Abstract
We investigated whether the affinity of tissue inhibitor of metalloproteinases (TIMP)-3 for adamalysins with thrombospondin motifs (ADAMTS)-4 and ADAMTS-5 is affected by the non-catalytic ancillary domains of the enzymes. For this purpose, we first established a novel method of purifying recombinant FLAG-tagged TIMP-3 and its inhibitory N-terminal domain (N-TIMP-3) by treating transfected HEK293 cells with sodium chlorate to prevent heparan sulfate proteoglycan-mediated TIMP-3 internalization. TIMP-3 and N-TIMP-3 affinity for selected matrix metalloproteinases and forms of ADAMTS-4 and -5 lacking sequential C-terminal domains was determined. TIMP-3 and N-TIMP-3 displayed similar affinity for various matrix metalloproteinases as has been previously reported for E. coli-expressed N-TIMP-3. ADAMTS-4 and -5 were inhibited more strongly by N-TIMP-3 than by full-length TIMP-3. The C-terminal domains of the enzymes enhanced interaction with N-TIMP-3 and to a lesser extent with the full-length inhibitor. For example, N-TIMP-3 had 7.5-fold better Ki value for full-length ADAMTS-5 than for the catalytic and disintegrin domain alone. We propose that the C-terminal domains of the enzymes affect the structure around the active site, favouring interaction with TIMP-3.
Keywords: aggrecanase, inhibition kinetics, MMPs
1. Introduction
In addition to their catalytic domains, proteolytic enzymes often have non-catalytic ancillary domains that modulate interaction of the enzyme with substrates or inhibitors. Indeed, almost all members of the metzincin family of metalloproteinases have such ancillary domains, and they have been shown to mediate recognition and cleavage of numerous substrates. For example, the hemopexin domain of matrix metalloproteinase 1 (MMP-1) is required for cleavage of collagen (Clark and Cawston 1989). Among the related adamalysins with thrombospondin motifs (ADAMTSs), the C-terminal domains of ADAMTS-4 and ADAMTS-5 have been shown to promote aggrecan cleavage (Kashiwagi et al., 2004; Gendron et al., 2007; Fushimi et al., 2008), the C-terminal spacer domain of ADAMTS13 promotes cleavage of von Willebrand factor (Gao et al., 2008) and C-terminal domains of ADAMTS-2 modulate processing of the N-terminal propeptide of procollagen (Colige et al., 2005). These ancillary domains have also been shown to modulate interaction of some metzincins with the endogenous tissue inhibitors of metalloproteinases (TIMPs). For example, the MMP-2 hemopexin domain interacts strongly with TIMP-2 (Murphy et al., 1992; Butler et al., 1999; Morgunova et al., 2002). While the MMPs are inhibited by all four of the mammalian TIMPs, most adamalysins and ADAMTSs are inhibited only by TIMP-3. Compared to the MMPs, the adamalysins and ADAMTSs have a greater number and diversity of C-terminal ancillary domains and the role of these in modulating interactions with TIMP-3 is largely unknown, as a lack of sensitive substrates has hampered in-depth kinetic analysis. Good substrates are available for ADAM17, or tumour necrosis factor-α converting enzyme (TACE) and the C-terminal domains of this enzyme have been shown to reduce the affinity of the enzyme for both full-length TIMP-3 and N-TIMP-3 by 10-fold (Lee et al., 2002).
ADAMTS-4 and -5 cleave the cartilage matrix proteoglycan aggrecan at various sites, releasing the chondroitin and keratan sulfate-bearing region of the molecule from the tissue. This is an early and crucial step in the development of osteoarthritis as it reduces the ability of the tissue to resist compressive loads. Both enzymes are readily proteolyzed to smaller isoforms, which have altered proteolytic activity (Gao et al., 2004). Here we investigate TIMP-3 inhibition of the isoforms, with an aim to understanding whether, as in the case of ADAM17, the C-terminal ancillary domains of the enzymes regulate TIMP-3 binding to the active site. To do this, we developed a novel method of purifying sufficient soluble full-length TIMP-3 and its inhibitory N-terminal domain (N-TIMP-3) for kinetic analysis.
N-TIMP-3 can be successfully expressed in E. coli and refolded in vitro (Kashiwagi et al., 2001), but we have not been able to refold full-length TIMP-3 using this system. Unlike the other TIMPs, TIMP-3 binds tightly to the extracellular matrix and is not readily soluble (Lee et al., 2007; Yu et al., 2000). It is thus difficult to recombinantly express TIMP-3 in mammalian cells, and to date full-length TIMP-3 has only been recombinantly produced in NSO mouse myeloma cells, which do not produce an extracellular matrix (Apte et al., 1995). Here we describe successful purification of full-length TIMP-3 and N-TIMP-3 recombinantly expressed in human embryonic kidney HEK293 cells. Recently developed synthetic fluorescent quenched substrates allowed us to determine the inhibition constants of both TIMP-3 and N-TIMP-3 for various forms of ADAMTS-4 and ADAMTS-5.
2. Results
2.1. Purification of recombinant TIMP-3 and N-TIMP-3
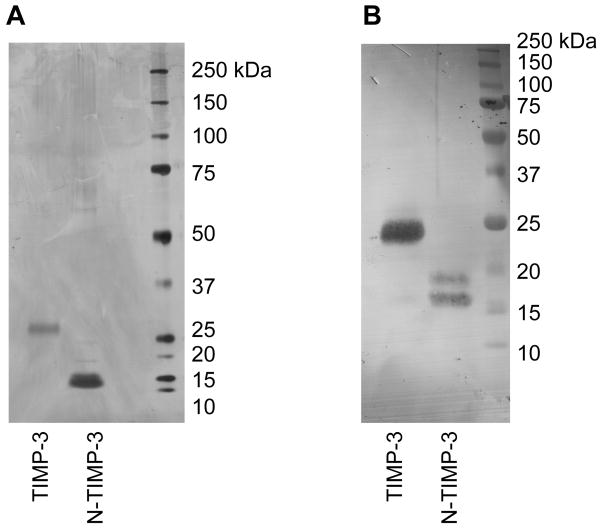
This study describes a novel method of purifying soluble full-length TIMP-3. No TIMP-3 is observed in the conditioned medium of transfected HEK293 cells, and we have previously shown that TIMP-3 is rapidly endocytosed after secretion from the cell by a scavenger endocytic receptor of the low-density lipoprotein receptor-related protein (LRP) family (Troeberg et al., 2008). TIMP-3 accumulates in the conditioned medium if this endocytic pathway is blocked, for example by the addition of receptor-associated protein (RAP), an inhibitor of LRP-mediated endocytosis (Troeberg et al., 2008). Heparin also causes an accumulation of TIMP-3, indicating that a heparan sulfate proteoglycan co-receptor is possibly required for LRP-mediated endocytosis as has been reported for other LRP ligands (Godyna et al., 1995; Sarafanov et al., 2001). However, heparin binds to TIMP-3 tightly and purification of TIMP-3 from heparin-treated cells proved difficult. To keep recombinantly expressed TIMP-3 in the medium, we treated transfected cells with sodium chlorate (NaClO3), which blocks sulfation of cell surface glycosaminoglycans (Baeuerle and Huttner 1986; Safaiyan et al., 1999). This resulted in the accumulation of FLAG-tagged TIMP-3 and N-TIMP-3 in the conditioned medium of transfected cells. Both proteins were purified from the conditioned medium by anti-FLAG affinity chromatography (Fig. 1A). As previously reported for full-length ADAMTS-4 (Kashiwagi et al., 2004), substantial processing of the C-terminal FLAG-tag occurred, reducing the yield to approximately 50 μg of purified protein per litre of conditioned medium. Both TIMP-3 and N-TIMP-3 expressed in HEK293 were confirmed to have the expected N-terminal sequence 1CTCSPSHPQD and to react with a polyclonal anti-TIMP-3 antibody (Fig 1B). Titration against a known concentration of MMP-1 showed all preparations of TIMP-3 to be 100% active.
Fig. 1.
Purification of TIMP-3 and N-TIMP-3. FLAG-tagged glycosylated human TIMP-3 and N-TIMP-3 were expressed by treating transfected HEK293 with 30 mM sodium chlorate in serum-free DMEM and purified from the conditioned medium by FLAG affinity chromatography and cation exchange chromatography. A, Silver-stained 10% polyacrylamide SDS-PAGE gel of TIMP-3 (100 ng) and N-TIMP-3 (100 ng). B, Immuoblot of 12% polyacrylamide SDS-PAGE gel of TIMP-3 (100 ng) and N-TIMP-3 (100 ng) stained using a polyclonal rabbit anti-TIMP-3 antibody.
TIMP-3 contains a single N-glycosylation site in its C-terminal domain (Apte et al., 1995) and various possible O-glycosylation sites. Various cell types have been shown to express both a 27 kDa glycosylated and a 24 kDa non-glycosylated form (Apte et al., 1995; Fabunmi et al., 1996; Langton et al., 1998). Transfected HEK293 cells also synthesized both glycosylated and non-glycosylated forms, but only the glycosylated form remained after purification, with the non-glycosylated form possibly lost due to increased “stickiness”. Indeed, we obtained lower yields when we treated cells with tunicamycin to obtain only the non-N-glycosylated form. N-TIMP-3 was expressed primarily as a non-glycosylated form, with a lower amount of a higher molecular mass glycosylated form present. Since N-TIMP-3 contains no N-glycosylation sites, this possibly reflects O-glycosylation.
2.2. Ki(app) of TIMP-3 and N-TIMP-3 for selected MMPs
We compared the inhibitory activity of our mammalian-expressed TIMP-3 and N-TIMP-3 with the previously characterized E. coli-expressed N-TIMP-3 with a C-terminal His-tag (Kashiwagi et al., 2001). Ki(app) values determined in the current study for E. coli-expressed N-TIMP-3 agreed well with previously published values for this protein (Kashiwagi et al., 2001). Although we determined a 6-fold lower Ki(app) value for N-TIMP-3 inhibition of the catalytic domain of MMP-3 (MMP-3cat), our results agree with that found in the previous study that N-TIMP-3 is a weaker inhibitor of MMP-3cat than of the catalytic domain of MMP-1 (MMP-1cat) or MMP-2 (Table 1).
Table 1.
Ki(app) values a (nM) for TIMP-3 and N-TIMP-3 inhibition of selected MMPs.
| Kashiwagi et al. E. coli-expressed N-TIMP-3 |
Current study E. coli-expressed N-TIMP-3 |
Mammalian expressed N-TIMP-3 |
Mammalian expressed TIMP-3 |
|
|---|---|---|---|---|
| MMP-1cat | 1.2 | 0.8 ± 0.4 | 0.9 ± 0.6 | 1. 1 ± 0.6 |
| MMP-2 | 4.3 | 1.3 ± 0.5 | 0.9 ± 0.3 | <0.1 |
| MMP-2cat | ND | 0.8 ± 0.1 | 1.0 ± 0.3 | 0.9 ± 0.3 |
| MMP-3cat | 66.9 | 11.1 ± 5.0 | 9.1 ± 3.3 | 1.0 ± 0.4 |
Ki(app) given as mean ± standard deviation (n=4-6).
We found that mammalian expressed N-TIMP-3 had essentially identical inhibitory properties to the E. coli-expressed N-TIMP-3, being a strong inhibitor of MMP-1 cat and MMP-2, and a 10-fold weaker inhibitor of MMP-3cat.
MMP-1cat was equally well inhibited by N-TIMP-3 and full-length TIMP-3. MMP-3cat, however, was more strongly inhibited by full-length TIMP-3 than by N-TIMP-3, implying that the C-terminal domain of the inhibitor contributes to binding. The MMP-2 catalytic domain (MMP-2cat) was equally well inhibited by N-TIMP-3 and full-length TIMP-3, but the full-length enzyme was more strongly inhibited by the full-length TIMP-3 than by N-TIMP-3. This indicates that the C-terminal domain of TIMP-3 interacts with the C-terminal hemopexin domain of MMP-2.
2.3. Ki of TIMP-3 and N-TIMP-3 for ADAMTS-4 and ADAMTS-5
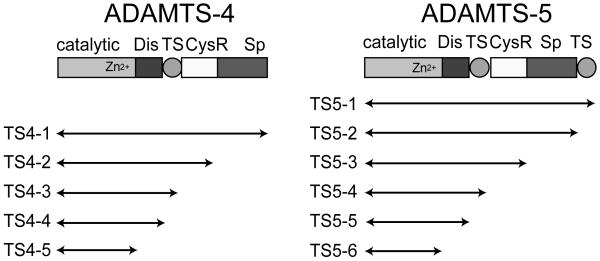
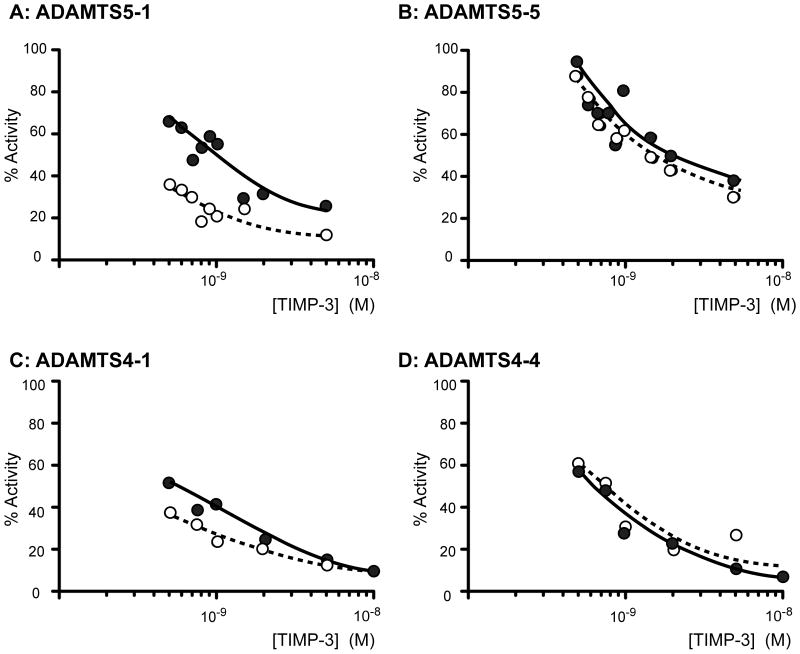
We analyzed TIMP-3 and N-TIMP-3 inhibition of various isoforms (Fig. 2) of ADAMTS-4 and -5 lacking C-terminal ancillary domains. Fig. 3 shows representative data for four enzyme isoforms fitted to the tight-binding equation (Bieth, 1995). All isoforms of ADAMTS-4 and -5 were effectively inhibited by TIMP-3 and N-TIMP-3, with Ki values in the sub-nanomolar range. All ADAMTS-5 isoforms were inhibited more strongly by N-TIMP-3 than by full-length TIMP-3 (Table 2). The C-terminal domains of ADAMTS-5 enhanced inhibition by N-TIMP-3, with full-length ADAMTS-5 (ADAMTS5-1) having a 7.5-fold better Ki value than ADAMTS5-5, which consists of the catalytic and disintegrin domain. The C-terminal domains of the enzyme had a similar, although less marked, effect on association with the full-length inhibitor. The C-terminal domains of ADAMTS-4 also had a similar effect on association with full-length and N-TIMP-3 (Table 2).
Fig. 2.
ADAMTS-4 and ADAMTS-5 isoforms analysed. Each isoform has a FLAG-tag at its C-terminus and was purified from the conditioned medium of transfected HEK293 cells by FLAG affinity chromatography. Dis, disintegrin domain; TS, thrombospondin domain; CysR, cysteine-rich domain; Sp, spacer domain.
Fig. 3.
Inhibition of ADAMTS-4 and ADAMTS-5 by TIMP-3 and N-TIMP-3. ADAMTS5-1 (A, 0.5 nM) and ADAMTS5-5 (B, 0.5 nM) were incubated with 0.5-5 nM TIMP-3 ( ) or N-TIMP-3 (➃) for 1 h at 37°C and residual activity against Abz-TESE∼SRGAIY-Dpa-KK determined (18 h, 37°C). TIMP-3 and N-TIMP-3 had Ki(app) values of 0.70 ± 0.04 nM and 0.14 ± 0.02 nM respectively for ADAMTS5-1, and 1.89 ± 0.06 nM and 1.35 ± 0.04 nM respectively for ADAMTS5-5. ADAMTS4-1 (C, 0.5 nM) and ADAMTS4-4 (D, 0.5 nM) were incubated with 0.25-10 nM TIMP-3 ( ) or N-TIMP-3 (➃) for 1 h at 37°C and residual activity against FAM-AE∼LQGRPISIAK-TAMRA (1 μM) determined (18 h, 37°C). TIMP-3 and N-TIMP-3 had Ki(app) values of 0.39 ± 0.03 nM and 0.19 ± 0.04 nM respectively for ADAMTS4-1, and 0.38 ± 0.04 nM and 0.55 ± 0.05 nM respectively for ADAMTS4-4. The error values given are the standard errors on the fit of the data to the tight binding equation (equation 2) (Bieth, 1995).
Table 2.
Ki values (nM) a for mammalian-expressed TIMP-3 and N-TIMP-3 inhibition of ADAMTS-4 and ADAMTS-5.
| N-TIMP-3 | TIMP-3 | N-TIMP-3 | TIMP-3 | ||
|---|---|---|---|---|---|
| ADAMTS5-1 | 0.13 ± 0.06 | 0.63 ± 0.17 | |||
| ADAMTS5-2 | 0.35 ± 0.03 | 0.72 ± 0.17 | ADAMTS4-1 | 0.13 ± 0.06 | 0.22 ± 0.15 |
| ADAMTS5-3 | 0.29 ± 0.08 | 1.00 ± 0.39 | ADAMTS4-2 | 0.18 ± 0.02 | 0.22 ± 0.07 |
| ADAMTS5-4 | 0.40 ± 0.18 | 1.13 ± 0.37 | ADAMTS4-3 | 0.10 ± 0.01 | 0.40 ± 0.07 |
| ADAMTS5-5 | 1.01 ± 0.45 | 1.17 ± 0.33 | ADAMTS4-4 | 0.64 ± 0.15 | 0.51 ± 0.21 |
Ki values given as mean ± standard deviation (n=4-6).
Glycosylation had little effect on TIMP-3 or N-TIMP-3 inhibition of ADAMT5-2 or ADAMTS4-2 (Table 3).
Table 3.
Effect of glycosylation on TIMP-3 and N-TIMP-3 inhibition of ADAMTS4-2 and ADAMTS5-2.
| N-TIMP-3 Kia | TIMP-3 Kia | |||
|---|---|---|---|---|
| glycosylated b | non-glycosylated c | glycosylated b | non-glycosylated d | |
| ADAMTS5-2 | 0.35 ± 0.03 | 0.32 ± 0.12 | 0.72 ± 0.17 | 1.37 ± 0.4 |
| ADAMTS4-2 | 0.18 ± 0.02 | 0.79 ± 0.02 | 0.22 ± 0.07 | 0.20 ± 0.08 |
Ki values (nM) are expressed as mean ± standard deviation (n=4).
Glycosylated TIMP-3 and N-TIMP-3 were expressed in HEK293 cells.
Non-glycosylated N-TIMP-3 was expressed in E. coli.
Non-glycosylated TIMP-3 was expressed in HEK293 cells treated with 5 μg/ml tunicamycin.
The isolated catalytic domains of ADAMTS-4 and -5 (ADAMTS4-5 and ADAMTS5-6) had only minimal activity on natural substrates and the synthetic substrates used, so their inhibition by TIMP-3 could not be analyzed by enzyme inhibition kinetics.
3. Discussion
Here we report a novel method to generate recombinant TIMP-3 protein by treating transfected HEK293 cells with sodium chlorate. Our initial attempts to express TIMP-3 in HEK293 or HTB94 chondrosarcoma cells were hampered by the observation that, although these cells transcribe a considerable amount of TIMP-3 mRNA, no TIMP-3 accumulates in the conditioned medium. We have previously reported that TIMP-3 is normally internalized but accumulates in medium in the presence of an antagonist of the LRP endocytic receptor or heparin (Troeberg et al., 2008). We thus treated cells with sodium chlorate, which blocks sulfation of cell surface glycosaminoglycans (Baeuerle and Huttner 1986; Safaiyan et al., 1999) and observed accumulation of soluble TIMP-3 in the medium. TIMP-3 has previously been purified from the conditioned medium of transfected NSO mouse myeloma cells (Apte et al., 1995), which appear to lack this endocytic pathway. Our transfected cells showed no signs of TIMP-3-induced apoptosis (Bond et al., 2000) in the absence of sodium chlorate, but exhibited increased cell death once chlorate was added and TIMP-3 accumulated in the medium.
Our values for TIMP-3 inhibition of ADAMTS-5 (Ki = 0.1-1.2 nM, Table 2) are in agreement with the previously reported value of 0.66 nM (Kashiwagi et al., 2001). Our Ki values for TIMP-3 inhibition of ADAMTS-4 (Ki = 0.1-0.7 nM) are also in agreement with that reported by Kashiwagi et al. (2001), although higher values of 4-8 nM have been reported in other studies (Hashimoto et al., 2001; Wayne et al., 2007).
C-terminal domains distal to the disintegrin domain are required for ADAMTS-4 and -5 activity on aggrecan and various other protein substrates, but these domains had little effect on hydrolysis of the synthetic substrates used in this study (data not shown). The isolated catalytic domains of ADAMTS-4 and -5 had negligible activity on these synthetic substrates. To date, the catalytic domain alone has not been shown to effectively cleave protein substrates such as aggrecan, suggesting that the disintegrin domain may be part of the minimal catalytic unit. This view is supported by the crystal structures of ADAMTS-4 and -5, which show that the disintegrin domain is located close to the catalytic domain and may act as an extension of the catalytic cleft (Mosyak et al., 2008).
The inhibitory machinery of TIMPs is contained in their N-terminal domains, and in most cases, the C-terminal domains of both MMPs and TIMPs have a minimal effect on complex formation (Brew et al., 2000). There are some notable exceptions to this, in which interactions other than binding of the N-terminal TIMP domain to the catalytic domain of the enzyme contribute substantially to the overall binding energy. In particular, the C-terminal domains of TIMP-2, TIMP-3 and TIMP-4 bind strongly to the MMP-2 hemopexin domain (Butler et al., 1999; Lee et al., 2001; Morgunova et al., 2002; Murphy et al., 1992; Troeberg et al., 2002) and the C-terminal domain of TIMP-1 binds to the hemopexin domain of MMP-9 (Bodden et al., 1994).
While the C-terminal domains of MMPs either have little effect or strengthen binding to TIMPs, the C-terminal domains of ADAM17 weaken interaction with both full-length TIMP-3 and N-TIMP-3 by 10-fold (Lee et al., 2002). Also, the ADAM17 catalytic domain interacts more strongly with the prodomain than the enzyme with disintegrin and cysteine-rich domain domains attached to the catalytic domain (Milla et al., 1999). Lee et al. (2002) postulated that the C-terminal domains of the enzyme may sterically hinder access to the catalytic site. The spatial orientation of the ADAM17 C-terminal domains is not known, as crystal structures are only available for the catalytic domain in complex with either a hydroxamate inhibitor or N-TIMP-3 (Maskos et al., 1998; Wisniewska et al., 2008). However, for the related snake venom metalloproteinases, called vascular apoptosis-inducing proteins (VAPs), crystal structures are available showing the catalytic, disintegrin and cysteine-rich domains (Igarashi et al., 2007; Igarashi et al., 2006; Takeda 2008). These structures support Lee et al.'s proposal that the C-terminal domains of ADAM17 may sterically hinder TIMP-3 access to the active site (Lee et al., 2002) as the C-terminal VAP domains adopt a C-shaped conformation, with the concave surface of the cysteine-rich domain being located near to and facing the catalytic domain (Igarashi et al., 2007). Additionally, there is a “hinge” region between the catalytic and disintegrin domains and the 6 available VAP structures indicate flexibility in the orientation of the catalytic domain relative to the C-terminal domains.
In contrast to ADAM17, we found that the ADAMTS-4 and -5 C-terminal domains increase association with N-TIMP-3 by 5-7-fold (Table 2). The C-terminal domains also increase association with full-length TIMP-3, albeit to a lesser extent (Ki for ADAMTS-4 and -5 improves 2-fold). This is in agreement with the report of Wayne et al. (2007). In particular, our data suggest that the TS domains of ADAMTS-4 and -5 are involved in interaction with TIMP-3. Deletion of the C-terminal TS domain of ADAMTS-5 increases Ki for N-TIMP-3 by 3-fold. Further deletion of the Sp and CysR domains has minimal effect on Ki, but deletion of the first TS domain results in a further increase in Ki for N-TIMP-3. Ki values for ADAMTS-4 deletion mutants show the same result, with deletion of the TS domain increasing Ki for N-TIMP-3 by 6-fold, while deletion of the Sp and CysR domains had little effect. Interestingly, the TS domains have also been shown to play a role in substrate recognition and cleavage (Tortorella et al., 2000; Fushimi et al., 2008).
The domain architecture of the full-length ADAMTSs is not known, as three-dimensional structures are only available for the catalytic and disintegrin domains of ADAMTS-1, -4 and -5 (Gerhardt et al., 2007; Mosyak et al., 2008; Shieh et al., 2008). Although the ADAMTS and VAP disintegrin domains share sequence homology, it is not possible to extrapolate to the ADAMTSs from the available VAP structures as their domain arrangements are dissimilar and may adopt a different orientation relative to the catalytic domain (Gerhardt et al., 2007; Takeda 2008). The crystal structures of the catalytic and disintegrin domains of ADAMTS-4 with and without a low molecular weight inhibitor determined by Moysak et al. (2008) indicated an ‘open’ and a ‘closed’ form, respectively. In the latter form, the residues Asp328 and Thr329 in the so-called S2′ loop of 322CGXXXCDTL330 are around the catalytic zinc and the side chain of Asp328 chelates the Zn2+ ion and that of Thr329 fills the space at the mouth of the S1′ pocket. Thus, the S2′ loop remains as an auto-inhibitor unless structural re-arrangements occur around this region and disrupt the interaction between Asp328 and the Zn2+ ion. However, as proposed by Moysak et al. (2008), the active open form and the inactive closed form may exist in equilibrium. Full-length ADAMTS-4 and ADAMTS-5 are highly active against a natural substrate, aggrecan, but deletion of the C-terminal non-catalytic domains of the enzymes greatly reduces their activity (Kashiwagi et al., 2004; Gendron et al., 2007; Fushimi et al., 2008). This suggests that these domains alone or substrate binding to these non-catalytic domains may shift the equilibrium to the more open form. Our data support this possibility, since the presence of the C-terminal domains enhances association with N-TIMP-3. Also, agents that interact with the ADAMTS C-terminal domains, such as calcium pentosan polysulfate and aggrecan, improve affinity for TIMP-3 (Troeberg et al., 2008; Wayne et al., 2007).
It is unclear why the C-terminal domains favour association with N-TIMP-3 more than with full-length TIMP-3. Given that N-TIMP-3 is a slightly better inhibitor of full-length ADAMTS-4 and -5 than full-length TIMP-3 (Table 2), steric clashes may occur between the C-terminal domain of TIMP-3 and the ADAMTS non-catalytic domains. Alternatively, a surface available in N-TIMP-3 but not full-length TIMP-3 may enhance interaction with the non-catalytic domains of the enzyme, which in turn, could increase the affinity of binding to the active site. Interestingly, N-TIMP-4 is also a better inhibitor of ADAM17 than full-length TIMP-4 (Lee et al., 2005). The opposite is true for ADAM10, which is not inhibited by N-TIMP-1 or N-TIMP-3, but is effectively inhibited by full-length TIMP-1 and TIMP-3 (Rapti et al., 2008). The ADAMs and the ADAMTSs analyzed to date thus vary considerably in their affinity for N-TIMP-3 relative to full-length TIMP-3, possibly reflecting conformational diversity of their C-terminal domains. Structural studies of the complex formed between full-length ADAMTS-4 or -5 and TIMP-3 are necessary to obtain further information on these molecular interactions.
4. Experimental procedures
4. 1. Preparation of TIMP-3 and N-TIMP-3 from HEK293
Recombinant human C-terminally FLAG-tagged TIMP-3 and N-TIMP-3 were expressed using pCEP4-based expression vectors (Invitrogen, Paisley, UK) constructed by the PCR method. The TIMP-3 expression vector was constructed using the sense primer 5′-CTCGAGTCCCTTTTTTTTCCACAGGAGCTCGCCGCCACCATGACCCCTTGGCT CGGGCTCATCGTGCTC-3′ containing an XhoI site (underlined), Kozak consensus sequence (in italics) and TIMP-3 N-terminal sequence, and the antisense primer 5′-CTCGAGCTACTTATCGTCGTCATCCTTGTAATCGGGGTCTGTGGCATTGATGATG CTTTTATCCGG-3′ containing an XhoI site (underlined), FLAG epitope (Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys-stop) (in italics) and the C-terminal sequence of TIMP-3, with a TIMP-3 vector kindly provided by Prof M. Seiki (University of Tokyo, Japan) as a template. The N-TIMP-3 (residues 1-121, from 1CTCSPS to HLGCN121) vector was constructed using the same sense primer and the anti-sense primer 5′-CTCGAGCTACTTATCGTCGTCATCCTTGTAATCGTTACAACCCAGGTGATACCGA TAGTTCAGCC-3′ containing an XhoI site (underlined), FLAG epitope (in italics) and the C-terminal sequence of N-TIMP3. PCR fragments were ligated into pCEP4 at the XhoI site.
Human embryonic kidney HEK293 cells were transfected with the expression plasmids by lipofection with FuGENE6 (Roche Applied Science, Basel, Switzerland) and transfected cells selected by treatment with 800 μg/ml hygromycin B (PAA Laboratories GmbH, Pasching, Austria) over 3 weeks. Transfected cell lines were maintained under standard cell culture conditions in Dulbecco's modified Eagle's medium (DMEM) containing 10% (v/v) fetal calf serum, penicillin (100 units/ml), streptomycin (100 units/ml), 250 μg/ml G418 (PAA Laboratories GmbH, Pasching, Austria) and 800 μg/ml hygromycin B.
For expression of recombinant FLAG-tagged TIMP-3 and N-TIMP-3, transfected cells were treated with 30 mM sodium chlorate (NaClO3, Sigma-Aldrich, Dorset, UK) in serum-free DMEM. Non-glycosylated full-length human TIMP-3 was prepared by treating transfected HEK293 cells with 5 μg/ml tunicamycin in addition to sodium chlorate. In all cases, medium was changed every 2 days to reduce TIMP-3-induced apoptosis (Bond et al., 2000). Conditioned media (0.5-1 l) were centrifuged to remove cell debris and applied to a column of anti-FLAG M2-agarose (1.5 ml, Sigma-Aldrich, Dorset, UK). The column was washed with 50 mM Tris-HCl, pH 7.5, 1 M NaCl, 10 mM CaCl2, 0.05% Brij-35, 0.02% NaN3. Bound proteins were eluted with 200 μg/ml FLAG peptide (Sigma-Aldrich, Dorset, UK) in TNC buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM CaCl2, 0.05% Brij-35 and 0.02% NaN3). To remove the FLAG peptide, the eluate was applied to a Macro-Prep 25 S resin (400 μl, BioRad, Hemel Hempstead, UK) equilibrated in TNC buffer. Recombinant protein was eluted with 50 mM Tris-HCl, pH 7.5, 1 M NaCl, 10 mM CaCl2, 0.05% Brij-35 and 0.02% NaN3. Finally, the buffer was exchanged for TNC buffer using a PD-10 desalting column (GE Healthcare, Bucks, UK).
The purity of recombinant TIMP-3 and N-TIMP-3 was confirmed by reducing SDS-PAGE (Bury, 1981) and their active concentrations determined by titration with known concentrations of the catalytic domain of MMP-1. The identity of the purified proteins was confirmed using a commercial rabbit polyclonal antibody (AbCam, Cambridge, UK, catalogue number ab39184) and M2 anti-FLAG antibody (Sigma-Aldrich, Dorset, UK).
N-terminal analysis was carried out on TIMP-3 and N-TIMP-3 prepared from HEK293. Samples were denatured in SDS sample buffer at 80°C in the presence of 50 mM dithiothreitol. SDS-PAGE was performed on a 10% gel (10 cm × 10 cm × 0.15 cm) according to Bury (1981). The gel was electroblotted to Immobilon-P membrane (Millipore) in 10 mM CAPS 10% methanol pH 11 (Matsudaira, 1987) and stained for protein using Coomassie Brilliant Blue. Bands of interest were excised and placed directly onto a Polybrene-treated glass filter. Samples were analyzed by automated Edman degradation using an Applied Biosystems Procise 494HT sequencer with on-line phenylthiohydantoin HPLC analysis. The instruments were operated according to the manufacturers' instructions.
4.2. Preparation of E.coli-expressed N-TIMP-3, ADAMTSs and MMPs
The N-terminal domain of human TIMP-3 with C-terminal His-tag was expressed in Escherichia coli, purified and folded in vitro (Kashiwagi et al., 2001). Human ADAMTS-4 and ADAMTS-5 deletion mutants (Fig. 2) were expressed and purified as previously described (Fushimi et al., 2008). The catalytic domains of human MMP-1 (MMP-1cat) and MMP-3 (MMP-3cat) were prepared as described (Chung et al., 2000; Suzuki et al., 1998). Human proMMP-2 and the pro-form of the MMP-2 catalytic domain (proMMP-2ΔC, lacking hemopexin-domain residues 466 to 660) were expressed in HEK293 cells using pCEP4-based expression vectors. The proMMP-2ΔC expression plasmid was constructed from the proMMP-2/pCEP4 plasmid by PCR mutagenesis using the sense primer 5′-CAGTACATCAAGTGTATCATATGCCAAGTCCGCCCC-3′ and the anti-sense primer 5′-GCCAAGGTCAATAAGCTTTCATCACCCATAGAGCTC-3′. The resulting PCR product was digested with KpnI and HindIII and ligated into similarly cut pCEP4. Both vectors were transfected into HEK293 cells and transfected cells selected by treatment with 1000 μg/ml hygromycin B as described for TIMP-3 above. Recombinant proMMP-2 and proMMP-2cat were purified from the conditioned medium by gelatin affinity chromatography and gel filtration chromatography (Itoh et al., 1995). Active forms of the enzymes were prepared by treatment with 1 mM 4-aminophenylmercuric acetate (ICN Biochemicals, Solon, OH, USA) for 60 min at 37°C, after which the buffer was exchanged for TNC buffer using a PD-10 desalting column (GE Healthcare, Bucks, UK). Active concentrations of all enzymes were determined by titration against a known concentration of E. coli-expressed N-TIMP-3 (for ADAMTS-4 and -5) or TIMP-1 (for MMPs).
4.3. Measurement of Inhibition Constant, Ki
All enzyme assays were conducted in TNC buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM CaCl2, 0.05% Brij-35 and 0.02% NaN3) at 37°C, using a Gemini microplate spectrofluorometer (Molecular Devices, Sunnyvale, CA, USA).
The activity of ADAMTS-4 was monitored using the fluorescent peptide substrate carboxyfluorescein-Ala-Glu∼Leu-Asn-Gly-Arg-Pro-Ile-Ser-Ile-Ala-Lys-N,N,N′,N′-tetramethyl-6-carboxyrhodamine (FAM-AE∼LQGRPISIAK-TAMRA) at a final concentration of 0.5 μM with an excitation wavelength of 485 nm and an emission wavelength of 538 nm (495 nm cut-off) (Wayne et al., 2007).
The activity of ADAMTS-5 was monitored using the fluorescent peptide substrate ortho-aminobenzoyl-Thr-Glu-Ser-Glu∼Ser-Arg-Gly-Ala-Ile-Tyr-(N-3-[2,4-dinitrophenyl]-L-2,3-diaminopropionyl)-Lys-Lys-NH2 [Abz-TESE∼SRGAIY-Dpa-KK] (kindly provided by Dr. Andrew Parker, AstraZeneca, Macclesfield, UK). This peptide is based on the ADAMTS-4 cleavage site in rat brevican (Nakamura et al., 2000; Matthews et al., 2000). ADAMTS-5 has been predicted to also cleave at this site (Nakada et al., 2005) and we found that recombinant ADAMTS-5 cleaved this substrate more readily than ADAMTS-4 in vitro. The substrate was used at a final concentration of 20 μM with an excitation wavelength of 300 nm and an emission wavelength of 430 nm (420 nm cutoff). Substrate hydrolysis by 0.5 nM ADAMTS-5 was confirmed to be linear for 18 h at 37 °C.
Activities of MMP-1 and MMP-2 were measured using the fluorescent quenched peptide substrate (7-methoxycoumarin-4-yl)acetyl-Pro-Leu-Gly-Leu-(N-3-[2,4-dinitrophenyl]-L-2,3-diaminopropionyl)-Ala-Arg-NH2 (Mca-PLGL-Dpa-AR) at 1.5 μM final concentration (Knight et al., 1992). Activity of MMP-3cat was measured using the fluorescent quenched substrate NFF-3, Mca-Arg-Pro-Lys-Pro-Val-Glu-Nva-Trp-Arg-Lys(2,4-dinitrophenyl)-NH2 at 1.5 μM final concentration (Nagase et al., 1994).
Ki values of TIMP-3 for various isoforms of ADAMTS-4 and ADAMTS-5 were determined under equilibrium kinetic conditions (Bieth, 1995). Final enzyme concentrations for Ki determinations were as follows: ADAMTS-4 and -5 were used at 0.5 nM, MMP-1, MMP-1cat and MMP-3cat were used at 1 nM, and MMP-2 and MMP-2cat were used at 0.125 nM. Enzymes were pre-incubated (1 h, 37°C) with TIMP-3 (0.5-100 nM final concentration) and equilibrium rate of substrate hydrolysis (vs) determined (18 h, 37°C). Prism software (GraphPad, La Jolla, CA, USA) was used to fit the data to the tight-binding equation (Bieth, 1995):
| (1) |
where vo is equilibrium rate of substrate hydrolysis in the absence of inhibitor, Eo is the total enzyme concentration, Io is the total inhibitor concentration and Ki(app) is the apparent inhibition constant.
To determine Ki from Ki(app), the Km of the enzymes for the substrates used must be known (Bieth, 1995). Wayne et al. (2007) determined a Km value of 15 μM for ADAMTS-4 cleavage of FAM-AE∼LQGRPISIAK-TAMRA, which we used at 0.5 μM. We determined a Km value of 76 μM for ADAMTS-5 cleavage of Abz-TESE∼SRGAIY-Dpa-KK (data not shown), used at 20 μM. Ki was then calculated from the equation:
| (2) |
where Ki is the inhibition constant, [S] is the initial substrate concentration and Km is the Michaelis constant for the substrate used. Therefore, Ki(app) was divided by 1.033 to determine Ki for ADAMTS-4, and by 1.26 to determine Ki for ADAMTS-5.
Acknowledgments
We thank Dr Andrew Parker (AstraZeneca, Macclesfield, UK) for provision of the Abz-TESE∼SRGAIY-Dpa-KK fluorescent substrate and Prof. M. Seiki (University of Tokyo, Japan) for the TIMP-3 vector. This work was supported by the Wellcome Trust (grant 057473) and Award Number AR40994 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS). The content is solely the responsibility of the authors and does not necessarily represent the official views of NIAMS or NIH.
Abbreviations
- ADAM
adamalysin
- ADAMTS
adamalysin with thrombospondin motifs
- cat
catalytic domain
- CysR
cysteine-rich
- Dis
disintegrin
- LRP
low-density lipoprotein receptor-related protein
- MMP
matrix metalloproteinase
- N-TIMP
N-terminal domain of TIMP
- RAP
receptor-associated protein
- Sp
spacer
- TACE
tumour necrosis factor-α converting enzyme
- TIMP
tissue inhibitor of metalloproteinase
- TS
thrombospondin
- VAP
vascular apoptosis-inducing protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apte SS, Olsen BR, Murphy G. The gene structure of tissue inhibitor of metalloproteinases (TIMP)-3 and its inhibitory activities define the distinct TIMP gene family. J Biol Chem. 1995;270:14313–14318. doi: 10.1074/jbc.270.24.14313. [DOI] [PubMed] [Google Scholar]
- Baeuerle PA, Huttner WB. Chlorate - a potent inhibitor of protein sulfation in intact cells. Biochem Biophys Res Commun. 1986;141:870–877. doi: 10.1016/s0006-291x(86)80253-4. [DOI] [PubMed] [Google Scholar]
- Bieth JG. Theoretical and Practical Aspects of Proteinase Inhibition Kinetics Meth. Enzymol. 1995;248:59–84. doi: 10.1016/0076-6879(95)48007-2. [DOI] [PubMed] [Google Scholar]
- Bodden MK, Windsor LJ, Caterina NC, Harber GJ, Birkedal-Hansen B, Birkedal-Hansen H. Human TIMP-1 binds to pro-M(r) 92K GL (gelatinase B, MMP-9) through the “second disulfide knot”. Ann N Y Acad Sci. 1994;732:403–407. doi: 10.1111/j.1749-6632.1994.tb24767.x. [DOI] [PubMed] [Google Scholar]
- Bond M, Murphy G, Bennett MR, Amour A, Knauper V, Newby AC, Baker AH. Localization of the death domain of tissue inhibitor of metalloproteinase-3 to the N terminus. Metalloproteinase inhibition is associated with proapoptotic activity. J Biol Chem. 2000;275:41358–41363. doi: 10.1074/jbc.M007929200. [DOI] [PubMed] [Google Scholar]
- Brew K, Dinakarpandian D, Nagase H. Tissue inhibitors of metalloproteinases: evolution, structure and function. Biochim Biophys Acta. 2000;1477:267–283. doi: 10.1016/s0167-4838(99)00279-4. [DOI] [PubMed] [Google Scholar]
- Bury AF. Analysis of protein and peptide mixtures. Evaluation of three sodium dodecyl sulfate -polyacrylamide gels electrophoresis systems. J Chromatography. 1981;213:491–500. [Google Scholar]
- Butler GS, Apte SS, Willenbrock F, Murphy G. Human tissue inhibitor of metalloproteinases 3 interacts with both the N- and C-terminal domains of gelatinases A and B. Regulation by polyanions. J Biol Chem. 1999;274:10846–10851. doi: 10.1074/jbc.274.16.10846. [DOI] [PubMed] [Google Scholar]
- Colige A, Ruggiero F, Vandenberghe I, Dubail J, Kesteloot F, Van Beeumen J, Beschin A, Brys L, Lapière CM, Nusgens B. Domains and maturation processes that regulate the activity of ADAMTS-2, a metalloproteinase cleaving the aminopropeptide of fibrillar procollagens types I-III and V. J Biol Chem. 2005;280:34397–34408. doi: 10.1074/jbc.M506458200. [DOI] [PubMed] [Google Scholar]
- Chung L, Shimokawa K, Dinakarpandian D, Grams F, Fields GB, Nagase H. Identification of the (183)RWTNNFREY(191) region as a critical segment of matrix metalloproteinase 1 for the expression of collagenolytic activity. J Biol Chem. 2000;275:29610–29617. doi: 10.1074/jbc.M004039200. [DOI] [PubMed] [Google Scholar]
- Clark IM, Cawston TE. Fragments of human fibroblast collagenase. Purification and characterization. Biochem J. 1989;263:201–206. doi: 10.1042/bj2630201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabunmi RP, Baker AH, Murray EJ, Booth RF, Newby AC. Divergent regulation by growth factors and cytokines of 95 kDa and 72 kDa gelatinases and tissue inhibitors or metalloproteinases-1, -2, and -3 in rabbit aortic smooth muscle cells. Biochem J. 1996;315(Pt 1):335–342. doi: 10.1042/bj3150335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushimi K, Troeberg L, Nakamura H, Lim NH, Nagase H. Functional differences of the catalytic and non-catalytic domains in human ADAMTS-4 and ADAMTS-5 in aggrecanolytic activity. J Biol Chem. 2008;283:6706–6716. doi: 10.1074/jbc.M708647200. [DOI] [PubMed] [Google Scholar]
- Gao G, Plaas A, Thompson VP, Jin S, Zuo F, Sandy JD. ADAMTS4 (aggrecanase-1) activation on the cell surface involves C-terminal cleavage by glycosylphosphatidyl inositol-anchored membrane type 4-matrix metalloproteinase and binding of the activated proteinase to chondroitin sulfate and heparan sulfate on syndecan-1. J Biol Chem. 2004;279:10042–10051. doi: 10.1074/jbc.M312100200. [DOI] [PubMed] [Google Scholar]
- Gao W, Anderson PJ, Sadler JE. Extensive contacts between ADAMTS13 exosites and von Willebrand factor domain A2 contribute to substrate specificity. Blood. 2008;112:1713–1719. doi: 10.1182/blood-2008-04-148759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron C, Kashiwagi M, Lim NH, Enghild JJ, Thogersen IB, Hughes C, Caterson B, Nagase H. Proteolytic activities of human ADAMTS-5: Comparative studies with human ADAMTS-4. J Biol Chem. 2007;282:18294–18306. doi: 10.1074/jbc.M701523200. [DOI] [PubMed] [Google Scholar]
- Gerhardt S, Hassall G, Hawtin P, McCall E, Flavell L, Minshull C, Hargreaves D, Ting A, Pauptit RA, Parker AE, Abbott WM. Crystal structures of human ADAMTS-1 reveal a conserved catalytic domain and a disintegrin-like domain with a fold homologous to cysteine-rich domains. J Mol Biol. 2007;373:891–902. doi: 10.1016/j.jmb.2007.07.047. [DOI] [PubMed] [Google Scholar]
- Godyna S, Liau G, Popa I, Stefansson S, Argraves WS. Identification of the low density lipoprotein receptor-related protein (LRP) as an endocytic receptor for thrombospondin-1. J Cell Biol. 1995;129:1403–1410. doi: 10.1083/jcb.129.5.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto G, Aoki T, Nakamura H, Tanzawa K, Okada Y. Inhibition of ADAMTS4 (aggrecanase-1) by tissue inhibitors of metalloproteinases (TIMP-1, 2, 3 and 4) FEBS Lett. 2001;494:192–195. doi: 10.1016/s0014-5793(01)02323-7. [DOI] [PubMed] [Google Scholar]
- Igarashi T, Araki S, Mori H, Takeda S. Crystal structures of catrocollastatin/VAP2B reveal a dynamic, modular architecture of ADAM/adamalysin/reprolysin family proteins. FEBS Lett. 2007;581:2416–2422. doi: 10.1016/j.febslet.2007.04.057. [DOI] [PubMed] [Google Scholar]
- Igarashi T, Oishi Y, Araki S, Mori H, Takeda S. Crystallization and preliminary X-ray crystallographic analysis of two vascular apoptosis-inducing proteins (VAPs) from Crotalus atrox venom. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:688–691. doi: 10.1107/S1744309106022548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Binner S, Nagase H. Steps involved in activation of the complex of pro-matrix metalloproteinase 2 (progelatinase A) and tissue inhibitor of metalloproteinases (TIMP)-2 by 4-aminophenylmercuric acetate. Biochem J. 1995;308(Pt 2):645–651. doi: 10.1042/bj3080645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi M, Enghild JJ, Gendron C, Hughes C, Caterson B, Itoh Y, Nagase H. Altered proteolytic activities of ADAMTS-4 expressed by C-terminal processing. J Biol Chem. 2004;279:10109–10119. doi: 10.1074/jbc.M312123200. [DOI] [PubMed] [Google Scholar]
- Kashiwagi M, Tortorella M, Nagase H, Brew K. TIMP-3 is a potent inhibitor of aggrecanase 1 (ADAM-TS4) and aggrecanase 2 (ADAM-TS5) J Biol Chem. 2001;276:12501–12504. doi: 10.1074/jbc.C000848200. [DOI] [PubMed] [Google Scholar]
- Knight CG, Willenbrock F, Murphy G. A novel coumarin-labelled peptide for sensitive continuous assays of the matrix metalloproteinases. FEBS Lett. 1992;296:263–266. doi: 10.1016/0014-5793(92)80300-6. [DOI] [PubMed] [Google Scholar]
- Langton KP, Barker MD, McKie N. Localization of the functional domains of human tissue inhibitor of metalloproteinases-3 and the effects of a Sorsby's Fundus Dystrophy mutation. J Biol Chem. 1998;273:16778–16781. doi: 10.1074/jbc.273.27.16778. [DOI] [PubMed] [Google Scholar]
- Lee MH, Atkinson S, Murphy G. Identification of the extracellular matrix (ECM) binding motifs of tissue inhibitor of metalloproteinases (TIMP)-3 and effective transfer to TIMP-1. J Biol Chem. 2007;282:6887–6898. doi: 10.1074/jbc.M610490200. [DOI] [PubMed] [Google Scholar]
- Lee MH, Knäuper V, Becherer JD, Murphy G. Full-length and N-TIMP-3 display equal inhibitory activities toward TNF-alpha convertase. Biochem Biophys Res Commun. 2001;280:945–950. doi: 10.1006/bbrc.2000.4192. [DOI] [PubMed] [Google Scholar]
- Lee MH, Rapti M, Murphy G. Total conversion of tissue inhibitor of metalloproteinase (TIMP) for specific metalloproteinase targeting: fine-tuning TIMP-4 for optimal inhibition of tumor necrosis factor-(alpha)-converting enzyme. J Biol Chem. 2005;280:15967–15975. doi: 10.1074/jbc.M500897200. [DOI] [PubMed] [Google Scholar]
- Lee MH, Verma V, Maskos K, Becherer JD, Knäuper V, Dodds P, Amour A, Murphy G. The C-terminal domains of TACE weaken the inhibitory action of N-TIMP-3. FEBS Lett. 2002;520:102–106. doi: 10.1016/s0014-5793(02)02776-x. [DOI] [PubMed] [Google Scholar]
- Maskos K, Fernandez-Catalan C, Huber R, Bourenkov GP, Bartunik H, Ellestad GA, Reddy P, Wolfson MF, Rauch CT, Castner BJ, Davis R, Clarke HR, Petersen M, Fitzner JN, Cerretti DP, March CJ, Paxton RJ, Black RA, Bode W. Crystal structure of the catalytic domain of human tumor necrosis factor-alpha-converting enzyme. Proc Natl Acad Sci USA. 1998;95:3408–3412. doi: 10.1073/pnas.95.7.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- Matthews RT, Gary SC, Zerillo C, Pratta M, Solomon K, Arner EC, Hockfield S. Brain-enriched hyaluronan binding (BEHAB)/brevican cleavage in a glioma cell line is mediated by a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) family member. J Biol Chem. 2000;275:22695–22703. doi: 10.1074/jbc.M909764199. [DOI] [PubMed] [Google Scholar]
- Milla ME, Leesnitzer MA, Moss ML, Clay WC, Carter HL, Miller AB, Su JL, Lambert MH, Willard DH, Sheeley DM, Kost TA, Burkhart W, Moyer M, Blackburn RK, Pahel GL, Mitchell JL, Hoffman CR, Becherer JD. Specific sequence elements are required for the expression of functional tumor necrosis factor-alpha-converting enzyme (TACE) J Biol Chem. 1999;274:30563–30570. doi: 10.1074/jbc.274.43.30563. [DOI] [PubMed] [Google Scholar]
- Morgunova E, Tuuttila A, Bergmann U, Tryggvason K. Structural insight into the complex formation of latent matrix metalloproteinase 2 with tissue inhibitor of metalloproteinase 2. Proc Natl Acad Sci USA. 2002;99:7414–7419. doi: 10.1073/pnas.102185399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosyak L, Georgiadis K, Shane T, Svenson K, Hebert T, McDonagh T, Mackie S, Olland S, Lin L, Zhong X, Kriz R, Reifenberg EL, Collins-Racie LA, Corcoran C, Freeman B, Zollner R, Marvell T, Vera M, Sum PE, LaVallie ER, Stahl M, Somers W. Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5. Protein Sci. 2008;17:16–21. doi: 10.1110/ps.073287008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy G, Willenbrock F, Ward RV, Cockett MI, Eaton D, Docherty AJ. The C-terminal domain of 72 kDa gelatinase A is not required for catalysis, but is essential for membrane activation and modulates interactions with tissue inhibitors of metalloproteinases. Biochem J. 1992;283(Pt 3):637–641. doi: 10.1042/bj2830637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase H, Fields CG, Fields GB. Design and characterization of a fluorogenic substrate selectively hydrolyzed by stromelysin 1 (matrix metalloproteinase-3) J Biol Chem. 1994;269:20952–20957. [PubMed] [Google Scholar]
- Nakada M, Miyamori H, Kita D, Takahashi T, Yamashita J, Sato H, Miura R, Yamaguchi Y, Okada Y. Human glioblastomas overexpress ADAMTS-5 that degrades brevican. Acta Neuropathol. 2005;110:239–246. doi: 10.1007/s00401-005-1032-6. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Fujii Y, Inoki I, Sugimoto K, Tanzawa K, Matsuki H, Miura R, Yamaguchi Y, Okada Y. Brevican is degraded by matrix metalloproteinases and aggrecanase-1 (ADAMTS4) at different sites. J Biol Chem. 2000;275:38885–38890. doi: 10.1074/jbc.M003875200. [DOI] [PubMed] [Google Scholar]
- Rapti M, Atkinson SJ, Lee MH, Trim A, Moss M, Murphy G. The isolated N-terminal domains of TIMP-1 and TIMP-3 are insufficient for ADAM10 inhibition. Biochem J. 2008;411:433–439. doi: 10.1042/BJ20071430. [DOI] [PubMed] [Google Scholar]
- Safaiyan F, Kolset SO, Prydz K, Gottfridsson E, Lindahl U, Salmivirta M. Selective effects of sodium chlorate treatment on the sulfation of heparan sulfate. J Biol Chem. 1999;274:36267–36273. doi: 10.1074/jbc.274.51.36267. [DOI] [PubMed] [Google Scholar]
- Sarafanov AG, Ananyeva NM, Shima M, Saenko EL. Cell surface heparan sulfate proteoglycans participate in factor VIII catabolism mediated by low density lipoprotein receptor-related protein. J Biol Chem. 2001;276:11970–11979. doi: 10.1074/jbc.M008046200. [DOI] [PubMed] [Google Scholar]
- Shieh HS, Mathis KJ, Williams JM, Hills RL, Wiese JF, Benson TE, Kiefer JR, Marino MH, Carroll JN, Leone JW, Malfait AM, Arner EC, Tortorella MD, Tomasselli A. High resolution crystal structure of the catalytic domain of ADAMTS-5 (aggrecanase-2) J Biol Chem. 2008;283:1501–1507. doi: 10.1074/jbc.M705879200. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kan CC, Hung W, Gehring MR, Brew K, Nagase H. Expression of human pro-matrix metalloproteinase 3 that lacks the N-terminal 34 residues in Escherichia coli: autoactivation and interaction with tissue inhibitor of metalloproteinase 1 (TIMP-1) Biol Chem. 1998;379:185–191. doi: 10.1515/bchm.1998.379.2.185. [DOI] [PubMed] [Google Scholar]
- Takeda S. Three-dimensional domain architecture of the ADAM family proteinases. Seminars in Cell & Developmental Biology. 2008 doi: 10.1016/j.semcdb.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Tortorella M, Pratta M, Liu RQ, Abbaszade I, Ross H, Burns T, Arner E. The thrombospondin motif of aggrecanase-1 (ADAMTS-4) is critical for aggrecan substrate recognition and cleavage. J Biol Chem. 2000;275:25791–25797. doi: 10.1074/jbc.M001065200. [DOI] [PubMed] [Google Scholar]
- Troeberg L, Fushimi K, Khokha R, Emonard H, Ghosh P, Nagase H. Calcium pentosan polysulfate is a multifaceted exosite inhibitor of aggrecanases. FASEB J. 2008;22:3515–3524. doi: 10.1096/fj.08-112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troeberg L, Tanaka M, Wait R, Shi YE, Brew K, Nagase H. E. coli expression of TIMP-4 and comparative kinetic studies with TIMP-1 and TIMP-2: insights into the interactions of TIMPs and matrix metalloproteinase 2 (gelatinase A) Biochemistry. 2002;41:15025–15035. doi: 10.1021/bi026454l. [DOI] [PubMed] [Google Scholar]
- Wayne GJ, Deng SJ, Amour A, Borman S, Matico R, Carter HL, Murphy G. TIMP-3 inhibition of ADAMTS-4 (Aggrecanase-1) is modulated by interactions between aggrecan and the C-terminal domain of ADAMTS-4. J Biol Chem. 2007;282:20991–20998. doi: 10.1074/jbc.M610721200. [DOI] [PubMed] [Google Scholar]
- Wisniewska M, Goettig P, Maskos K, Belouski E, Winters D, Hecht R, Black R, Bode W. Structural determinants of the ADAM inhibition by TIMP-3: crystal structure of the TACE-N-TIMP-3 complex. J Mol Biol. 2008;381:1307–1319. doi: 10.1016/j.jmb.2008.06.088. [DOI] [PubMed] [Google Scholar]
- Yu WH, Yu S, Meng Q, Brew K, Woessner JF. TIMP-3 binds to sulfated glycosaminoglycans of the extracellular matrix. J Biol Chem. 2000;275:31226–31232. doi: 10.1074/jbc.M000907200. [DOI] [PubMed] [Google Scholar]