legend to Fig. 3 or by use of the Wilcoxon rank-sum test comparing two genotype groups. Bottom, FoxA2 Luciferase reporters containing −1945/+1204 of the FoxA2 gene with the respective (CCT) n = 13, 14, or 15 repeat genotypes were cotransfected into HepG2 cells with a CMX-LacZ reporter. Luciferase activities, normalized to β-galactosidase activity, were graphed as fold change for each variant genotype relative to the baseline activity of the empty pGL3-basic plasmid (set as one). Values represent the mean ± S.D. measured in triplicate in at least two independent transfections, and results for each genotype reporter were aligned underneath the corresponding results for FoxA2 genotype associated with hepatic mRNA expression. The transcriptional activity of the FoxA2 (CCT)14 reporter differed significantly from the (CCT)13 and (CCT)15 reporters, p = 0.0028 and p = 0.0122, respectively.
Functional Analysis of the FoxA2 Polymorphic CCT Repeat.
We determined the effect of the −415 CCT repeat polymorphism on FoxA2 transcription by amplifying −1945/+1204 bp (relative to TSS.2; −3149/−1 bp relative to TSS.1) of the FoxA2 promoter from donors with wild-type and variant CCT repeat alleles and cloning it into a luciferase reporter. Transfection of the FoxA2 reporters into HepG2 cells revealed that the 14 CCT repeat had the highest transcriptional activity (14 > 13 > 15 > 19) (Fig. 4, bottom; and not shown). Moreover, the transcriptional activities of the FoxA2 promoters were concordant with expression of FoxA2 (and target genes PXR and CYP3A4) mRNA expression in human livers (Fig. 4, top).
Significant Correlations Are also Found among mRNA Levels of Additional Transcription Factors and Coregulated CYP3A4.
To determine whether any other hepatic transcription factors, besides PXR and FoxA2, were associated with CYP3A4 expression, we quantified the relative mRNA levels of HNF4α, FoxA2, FoxA3, C/EBPα, Nr1I2 (PXR), PAR.2 (an isoform of PXR with a longer amino terminal end), NR0B2 (SHP); PGC1α (Ppargc1α), Ncor1, and Ncor2 (SMRT) in the hepatic RNA samples from white subjects by use of TaqMan gene expression assays. In addition, we quantified CYP3A4, CYP3A5, CYP3A7, CYP1A2, CYP2E1, and ABCB1 (MDR1) (Table 2). Extensive interindividual variation in expression was observed. CYP3A4 mRNA expression in white males and females had a ∼13- and 8-fold range of variation, respectively. Sex had limited influence on gene expression, with the exception of CYP3A4 mRNA levels. CYP3A4 relative mRNA expression was significantly higher in females (n = 52; mean ± S.D. = 7.6 ± 2.43; median = 7.61) compared with males (n = 76; mean + S.D. = 6.00 + 2.97; median = 6.05) (p = 0.002).
TABLE 2.
Hepatic expression of the candidate genes in livers of white donors
| Gene | ABI Expression Assay | n | Min a | Med a | S.D. a | Range a | Min a | Max a |
|---|---|---|---|---|---|---|---|---|
| CYP3A4 | ||||||||
| Males | Hs00430021_m1 | 76 | 6.00 | 6.06 | 2.97 | 13.26 | −1.19 | 12.07 |
| Females | 52 | 7.61 | 7.61 | 2.43 | 8.37 | 2.88 | 11.25 | |
| CYP1A2 | Hs00167927_m1 | 40 | 3.15 | 3.45 | 2.54 | 8.84 | −1.91 | 6.93 |
| CYP2E1 | Hs00559368_m1 | 40 | −0.11 | −0.06 | 0.86 | 3.46 | −1.87 | 1.59 |
| CYP3A5 | Hs00241417_m1 | 40 | −0.07 | −0.30 | 2.26 | 8.94 | −4.28 | 4.66 |
| CYP3A7 | Hs00426361_m1 | 39 | 5.03 | 4.85 | 3.15 | 11.50 | −0.56 | 10.95 |
| PXR (Nr1I2) | Hs00243666_m1 | 128 | 4.76 | 5.08 | 2.15 | 10.44 | −1.83 | 8.61 |
| PAR0.2 | Hs00254365_m1 | 127 | 6.07 | 6.20 | 2.32 | 9.98 | 0.58 | 10.56 |
| SHP (NR0B2) | Hs00222677_m1 | 40 | 0.19 | 0.26 | 1.23 | 6.66 | −3.81 | 2.85 |
| FoxA2 (HNF3β) | Hs00232764_m1 | 128 | −0.64 | −0.38 | 2.42 | 11.95 | −7.88 | 4.08 |
| FoxA3 (HNF3γ) | Hs00270130_m1 | 112 | 0.37 | 0.72 | 1.78 | 10.09 | −6.15 | 3.94 |
| HNF4α | Hs01023294_m1 | 91 | 0.36 | 0.75 | 2.03 | 11.07 | −7.20 | 3.87 |
| C/EBPα | Hs00269972_s1 | 40 | −1.52 | −1.05 | 1.62 | 6.53 | −5.69 | 0.84 |
| Ncor2 (SMRT) | Hs00196955_m1 | 100 | −0.74 | −0.64 | 1.16 | 6.39 | −4.74 | 1.66 |
| Ncor1 | Hs00196920_m1 | 40 | −0.84 | −0.60 | 1.57 | 6.57 | −4.93 | 1.64 |
| PGC1α (Ppargc1α) | Hs00173304_m1 | 40 | −0.04 | 0.31 | 1.63 | 7.09 | −4.27 | 2.81 |
| MDR1 (ABCB1) | Hs00184491_m1 | 40 | −0.02 | 0.17 | 0.89 | 3.93 | −2.55 | 1.38 |
The expression data were log transformed (to base 2) before analysis.
The Pearson correlation (Table 3) was calculated to describe the correlation between CYP3A mRNAs and the liver transcription factors, and p values were calculated for the observed correlations. As expected, CYP3A4 mRNA levels were significantly correlated with CYP3A5 and CYP3A7 mRNA levels. PAR.2 is an isoform of PXR, and although it is transcribed from an alternate promoter, its expression was found to be highly correlated with PXR and CYP3A4 mRNA levels in all whites (Table 3). Both PXR and PAR.2 were correlated with HNF4α, HNF3γ, FoxA2, and C/EBPα. The only mRNA demonstrating a negative correlation with CYP3A4 and other mRNAs was SMRT (Ncor2), a nuclear receptor corepressor. Similar results were seen for Spearman's correlation (ρ) of colinearity of rank between CYP3A4 and liver transcription factor mRNA expression levels (not shown).
TABLE 3.
Statistical significance of linear correlations among mRNA levels of transcription factors and coregulated CYP3A genes (Pearson's correlation coefficient and p values)
| r (Pearson) | MDR1 | PGC1α | Ncor1 | Ncor2 | CEBPα | HNF4α | HNF3γ | Foxa2 | SHP | Par2 | PXR | CYP3A7 | CYP3A5 | CYP2E1 | CYP1A2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CYP3A4 | |||||||||||||||
| r | 0.246 | 0.054 | 0.242 | −0.121 | 0.249 | 0.157 | 0.053 | 0.235 | 0.210 | 0.449 | 0.471 | 0.674 | 0.498 | 0.276 | 0.240 |
| sig. | 0.126 | 0.740 | 0.132 | 0.230 | 0.121 | 0.137 | 0.580 | 0.008 | 0.193 | 0.000 | 0.000 | 0.000 | 0.001 | 0.084 | 0.136 |
| CYP1A2 | |||||||||||||||
| r | 0.278 | 0.374 | 0.380 | −0.235 | 0.569 | 0.633 | 0.571 | 0.380 | 0.218 | 0.428 | 0.569 | −0.008 | 0.272 | 0.318 | |
| sig. | 0.083 | 0.018 | 0.016 | 0.145 | 0.000 | 0.000 | 0.000 | 0.015 | 0.177 | 0.006 | 0.000 | 0.960 | 0.089 | 0.046 | |
| CYP2E1 | |||||||||||||||
| r | 0.563 | 0.437 | 0.467 | −0.219 | 0.255 | 0.306 | 0.351 | 0.440 | 0.323 | 0.488 | 0.492 | 0.180 | 0.295 | ||
| sig. | 0.000 | 0.005 | 0.002 | 0.174 | 0.112 | 0.054 | 0.027 | 0.005 | 0.042 | 0.001 | 0.001 | 0.272 | 0.064 | ||
| CYP3A5 | |||||||||||||||
| r | 0.225 | 0.208 | 0.329 | −0.191 | 0.227 | 0.200 | 0.262 | 0.278 | 0.251 | 0.353 | 0.366 | 0.353 | |||
| sig. | 0.162 | 0.197 | 0.038 | 0.238 | 0.159 | 0.215 | 0.102 | 0.082 | 0.118 | 0.025 | 0.020 | 0.027 | |||
| CYP3A7 | |||||||||||||||
| r | 0.519 | 0.250 | 0.346 | −0.153 | 0.338 | 0.272 | 0.245 | 0.359 | 0.350 | 0.409 | 0.294 | ||||
| sig. | 0.001 | 0.124 | 0.031 | 0.354 | 0.035 | 0.093 | 0.133 | 0.025 | 0.029 | 0.010 | 0.070 | ||||
| PXR | |||||||||||||||
| r | 0.610 | 0.571 | 0.713 | 0.179 | 0.667 | 0.702 | 0.602 | 0.682 | 0.619 | 0.903 | |||||
| sig. | 0.000 | 0.000 | 0.000 | 0.075 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |||||
| Par2 | |||||||||||||||
| r | 0.565 | 0.534 | 0.636 | 0.072 | 0.582 | 0.647 | 0.557 | 0.659 | 0.561 | ||||||
| sig. | 0.000 | 0.000 | 0.000 | 0.479 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | ||||||
| SHP | |||||||||||||||
| r | 0.460 | 0.403 | 0.438 | −0.246 | 0.292 | 0.247 | 0.445 | 0.418 | |||||||
| sig. | 0.003 | 0.010 | 0.005 | 0.127 | 0.068 | 0.125 | 0.004 | 0.007 | |||||||
| Foxa2 | |||||||||||||||
| r | 0.715 | 0.603 | 0.771 | 0.206 | 0.671 | 0.711 | 0.744 | ||||||||
| sig. | 0.000 | 0.000 | 0.000 | 0.040 | 0.000 | 0.000 | 0.000 | ||||||||
| HNF3γ | |||||||||||||||
| r | 0.506 | 0.510 | 0.571 | 0.137 | 0.852 | 0.778 | |||||||||
| sig. | 0.001 | 0.001 | 0.000 | 0.186 | 0.000 | 0.000 | |||||||||
| HNF4α | |||||||||||||||
| r | 0.503 | 0.449 | 0.629 | 0.255 | 0.895 | ||||||||||
| sig. | 0.001 | 0.004 | 0.000 | 0.015 | 0.000 | ||||||||||
| CEBPα | |||||||||||||||
| r | 0.511 | 0.416 | 0.599 | −0.051 | |||||||||||
| sig. | 0.001 | 0.008 | 0.000 | 0.755 | |||||||||||
| Ncor2 | |||||||||||||||
| r | 0.085 | 0.025 | 0.306 | ||||||||||||
| sig. | 0.604 | 0.880 | 0.055 | ||||||||||||
| Ncor1 | |||||||||||||||
| r | 0.777 | 0.621 | |||||||||||||
| sig. | 0.000 | 0.000 | |||||||||||||
| PGC1α | |||||||||||||||
| r | 0.640 | ||||||||||||||
| sig. | 0.000 |
Note: Significant (sig.) correlations (p < 0.05) are in bold. A value of 0 indicates that P < 0.0005.
Prediction of CYP3A4 Expression by Multiple Linear Regression Using mRNA Expression of Transcription Factors as Predictors.
Using mRNA expressions of transcription factors as predictors/variables in a multivariable model, we determined the percentage variability (r2) of CYP3A4 mRNA levels that could be explained by a subset of the transcription factors (PXR, PAR.2, FoxA2, HNF4α, FoxA3, and SMRT), because these genes had the largest dataset available (n = 88 livers), making this particular model more stable. By use of a factor analysis, the best predictors of CYP3A4 mRNA expression were: 1) PXR/PAR2 mRNA levels, which together explained as much as 19.0%; 2) sex, which explained 5.69%; and 3) SMRT (NCor2) mRNA level, which explained 3.73% CYP3A4 mRNA variability.
Resequencing and Genotyping of Additional Transcription Factors Coregulating CYP3A4, and of Select Regions of the CYP3A Locus.
We resequenced selected regions of HNF4α, FoxA3, and SMRT (Ncor2) in DNA from the same mRNA phenotype livers and identified polymorphisms in each gene (Table 4). In addition, three SNPs in MDR1/ABCB1 were genotyped because we have previously shown that expression of the mdr1 efflux transporter can influence hepatic expression of Cyp3a (Schuetz et al., 2000), presumably by regulating the intracellular concentration of an endogenous CYP3A physiologic regulator, and because we previously found that SNPs (2677G>T and 3435C>T) in ABCB1 were significantly associated with basal CY3A4 expression and activity in a smaller liver cohort. We also genotyped five SNPs in the CYP3A locus that some have found to be associated with CYP3A4 expression: CYP3A5*3, CYP3A7*1C, the CYP3A4 3′-UTR 27674A>T (rs12333983), an intron 7 SNP (rs2246709), and CYP3A4*1B. In addition, three regions of the CYP3A4 5′ region were selected for resequencing analysis: −6, −12, and −19 kb upstream of the CYP3A4, because they had not been resequenced previously. They demonstrated clustering of LETF binding sites and regulation by LETFS such as C/EBPβ (Martínez-Jiménez et al., 2005), or they had high multispecies conservation or high regulatory potential. Three polymorphisms were identified: −11833C>G, −18440A>T (rs12705060), and −18277C>G (rs11766150).
TABLE 4.
SNPs identified and genotyped in CYP3A4 and transcription factors
| Position in Ref Seq | Position Relative to ATG | rs Number | SNP and Flanking Sequence | Str a | MAF(MA) All | Het F | Het M | Hom Var F | Hom Var M |
|---|---|---|---|---|---|---|---|---|---|
| NC_000019 b | FoxA3 (HNF3γ) | ||||||||
| 51067448 | 7894 | ccctggcac(C>T)gccaagccac | (+) | 0.01(T) | 1 | 2 | 0 | 0 | |
| 51067619 | 8065 | rs16980091 | tgtctttca(C>T)gactgcttcg | (+) | 0.13(T) | 12 | 17 | 1 | 1 |
| 51068057 | 8503 | rs3810327 | cagcacctcc(C>A)aaactggacg | (+) | 0.1(A) | 10 | 15 | 0 | 0 |
| NT_011362 c | HNF4α | ||||||||
| 8036502 | −858 | rs11508796 | ccagaggag(C>T)gccagacagg | (+) | 0.13(T) | 12 | 15 | 2 | 0 |
| 8038170 | 811 | ttcttcagtg(T>G)accgttttca | (−) | 0.03(G) | 2 | 6 | 0 | 0 | |
| 8038270 | 911 | rs6031546 | tattgtgtc(T>C)tccacgaatt | (−) | 0.21(C) | 17 | 24 | 3 | 1 |
| 8038389 | 1030 | tgtgtgattt(G>T)aacaggttta | (−) | 0.004(T) | 1 | 0 | 0 | 0 | |
| 8038632 | 1273 | rs2144908 | attccctggc(C>T)ctctgtcctc | (−) | 0.18(T) | 15 | 18 | 3 | 2 |
| 8052201 | 14842 | taggctgtct(T>C)gtcccctgct | (+) | 0.02(C) | 1 | 3 | 0 | 0 | |
| 8052545 | 15186 | rs6103716 | gaaagttgtc(A>C)aaacaggcatga | (+) | 0.33(C) | 23 | 37 | 3 | 6 |
| 8052558 | 15199 | rs6031558 | acaggcatg(G>C)gaagggcaga | (+) | 0.31(C) | 27 | 31 | 4 | 4 |
| 8078452 | 41093 | gaataaacc(C>T)agaccttgtg | (−) | 0.004(T) | 1 | 0 | 0 | 0 | |
| 8078520 | 41161 | rs717248 | tgtcgtgaag(A>G)ttaaatgaga | (−) | 0.06(G) | 3 | 11 | 0 | 0 |
| 8078699 | 41340 | rs717247 | aaaaatatt(A>G)gcttctttct | (−) | 0.31(G) | 23 | 29 | 4 | 9 |
| 8115620 | 78261 | atgcctacat(A>G)tggggacatc | (+) | 0.005(G) | 1 | 0 | 0 | 0 | |
| 8115852 | 78493 | gagagctccc(G>A)ttacatgagg | (+) | 0.005(A) | 1 | 0 | 0 | 0 | |
| 8115972 | 78613 | ggccgctccc(G>A)ccttcccctg | (+) | 0.28(A) | 21 | 21 | 2 | 7 | |
| 8115985 | 78626 | ttcccctgtg(C>T)cttccctcca | (+) | 0.005(T) | 0 | 1 | 0 | 0 | |
| 8116201 | 78842 | rs6017341 | agggcaaagg(G>C)ctgatgaggt | (−) | 0.04(C) | 5 | 4 | 0 | 0 |
| NC_000012 d | SMRT (Ncor2) | ||||||||
| 123568072 | −22322 | rs6488932 | gcgtgggctc(A>G)ggcaggggag | (−) | 0.02(G) | 1 | 3 | 0 | 0 |
| 123568276 | −22526 | agcctcaccc(C>T)ggcctgcccg | (+) | 0.02(T) | 1 | 4 | 0 | 0 | |
| 123568478 | −22728 | rs837948 | cgacagaact(G>A)gggctctttc | (+) | 0.37(A) | 15 | 39 | 9 | 10 |
| 123568845 | −23095 | gcagaggggc(G>A)gccccagagg | (+) | 0.004(A) | 0 | 1 | 0 | 0 | |
| 123568894 | −23144 | ctcccatccc(G>A)gccctggagg | (+) | 0.01(A) | 1 | 1 | 0 | 0 | |
| 123569005 | −23255 | rs12426064 | ccgcctgacc(C>T)cacggctggg | (+) | 0.01(T) | 1 | 1 | 0 | 0 |
| NG_000004 e , f | CYP3A4 | ||||||||
| 135607 | −392 | rs2740574 | gacaagggc(A>G)gagagaggcg | (+) | 0.07(G) | 4 | 11 | 0 | 1 |
| 124166 | −11833 | cagtgtgtg(C>G)gttcccctcc | (−) | 0.18(G) | 16 | 21 | 1 | 3 | |
| 117722 | −18277 | rs11766150 | ctaagcttct(C>G)ttctcgcttc | (−) | 0.07(G) | 4 | 12 | 0 | 1 |
| 117559 | −18440 | rs12705060 | caaatggct(T>A)actagaataa | (+) | 0.49(A) | 21 | 35 | 14 | 17 |
| 151975 | 15977 | rs2246709 | aatccatag(A/G)gcagaaagtt | (−) | 0.32(G) | 17 | 35 | 7 | 7 |
| 163672 | 27674 | rs12333983 | atatacacgg(T>A)tacatccatt | (−) | 0.14(A) | 10 | 19 | 1 | 2 |
| CYP3A5 | |||||||||
| 247167 | 6986 | rs776746 | ttgtctttc(A>G)tatctcttc | (+) | 0.13(A) | 9 | 17 | 1 | 2 |
| AF364606 g | PXR (NR1I2) | ||||||||
| 63396 | −6993 | rs2472677 | catatttttt(T>C)tgattaaaaa | (+) | 0.35(C) | 27 | 25 | 4 | 10 |
| 46370 | −24019 | rs3842689 | atcaccacag(-/GAGAAG)ccttaactac | (+) | 0.34(Del) | 26 | 30 | 3 | 12 |
| 44477 | −25912 | rs1523130 | tattggaaag(G>A)aaaagagtaa | (+) | 0.38(A) | 24 | 30 | 6 | 14 |
| NT_007933 h | MDR1 (ABCB1) | ||||||||
| 12455162 | 8615 | rs3789243 | acaacgacgc(C>T)ccataaatta | (−) | 0.47(T) | 32 | 31 | 9 | 18 |
| 12394894 | 68883 | rs2032582 | actagaaggt(G>T)ctgggaaggt | (−) | 0.49(T) | 24 | 24 | 11 | 24 |
| 12372921 | 90856 | rs1045642 | aggaagagat(T>C)gtgagggcag | (−) | 0.42(C) | 30 | 24 | 7 | 19 |
MA, minor allele; MAF, minor allele frequency; Gene and ATG positions in the Reference Sequence (Ref Seq): Het, heterozygous; Hom, homozygous; Var, variant.
Strand orientation with respect to GenBank sequence.
FoxA3 = 51059358–51068895, ATG = 51059555–51059557.
HNF4α = 8037356–8112945, ATG = 8037360–8037362.
Ncor2 = 123374914–123568793, ATG = 123545750-48 (compliment).
CYP3A4 = 135895–163090, ATG = 135999–136001.
CYP3A5 = 240080–271889, ATG = 240182–240184.
PXR = 45837–80887, ATG = 70390–70392.
ABCB1 = 12367224–12576840, ATG = 12463776-4 (compliment).
Univariate Analysis of cis- and trans-Genetic Variation for Association with CYP3A4 mRNA Expression.
In the combined population, CYP3A4 expression was significantly associated with 1) PXR 44477C>T (rs1523130) (CC versus CT+TT; 7.4 ± 2.75 versus 6.13 ± 2.87, p = 0.01) and 2) the MDR1 exon 26 polymorphism at 3435C>T. Livers homozygous for the T allele had significantly higher CYP3A4 mRNA expression compared with livers with CT or CC genotypes (7.32 ± 2.77 versus 6.66 ± 2.48 versus 5.35 ± 3.41; p = 0.02) (Supplemental Table 3). None of the polymorphisms in HNF4α, FoxA3, or SMRT was predictive of CYP3A4 mRNA expression when all whites were combined or after stratification by sex.
Multivariable Analysis of Variation in the CYP3A4 Regulatory Region and in Transcription Factors for Association with CYP3A4 mRNA Expression.
Because univariate analysis identified multiple transcription factor polymorphisms associated with CYP3A4 mRNA levels in liver, the data were further analyzed by use of multiple linear regression (MLR) to explain the variability observed in CYP3A4 mRNA levels (Table 5). MLR analysis was performed by use of different filtering stringencies that either included all the SNPs (without any filters) or included only those SNPs that fulfilled the following criteria: 1) SNPs with p value of <0.1 in univariate analysis; 2) SNPs with more than three samples for each genotype category (wild-type, heterozygous, or variant). With use of the more stringent filtering criteria, 19.5% of the variation in CYP3A4 expression could be explained by the combined effect of sex and polymorphisms in ABCB1/MDR1, FoxA2, HNF3γ, and the PXR promoter and by the interaction between sex and FoxA2 SNP rs1203910. Using less stringent filtering criteria, sex, polymorphisms in MDR1 exon 26, FoxA2, and interactions of sex with SNPs in FoxA2 and HNF4α explained 24.6% of the variation observed in CYP3A4 mRNA levels. The CYP3A4 locus SNP (rs12705060) at −19 kb upstream of CYP3A4, close to a cluster of LETF sites, also came up as a predictor in the MLR analysis and explained an additional 2.3% CYP3A4 mRNA variability. To check concordance of the MLR results, we also used classification and regression tree analysis and found very similar results (data not shown).
TABLE 5.
Results of multiple linear regression analysis to explain the variability observed in CYP3A4 mRNA levels
| Gene | Effect on CYP3A4 Expression | % of Total Variability Explained with Addition of Factor | p Value | BIC |
|---|---|---|---|---|
| More filtering | ||||
| Base a | 225 | |||
| rs1045642 (MDR1) | Increased in CT and TT | 6.6 | 0.01 | 219 |
| Sex | Increased in females | 9.5 | 0.04 | 217 |
| rs1212275 (FoxA2) | Increased in CT and TT | 12.8 | 0.03 | 214 |
| rs16980091(HNF3γ) | Increased in CT and TT | 15.3 | 0.05 | 212 |
| rs1523130 (PXR) | Decreased in CT and TT | 17.0 | 0.08 | 210 |
| Interaction: sex and rs1203910 (FoxA2) b | Decreased in TT, increased in males and TT | 19.5 | 0.08 | 209 |
| Using less filtering | ||||
| Base a | 191 | |||
| rs1045642 (MDR1) | Increased in CT and TT | 11.0 | 0.002 | 182 |
| rs1212275 (FoxA2) | Increased in CT and TT | 14.7 | 0.03 | 179 |
| Sex | Increased in females | 16.9 | 0.02 | 177 |
| Interaction: sex and rs1203910 (FoxA2) b | Decreased in TT, increased in males and TT | 17.9 | 0.2 | 178 |
| rs6031546 (HNF4α) | Increased in TT | 20.0 | 0.08 | 177 |
| Interaction: sex and rs11508796 (HNF4α) b | Increased in TT, decreased in males and TT | 22.3 | 0.1 | 176 |
| rs12705060 (CYP3A4) | Increased in TT | 24.6 | 0.1 | 175 |
BIC, Bayesian Information Criterion.
Base is the base model without the addition of any factor or variable.
Interaction denotes a significant difference in effect of a SNP in males and females.
Discussion
CYP3A4 is known to be under combinatorial transcriptional control. We determined how differential expression of CYP3A4 regulators contributes to variable CYP3A4 expression. Our data suggest a pattern of correlated expression of these transcription factor mRNAs with CYP3A4 mRNA. We showed for the first time that CYP3A4 hepatic expression was correlated significantly with the FoxA2 mRNA expression in white females, and, as previously shown in smaller cohorts (Wolbold et al., 2003; Lamba et al., 2004), CYP3A4 mRNA was correlated with PXR mRNA in all livers. PXR expression, in turn, was significantly related with expression of HNF4α, HNF3γ, and C/EBPα. The data also revealed a negative correlation of CYP3A4 expression with SMRT (Ncor2) expression. This relationship is biologically plausible because Ncor2 is a known PXR corepressor. The association of CYP3A4 positively with PXR and negatively with Ncor2 suggests that PXR may reside at the CYP3A4 promoter directing some level of basal transcriptional activity.
Because CYP3A4 expression can be considered a complex trait under multifactorial control, we used multiple linear regression analysis to determine which SNPs affected/predicted CYP3A4 expression. Polymorphisms in PXR, FoxA2, FoxA3 (HNF3γ) (a nonsynonymous SNP), and HNF4α (a 5′-SNP), combined with sex, together explained as much as 24.6% of the variation in CYP3A4 expression. As previously found in a smaller liver cohort, individuals with the MDR1 rs1045642 SNP have higher levels of hepatic CYP3A4, presumably because of higher intracellular concentrations of endogenous CYP3A4 regulators.
Despite the significant correlations between PXR and FoxA2 mRNA expression levels, CYP3A4 expression was more highly associated with PXR mRNA expression and less with FoxA2 expression. This evidence suggests one hypothetical model in which FoxA2 could be involved in regulating the expression of PXR, which in turn regulates CYP3A4 transcription. Indeed, FoxA2 has been shown to bind to the proximal mouse PXR promoter (Kyrmizi et al., 2006), to regulate mouse liver PXR expression (Bochkis et al., 2008), and to amplify this biological network by directly regulating the PXR coregulator Ncoa2 (SRC-2) (Bochkis et al., 2009). However, whether FoxA2 is an equally important regulator of human CYP3A4 and PXR was not known, in particular, because Odom et al. (2007) previously determined that Foxa2 binding regions within target genes in mouse and human genomes have diverged substantially. We had previously suggested that FoxA2 might be a human PXR regulator because we found that multiple SNPs in putative FoxA2 binding sites in the PXR gene were associated with PXR mRNA expression and with target genes CYP3A4 and MDR1 (Lamba et al., 2008). In addition, FoxA2 is probably a direct regulator of CYP3A4 as well, because Cyp3a is a direct FoxA2 target in mice (Bochkis et al., 2008), and ectopic overexpression of FoxA2 induces CYP3A4 mRNA in HepG2 cells and LS180 cells (E. Schuetz unpublished observation).
Although our study analyzed constitutive CYP3A4 expression in human livers, there is evidence that FoxA2 is also important for CYP3A4 inducibility because 1) we previously showed that SNPs in putative FoxA2 binding sites in PXR were also associated with the magnitude of PXR-mediated rifampin induction of CYP3A4 in primary human hepatocytes (Lamba et al., 2008), and 2) Kaestner (Bochkis et al., 2008) found that Cyp3a was less inducible by bile acids in mice with conditional hepatic FoxA2 ablation. Although decreased expression of PXR in Foxa2−/− mice contributes to this phenotype, it is also possible that Foxa2 facilitates PXR access to the Cyp3a chromatin because Foxa2 binding relieves chromatin compaction and allows binding of other transcription factors (Lee et al., 2005). Indeed, Foxa proteins have been suggested to act as “pioneer factors” facilitating nuclear hormone receptor binding to target genes (Friedman and Kaestner, 2006). Foxa2 binding sites are predicted in the CYP3A4 promoter at (−8237 and −8333bp) near distal PXR elements (−7733/−7898 bp) and at −10787, 10837, and −11265 bp in or adjacent to the HNF1 and HNF4 elements in the CLEM-4 regulatory region. There is at least one additional implication of the inter-relationship of FoxA2, PXR, and CYP3A. Insulin was first reported to repress drug metabolism in 1961 (Dixon et al., 1961), and Omiecinski reported that insulin treatment significantly attenuated Cyp3a induction by dexamethasone in rat hepatocytes (Sidhu and Omiecinski, 1999). Because insulin inactivates FoxA2 through phosphorylation, it will be interesting to determine whether insulin-inactivated FoxA2 contributes to decreased Cyp3a induction.
The identification of common FoxA2 polymorphisms predictive of FoxA2 and target gene PXR and CYP3A4 expression is of additional interest because FoxA2 is important to bile acid and glucose homeostasis (Rausa et al., 2000) and is a candidate gene associated with maturity onset diabetes of the young (MODY). An earlier effort to resequence FoxA2 in the French population identified three of the polymorphisms found in this study, A97A, G279G, and Q396Q (Abderrahmani et al., 2000), but none was associated with MODY in that study or with hepatic FoxA2 expression in our study. Therefore, several polymorphisms that we identified are of interest as candidate MODY polymorphisms, in particular, because none of these was in LD with FoxA2 Tag SNPs. First are the rare nonsynonymous SNPs that result in M87V and M121V amino acid substitutions, because these changes are predicted by Polyphen to be damaging to the FoxA2 protein. Second are the two polymorphisms that are associated with FoxA2 and PXR mRNAs: 1) the common rs1212275 (Q396Q) SNP [although this is a synonymous change, there is precedence for silent polymorphisms, such as the MDR1 exon 26 SNP, to alter mRNA stability and conformation] (Kimchi-Sarfaty et al., 2007); and 2) the (CCT)n trinucleotide repeat polymorphism. This repeat is located in intron 1 of the FoxA2 mRNA encoding the 457-aa FoxA2 protein but in the promoter (−212 bp upstream) of TSS.1 of the alternative FoxA2 mRNA. Any deviation from 14 repeats was associated with lower FoxA2, PXR, and CYP3A4 mRNAs. Likewise, there was a corresponding decrease in the transcriptional activity of FoxA2 reporter plasmids with the polymorphic repeats in transfected liver cells. The FoxA2 repeat polymorphism is also of interest because expansion of trinucleotide repeats is responsible for multiple human diseases, although those expansions are typically much larger in number than observed here. Nevertheless, there are numerous accounts in the literature reporting the involvement of polymorphic repeats with variability in mRNA expression. For example, a polymorphic CA repeat in intron 1 of the epidermal growth factor receptor is associated with its expression in breast cancer (Gebhardt et al., 1999); a repeat polymorphism in the ERβ gene has also been implicated in menopausal and premenstrual symptoms (Takeo et al., 2005); and a shorter CAG repeat in the androgen receptor gene has been shown to be associated with atypical hyperplasia and breast carcinoma (De Abreu et al., 2007). Hence, the results from our study and the report that the CCT trinucleotide repeat polymorphism was associated with risk of type 2 diabetes in North Indians (Tabassum et al., 2008) make this an interesting polymorphism for further study in diabetes risk and for association with altered CYP3A-mediated drug metabolism in diabetes.
Supplementary Material
Acknowledgments
We thank the Hartwell Center at St. Jude Children's Research Hospital for the DNA sequencing and oligonucleotide synthesis.
This work was supported in part by the National Institutes of Health [Grant GM60346]; National Institutes of Health National Institute of General Medical Sciences Pharmacogenetics Research Network and Database [Grants U01-GM61374, U01-GM61393]; the National Institutes of Health Cancer Center Support [Grant P30-CA21765]; and by the American Lebanese Syrian Associated Charities.
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.109.160804.
The online version of this article (available at http://jpet.aspetjournals.org) contains supplemental material.
- LETF
- liver enriched transcription factor
- PXR
- pregnane X receptor
- FoxA2, HNF3β
- hepatic nuclear factor-3-β
- FoxA3, HNF3γ
- hepatic nuclear factor-3-γ
- C/EBP
- CCAAT/enhancer binding protein
- SMRT
- silencing mediator for retenoid or thyroid hormone receptor
- Ncor2
- nuclear receptor corepressor 2
- SNP
- single-nucleotide polymorphism
- MDR1/ABCB1
- multidrug resistance protein 1/ATP-binding cassette subfamily B member 1
- MLR
- multiple linear regression
- PCR
- polymerase chain reaction
- UTR
- untranslated region
- LD
- linkage disequilibrium.
References
- Abderrahmani A, Chèvre JC, Otabe S, Chikri M, Hani EH, Vaxillaire M, Hinokio Y, Horikawa Y, Bell GI, Froguel P. (2000) Genetic variation in the hepatocyte nuclear factor-3beta gene (HNF3B) does not contribute to maturity-onset diabetes of the young in French Caucasians. Diabetes 49:306–308 [DOI] [PubMed] [Google Scholar]
- Bochkis IM, Rubins NE, White P, Furth EE, Friedman JR, Kaestner KH. (2008) Hepatocyte-specific ablation of Foxa2 alters bile acid homeostasis and results in endoplasmic reticulum stress. Nat Med 14:828–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkis IM, Schug J, Rubins NE, Chopra AR, O'Malley BW, Kaestner KH. (2009) Foxa2-dependent hepatic gene regulatory networks depend on physiological state. Physiol Genomics 38:186–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Abreu FB, Pirolo LJ, Canevari Rde A, Rosa FE, Moraes Neto FA, Caldeira JR, Rainho CA, Rogatto SR. (2007) Shorter CAG repeat in the AR gene is associated with atypical hyperplasia and breast carcinoma. Anticancer Res 27:1199–1205 [PubMed] [Google Scholar]
- Dixon RL, Hart LG, Fouts JR. (1961) The metabolism of drugs by liver microsomes from alloxan-diabetic rats. J Pharmacol Exp Ther 133:7–11 [PubMed] [Google Scholar]
- Friedman JR, Kaestner KH. (2006) The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci 63:2317–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuen S, Fukuda T, Maune H, Ikenaga Y, Yamamoto I, Inaba T, Azuma J. (2002) Novel detection assay by PCR-RFLP and frequency of the CYP3A5 SNPs, CYP3A5*3 and *6, in a Japanese population. Pharmacogenetics 12:331–334 [DOI] [PubMed] [Google Scholar]
- Gebhardt F, Zänker KS, Brandt B. (1999) Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 14. J Biol Chem 274:13176–13180 [DOI] [PubMed] [Google Scholar]
- Jover R, Bort R, Gómez-Lechón MJ, Castell JV. (2002) Down-regulation of human CYP3A4 by the inflammatory signal interleukin-6: molecular mechanism and transcription factors involved. FASEB J 16:1799–1801 [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. (2007) A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315:525–528 [DOI] [PubMed] [Google Scholar]
- Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, Talianidis I. (2006) Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev 20:2293–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba J, Lamba V, Strom S, Venkataramanan R, Schuetz E. (2008) Novel single nucleotide polymorphisms in the promoter and intron 1 of human pregnane X receptor/NR1I2 and their association with CYP3A4 expression. Drug Metab Dispos 36:169–181 [DOI] [PubMed] [Google Scholar]
- Lamba J, Strom S, Venkataramanan R, Thummel KE, Lin YS, Liu W, Cheng C, Lamba V, Watkins P, Schuetz E. (2006) MDR1 genotype is associated with Hepatic CYP3A4 basal and induction phenotype.Clin Pharmacol Therap 79:325–338 [DOI] [PubMed] [Google Scholar]
- Lamba JK, Lin YS, Schuetz EG, Thummel KE. (2002) Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev 54:1271–1294 [DOI] [PubMed] [Google Scholar]
- Lamba V, Yasuda K, Lamba JK, Assem M, Davila J, Strom S, Schuetz EG. (2004) PXR (NR1I2): splice variants in human tissues, including brain, and identification of neurosteroids and nicotine as PXR activators. Toxicol Appl Pharmacol 199:251–265 [DOI] [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, Kaestner KH. (2005) The initiation of liver development is dependent on Foxa transcription factors. Nature 435:944–947 [DOI] [PubMed] [Google Scholar]
- Li Z, White P, Tuteja G, Rubins N, Sackett S, Kaestner KH. (2009) Foxa1 and Foxa2 regulate bile duct development in mice. J Clin Invest 119:1537–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YP, Huang JD. (2008) Interplay of pregnane X receptor with other nuclear receptors on gene regulation. Drug Metab Pharmacokinet 23:14–21 [DOI] [PubMed] [Google Scholar]
- Lin YS, Dowling AL, Quigley SD, Farin FM, Zhang J, Lamba J, Schuetz EG, Thummel KE. (2002) Co-regulation of CYP3A4 and CYP3A5 and contribution to hepatic and intestinal midazolam metabolism. Mol Pharmacol 62:162–172 [DOI] [PubMed] [Google Scholar]
- Martínez-Jiménez CP, Gómez-Lechón MJ, Castell JV, Jover R. (2005) Transcriptional regulation of the human hepatic CYP3A4: identification of a new distal enhancer region responsive to CCAAT/enhancer-binding protein beta isoforms (liver activating protein and liver inhibitory protein). Mol Pharmacol 67:2088–2101 [DOI] [PubMed] [Google Scholar]
- Martínez-Jiménez CP, Jover R, Donato MT, Castell JV, Gómez-Lechón MJ. (2007) Transcriptional regulation and expression of CYP3A4 in hepatocytes. Curr Drug Metab 8:185–194 [DOI] [PubMed] [Google Scholar]
- Nakamura K, Moore R, Negishi M, Sueyoshi T. (2007) Nuclear pregnane X receptor cross-talk with FoxA2 to mediate drug-induced regulation of lipid metabolism in fasting mouse liver. J Biol Chem 282:9768–9776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DA, Tobe VO, Taylor SL. (1997) PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res 25:2745–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E. (2007) Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet 39:730–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ourlin JC, Jounaidi Y, Maurel P, Vilarem MJ. (1997) Role of the liver-enriched transcription factors C/EBP alpha and DBP in the expression of human CYP3A4 and CYP3A7. J Hepatol 26 (Suppl 2):54–62 [DOI] [PubMed] [Google Scholar]
- Ozdemir V, Kalow W, Tang BK, Paterson AD, Walker SE, Endrenyi L, Kashuba AD. (2000) Evaluation of the genetic component of variability in CYP3A4 activity: a repeated drug administration method. Pharmacogenetics 10:373–388 [DOI] [PubMed] [Google Scholar]
- Rausa FM, Tan Y, Zhou H, Yoo KW, Stolz DB, Watkins SC, Franks RR, Unterman TG, Costa RH. (2000) Elevated levels of hepatocyte nuclear factor 3beta in mouse hepatocytes influence expression of genes involved in bile acid and glucose homeostasis. Mol Cell Biol 20:8264–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Antona C, Bort R, Jover R, Tindberg N, Ingelman-Sundberg M, Gómez-Lechón MJ, Castell JV. (2003) Transcriptional regulation of human CYP3A4 basal expression by CCAAT enhancer-binding protein alpha and hepatocyte nuclear factor-3 gamma. Mol Pharmacol 63:1180–1189 [DOI] [PubMed] [Google Scholar]
- Schirmer M, Rosenberger A, Klein K, Kulle B, Toliat MR, Nürnberg P, Zanger UM, Wojnowski L. (2007) Sex-dependent genetic markers of CYP3A4 expression and activity in human liver microsomes. Pharmacogenomics 8:443–453 [DOI] [PubMed] [Google Scholar]
- Schuetz EG. (2004) Lessons from the CYP3A4 promoter. Mol Pharmacol 65:279–281 [DOI] [PubMed] [Google Scholar]
- Schuetz EG, Umbenhauer DR, Yasuda K, Brimer C, Nguyen L, Relling MV, Schuetz JD, Schinkel AH. (2000) Altered expression of hepatic cytochromes P-450 in mice deficient in one or more mdr1 genes. Mol Pharmacol 57:188–197 [PubMed] [Google Scholar]
- Shimada T, Iwasaki M, Martin MV, Guengerich FP. (1989) Human liver microsomal cytochrome P-450 enzymes involved in the bioactivation of procarcinogens detected by umu gene response in Salmonella typhimurium TA 1535/pSK1002. Cancer Res 49:3218–3228 [PubMed] [Google Scholar]
- Sidhu JS, Omiecinski CJ. (1999) Insulin-mediated modulation of cytochrome P450 gene induction profiles in primary rat hepatocyte cultures. J Biochem Mol Toxicol 13:1–9 [DOI] [PubMed] [Google Scholar]
- Tabassum R, Chavali S, Dwivedi OP, Tandon N, Bharadwaj D. (2008) Genetic variants of FOXA2: risk of type 2 diabetes and effect on metabolic traits in North Indians. J Hum Genet 53:957–965 [DOI] [PubMed] [Google Scholar]
- Takeo C, Negishi E, Nakajima A, Ueno K, Tatsuno I, Saito Y, Amano K, Hirai A. (2005) Association of cytosine-adenine repeat polymorphism of the estrogen receptor-beta gene with menopausal symptoms. Gend Med 2:96–105 [DOI] [PubMed] [Google Scholar]
- Tirona RG, Lee W, Leake BF, Lan LB, Cline CB, Lamba V, Parviz F, Duncan SA, Inoue Y, Gonzalez FJ, et al. (2003) The orphan nuclear receptor HNF4 alpha determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nat Med 9:220–224 [DOI] [PubMed] [Google Scholar]
- Uetzmann L, Burtscher I, Lickert H. (2008) A mouse line expressing Foxa2-driven Cre recombinase in node, notochord, floorplate, and endoderm. Genesis 46:515–522 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Kumai T, Matsumoto N, Tanaka M, Suzuki S, Satoh T, Kobayashi S. (2004) Expression of CYP3A4 mRNA is correlated with CYP3A4 protein level and metabolic activity in human liver. J Pharmacol Sci 94:459–462 [DOI] [PubMed] [Google Scholar]
- Wolbold R, Klein K, Burk O, Nüssler AK, Neuhaus P, Eichelbaum M, Schwab M, Zanger UM. (2003) Sex is a major determinant of CYP3A4 expression in human liver. Hepatology 38:978–988 [DOI] [PubMed] [Google Scholar]
- Wolfrum C, Asilmaz E, Luca E, Friedman JM, Stoffel M. (2004) Foxa2 regulates lipid metabolism and ketogenesis in the liver during fasting and in diabetes. Nature 432:1027–1032 [DOI] [PubMed] [Google Scholar]
- Wortham M, Czerwinski M, He L, Parkinson A, Wan YJ. (2007) Expression of constitutive androstane receptor, hepatic nuclear factor 4 alpha, and P450 oxidoreductase genes determines interindividual variability in basal expression and activity of a broad scope of xenobiotic metabolism genes in the human liver. Drug Metab Dispos 35:1700–1710 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.