Abstract
Circadian rhythms in physiology and behavior are known to be influenced by the estrous cycle in female rodents. The clock genes responsible for the generation of circadian oscillations are widely expressed both within the central nervous system and peripheral tissues, including those that comprise the reproductive system. To address whether the estrous cycle affects rhythms of clock gene expression in peripheral tissues, we first examined rhythms of clock gene expression (Per1, Per2, Bmal1) in reproductive (uterus, ovary) and non-reproductive (liver) tissues of cycling rats using quantitative real-time PCR (in vivo) and luminescent recording methods to measure circadian rhythms of PER2 expression in tissue explant cultures from cycling PER2::LUCIFERASE (PER2::LUC) knockin mice (ex vivo). We found significant estrous variations of clock gene expression in all three tissues in vivo, and in the uterus ex vivo. We also found that exogenous application of estrogen and progesterone altered rhythms of PER2::LUC expression in the uterus. In addition, we measured the effects of ovarian steroids on clock gene expression in a human breast cancer cell line (MCF-7 cells) as a model for endocrine cells that contain both the steroid hormone receptors and clock genes. We found that progesterone, but not estrogen, acutely up-regulated Per1, Per2, and Bmal1 expression in MCF-7 cells. Together, our findings demonstrate that the timing of the circadian clock in reproductive tissues is influenced by the estrous cycle and suggest that fluctuating steroid hormone levels may be responsible, in part, through direct effects on the timing of clock gene expression.
Keywords: Estrogen, Progesterone, Uterus, PER2::LUC, MCF-7 cell line, Circadian Rhythm
1. Introduction
The circadian and reproductive systems communicate bi-directionally through the brain and periphery to coordinate the timing of important reproductive events. The central circadian clock located in the suprachiasmatic nucleus (SCN) of the hypothalamus generates daily oscillations [1] responsible for organizing the timing of most behaviors and physiological events in mammals, including the luteinizing hormone (LH) surge during the estrous cycle that triggers ovulation [2, 3]. In cycling female rodents, the LH surge occurs on the day of proestrus at a specific “critical period” in the late afternoon, approximately 2 – 4 h before lights off [4, 5]. In female rats, SCN-lesions abolish this LH surge and prevent ovulation [6].
Reproductive events, in turn, have both direct and indirect feedback effects on the temporal organization of behavior and physiology. For example, in humans, menstruation, pregnancy and menopause have all been associated with sleep disorders and changes in body temperature rhythms [7–10]. Similarly, circadian rhythms of activity significantly change during the estrous cycle in rodents [11–14]: female rats show phase advances of locomotor activity and higher total activity during stages of proestrus and estrus relative to diestrus [11]. Estrogen itself alters circadian rhythms, as implants shorten the period of locomotor activity of female rats and hamsters while increasing both the amplitude and activity bout length [12–14]. Interestingly, these estrogenic effects are alleviated in the presence of progesterone, suggesting that complex regulation of the hormonal milieu across the estrous cycle is necessary for the observed behavioral effects.
The specific role of the SCN in both the timing of reproductive events and in the temporal reorganization during these events has been bolstered by our increased understanding of the molecular clock and its constituent clock genes, which drive circadian rhythms of gene expression and physiology with a 24 h period. These genes and their protein products are organized into interlocking positive and negative transcriptional and translational feedback loops that regulate circadian rhythm generation in the SCN [15]. A recent and important finding revealed that the same or similar molecular feedback loops found in SCN cells, are also present in a majority of peripheral organs [16]. Furthermore, evidence suggests that several of these peripheral clocks can continue to oscillate in the absence of the SCN [17]. Therefore, it is possible that peripheral oscillators may locally regulate physiology while also receiving modulatory timing cues from the SCN. Although numerous studies have described the expression of circadian clock genes in reproductive tissues, including the ovary [18, 19] and uterus [20, 21], the physiological function of clock genes in tissues of the reproductive system remains largely unknown. Further, the very nature of the temporal cues carrying timing information from the SCN to peripheral oscillators, and perhaps in the reverse direction, are also poorly understood. Several reports demonstrate reproductive dysfunctions in mice with clock gene mutations [22, 23]. For example, female Clock-mutant mice, which carry a 51-amino acid deletion in the transcriptional activation domain of the CLOCK protein gene, have irregular estrous cycles, do not have a normal LH surge on the day of proestrus [23] and fail to show circadian rhythms of clock gene expressions in the uterus [22].
In the present study, our goal was to address two hypotheses: 1) that the estrous cycle drives changes in the timing of the circadian clocks in reproductive tissues and 2) that fluctuating levels of circulating ovarian steroid hormones [17β-estradiol (E2) and progesterone (P4)] during the cycle modulate the timing of clock gene expression in target oscillators. To address these questions, we have employed four sets of experiments using multiple levels of analysis. First, using cycling rats, we examined diurnal rhythms of Per1, Per2, and Bmal1 mRNA as a function of estrous cycle stage in both reproductive (uterus and ovary) and non-reproductive tissues (liver). Second, using tissue explants from PER2::LUCIFERASE (PER2::LUC) knockin mice, we compared rhythmic properties of both central and peripheral clocks at different stages of the estrous cycle. To examine the putative role for circulating ovarian steroid hormones, we measured their lasting effects on the period of PER2::LUC expression rhythms in uterine explants from mice. Finally, using the MCF-7 human cancer cell line as a model for endocrine cells containing both steroid hormone receptors and circadian clock genes, we examined the effects of acute E2 or P4 treatment on the level of clock gene expression.
2. Experimental
2.1. Experiment 1: Effects of the Estrous Cycle on Diurnal Rhythms of Clock Gene Expression in Peripheral Tissues of rats
Intact female Wistar rats were obtained from Charles River Laboratories (Yokohama, Japan) at 7 – 8 weeks of age, and were maintained under controlled environmental conditions (temperature, 24 ± 1 °C; lights on at 0600 – 1800 h) with food and water available ad libitum for at least 2 weeks. Vaginal smears were taken every morning and those rats that exhibited at least two consecutive 4-day estrous cycles were used in the present study. Animals were sacrificed in groups on the same day of the estrous cycle by deep ether anesthesia every 4 h beginning at zeitgeber time 0 (ZT 12 is defined as the time of lights off; n = 4 for each time point). After the uterus, ovary, and liver were rapidly removed, and tissues were frozen in dry ice and stored at − 80 °C. All animal housing and surgical procedures were in accordance with the guidelines of the Japanese Physiological Society and were approved by the Institutional Animal Care and Use Committee of Graduate School of Bioagricultural Sciences in Nagoya University.
RNA from the uterus, ovary, and liver was extracted with TRIzol solution (Invitrogen, Carlsbad, CA) and digested with DNase (Invitrogen) to remove possible DNA contaminants. The samples were dissolved in 20 µl of diethyl pyrocarbonate-treated water and stored at − 80 °C until use. Briefly, 1 µg of total RNA was reverse transcribed to cDNA with random primers using SuperScript II RT (Invitrogen). Real-time PCR was performed at 95 °C for 10 min and then run for 35 cycles at 95 °C for 15 sec, 60 °C for 1 min using SYBR® Green (Applied Biosystems, Foster City, CA) and the ABI PRISM® 7000 real-time PCR system (Applied Biosystems). PCR primers were designed using Primer Express V1.0 Software (Applied Biosystems) and the sequences of the primers were as follows: Per1 (GenBank accession number: AB092976), 5’-GCTTGTGTGGACTGTGGTAGCA-3’ and 5’- GCCCCAATCCATCCAGTTCT-3’; Per2 (GenBank accession number: AB016532), 5’-AAGCATCCAGTCCTGTTTTCTTTAGA-3’ and 5’-CATCGCACAAACTCTGAGAGCTTA-3’; Bmal1 (GenBank accession number: AB012600), 5’-GGAGAAGGTGGCCCAAAGAG-3’ and 5’- TCGGCAATCATTCGACCTATT-3’; cyclophilin (GenBank accession number: M19533), 5’- GGGTTCCTCCTTTCACAGAATTATT-3’ and 5’-TTGCCACCAGTGCCATTATG-3’. Relative standard curves were created with serially diluted cDNA product for each transcript (0, 102, 103, 104, 105 106, 107, 108 copies/µl). All cDNA samples (10 ng – 100 ng) were analyzed in duplicate and amounts of the target cDNA were calculated. A melt curve analysis was then performed to confirm there was no non-specific product amplification. In addition, to correct for variations in RNA and/or cDNA quality and quantity, average transcript levels of the target genes were normalized to the average level of the constitutively expressed housekeeping gene Cyclophilin.
2.2. Experiment 2: Effects of the Estrous Cycle on Circadian Rhythms of PER2::LUC Expression in Tissue Explants from mice
Three to four-month-old intact female PER2::LUC knockin mice [originally purchased from The Jackson Laboratory, Bar Harbor, ME; [17]] from our breeding colony were used for all experiments. Animals were maintained under controlled environmental conditions (temperature, 22 ± 2 °C; lights on at 0800 – 2000 h) with food and water available ad libitum. Vaginal smears were taken daily and those mice that exhibited at least two consecutive 4 or 5-day estrous cycles were used in the present study. All procedures and standards of care were approved by the UCLA Division of Laboratory Animals and were conducted according to the National Institutes of Health guidelines for the use of experimental animals. Mice identified at stages of proestrus or metestrus were euthanized 1 – 2 h before lights-off with deep isoflurane anesthesia, rapidly decapitated and tissue culture procedures were carried out as previously reported [24]. Briefly, for SCN cultures, brain tissue was removed, placed in chilled Hank's buffered salt solution (HBSS) and sliced in the coronal plane on a vibratome at a thickness of 300 µm. The bilateral SCN and a minimum of surrounding tissue were isolated from the slice using scalpels. The uterus, ovary, and liver were removed, placed in chilled HBSS, isolated from the surrounding adipose tissue and dissected into small pieces (≈ 1 mm3) with the aid of a dissecting microscope. SCN slices and peripheral tissue explants were placed at the liquid interface on membranes (Millicell-CM, PICM030-50; Millipore, Billerica, MA ) in 35 mm dishes (Nunc, Thermo Fisher Scientific, Rochester, NY) containing 1.0 ml recording medium [serum-free, no sodium bicarbonate, no phenol red, Dulbecco's Modified Eagle's Medium (D-2902, Sigma-Aldrich; St. Louis, MO)] supplemented with 0.35 g/l sodium bicarbonate, 10 mM HEPES (pH 7.2), B27 supplement (2%; 17504-010, Invitrogen) and 0.1 mM luciferin (beetle luciferin, potassium salt; Promega, Madison, WI) and antibiotics (25 U/ml penicillin, 25 mg/ml streptomycin; Invitrogen). Although serum-free, our cell culture medium for uterine explants does contain a B27 supplement that is known to contain a small amount of P4 [25]. The dishes were sealed using vacuum grease and placed into a LumiCycle (Actimetrics, Wilmette, IL) inside a light-tight 36.5 °C environmental chamber.
The bioluminescence signal was counted every ten minutes for at least 4 days and data were normalized by subtraction of the 24 h running average from the raw data and then smoothed with a 2 h running average (Origin Lab 7.0; Origin Labs Inc., Northampton, MA). The peak was calculated as the highest point of smoothed data during the interval between 12 h and 36 h in culture. The amplitude was summed by the first lowest point and the next highest point of the data.
2.3. Experiment 3: Effects of Estrogen and Progesterone on Circadian Rhythms of PER2::LUC Expression in Uterine Explants from mice
Procedures for animal and tissue culture are identical to those in Experiment 2. Mice identified at stages of metestrus were euthanized 1 – 2 h before lights-off with deep isoflurane anesthesia, rapidly decapitated and tissue culture procedures were carried out. Hormone treatment was carried out 2 days after the onset of recording. This delay allows us to monitor at least one full cycle of luminescence before the start of treatment and allows us to simultaneously mimic the timing of the steroid hormone background on proestrus. Culture dishes were temporarily removed from the LumiCycle and stock solutions of E2 and P4 were directly applied to culture media at projected ZT 1 (for E2; in the morning) and ZT 7 (for P4; in the afternoon). The dishes were back to LumiCycle and continued recording. The peak and amplitude were calculated from smoothed data after the treatments.
2.4. Experiment 4: Effects of Estrogen or Progesterone on the Expression of Per1, Per2, and Bmal1 mRNA in MCF-7 Human Cancer Cells
The human breast cancer MCF-7 cells (ER and PR positive; ATCC HTB-22) were cultured at 37 °C (5% CO2) in DMEM (Invitrogen) supplemented with 10 % FBS (Omega Scientific, Inc., Tarzana, CA) containing 50 U/ml penicillin and 50 µg/ml streptomycin. Approximately 5 × 104 cells were seeded in a 24-well dish at least 3 days before the experiment. When the cells reached 90 – 100 % confluency, they were gently washed with PBS and treated with media containing either compound or vehicle. At the end of treatment, the medium was removed and the cells were washed with PBS once, and TRIzol solution (Invitrogen) was added for RNA extraction. The cells were transferred into 1.5 ml tubes and stored at −80 °C until RNA extraction. Stock solutions of drugs were directly added to the media at a volume of 0.1 %. RNA extractions and real-time PCR procedures were identical to those in Experiment 1, with the modification that an iQ5 Real-Time PCR Detection System (Bio-Rad Laboratories; Hercules, CA) was used instead, and the PCR primers were designed to span at least an exon/intron junction to avoid amplification of DNA sequences using Primer3 Software. The sequences of the primers were as follows: Per1 (GenBank accession number: AF022991), 5’-ACTTTACCCAGGAGAAGTCCG-3’ and 5’-CTTGGTCACATACGGGGTTAG-3’; Per2 (GenBank accession number: NM_022817), 5’-CCAGATACCTTTAGCCTGATGA-3’ and 5’-GGACCTTCAGCTCCTTTAGTG-3’; Bmal1 (GenBank accession number: NM_001030272), 5’-TGTGACCGAGGGAAGATACTC-3’ and 5’-GGATGCAGGTAGTCAAACAAAC-3’; cyclophilin (GenBank accession number: NM_000942), 5’-GTGATCTTTGGTCTCTTCGG-3’ and 5’-GAAGTCCTTGATTACACGATGG-3’.
2.5.Hormones
All hormone compounds were acquired from Sigma-Aldrich. Water-soluble E2 and P4 were diluted in sterile PBS and the progesterone receptor antagonist (RU486) was diluted in ethanol (EtOH).
2.6. Statistics
In Experiment 1, data were analyzed by one-way ANOVA to determine diurnal variations of clock gene expression in each tissue as a function of estrous stage, and two-way ANOVA to determine estrous variations of clock gene expression as a function of both time of day (ZT) and estrous stage. In Experiment 2, the Student's t-test was used to compare the peak timing and amplitude differences between the proestrus and metestrus groups. In other experiments, statistical significances between groups were determined by one-way ANOVA followed by Tukey’s post-hoc analyses. All results are presented as the mean ± SEM, and were considered significant at P < 0.05.
3. Results
3.1. Experiment 1: Effects of the Estrous Cycle on Diurnal Rhythms of Clock Gene Expression in Peripheral Tissues of rats
To determine whether fluctuating hormone levels during the estrous cycle affect the diurnal rhythm of clock gene expression in tissues of the reproductive system, we examined rhythms of Per1, Per2, and Bmal1 mRNA expression in the uterus, ovary, and liver of female rats at different stages of estrous using quantitative real-time PCR. Although not considered a tissue of the reproductive system, the liver is a well established peripheral oscillator that displays robust circadian rhythms of clock gene expression.
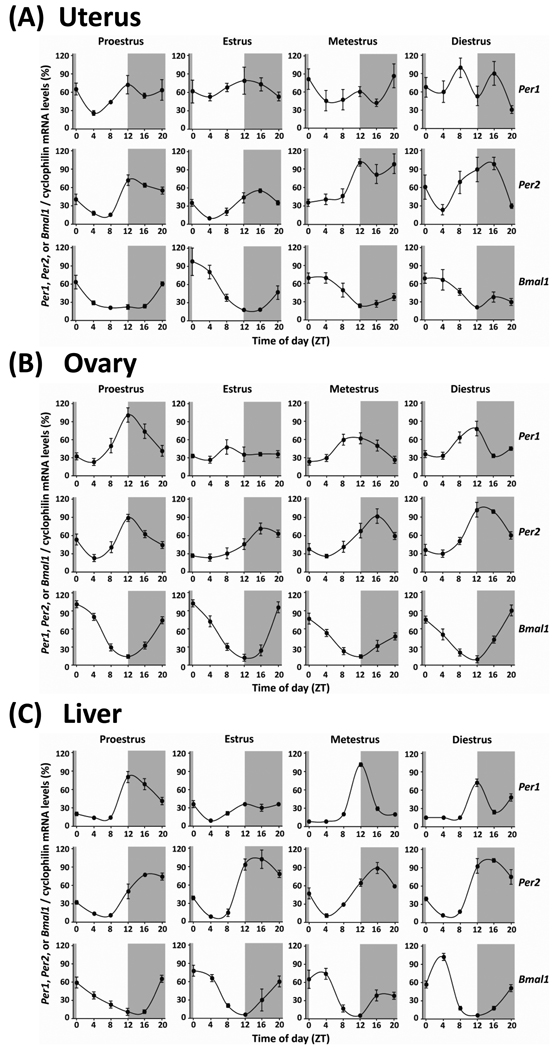
Figure 1 shows the patterns of Per1, Per2, and Bmal1 mRNA rhythm in the uterus, ovary, and liver. In the uterus (Fig. 1A), Per1 mRNA expression was observed statistically rhythmic only in the stage of proestrus (see Table 1 for details of statistical analysis). At other stages of the estrous cycle, Per1 expression exhibits a bimodal pattern with two peaks per 24-h cycle. Per2 and Bmal1 mRNA levels in the uterus were found to exhibit significant diurnal variations in all stages of the estrous cycle (Table 1). Two-way ANOVA revealed a main effect of cycle stage on the rhythm of Per2 expression, but not Per1 and Bmal1 expression, in the uterus (Table 1). In addition, the peaks of Per2 mRNA levels fluctuated through the estrous cycle such that peak expression was gradually delayed from ZT 12 on proestrus to ZT 16 on diestrus. The F value from the one-way ANOVA analysis indicates that the strongest Per2 mRNA rhythm is on proestrus while the daily rhythms steadily weakened on estrus through diestrus (Table 1).
Fig. 1. Rhythms of Clock Gene Expressions in Peripheral Tissues from Cycling Rats.
Diurnal rhythms of Per1, Per2 and Bmal1 genes expression in the uterus (A), ovary (B), and liver (C) of intact cycling rats throughout the estrous cycle. Cyclophilin mRNA was used as internal controls for transcripts the expression of which was constant throughout the day. Data are normalized by setting the highest point during the estrous cycle to 100%. Values represent the mean ± SEM (n=3 – 4, in each time point). The gray shades indicate dark phases.
Table 1.
Statistical analyses for rhythmic expressions of Per1, Per2, and Bmal1 in the liver, ovary, and uterus of rats throughout the estrus cycle.
| Uterus | |||||||
| One-way ANOVA | |||||||
| Two-way ANOVA | |||||||
| Proestrus | Estrus | Metestrus | |||||
| Diestrus | (ZT × estrus cycle) | ||||||
| Per1 | F(5,18) = 2.86 | F(5,18) = 0.32 | F(5,18) = 1.60 | F(5,16) = 2.32 | |||
| F(15,71) = 1.55 | |||||||
| P<0.05 | N.S. | N.S. | |||||
| N.S. | N.S. | ||||||
| Per2 | F(5,18) = 10.36 | F(5,18) = 8.28 | F(5,18) = 4.98 | F(5,17) = 3.62 | |||
| F(15,71) = 2.28 | |||||||
| P<0.01 | P<0.01 | P<0.01 | |||||
| P<0.05 | P<0.05 | ||||||
| Bmal1 | F(5,18) = 8.83 | F(5,18) = 5.48 | F(5,18) = 4.35 | F(5,17) = 3.07 | |||
| F(15,71) = 1.64 | |||||||
| P<0.01 | P<0.01 | P<0.01 | |||||
| P<0.05 | N.S. | ||||||
| Ovary | |||||||
| One-way ANOVA | |||||||
| Two-way ANOVA | |||||||
| Proestrus | Estrus | Metestrus | |||||
| Diestrus | (ZT × estrus cycle) | ||||||
| Per1 | F(5,18) = 14.37 | F(5,18) = 1.45 | F(5,18) = 11.61 | F(5,18) = 15.20 | |||
| F(15,72) = 4.82 | |||||||
| P<0.01 | N.S. | P<0.01 | |||||
| P<0.01 | P<0.001 | ||||||
| Per2 | F(5,18) = 26.49 | F(5,18) = 18.04 | F(5,18) = 14.14 | F(5,18) = 32.34 | |||
| F(15,72) = 5.11 | |||||||
| P<0.01 | P<0.01 | P<0.01 | |||||
| P<0.01 | P<0.001 | ||||||
| Bmal1 | F(5,18) = 88.01 | F(5,18) = 70.23 | F(5,18) = 26.21 | F(5,18) = 40.76 | |||
| F(15,72) = 7.40 | |||||||
| P<0.01 | P<0.01 | P<0.01 | |||||
| P<0.01 | P<0.001 | ||||||
| Liver | |||||||
| One-way ANOVA | |||||||
| Two-way ANOVA | |||||||
| Proestrus | Estrus | Metestrus | |||||
| Diestrus | (ZT × estrus cycle) | ||||||
| Per1 | F(5,18) = 27.80 | F(5,18) = 9.02 | F(5,18) = 273.23 | F(5,18) = 35.35 | |||
| F(15,72) = 15.85 | |||||||
| P<0.01 | P<0.01 | P<0.01 | |||||
| P<0.01 | P<0.001 | ||||||
| Per2 | F(5,18) = 23.66 | F(5,18) = 22.07 | F(5,18) = 26.05 | F(5,18) = 26.82 | |||
| F(15,72) = 2.02 | |||||||
| P<0.01 | P<0.01 | P<0.01 | |||||
| P<0.01 | P<0.05 | ||||||
| Bmal1 | F(5,18) = 12.68 | F(5,18) = 6.26 | F(5,18) = 8.99 | F(5,18) = 45.37 | |||
| F(15,72) = 2.60 | |||||||
| P<0.01 | P<0.01 | P<0.01 | |||||
| P<0.01 | P<0.01 | ||||||
N.S. = no significance
Fig. 1B displays estrous variations of Per1, Per2, and Bmal1 mRNA rhythm in the ovary. Statistical analysis shows significant daily rhythms of Per1 mRNA expression peaking at ZT 12 on proestrus, metestrus and diestrus, but not on estrus (Table 1). Per2 and Bmal1 mRNA levels also exhibited diurnal rhythms in all stages of the estrous cycle, peaking between ZT 12 – 16 and ZT 20 - 0, respectively. Two-way ANOVA supported a main effect of the estrous cycle on rhythms of Per1, Per2, and Bmal1 mRNA rhythms in the ovary (Table 1). Notably, the peak of Per2 mRNA appears slightly advanced on diestrus and proestrus (peaks between ZT 12 – 14) compared with metestrus and estrus (peaks at ZT 16).
In the liver (Fig. 1C), Per1 mRNA expression levels displayed diurnal rhythms with peak levels at the transition between the light and dark period (ZT 12) throughout the estrous cycle. The F value from the one-way ANOVA analysis indicates that the Per1 mRNA rhythm in the stage of estrus was much lower than other stages (Table 1). The expression levels of Per2 and Bmal1 in the liver also showed diurnal rhythms, peaking in the middle of the night (ZT 16) and early morning (ZT 0 – 4), respectively. Two-way ANOVA revealed a significant main effect of the estrous cycle on all there clock genes mRNA expression rhythms in the liver (Table 1).
3.2. Experiment 2: Effects of the Estrous Cycle on Circadian Rhythms of PER2::LUCIFERASE Expression in Tissue Explants from mice
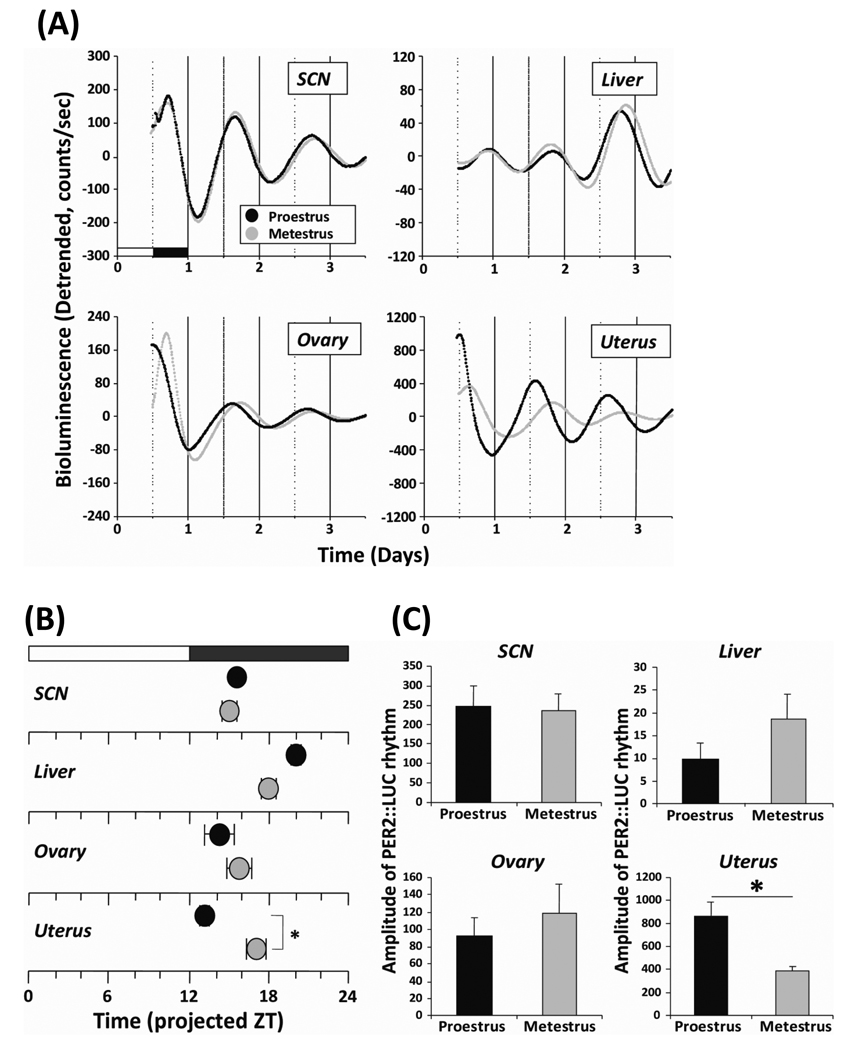
To closely assess the influence of the estrous cycle on rhythms of clock gene expression in peripheral tissues, we conducted bioluminescence recording of tissue explants from cycling PER2::LUC mice. As we are interested in the effects of fluctuating hormone levels on the timing of the molecular clock in target tissues of the reproductive system, we chose to examine tissue culture explants from animals at different stages of the cycle. Given that hormone levels show their largest variations on proestrus and are relatively stable and quiescent on metestrus, we compared tissue explants taken from animals at these two divergent stages of the estrous cycle. We analyzed both the peak time and amplitude on the first complete cycle in culture as a measure of the lasting in vitro effects of the in vivo hormonal milieu prior to euthanasia and tissue culture. We found that cultured SCN, liver, ovary, and uterus from both proestrus and metestrus demonstrated robust circadian rhythms of PER2::LUC expression (Fig. 2A). In the SCN and ovary, we did not observe a significant effect of estrous stage on the peak phase and amplitude of PER2::LUC expression (Fig. 2B and C). Although there was a trend suggesting that the estrous stage influenced the peak phase and amplitude of the PER2::LUC rhythm in the liver, this effect was not significant (Fig. 2B and C; P = 0.11, Student's t-test). Notably, the estrous cycle did have a significant effect on the peak phase and amplitude of PER2::LUC expression in the uterus. The rhythm on proestrus displayed an earlier peak time (≈ 4 h) and significantly larger amplitude (≈ 2-fold) relative to the rhythm on metestrus (Fig. 2B and C; P < 0.01, Student's t-test).
Fig. 2. Circadian rhythms of PER2::LUC expression in both reproductive and non-reproductive tissues from female mice at different stages of estrus.
(A) Representative records of bioluminescence showing circadian profiles of PER2::LUC expression rhythms in cultured SCN, liver, ovary, and uterus explants from female mice. Tissues were explanted just before lights off on proestrus (black trace) and metestrus (gray trace). Bioluminescence counts are plotted relative to the light: dark cycle prior to sacrifice, with light onset = 0. (B) Phase map of peak PER2::LUC expression in the SCN, liver, ovary, and uterus. Each point represents the average (± SEM; n = 5 for each group) peak of PER2::LUC expression during the first full 24 h in vitro (interval between 12 and 36 h in culture) plotted relative to the light: dark cycle prior to sacrifice. Black circle, data from proestrus; gray circle, data from metestrus. (C) Amplitude of PER2::LUC expression in the SCN, ovary, uterus, and liver. The amplitude of each oscillation was determined to sum the lowest and points (counts) during the first full 24 h in vitro. The average counts (± SEM) of amplitude are shown (n = 5 for each group). *, Statistically significant (P<0.05, Student’s t-test)
3.3. Experiment 3: Effects of Estrogen and Progesterone on Circadian Rhythms of PER2::LUC Expression in Uterine Explants from mice
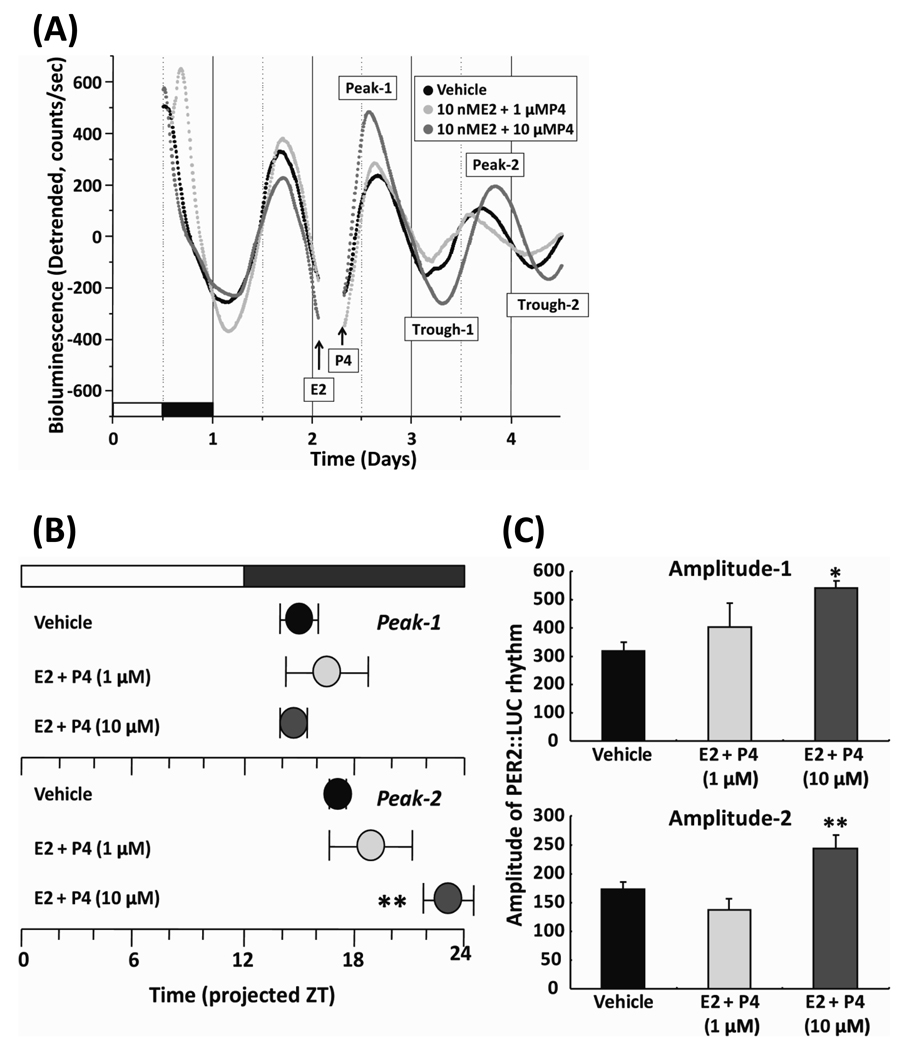
Since we observed significant estrous variations of Per2 rhythm in the uterus both in vivo and ex vivo, we sought to determine whether ovarian steroid hormones directly modulate circadian rhythms of clock gene expression in the steroid responsive tissue. We examined the effect of exogenous application of E2 and P4 on rhythms of PER2::LUC expression in uterine explants from cycling PER2::LUC mice. Uterine explants were made on the day of metestrus and after 2 days in culture, E2 was applied at projected ZT 1 (morning of proestrus) and P4 was applied at projected ZT 7 (afternoon of proestrus). The uterine explants remained in culture for 2 days after treatment to allow us to measure the peak phase and amplitude for two full circadian cycles (Fig. 3A). Treatment with 10 nM E2 and 1 µM P4 did not have a significant effect on the rhythms of PER2::LUC expression in the uterus. Treatment with higher concentrations of P4 (10 µM), in the presence of the same concentration of E2 (10 nM), significantly delayed the second peak (P < 0.01 vs. vehicle, Tukey’s test) and increased both first (P < 0.05 vs. vehicle, Tukey’s test) and second (P < 0.01 vs. vehicle, Tukey’s test) amplitudes (Fig. 3B and C).
Fig. 3. Effect of exogenous treatment with estrogen and progesterone on circadian rhythms of PER2::LUC expression in uterine explants.
(A) Representative records of bioluminescence showing effect of E2 and P4 on rhythmic expression of PER2::LUC in cultured uterus. Tissues were explanted just before lights off on metstrus. After 2 days, E2 at projected ZT 1 and P4 at projected ZT 7 were applied to the explant culture. Bioluminescence counts are plotted relative to the light: dark cycle prior to sacrifice, with light onset = 0. (B) Phase map of peak PER2::LUC expression rhythms after the treatment. Each point represents the average (± SEM; n = 6 for each group) first and second peak phases of PER2: LUC expression plotted relative to the light: dark cycle. (C) Amplitude of PER2::LUC expression rhythms after the treatment. Amplitude-1 and 2 represent the sum of peak-1 and trough-1, and peak-2 and trough-2, respectively. The average counts (± SEM) of amplitude are shown (n = 6 for each group). * P<0.05, ** P<0.01, vs. vehicle (Tukey’s test).
3.4. Experiment 4: Effects of Estrogen or Progesterone on the Expression of Per1, Per2, and Bmal1 mRNA in MCF-7 Human Cancer Cells
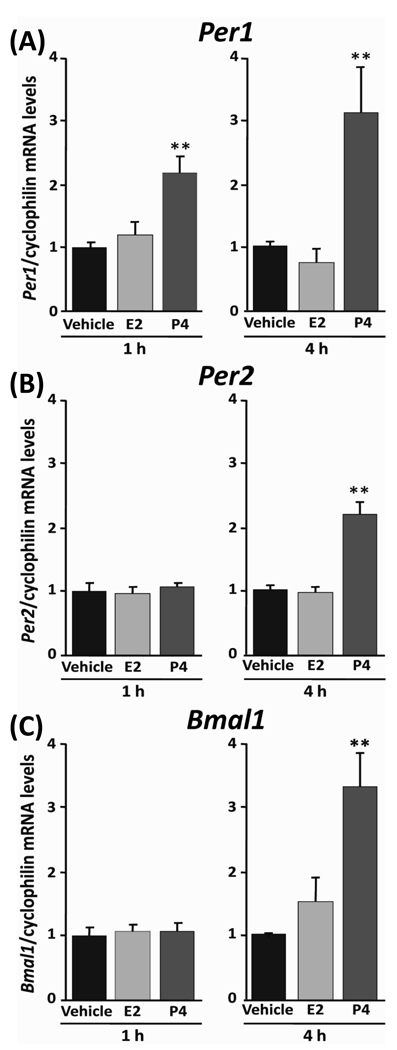
In experiment 3, the treatment with E2 and P4 increased the amplitude of PER2::LUC expression rhythm in the uterus. To better assess the acute effect of E2 or P4 on clock gene expression in target cells of the uterus, we examined the response of Per1, Per2, and Bmal1 mRNA expression in MCF-7 cells following exposure (1 or 4 h) to E2 or P4. Previous work has established that MCF-7 cells express estrogen (ERα, β) and progesterone (PRA, B) receptors and show appropriate responses following treatment with either hormone [26]. We initially treated MCF-7 cells with a single dose of E2 (10 nM) or P4 (1 µM), which approximate physiologically relevant concentrations [27] that induce Per1 mRNA expression in the rat uterus [28]. We did not observe a significant effect of 10 nM E2 on Per1, Per2, and Bmal1 mRNA expression in MCF-7 cells (Fig. 4). However, treatment with 1 µM P4 (P < 0.01 vs. vehicle, Tukey’s test) resulted in a significant increase in Per1 mRNA within 1 h (Fig. 4A) which was further potentiated after 4 h (P < 0.01 vs. vehicle, Tukey’s test). Further, treatment with 1 µM P4 for 4 h, but not 1 h, induced a 2-fold increase in Per2 mRNA expression (Fig. 4B; P < 0.01 vs. vehicle, Tukey’s test). In addition, treatment for 4 h with 1 µM P4 significantly increased the level of Bmal1 mRNA expression in MCF-7 cells (Fig. 4C; P < 0.01 vs. vehicle, Tukey’s test).
Fig. 4. Effect of exogenous treatment with estrogen or progesterone on the level of Per1, Per2, and Bmal1 mRNA expression in MCF-7 cells.
The transcript levels of Per1 (A), Per2 (B), and Bmal1 (C) in MCF-7 cells were stimulated by 1 or 4 h treatment of 10 nM E2 or 1 µM P4. Data were normalized as vehicle (PBS) to 1 and shown mean ± SEM (n = 5 for each group). **P<0.01, ***P<0.001, when compare with vehicle group (Tukey’s test).
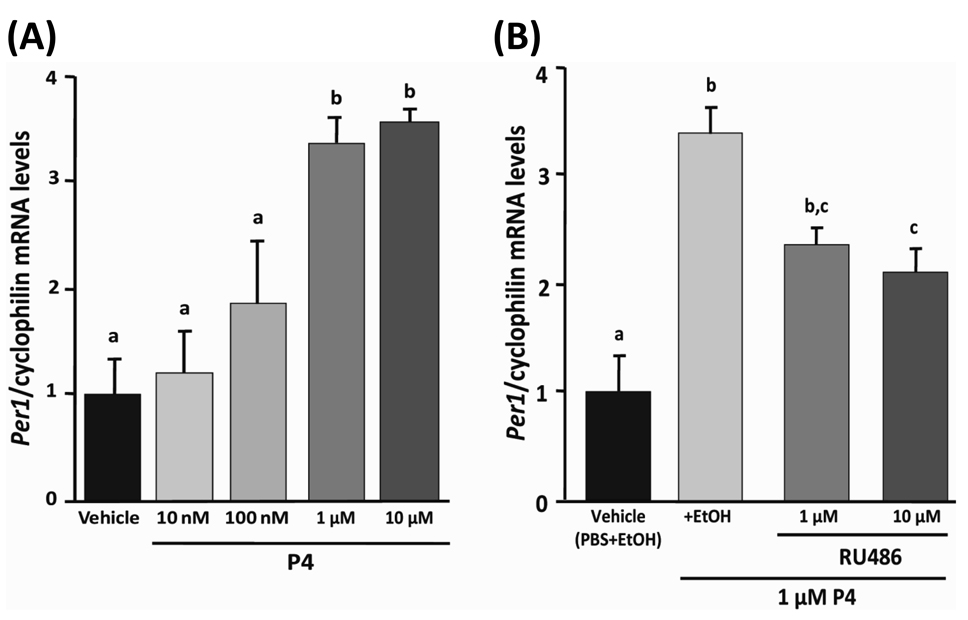
We decided to examine the dose- and receptor- dependent effects of P4 on the molecular clock in MCF-7 cells. To this end, we assessed the effects of P4-treatment at several concentrations in the presence or absence of the progesterone receptor antagonist RU486. We treated MCF-7 cells with 10 nM, 100 nM, 1 µM, or 10 µM P4 for 4 h. We observed a dose dependent increase in Per1 mRNA expression in MCF-7 cells following treatment with P4. P4 at concentrations of 10 nM and 100 nM did not alter the expression levels of Per1, whereas 1 µM and 10 µM P4 significantly increased the expression of Per1 mRNA in MCF-7 cells (Fig. 5A; P < 0.01 vs. vehicle for 1 and 10 µM, Tukey’s test). Treatment of MCF-7 cells with RU486 at concentration of either 1 µM or 10 µM in the presence of 1 µM P4 attenuated, but did not abolish, the effects of P4 on the level of Per1 mRNA in MCF-7 cells (Fig. 5B; P < 0.05, EtOH control and 1 µM P4 vs. 1 µM P4 and 10 µM RU486, Tukey’s test).
Fig. 5. Dose-dependent and receptor-mediated effects of P4 on the expression of Per1 in MCF-7 cells.
(A) Per1 transcript levels in MCF-7 cells treated with increasing concentrations of P4 (10 nM, 100 nM, 1 µM, and 10 µM) or PBS vehicle for 4 h. (B) Per1 transcript levels in MCF-7 treated with vehicle containing both PBS and EtOH, EtOH + 1 µM P4, 1 µM P4 + 1 µM RU486, or 1 µM P4 + 10 µM RU486. Data were normalized for comparisons by setting to vehicle treatment (PBS + EtOH) to 1. Data are plotted as mean ± SEM (n = 5 for each group). Differing letters between groups indicate significant differences (P<0.05, Tukey’s test).
4. Discussion
We observed that the rhythms of Per1, Per2, and Bmal1 expression varied with the stage of the estrous cycle in both reproductive and non-reproductive tissues of female rats. Focusing on Per2 rhythms in the uterus, more robust daily rhythms with an advanced peak phase were observed on proestrus compared with other stages. This in vivo result was also seen in the ex vivo experiment as uterine explants exhibited a larger amplitude and phase advanced rhythm in PER2::LUC expression when cultures were prepared on proestrus relative to metestrus. These data strongly support our hypothesis that the estrous cycle drives changes in the timing of the circadian clocks in reproductive tissues. However, the impact of the estrous cycle varied across tissues. In previous work, we found evidence that estrogen may be an important regulator of clock gene expression. Eliminating the estrous-driven changes in estrogen and progesterone by ovariectomy and E2 implants altered circadian rhythms of Per1 and Per2 mRNA expression in peripheral tissues including the liver, kidney, and uterus. These data suggest that E2 differentially regulates the rhythm of clock gene expression in these tissues [21]. In the central nervous system, there is evidence that PER2 expression in the bed nucleus of the stria terminalis and central nucleus of the amygdala, but not the SCN, varies across the estrous cycle in female rats [29]. In the present study, we also found that the rhythms of PER2::LUC expression in SCN explants were unaffected by the estrous stage. These data indicate significant tissue-specific effects of the estrous cycle on clock gene expression in both neural and non-neural oscillators, but not the SCN. It may be that these tissue-specific effects of the estrous cycle are mediated by cycle dependent changes in the distribution of hormone receptors and other proteins, such as coactivators and corepressors in the target tissue. For instance, in situ hybridization, immunocytochemistry, and RT-PCR analyses indicate that the SCN contains ERβ [30–32], whereas the liver and the kidney express abundant levels of ERα [33] and ERα is predominantly expressed in the uterus [34–36]. Further studies using additional molecular techniques will be required to determine how the estrous cycle modifies clock gene expression at the cellular level, with a focus on both steroid hormone binding to consensus sequences on target genes and pituitary peptide hormone receptor signaling mechanisms within target tissues.
Treatment of uterine explants with estrogen and progesterone concentrations intended to mimic the steroid hormone background of proestrus increased the amplitude and delayed the peak of PER2::LUC expression in the uterus. This effect is consistent with the response of the uterine rhythm of Per2 expression on proestrus that we observed in our in vivo and ex vivo experiments. Further, the phase-delay observed on the next treatment day matches the phase-delay of Per2 mRNA rhythm on estrus in vivo. These data support our second hypothesis, which suggests that fluctuating levels of circulating ovarian steroid hormones during the estrous cycle modulate the timing of clock gene expression in target oscillators.
We also found that P4, but not E2, acutely induces clock gene expression in MCF-7 human cancer cells. The MCF-7 expresses functional ERs and PRs [26] and has been extensively used as a model to study steroid hormone dependent signaling mechanisms. While they do not intrinsically represent a model for the direct effects of the estrus cycle on clock gene expression, the fact that they express functional steroid receptors and clock genes makes them an excellent model for the study of steroid effects on the molecular clock. Therefore, by extension, we propose that fluctuating steroid hormone levels in the cycling female can affect the molecular clock in target cells (e.g. uterus, ovary, etc.) that are known to express the components of the molecular clock. We observed a dose dependent stimulatory effect of P4 on Per1 expression that was attenuated, but not abolished, by co-treatment with the P4-receptor antagonist RU-486. These data suggest that the molecular clock can be modulated by progesterone, acting via the PR in target cells. Further, 1 h treatment with P4 induced Per1 mRNA expression, but failed to stimulate either Per2 or Bmal1 mRNA expression in MCF-7 cells. P4 and E2 are known to exert their effects by binding to cytoplasmic nuclear hormone receptors that in turn bind to unique DNA sequences referred to as PRE (progesterone response elements) or ERE (estrogen response elements) [37]. There is a PRE-half sequence in the first intron and some ERE-half sites in the 5’ flanking regulatory regions of mouse Per1 gene (GeneBank accession number: AB030818). Taken together, it is possible that P4 directly enhances Per1 transcription following direct binding and activation of these target elements in the Per1 promoter. Although further studies using ER and PR positive or negative-cell lines will be required to determine how the classical nuclear steroid receptors regulate the molecular circadian clock, our results indicate that MCF-7 cells are a good model to analyze how the signaling from steroid hormones affects molecular clocks. Unlike in previous reports [38], we did not observe a significant effect of E2 on clock gene expression in these cells. This difference is likely to due treatment parameters as in the previous work. That is, E2 was applied at a considerably higher concentration (1 µM) and duration (16 h) in that study relative to our present work (10 nM, 1 or 4 h). Another report also showed that 6 h of stimulation with 10 nM E2 induced Per1 mRNA expression in cultured rat uterine stroma cells [28], the induction level in that study was ≤ 1.5-fold. Our previous ex vivo study revealed that 10 nM – 10 µM E2 significantly shortened the period of PER2::LUC expression in cultured uterine explants, whereas P4 did not [24]. These data support the notion that E2 may directly affect the period and phase of the circadian clock in ER-positive tissues, without affecting the amplitude of clock gene expression. Further, it is reasonable to speculate, based on the current data and previous reports, that E2 and P4 act differentially but in concert to modulate circadian rhythms of clock gene expression within target tissues.
In summary, we have identified estrous cycle dependent changes in clock gene expression rhythms in both reproductive and non-reproductive tissues. The influence of fluctuating hormone levels during the estrous cycle on rhythms of clock gene expression in reproductive tissues was confirmed by bioluminescence recordings of cultured uterine explants. We have also shown that progesterone treatment causes acute up-regulation of Per1, Per2, and Bmal1 expression in human breast cancer MCF-7 cells that are known to express nuclear hormone receptors. The effect of P4 in these cells was attenuated by the addition of a selective receptor antagonist, implicating classic PR-dependent signaling in the response of the clock to P4-treatment. Taken together, our results strongly support our hypotheses: 1) that the estrous cycle drives changes in the timing of the circadian clocks in reproductive tissues and 2) that fluctuating levels of circulating ovarian steroid hormones during the cycle modulate the timing of clock gene expression in target oscillators. Finally, our present and previous data strongly suggest that ovarian steroid hormones mediate these effects by acting on their cognate receptors. By adjusting the timing of the circadian clock in tissues such as the uterus across the days of the estrous cycle, ovarian steroid hormones may play a critical role in the stable homeostasis of female reproductive function.
Acknowledgements
We thank Drs. Dawn Loh, Tsuyoshi Watanabe, Shigeru Tomida, and Mayumi Kojima for their help in carrying out the experiments. We would like to express our gratitude to Mr. Andy Vosko for critical reading and comments on the manuscript. This work was supported by NIH Grants RO1 MH062517 (to G.D.B.), Grants-in-Aid for the Promotion of Science for Young Scientists from Japanese Ministry of Education, Science and Culture 14011687 (to T.J.N), and the Friends of Semel Institute for Neuroscience at UCLA funded by the Joseph Drown Foundation (to T.J.N).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 2.Alleva JJ, Waleski MV, Alleva FR. A biological clock controlling the estrous cycle of the hamster. Endocrinology. 1971;88:1368–1379. doi: 10.1210/endo-88-6-1368. [DOI] [PubMed] [Google Scholar]
- 3.Funabashi T, Mitsushima D, Nakamura TJ, Uemura T, Hirahara F, Shinohara K, et al. Gonadotropin-releasing hormone (GnRH) surge generator in female rats. Prog Brain Res. 2002;141:165–173. doi: 10.1016/S0079-6123(02)41091-6. [DOI] [PubMed] [Google Scholar]
- 4.Chappell PE, Lee J, Levine JE. Stimulation of gonadotropin-releasing hormone surges by estrogen. II. Role of cyclic adenosine 3'5'-monophosphate. Endocrinology. 2000;141:1486–1492. doi: 10.1210/endo.141.4.7427. [DOI] [PubMed] [Google Scholar]
- 5.Everett JW, Sawyer CH. A 24-hour periodicity in the "LH-release apparatus" of female rats, disclosed by barbiturate sedation. Endocrinology. 1950;47:198–218. doi: 10.1210/endo-47-3-198. [DOI] [PubMed] [Google Scholar]
- 6.Wiegand SJ, Terasawa E. Discrete lesions reveal functional heterogeneity of suprachiasmatic structures in regulation of gonadotropin secretion in the female rat. Neuroendocrinology. 1982;34:395–404. doi: 10.1159/000123335. [DOI] [PubMed] [Google Scholar]
- 7.Cagnacci A, Soldani R, Laughlin GA, Yen SS. Modification of circadian body temperature rhythm during the luteal menstrual phase: role of melatonin. J Appl Physiol. 1996;80:25–29. doi: 10.1152/jappl.1996.80.1.25. [DOI] [PubMed] [Google Scholar]
- 8.Moline ML, Broch L, Zak R, Gross V. Sleep in women across the life cycle from adulthood through menopause. Sleep Med Rev. 2003;7:155–177. doi: 10.1053/smrv.2001.0228. [DOI] [PubMed] [Google Scholar]
- 9.Shinohara K, Uchiyama M, Okawa M, Saito K, Kawaguchi M, et al. Menstrual changes in sleep, rectal temperature and melatonin rhythms in a subject with premenstrual syndrome. Neurosci Lett. 2000;281:159–162. doi: 10.1016/s0304-3940(00)00826-0. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki H, Uchiyama M, Shibui K, Kim K, Tagaya H, Shinohara K. Long-term rectal temperature measurements in a patient with menstrual-associated sleep disorder. Psychiatry Clin Neurosci. 2002;56:475–478. doi: 10.1046/j.1440-1819.2002.01039.x. [DOI] [PubMed] [Google Scholar]
- 11.Wollnik F, Turek FW. Estrous correlated modulations of circadian and ultradian wheel-running activity rhythms in LEW/Ztm rats. Physiol Behav. 1988;43:389–396. doi: 10.1016/0031-9384(88)90204-1. [DOI] [PubMed] [Google Scholar]
- 12.Albers HE, Gerall AA, JF A. Effect of reproductive state on circadian periodicity in the rat. Physiol Behav. 1981;26:21–25. doi: 10.1016/0031-9384(81)90073-1. [DOI] [PubMed] [Google Scholar]
- 13.Morin LP. Effect of ovarian hormones on synchrony of hamster circadian rhythms. Physiol Behav. 1980;24:741–749. doi: 10.1016/0031-9384(80)90406-0. [DOI] [PubMed] [Google Scholar]
- 14.Morin LP, Fitzgerald KM, Zucker I. Estradiol shortens the period of hamster circadian rhythms. Science. 1977;196:305–307. doi: 10.1126/science.557840. [DOI] [PubMed] [Google Scholar]
- 15.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(Spec No 2):R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 16.Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 17.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fahrenkrug J, Georg B, Hannibal J, Hindersson P, Gras S. Diurnal rhythmicity of the clock genes Per1 and Per2 in the rat ovary. Endocrinology. 2006;147:3769–3776. doi: 10.1210/en.2006-0305. [DOI] [PubMed] [Google Scholar]
- 19.Karman BN, Tischkau SA. Circadian clock gene expression in the ovary: Effects of luteinizing hormone. Biol Reprod. 2006;75:624–632. doi: 10.1095/biolreprod.106.050732. [DOI] [PubMed] [Google Scholar]
- 20.Johnson MH, Lim A, Fernando D, Day ML. Circadian clockwork genes are expressed in the reproductive tract and conceptus of the early pregnant mouse. Reprod Biomed Online. 2002;4:140–145. doi: 10.1016/s1472-6483(10)61931-1. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura TJ, Moriya T, Inoue S, Shimazoe T, Watanabe S, Ebihara S, et al. Estrogen differentially regulates expression of Per1 and Per2 genes between central and peripheral clocks and between reproductive and nonreproductive tissues in female rats. J Neurosci Res. 2005;82:622–630. doi: 10.1002/jnr.20677. [DOI] [PubMed] [Google Scholar]
- 22.Dolatshad H, Campbell EA, O'Hara L, Maywood ES, Hastings MH, Johnson MH. Developmental and reproductive performance in circadian mutant mice. Hum Reprod. 2006;21:68–79. doi: 10.1093/humrep/dei313. [DOI] [PubMed] [Google Scholar]
- 23.Miller BH, Olson SL, Turek FW, Levine JE, Horton TH, Takahashi JS. Circadian clock mutation disrupts estrous cyclicity and maintenance of pregnancy. Curr Biol. 2004;14:1367–1373. doi: 10.1016/j.cub.2004.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura TJ, Sellix MT, Menaker M, Block GD. Estrogen directly modulates circadian rhythms of PER2 expression in the uterus. Am J Physiol Endocrinol Metab. 2008;295:E1025–E1031. doi: 10.1152/ajpendo.90392.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res. 1993;35:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- 26.Horwitz KB, Costlow ME, McGuire WL. MCF-7; a human breast cancer cell line with estrogen, androgen, progesterone, and glucocorticoid receptors. Steroids. 1975;26:785–795. doi: 10.1016/0039-128x(75)90110-5. [DOI] [PubMed] [Google Scholar]
- 27.Sherman BM, Korenman SG. Hormonal characteristics of the human menstrual cycle throughout reproductive life. J Clin Invest. 1975;55:699–706. doi: 10.1172/JCI107979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He PJ, Hirata M, Yamauchi N, Hattori MA. Up-regulation of Per1 expression by estradiol and progesterone in the rat uterus. J Endocrinol. 2007;194:511–519. doi: 10.1677/JOE-07-0172. [DOI] [PubMed] [Google Scholar]
- 29.Perrin JS, Segall LA, Harbour VL, Woodside B, Amir S. The expression of the clock protein PER2 in the limbic forebrain is modulated by the estrous cycle. Proc Natl Acad Sci U S A. 2006;103:5591–5596. doi: 10.1073/pnas.0601310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gundlah C, Kohama SG, Mirkes SJ, Garyfallou VT, Urbanski HF, Bethea CL. Distribution of estrogen receptor beta (ERbeta) mRNA in hypothalamus, midbrain and temporal lobe of spayed macaque: continued expression with hormone replacement. Brain Res Mol Brain Res. 2000;76:191–204. doi: 10.1016/s0006-8993(99)02475-0. [DOI] [PubMed] [Google Scholar]
- 31.Hileman SM, Handa RJ, Jackson GL. Distribution of estrogen receptor-beta messenger ribonucleic acid in the male sheep hypothalamus. Biol Reprod. 1999;60:1279–1284. doi: 10.1095/biolreprod60.6.1279. [DOI] [PubMed] [Google Scholar]
- 32.Shughrue PJ, Lane MV, Merchenthaler I. Comparative distribution of estrogen receptor-alpha and -beta mRNA in the rat central nervous system. J Comp Neurol. 1997;388:507–525. doi: 10.1002/(sici)1096-9861(19971201)388:4<507::aid-cne1>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 33.Kuiper GG, Carlsson B, Grandien K, Enmark E, Haggblad J, Nilsson S, et al. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology. 1997;138:863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- 34.Hiroi H, Inoue S, Watanabe T, Goto W, Orimo A, Momoeda M, et al. Differential immunolocalization of estrogen receptor alpha and beta in rat ovary and uterus. J Mol Endocrinol. 1999;22:37–44. doi: 10.1677/jme.0.0220037. [DOI] [PubMed] [Google Scholar]
- 35.Pelletier G. Localization of androgen and estrogen receptors in rat and primate tissues. Histol Histopathol. 2000;15:1261–1270. doi: 10.14670/HH-15.1261. [DOI] [PubMed] [Google Scholar]
- 36.Pelletier G, El-Alfy M. Immunocytochemical localization of estrogen receptors alpha and beta in the human reproductive organs. J Clin Endocrinol Metab. 2000;85:4835–4840. doi: 10.1210/jcem.85.12.7029. [DOI] [PubMed] [Google Scholar]
- 37.Muramatsu M, Inoue S. Estrogen receptors: how do they control reproductive and nonreproductive functions? Biochem Biophys Res Commun. 2000;270:1–10. doi: 10.1006/bbrc.2000.2214. [DOI] [PubMed] [Google Scholar]
- 38.Gery S, Virk RK, Chumakov K, Yu A, Koeffler HP. The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene. 2007;26:7916–7920. doi: 10.1038/sj.onc.1210585. [DOI] [PubMed] [Google Scholar]