Abstract
Mucins are high-molecular-weight glycoproteins and are implicated in diverse biological functions. MUC4, a member of transmembrane mucin family, is expressed in airway epithelial cells and body fluids like saliva, tear film, ear fluid, and breast milk. In addition to its normal expression, an aberrant expression of MUC4 has been reported in a variety of carcinomas. Among various potential domains of MUC4, epidermal growth factor (EGF) -like domains are hypothesized to interact with and activate the ErbB2 receptors, suggesting an intramembrane-growth factor function for MUC4. The heavily glycosylated tandem repeat domain provides the structural rigidity to the extended extracellular region. MUC4, by virtue of its extended structure, serves as a barrier for some cell-cell and cell-extracellular matrix interactions and as a potential reservoir for certain growth factors. An intricate relationship between MUC4 and growth factor signaling is also reflected in the transcriptional regulation of MUC4. The MUC4 promoter has binding sites for different transcription factors, which are responsible for the regulation of its expression in different tissues. The interferon-γ, retinoic acid, and transforming growth factor-β signaling pathways regulate MUC4 expression in a partially interdependent manner. Taken together, all of these features of MUC4 strongly support its role as a potential candidate for diagnostic and therapeutic applications in cancer and other diseases.
Keywords: cell-cell matrix interactions, cell-extracellular matrix interactions, metastasis, receptor tyrosine kinases, HER2, cell signaling, tumor microenvironment
Mucins are high-molecular-weight glycoproteins expressed by the epithelial cells and in some cases by the endothelial cells (1). The major function of mucins is to protect and lubricate the ducts and the lumens within the human body (1, 2). In addition, mucins are involved in the differentiation and renewal of the epithelium and modulation of cell adhesion, immune response, and cell signaling (2-4). Mucins contain a polymorphic central domain, which is composed of a variable number of tandem repeats (VNTRs) rich in serine, threonine, and proline residues, a hallmark of the mucin family (2, 5-7). The biochemical and biophysical properties of mucins are largely governed by the extent and the nature of their glycosylation (8, 9). N-acetylgalactosamine is linked to a serine or threonine residue during the O-glycan chain synthesis and is present in all of the mucins. The subsequent carbohydrate moieties, however, may vary depending on the mucin type, the site of mucin expression, and the physiological or pathological conditions (10, 11). The glycosylation of mucins provides a potential basis for tissue-specific interactions with the milieu. By their characteristic pattern of glycosylation, mucins can modulate immunological response, can facilitate cell adhesion during tumor metastasis, and can also alter the functions of proteins interacting with the mucin carbohydrate moieties (12, 13).
On the basis of their structural properties, mucins are classified into three different categories: the gelforming/secreted mucins, the soluble mucins, and the membrane-bound mucins. Each mucin type has a characteristic structure-function relationship in a given tissue environment. The secreted mucins are exclusively expressed by specialized epithelial cells (secretory epithelium) and exhibit a restricted pattern of expression, whereas the membrane-bound mucins demonstrate a wide expression pattern within the human body. The main distinction between the members of these classes can be attributed to the presence of a transmembrane domain, which is responsible for anchoring mucins to the plasma membrane (14-16). Table 1 summarizes different types of mucins, their genomic localization, and the unique domains present in the mucin protein backbone. Some of the mucins share a common chromosomal locus. MUC4 and MUC20 are confined on chromosome locus 3q29, while MUC3A, MUC3B, MUC12, and MUC17 are clustered on the chromosome locus 7q22. MUC2, MUC5AC, MUC5B, and MUC6 are clustered together on the 11p15.5 chromosome locus (Table 1). In terms of domain architecture, the gelforming mucins contain various cysteine-rich domains (D domains) located both in the N terminus and the C terminus to the mucin backbone. These D domains are involved in the formation of disulfide-linked oligomers (17, 18). The only soluble mucin known is MUC7, which does not have sequence homology to any of the known mucins (19). The extracellular domain of the membrane-bound mucin family harbors at least one of the three putatively functional domains: the epidermal growth factor (EGF) -like domain; the sea-urchin sperm protein, the enterokinase, and agrin (SEA) module; and the cysteine-rich domain (1, 2, 20). MUC1 and MUC4 membrane-bound mucins have been extensively studied in relation to their normal and pathophysiological functions, differential expression in cancer and other diseases, and regulation. This review focuses on the MUC4 mucin, its structural features, the evolution of its domains, and its functional significance in normal biological processes and malignant states.
TABLE 1. Classification, genomic localization, and unique domains of different MUCs.
| MUC | Type | Location | Tandem repeat domaina | Other domains |
|---|---|---|---|---|
| MUC1 | TM | 1q21 | PDTRPAPGSTAPPAHGVTSA | SEA |
| MUC2 | Gel | 11p15 | PTTTPITTTTTVTPTPTPTGTQT | VWD, CTCK |
| MUC3A | TM | 7q22 | HSTPSFTSSITTTETTS | EGF |
| MUC3B | TM | 7q22 | HSTPSFTSSITTTETTS | EGF |
| MUC4 | TM | 3q29 | TSSASTGHATPLPVTD | EGF |
| MUC5AC | Gel | 11p15 | TTSTTSAP | VWD, CTCK |
| MUC5B | Gel | 11p15 | AT(G/S)STATPSS(T/S)PGT(T/A)(H/W)T(P/L) (P/T)VL(T/S)(T/S)T(A/T)TT(P/T)T |
VWD, CTCK |
| MUC6 | Gel | 11p15.5-p15.4 | SPFSSTGPMTATSFQTTTTYPTPSHPQTTLPTHVP PFSTSLVTPSTGTYITPTHAQMATSASIHSTPT GTIPPPTTLKATGSTHTAPPMTPTTSGTSQAH SSFSTAKSTSLHSHTSSTHHPEVTPTSTTTITP NPTSTPVAHTTSATSSRLPTPFTTHSPPTGS |
CTCK |
| MUC7 | Soluble | 4q13–q21 | TTAAPPTPSATTPAPPSSSAPPG | N.D |
| MUC8 | N.D | 12q24.3 | TSCPRPLQEGTRVANDTSCPRPLQEGTPGSR AAHALSRRGHRVHELPTSSPGGDTGF |
N.D |
| MUC10 | N.D | N.D | PTTDSTTPAPTTK | N.D |
| MUC11 | N.D | 7q22 | SGLSEESTT(S/F)HSSPGSTHTTLSPASTTT | N.D |
| MUC12 | TM | 7q22 | SGLSQESTTFHSSPGST(E/H)TTLSPAS(T/S)TT | EGF and SEA |
| MUC13 | TM | 3q13.3 | TTXXSXSPGSXSPXSTTT | EGF and SEA |
| MUC15 | TM | 11p14.3 | Lack of tandem repeat | N.D |
| MUC16 | TM | 19p13.2 | FNPWSSVPTTSTPGTSTPGTSTVHLATSGTP SSLPGHTAPVPLLIPFTLNFTITNLHYEENM QHPGSRKFNTTERVLQGLLKPLFKSTSVGPL YSGCRLTLLRPEKHGAATGVDAICTLRLDPTGP GLDRERLYWELSQLTNSVTELGPYTLDRDS LYVNG |
SEA |
| MUC17 | TM | 7q22 | LSTTPVASSEASTLSTSPVDTSTPVTNSSPT NSSPTTAEVTSMPTSTAGEGSTPLTNMP |
EGF-like domains, SEA |
| MUC19 | Gel | 12q12 | TTVAPGS | VWC |
| MUC20 | TM | 3q29 | SESSASSDGPHPVITPSRA | N.D |
CTCK, C-terminal cystine knot-like domain; N.D., not determined.
Most common repeat sequences.
MUC4 MUCIN: GENOMIC ORGANIZATION, STRUCTURAL FEATURES, AND SPLICE VARIANTS
MUC4 was cloned from the human tracheobranchial cDNA library and a human pancreatic tumor cell line (16, 21, 22). It was mapped on chromosome 3 in the region q29 (23). The genomic organization of MUC4 and the various domains encoded by different exons are illustrated in Fig. 1. The MUC4 gene contains 26 exons; the first encompasses the 5′-untranslated region (UTR) and codes for the leader peptide, the second exon codes for the large central tandem-repeat domain, and the 24 other exons code for the rest of the extracellular, transmembrane, and carboxyl tail of the MUC4 apoprotein. The exon sizes vary from 65 bp to 22 kb (exon-2 being the largest and polymorphic), while the intron size varies from 94 bp to 16.5 kb (intron-1 being the largest; refs. 24, 25). The size of the tandem repeat region varies from 7.5 to 19 kb due to a VNTR polymorphism (25). The MUC4 mucin is synthesized as a single polypeptide with the protein size varying from 550 to 930 kDa (26) depending on the size of polymorphic central tandem-repeat domain. Its amino-terminal is comprised of a 27 residue signal peptide, followed by three imperfect repetition motifs varying in size from 126 to 130 residues and a unique sequence of 554 residues. These sequences are followed by a central domain comprised of a variable number of 16 residue perfect tandem repetition motif, a unique feature of the human MUC4 mucin. The carboxy-terminal of MUC4 is composed of 12 distinct domains (CT1–CT12; Fig. 1B). Due to its sequence homology with rMuc4 (16), the Gly-Asp-Pro-His (GDPH) site in MUC4 is predicted to be the proteolytic cleavage site, generating two functional subunits (MUC4α and MUC4β; Fig. 1B). MUC4α is the mucin-like subunit, while MUC4β is the transmembrane growth factor-like subunit. The various functional domains present in the MUC4 protein are as follows: a centrally located large tandem repeat (TR) domain, which is followed by nidogen (NIDO) -like, adhesion-associated domain (AMOP; present in MUC4 and other proteins), von Willebrand factor (vWD; type D domain), and three EGF domains (16, 24). The NIDO domain present in MUC4 shows homology to the G1 domain of nidogen, a sulfated glycoprotein and an important constituent of the basement membrane known to be associated with laminin (27, 28). The AMOP domain is rich in cysteine residues that are probably involved in disulfide bridge formation (29). Both of the domains are hypothesized to play important roles in cell-cell interaction and adhesion to the extracellular matrix. The vWD domain, carrying the putative GDPH cleavage site in its N-terminal region, is found in various plasma proteins and is known to be involved in signal transduction events regulating numerous biological processes like cell adhesion, migration, and oligomer formation (30, 31). The vWD domain present in MUC4, however, is not rich in cysteine residues, which is usually the case with other vWD domain-containing proteins (32). The EGF-like domains present in MUC4 contain conserved cysteine residues. These domains are present in a variety of extracellular proteins (33-35). These cysteine rich domains are speculated to serve in homodimerization or oligomerization of the MUC4 mucin with itself as well as with other members of the mucin family. MUC4 has a short cytoplasmic tail of 22 amino acids that contains one tyrosine and three serine residues as potential phosphorylation sites and, hence, may participate in signal transduction events.
Figure 1.
MUC4 structure. A) MUC4 gene is encoded by 26 exons (E1–E26). E1 exon codes for amino-terminal of the protein. E2 is the largest. E2 is also polymorphic and codes for the central domain. Exons E3–E26 code for the carboxyl-terminal of the MUC4 protein, which includes various domains present in MUC4-like NIDO, AMOP, vWD, transmembrane region, and the cytoplasmic tail. B) MUC4 protein is divided in three regions: the N- terminal (NT), the C region (central domain), and the CT region. The C-terminal region codes for 12 domains (CT1–CT12). Different domains present in MUC4 protein are the central large tandem repeat domain, NIDO, AMOP, vWD, and 3 carboxyl-terminal located EGF domains. The MUC4 protein is hypothesized to be cleaved at GDPH, proteolytic site, generating two subunits: MUC4α and MUC4β. MUC4α is a mucin-like subunit that is heavily glycosylated, and MUC4β is a growth factor like subunit due to presence of EGF-like domains. MUC4 is anchored to the cell surface by the transmembrane region. MUC4 has a short cytoplasmic tail of 22 amino acids.
Alternative splicing of the MUC4 transcript has also been observed, which generates a series of splice variants (36). Of the 24 isoforms reported thus far, 22 are named as sv0 (the full-length MUC4) to sv21-MUC4, while the other 2 are named as MUC4/X and MUC4/Y. The majority of splice events occur downstream of the tandem repeat domain generating three distinct families of MUC4 variants: a secreted family comprised of 12 variants, a family of membrane-tethered variants having 5 members, and others that include 2 variants (MUC4/X and MUC4/Y) that are similar to the growth factor like membrane-bound form but lack the tandem repeat domain (24, 37). MUC4 splice variants are expressed in pancreatic carcinomas but not in the normal pancreas (25, 36, 37). Most of these variants lack the AMOP domain of MUC4 (29). The functional significance of the MUC4 splice variants is not yet known due to the lack of their expression data at the protein level. Evaluation of MUC4 splice variant expression at the protein level and understanding the regulation of splicing process will help us to elucidate the specificity of their expression as well as postulate their putative functions.
HOMOLOGY AND DOMAIN CONSERVATION AMONG DIFFERENT SPECIES
The evolutionary studies are helpful in defining the structure-function relationships of a protein and in identifying its putative roles in biological processes. Structural organization of human MUC4 and rat Muc4 shows that both the peptide sequences share >60% similarity (16). MUC4 protein sequences have been reported in humans (NP_060876.3) and mice (NP_536705.2) and predicted in dogs (XP_545147.2), rats (XP_221384.4), and chickens (XP_426704.2). However, some of these documented sequences have poorly characterized tandem-repeat regions. A homology analysis reveals the position of crucial amino acid residues conserved in all orthologues in different domains, including NIDO, AMOP, vWD, EGF-like domains, transmembrane domain (TM), and CT (Fig. 2A). The high degree of sequence similarity in different orthologues suggests that individual domains evolved from common ancestral domains. The sequence-based homology indicates the evolutionary closeness between human and dog, and rat and mouse sequences (Fig. 2B). All the homologues of MUC4 contain a fully conserved putative GDPH proteolytic cleavage site; however, the occurrence of cleavage event at this site has only been demonstrated in rMuc4. The GDPH cleavage occurs early during the transit of rMuc4 to the cell surface generating two subunits, a mucin subunit, ascitic sialo-glycoprotein-1 (ASGP1), and a transmembrane subunit, ASGP2 (38). The highest variability in MUC4 sequence is observed in the central tandem repeat domain across species. The sequence similarities between MUC4 proteins in different vertebrate species suggest that they might share some common functions.
Figure 2.
Evolution of the MUC4 mucin. A) Alignment of amino acid sequences of various protein domains of MUC4 from human, dog, mouse, rat, and chicken. The domains were defined by a simple modular architecture research tool (SMART) domain search and a Prosite search. The alignment was performed by using the ClustalW program of the European Bioinformatic Institute (EBI). “*” Indicates residues are identical in all sequences in the alignment. “:” Indicates conserved substitutions (within the amino acid subgroups). “.” Indicates semiconserved substitutions. According to the standard coloring scheme of EBI, red indicates small and hydrophobic residues. Blue and magenta indicate acidic and basic residues, respectively. Green represents amino acids containing hydroxyl and amine groups, while gray represents other residues. B) Phylogenetic tree of MUC4 orthologues from different species. Evolutionary relatedness of human and dog sequences and rat and mouse sequences was observed by sequence-based alignment. The phylogenetic tree was generated by the EBI ClustalW program by using complete protein sequences of different MUC4 orthologues. The branch lengths indicate the amount of evolutionary change. C) Evolution scheme of different domains of the MUC4 mucin. Each rectangle in the tandem repeat region represents a 16 aa motif that is repeated 146–500 times. The tandem repeat region probably evolved from duplications of Ser-Thr rich regions in ancestral protein A. The NIDO and EGF-like domains evolved from ancestral protein B, which is a common progenitor to the nidogen protein. Similarly, AMOP and vWD domains evolved from ancestral protein C, a common precursor to Susd2 protein. Domain structures of MUC4, nidogen, and Susd2 were analyzed using SMART. Nidogen_G2 = a β-barrel domain of nidogen; EGF_3 = EGF-like domain; THYROG = throglobulin type-1 domain; LDLRB = low-density lipoprotein-receptor class B; SMB_2 = somatomedin B domain; SUSHI = Sushi/SCP/SCR domain.
EVOLUTION OF MUC4 DOMAINS
MUC4 contains important domains like the tandem repeat domain, NIDO, AMOP, vWD, and EGF-like domains. All these domains are hypothesized to evolve from an ancestral protein harboring these domains. The tandem repeat domain varies in size and number in different species and is defined only for human, mouse and rat. The human MUC4 contains tandem repeat of 16 aa, which is repeated from 146–500 times; the mouse Muc4 contains tandem repeat of 124–126 aa, which is repeated only 20–21 times; while the rat muc4 contains tandem repeat of 117–124 aa but repeated only 12 times. The large exon coding for central repetitive domain is highly variable in size. The expansion of tandem repeat may be due to replication slippage, unequal sister chromatid exchanges, and gene conversion during the course of evolution (39). The variable size and number of tandem repeat in different species suggest that the evolution of tandem repeat is due to selective pressure to maintain the Ser and Thr codons corresponding to the O-glycosylation sites and through the internal successive duplications of the repeat region.
The other unique domains present in the MUC4 mucin but not found in other membrane-bound mucins are NIDO, AMOP, and vWD domains. A phylogenetic tree generated by Duraisamy et al. (40) by using the parsimony method using amino acid alignment of different domains of MUC4 revealed the origin of the NIDO domain from an ancestor common to the NIDO protein. The NIDO proteins also contained the EGF-like domains; however, AMOP and vWD were lacking in this protein. In similar studies, phylogenetic trees constructed using the AMOP and vWD domains showed the origin of these domains from an ancestor common to the Sushi-domain containing protein (Susd2; ref. 40). A hypothesized illustration of the evolution of MUC4 domains is indicated in Fig. 2C.
MUC4 EXPRESSION IN DIFFERENT TISSUES AND ORGANS
Mucin genes exhibit a spatiotemporal pattern of expression (41). MUC4 is present on the surface epithelium of the eye, vagina, ectocervix, trachea, and salivary gland, providing lubrication and protection to the ducts and lumen (42-46). It is expressed by a variety of tissue epithelia including simple epithelia, stratified squamous epithelia, and nonkeratinizing epithelia. The expression of both the transmembrane and secretory forms of MUC4 has been characterized in a variety of epithelial cells (47-52). Although the transmembrane forms of the MUC4 mucin are hypothesized to provide localized protection to the cell surface, the secretory forms are thought to be implicated in lubrication and provide broad protection to the luminal surfaces by trapping foreign particles and pathogens. Apart from its normal expression, an aberrant expression of MUC4 has been reported in various malignancies. The specialized expression of the MUC4 mucin at various tissue surfaces and its functions at those surfaces are discussed below.
Normal expression of MUC4
The expression of MUC4 has been reported in many normal epithelial tissues, both during development and in adults (7, 20, 41). MUC4 is the first mucin to be expressed in the lungs, even before organogenesis takes place (47). It is expressed 6.5 wk postgestation followed by MUC1 and MUC2 after 9.5 wk of gestation. Between weeks 8 and 12, MUC4 mRNA is present only in the trachea; however, its expression increases progressively in small bronchi and bronchioles after week 12 (48). In the digestive tract, MUC4 expression has been reported in the epithelial cell linings of the jejunum to colon axis as early as 6.5 wk postgestation (49). MUC4 is also expressed in squamous epithelial cells of the esophagus, and its expression correlates with the stages of squamous cell differentiation (50). In the embryonic stomach, MUC4 mRNA is expressed from 8 wk of gestation, while its expression in the fetal duodenum is not detected at any gestational stage (41, 51). The expression of MUC4 is also not detected in the liver, biliary tract, gallbladder, and pancreas during gestation (41).
Historically, two different types of milk mucins were described; one of the mucins was characterized as MUC1, which is now a well-known component of human fat globule membranes and as a soluble mucin. A second mucin, distinguished by its higher mass, could not be characterized and hence was called MUCX (53). MUC4, which has also been detected in milk and is a large sized mucin, could in fact represent the previously uncharacterized MUCX (54, 55). The presence of MUC4 has also been detected in lactating rat mammary glands, (56), which like MUC1 exists in both membrane-bound and soluble forms. A study performed by Ruvoen-Clouet et al. (54) using the anti-MUC4 monoclonal antibody (8G7) showed the presence of human MUC4 in milk in all the samples irrespective of the secretor character. Human breast milk contains various biologically active components that protect infants against microbes, viruses, and toxins. These include oligosaccharides, fucosylated oligosaccharides, hormones, growth factors, mucin, gangliosides, and endogenous peptides, which are present in secreted milk (53). In the recent study (54), the presence of MUC1 and MUC4 mucins was shown to strongly block the attachment of recombinant NV virus-like particles (rNV VLPs) to their carbohydrate ligands. The role of MUC4 in milk still remains poorly characterized. Nevertheless, MUC1 has been shown to bind to rotavirus and to enteropathogenic strains of Escherichia coli and is considered as an important component of innate immunity (57, 58). Mucins produced by breast milk also inhibit poxvirus activity by aggregating the poxviruses before their entry into host cells (59). It has also been suggested that mucins may inhibit other enveloped viruses such as HIV from entering into the host cells (59). Hence, mucins, including MUC4, in milk may play key roles in protection and immunity.
MUC4 expression is also detected in multiple tissues in adults, including salivary glands, reproductive tract, mammary epithelium, and body fluids like tears and saliva (43, 52, 60-62). MUC4 expression has been observed in salivary glands by immunohistochemical studies (52). A strong expression of MUC4 in striated ductal epithelial cells of parotid and submandibular glands is seen, while a weak staining is observed in serous acinar cells of both these glands. The extracellular portion of MUC4 has been detected in parotid secretions that might have resulted from shedding of membrane MUC4 or may represent alternatively spliced variants (52). MUC4 mRNA is also detected in the conjunctival stratified epithelium and in corneal epithelial cells in which it plays a crucial role in maintaining and stabilizing the tear film (63). In the reproductive tract, MUC4 is extensively expressed along with MUC1 (61). Expression of rMuc4 is reported in the vagina, cervix, and oviduct of the female rat reproductive tract (64). In rat mammary epithelial cells, rMuc4 is found in both soluble and membrane-associated forms (55, 65). Both these forms are also present at the luminal surface of the uterus acting in the same way as Muc1 in hindering the blastocyst implantation (66). However, in a recent study (67), no correlation between MUC4 allelic polymorphism and blastocyst implantation failure in women was observed.
MUC4 expression in malignancies and inflammatory diseases
An aberrant expression of MUC4 has been reported in various cancers and inflammatory diseases. MUC4 is up-regulated in high-grade dysplasia and adenocarcinoma of the esophagus (68). MUC4 is expressed at significantly lower levels in the intestinal epithelial cells in patients with Crohn’s disease (69, 70). Crohn’s disease is an inflammatory bowel condition characterized by mucosal ulcerations damaging the ileum and proximal colon (70, 71). An abnormal expression of MUC4 is observed in gallbladder carcinomas, whereas it remains undetectable under normal circumstances (41). MUC4 expression has also been reported in major and minor salivary gland mucoepidermoid carcinoma (72). High-grade salivary gland tumors have a trend for reduced MUC4 expression in comparison to the low-grade and intermediate-grade tumors. Also, MUC4-expressing salivary gland mucoepidermoid tumors are associated with improved patient survival and a longer time to recurrence as compared with patients whose tumors were diagnosed negative for MUC4 expression (60, 73, 74). MUC4 was reported to be overexpressed in 91.4% (32 of 35) cases of lung adenocarcinoma, while it was not expressed in malignant mesothelioma (n=41; ref. 75). Malignant mesothelioma has mixed morphological features and is often confused by the pleural infiltrations of lung adenocarcinomas. Therefore, MUC4 may be a clinically useful marker for the distinction between the two malignancies and for therapeutic and medicolegal purposes (75). Also, high MUC4 expression correlates with a short disease-free interval and a poor survival rate of small-sized lung adenocarcinomas, suggesting the potential role of MUC4 as a new independent factor for the prediction of disease outcome in lung adenocarcinoma (76).
In addition, MUC4 is known to be aberrantly expressed in pancreatic ductal adenocarcinoma with no detectable expression in the normal pancreas or chronic pancreatitis (20, 77, 78). The de novo expression of MUC4 is observed in precancerous pancreatic intraepithelial neoplasias (PanINs) and increases progressively with the disease advancement according to the pancreatic cancer progression model (78, 79). Changes in MUC4 expression level have also been reported in other inflammatory diseases of the airways like cystic fibrosis (CF) and chronic obstructive pulmonary disease (COPD) in humans (11, 80). An aberrant expression of MUC4 is also detected in human pancreatic tissues from cystic fibrosis patients as compared with the normal pancreas (81). In a recent study (82), transient up-regulation of Muc4 was observed in wood-smoke-treated rats during the recovery phase, suggesting yet another role of MUC4 in the renewal of tracheal epithelium. MUC4 is also aberrantly expressed in epithelial ovarian carcinomas, while exhibiting negligible expression in the normal ovary (83). In contrast to the aforementioned studies that have reported an overexpression of MUC4 in malignant conditions, a recent study (84) in prostate cancer has shown a down-regulation of MUC4 expression in prostate carcinomas as compared with the normal/benign prostrate region. Hence, MUC4 may serve diverse functions in a context-dependent manner. In this regard, aberrant biochemical modifications and alternative splicing of MUC4 may be important and, therefore, need to be examined.
MULTIFACETED FUNCTIONS OF THE MUC4 MUCIN
In general, all mucins play a role in the lubrication of epithelial surfaces and their protection from infections and injuries. Several lines of evidence implicate MUC4 in other important and more sophisticated biological processes as well, such as epithelial cell renewal and differentiation, cell signaling, cell adhesion, and cancer development (26, 82, 85-88).
Role of MUC4 in cancer cell signaling
In cancerous cells, MUC4 may gain novel functions due to its aberrant expression and biochemical modifications together with changes in cell polarity. These changes may allow MUC4 to interact with proteins that are otherwise cytoarchitecturally segregated. In our recent studies (26, 88), we have observed important roles of MUC4 in altered cell signaling. MUC4 colocalizes and physically interacts with the receptor tyrosine kinase HER2 in pancreatic cancer cells (unpublished data). HER2 is a member of the epidermal growth factor receptor (EGFR) family, which includes three other members, ErbB1 (also called as EGFR or HER1), ErbB2 (also called HER2), ErbB3 (or HER3), and ErbB4 (or HER4) (89). HER2 is often referred as an orphan receptor, as it does not have any soluble ligand. It, however, serves as a preferred heterodimerization partner for other members of the EGFR family (89, 90). An overexpression of HER2 is observed in many malignancies, where it activates intracellular signaling cascades responsible for cell proliferation, angiogenesis, metastatic spread, inhibition of apoptosis, and other processes important in cancer development (89, 91). These activation responses are mediated by the downstream effector molecules, including Ras, Raf, mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K), Akt, phospholipase-C, Rho, and signal transducer and activator of transcription (STATs; refs. 92-94). The rat orthologue of MUC4 (rMuc4) has been shown to specifically interact with ErbB2/HER2/neu via EGF-like domain in the transmembrane subunit (Fig. 3). The transmembrane subunit of human MUC4 also contains three EGF-like domains. The EGF-like domains of both rMuc4 and MUC4 have conserved residues similar to those of other EGF-like domains that are capable of binding and activating receptor tyrosine kinases of the ErbB-family. It is, however, not yet clear whether the complex formation of human MUC4 with HER2 is mediated by one of the EGF-like domain. The interaction of rMuc4 with ErbB2 leads to the phosphorylation of ErbB2/neu at a carboxyl terminal tyrosine residue (Tyr1248), which is implicated in cell transformation (95, 96). A recent study has shown that rMuc4 also facilitates phosphorylation of ErbB2 on tyrosine 1139, which being a Grb2-binding site recruits Grb2 to the apically localized ErbB2 receptor (97). The recruitment of Grb2 leads to phosphorylation of p38, which subsequently activates Akt. Studies in our MUC4-knock-down model have also revealed that MUC4 down-regulation correlates with the suppression of extracellular regulated kinase (ERK) signaling pathway (unpublished data). However, in contrast to observations in rMuc4, we found that the inhibition of MUC4 led to a decrease in the expression of HER2 and its active (pY1248-HER2) form (26). We hypothesize that MUC4 stabilizes HER2 either by decreasing the endocytosis or by increasing the recycling of the activated receptor. In fact, in a recent study (98), a similar mechanism for MUC1-mediated ErbB1/HER1 has been reported.
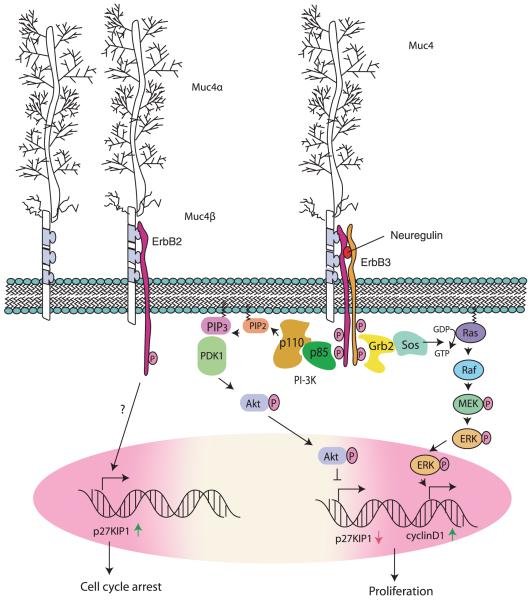
Figure 3.
rMuc4 and ErbB signaling. The interaction of rMuc4 with ErbB2 induces its limited phosphorylation and an up-regulation of p27Kip1, leading to cell cycle arrest. In the presence of neuregulin, rMuc4 is engaged in a quad-complex with ErbB2, ErbB3, and neuregulin and potentiate phosphorylation of ErbB2 leading to an Akt-mediated survival (via down-regulation of p27Kip1) and an ERK-mediated proliferative response. MEK = MAP-ERK kinase; PDK = phosphoinositide-dependent kinase; PIP = phosphatidylinositol phosphate.
Intriguingly, the interaction of rMuc4 with ErbB2 leads to context-dependent epithelial differentiation or cell proliferation. The binding of Muc4 alone with ErbB2 was shown to induce the limited phosphorylation of ErbB2 that leads to an up-regulation of the cyclin-dependent kinase inhibitor p27kip and did not activate mitogen-activated protein kinase or protein kinase B/Akt pathways (96). In contrast, in the presence of neuregulin, activation of both ErbB3 (neuregulin receptor) and ErbB2 was observed, which was further potentiated by rMuc4 and led to the down-regulation of the p27kip and an enhanced activation of ERK and Akt pathways. These changes in signaling pathways facilitated the cell cycle progression (96, 99). Another important finding was made in polarized epithelial cells where exogenous expression of rMuc4 resulted in the localization of ErbB2 from the basolateral to the apical surface (87, 97, 100). Furthermore, in a recent study (101), rMuc4 was also shown to enhance surface accumulation of ErbB2 and ErbB3 by suppressing their internalization. Thus, MUC4 plays important roles in cell signaling and can have differential effect on cell proliferation and differentiation.
Role of MUC4 in tumor growth and metastasis
An aberrant overexpression of MUC4 in a variety of carcinomas indicates its important role in the process of malignant progression. To understand the functional significance of MUC4 in pancreatic cancer, we silenced its expression in a MUC4 overexpressing pancreatic tumor cell line, CD18/HPAF. The resulting phenotype exhibited reduced tumor growth and metastasis (26). In subsequent studies (88), we showed that the MUC4-associated increase in tumor cell growth resulted from an enhanced proliferation and reduced apoptosis of pancreatic cancer cells. In other studies (102, 103), rMuc4 was also shown to potentiate tumorigenesis and metastasis via multiple mechanisms. Metastasis of the tumor cell is a multistep process that involves the detachment of tumor cells from the primary site, intravasation into the bloodstream, evasion of immune surveillance, adherence to vascular endothelial cells of distant organs, and finally extravasation into such tissues and subsequent secondary tumor formation. In the following subsections, we describe multiple attributes of MUC4 that can affect the metastatic progression of cancer cells.
MUC4 in adhesion and antiadhesion
Due to a high level of glycosylation, mucins attain an extended filamentous conformation (11, 104). MUC4 is a high-molecular-weight glycoprotein that is comprised of a heavily glycosylated large tandem-repeat region. The extracellular domain of MUC4 is predicted to extend >2 μm above the cell surface in the apical region (20). During the oncogenic transformation, cells lose their polarity, allowing MUC4 to be uniformly expressed all over the cell surface and interfere with the cell-cell interactions. In a similar way, MUC4 can also sterically reduce the accessibility of extracellular matrix ligands to the corresponding receptors. In a recent study (88), we observed enhanced tumor cell-extracellular matrix interactions after abrogation of MUC4 expression. MUC4 also reduced binding of the integrins (α2, α3, and α5) to their respective antibodies, suggesting the role of MUC4 in masking the accessibility of integrins to respective ligands (88). The antiadhesive effect (reduced cell-cell and cell-extracellular matrix interactions) of rMuc4 has also been demonstrated in A375 human melanoma cells, the extent of which was directly associated with the number of tandem repeats (105).
There is no direct evidence in the literature for the adhesive function of MUC4. Glycosylated mucins expressed on the tumor cell surface, however, can help in the adhesion of tumor cells to secondary sites via selectin-mediated mechanisms and hence promote metastasis (1). Moreover, it is also possible that MUC4 itself can establish novel interactions with extracellular matrix proteins through not yet functionally characterized AMOP and NIDO domains. We have preliminary evidence that MUC4 interacts with galectin-3 (unpublished data), which is ubiquitously present with in the cells, in the extracellular matrix, and on the endothelial surfaces (54, 106). Whether MUC4 can sequester various growth factors and cytokines and facilitate their binding to the corresponding receptors is yet to be established.
Resistance to apoptosis
Not all tumor cells that succeed to invade through the extracellular matrix and reach the bloodstream are able to thrive during the metastatic process. In fact, an appropriate cell-matrix interaction is imperative for cell survival. When this system is perturbed, cells undergo apoptotic cell death, a mechanism referred to as “anoikis.” In a normal cell, integrin-mediated cell signaling controls the apoptotic machinery. Cancer cells, however, develop resistance to apoptosis by alternate signaling mechanisms and are able to self-survive during the metastatic process. MUC4 overexpression has been shown to confer apoptotic resistance to tumor cells (88, 107). The rapid tumor growth in response to rMuc4 overexpression was significantly associated with decreased apoptosis in vitro (107). This indicates that rMuc4 can directly influence the ability of cells to undergo apoptosis without any requirement of crosstalk with the extracellular matrix, which is largely absent in any in vitro cell culture system. Similar evidence has been obtained for human MUC4 in an in vitro assay (88). The mechanism of suppression of apoptosis by MUC4 is not yet defined. It is possible that interaction of the MUC4 with ErbB2/HER2 plays a role in anoikis resistance. In fact, studies with rMuc4 have shown that the Muc4-ErbB2/neu interaction potentiates Akt-signaling, which is known to facilitate cell survival (95). However, there are studies that are not consistent with the role of MUC4 in apoptotic resistance (96, 108). MUC4 expression was shown to be enhanced in high-grade intraepithelial neoplasia (HGN) in Barrett’s esophagus (BO). In fact, MUC4 expression correlated with increased apoptosis (108). A role of rMuc4 in apoptosis was also demonstrated by up-regulating the expression of the cell cycle inhibitor p27kip (96). This up-regulation of p27kip was reversed in the presence of neuregulin, a ligand for ErbB3. Hence, the role of MUC4 in resistance to apoptotic cell death is seemingly regulated by a concerted signaling network that not only involves MUC4 and ErbB2 but also implicates other signaling components that are altered in a cancer cell.
Role in the reversal of contact inhibition
Epithelial cell proliferation is controlled by contact inhibition, a mechanism that is consistently active in normal cells to limit their proliferation (109-112). Cancer or even benign tumor cells, however, bypass this growth control mechanism and continue to proliferate, leading to the formation of a tumor mass (113). Cell-cell interactions are important in normal functioning of the growth control mechanisms, which are lost during malignant transformation. E-cadherins, which mediate cell-cell contact and associated catenins that are involved in modulation of the signaling machinery, are important in regulating cell proliferation (114). E-cadherin acts as an adhesion-activated cell receptor and thus participates in the cell signaling. The blockage of cell-cell interactions, however, can perturb this signaling. In a recent study (99), Muc4 was shown to abrogate the contact inhibition of breast cancer cells and this effect was correlated with the size of the tandem-repeat domain. rMuc4 directed the distribution of E-cadherin from the lateral surface of the cells to the apical cell surface and an activation of ERK leading to an enhanced expression of cyclinD1 (99). This study added another dimension to the antiadhesive effect of MUC4 that allows indirect changes in cell signaling. This finding may be significantly important in explaining the role of MUC4 in malignant progression by triggering the proliferation of contact-inhibited cancer cells, while simultaneously disrupting the cell-cell contact.
Suppression of immune surveillance
The immune system poses another barrier to combat the process of metastasis. Tumor cells must evade immune recognition and subsequent destruction to continue their malignant progression. The recognition of the tumor cells to the immune cells is made possible by the surface molecules present on the tumor cells. The presence of large proteins, such as mucins, on the surface of the tumor cells can mask these surface epitopes, and hence, tumor cells are able to dodge the immune surveillance (115-117). The overexpression of rMuc4 on tumor cells was shown to reduce the accessibility of the surface antigen to the cytotoxic immune cells such as cytotoxic-T lymphocytes or natural killer (NK) cells by steric-hindrance (102). The length of the tandem-repeat domain and the extent of glycosylation were important in the effective suppression of immune surveillance (102). It can be postulated that the human MUC4 can also exert a similar antirecognition effect and to a far greater extent than rMuc4 due to its substantially larger size.
REGULATION OF MUC4 EXPRESSION
Considering the multiple and diverse functions of MUC4 under normal and pathophysiological conditions, it has become important to define the molecular mechanisms that control its expression. The expression of MUC4 can be regulated at both the transcriptional and posttranscriptional levels (81, 118). Studies (118) on the MUC4 promoter have identified two highly active regions referred to as proximal and distal minimal promoters. These are located within a 2.8 kb 5′-UTR. The proximal promoter (−219/−1) does not contain a TATA-box, while it is present in the distal promoter (−2781/−2572). Four transcriptional initiation sites (3 with in the distal promoter at positions −2603, −2604, and −2605 and 1 in the proximal promoter at position −199) have been characterized. Various regulatory elements are present with in the 5′-UTR (118). A schematic illustration of various transcription factor binding elements in the MUC4 promoter is shown in Fig. 4A. The 5′-UTR is rich in GC-content at its 3′-end and contains various putative binding sites for specificity protein 1 (Sp1), activator protein (AP) −1/−2/−4, cAMP-response element binding (CREB) proteins, GATA, and STAT transcription factors. Sp1 and Sp3 are important regulators for the basal expression of MUC4 and are responsible, to some extent, for the expression of MUC4 in a variety of cell types (118). A large number of biologically active molecules, such as cytokines, bacterial products, growth factors, differentiation agents, and other factors, have been shown to regulate MUC4 synthesis (in vitro and/or in vivo) in various cell types (Fig. 4B; refs. 118-121).
Figure 4.
Regulation of MUC4 expression. A) Schematic of MUC4 promoter depicting the presence of various transcription factor binding sites AP-1, growth response element (GRE), hepatocyte nuclear factor 1α (HNF1α), STAT, retinoic acid receptor (RAR), SMAD, nuclear factor-κB (κB), retinoid receptor (RXR), and Sp1. The expression of MUC4 can be driven by an active proximal and a distal promoter region. The proximal promoter does not contain a TATA box, while it is present in the distal promoter region. B) Expression of MUC4 is transcriptionally regulated by a variety of growth factors, cytokines, and other components (IFNγ, RA, TGFβ, interleukins, and bile acids). IFNγ-induced expression of MUC4 is mediated by a novel mechanism that involves an up-regulation of STAT1. Retinoic acid-induced expression of MUC4 is mediated by TGFβ2. Induction of MUC4 expression by TGFβ occurs via both SMAD-dependent and -independent pathways. TGFβ-stimulation activates SMAD2/3, which in complex with SMAD4 gets translocated to the nucleus and facilitates the transcription of MUC4. In addition, TGFβ can drive MUC4 expression via MAPK, PI3K, and PKA pathways. Interleukins are known to regulate MUC4 expression via the STAT6 pathway in airway epithelial cells. Bile acids up-regulate MUC4 expression in esophageal cancer cells by involving the HNF1α transcription factor. RBP = retinoic acid binding protein; TCDC = taurochenodeoxycholate; TDC = taurodeoxycholate.
In our studies that focused on investigating the molecular mechanisms of MUC4 dysregulation in pancreatic cancer cells, we have identified two biomolecules, retinoic acid and interferon-γ (IFNγ), as regulators of MUC4. The expression of MUC4 is regulated by all trans-retinoic acid (ATRA) via a mechanism that implicates transforming growth factor-β2 (TGFβ2) as an interim mediator (Fig. 4B; 122). In another study (123), it was revealed that TGFβ could induce MUC4 expression via both Sma- and Mad-related protein (Smad) -dependent and Smad-independent pathways in pancreatic cancer cells. This activation is, however, negatively regulated by Smad7 and c-ski, both of which inhibit the activation of Smad4. Interestingly, in cells that have mutations in Smad4, such as CAPAN-1, TGFβ-induced expression of MUC4 could be regulated by MAPK, PI3K, and protein kinase A (PKA) signaling cascades (123). Regulation of MUC4 expression by IFNγ occurs via a STAT-1-dependent late response mechanism that requires an up-regulation of STAT-1. Furthermore, it has been shown that both retinoic acid (RA) and IFNγ synergistically up-regulate MUC4 expression in pancreatic cancer cells (120). The synergistic effect of RA and IFNγ involves a reprogramming of signaling pathways. The expression of MUC4 has also been shown to be regulated by tumor necrosis factor-α (TNFα), a proinflammatory cytokine, in synergism with other regulatory factors (118). The synergy between IFNγ and TNFα activates the STATs and the NF-κB transcription factors that on interaction with their cognate cis-element in the MUC4 promoter up-regulate its expression (118).
An increase in the MUC4 expression and the overall number of mucin-producing goblet cells is well reported in Barrett’s esophageal premalignant lesions (108). Barrett’s esophagus is caused by chronic reflux of bile acid from the stomach into the esophagus that was recently shown to up-regulate the expression of MUC4 at the transcriptional level (121). The bile acidmediated MUC4 up-regulation was rescued by the PI3K pathway inhibitor wartmannin, suggesting the role of PI3K signaling mechanisms in bile acid-mediated MUC4 regulation. Also, MUC4 expression by bile acids has been shown to be mediated by the HNF1α transcription factor that binds to the distal promoter region of MUC4 (124). Apart from the aforementioned pathways, interleukins (IL-4, IL-9) are also known to regulate MUC4 in respiratory epithelial cells in a time- and dose-dependent manner. Such up-regulation of MUC4 by different interleukins is mediated through the Janus kinase 3 (JAK3) pathway (125, 126). Furthermore, the IL-4-dependent increase in MUC4 expression involves the activation of the STAT-6 transcription factor (126).
CONCLUSION AND PERSPECTIVES
Based on the knowledge that we have gained on MUC4 so far, it is reasonable to assign it as a multifunctional protein (Fig. 5). Studies on the rat orthologue of MUC4 (95, 96) have indicated that MUC4 may perform diverse biological functions in a context-dependent manner (i.e., its role can differ significantly under normal and pathological conditions). Under normal conditions, there is a controlled interaction between neighboring cells and the cell and the extracellular matrix. During cancer development such interactions are interrupted, while novel interactions occur due to alterations in the cell surface proteins, extracellular matrix composition, and loss of cell polarity. All these molecular and cytoarchitectural changes create a favorable environment for tumor progression. MUC4 has many unique domains that suggest possible functions associated with growth factor signaling (via EGF-like domains in the membrane-bound MUC4β) and tumor cell interaction with the extracellular matrix (via NIDO and AMOP domains present the C-terminal region of the MUC4α-subunit). In addition, MUC4 can also modulate the function of other adhesion-associated signaling molecules via steric hindrance. It has been shown that rMuc4 physically interacts with the receptor tyrosine kinase ErbB2/neu, induces receptor phosphorylation, and potentiates downstream signaling. On the other hand, our recent studies (unpublished data) have demonstrated that MUC4 interacts with HER2/ErbB2 and modulates its expression. Therefore, it is imperative to understand the molecular mechanisms implicated in the MUC4-associated regulation of HER2 and to understand the biological significance of the MUC4-HER2 interaction.
Figure 5.

Multiple roles of MUC4 mucin in cancer development. MUC4 is engaged in complex formation with ErbB2 and/or ErbB3 (in presence of neuregulin) and initiates and/or potentiates downstream signaling and facilitates cell proliferation and cell survival. The large sized extracellular domain of MUC4 disrupts cell-cell and cell-extracellular matrix interactions via steric hindrance. MUC4 also increases cell motility via yet unknown mechanisms. The sialyl epitopes present on the heavily glycosylated tandem repeat domain of MUC4 may facilitate trans-interactions by binding to selectins or yet uncharacterized ligands on endothelial cells. The presence of MUC4 on the surface of the tumor cells can mask the surface epitopes to the cytotoxic immune cells such as cytotoxic-T lymphocytes or NK cells and, hence, escape from immune response.
There is a need to expedite efforts on studies related to the differential glycosylation and alternative splicing of MUC4 to understand its functional diversity. In addition, the MUC4 interactome needs to be explored to comprehend what defines the efficiency and specificity of MUC4 action in normal and pathophysiological conditions. The role of individual domains of MUC4 in initiating and/or potentiating cancer cell signaling is still being investigated. Once the cascades of signaling events activated by MUC4 are identified, the blockage of the pathways by using inhibitors may be helpful in controlling cancer progression. Also, delineating the signaling mechanisms implicated in the aberrant expression of MUC4 in cancer and defining the specific functions of MUC4 will lead to the formulations of better therapeutic strategies for the treatment of cancer.
Acknowledgments
This work was supported, in part, by grants from the U.S. National Institutes of Health (RO1 CA-78590 and C-A111294) and the U.S. Department of Defense (DOD OC040110 and PC040502). We also thank Kristi L.W. Berger (Eppley Institute, University of Nebraska Medical Center) for editorial assistance.
REFERENCES
- 1.Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat. Rev. Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 2.Moniaux N, Escande F, Porchet N, Aubert JP, Batra SK. Structural organization and classification of the human mucin genes. Front. Biosci. 2001;6:D1192–1206. doi: 10.2741/moniaux. [DOI] [PubMed] [Google Scholar]
- 3.Satoh S, Hinoda Y, Hayashi T, Burdick MD, Imai K, Hollingsworth MA. Enhancement of metastatic properties of pancreatic cancer cells by MUC1 gene encoding an anti-adhesion molecule. Int. J. Cancer. 2000;88:507–518. doi: 10.1002/1097-0215(20001115)88:4<507::aid-ijc1>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 4.Wesseling J, van der Valk SW, Vos HL, Sonnenberg A, Hilkens J. Episialin (MUC1) overexpression inhibits integrin-mediated cell adhesion to extracellular matrix components. J. Cell Biol. 1995;129:255–265. doi: 10.1083/jcb.129.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strous GJ, Dekker J. Mucin-type glycoproteins. Crit. Rev. Biochem. Mol. Biol. 1992;27:57–92. doi: 10.3109/10409239209082559. [DOI] [PubMed] [Google Scholar]
- 6.Corfield AP, Myerscough N, Gough M, Brockhausen I, Schauer R, Paraskeva C. Glycosylation patterns of mucins in colonic disease. Biochem. Soc. Trans. 1995;23:840–845. doi: 10.1042/bst0230840. [DOI] [PubMed] [Google Scholar]
- 7.Gendler SJ, Spicer AP. Epithelial mucin genes. Annu. Rev. Physiol. 1995;57:607–634. doi: 10.1146/annurev.ph.57.030195.003135. [DOI] [PubMed] [Google Scholar]
- 8.Carraway KL, Hull SR. O-glycosylation pathway for mucin-type glycoproteins. Bioessays. 1989;10:117–121. doi: 10.1002/bies.950100406. [DOI] [PubMed] [Google Scholar]
- 9.Hanisch FG. O-glycosylation of the mucin type. Biol. Chem. 2001;382:143–149. doi: 10.1515/BC.2001.022. [DOI] [PubMed] [Google Scholar]
- 10.Brockhausen I. Pathways of O-glycan biosynthesis in cancer cells. Biochim. Biophys. Acta. 1999;1473:67–95. doi: 10.1016/s0304-4165(99)00170-1. [DOI] [PubMed] [Google Scholar]
- 11.Lamblin G, Degroote S, Perini JM, Delmotte P, Scharfman A, Davril M, Lo-Guidice JM, Houdret N, Dumur V, Klein A, Rousse P. Human airway mucin glycosylation: a combinatory of carbohydrate determinants which vary in cystic fibrosis. Glycoconj. J. 2001;18:661–684. doi: 10.1023/a:1020867221861. [DOI] [PubMed] [Google Scholar]
- 12.Fukuda M. Possible roles of tumor-associated carbohydrate antigens. Cancer Res. 1996;56:2237–2244. [PubMed] [Google Scholar]
- 13.Fukuda M. Roles of mucin-type O-glycans in cell adhesion. Biochim. Biophys. Acta. 2002;1573:394–405. doi: 10.1016/s0304-4165(02)00409-9. [DOI] [PubMed] [Google Scholar]
- 14.Gendler SJ, Burchell JM, Duhig T, Lamport D, White R, Parker M, Taylor-Papadimitriou J. Cloning of partial cDNA encoding differentiation and tumor-associated mucin glycoproteins expressed by human mammary epithelium. Proc. Natl. Acad. Sci. U. S. A. 1987;84:6060–6064. doi: 10.1073/pnas.84.17.6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gendler SJ, Lancaster CA, Taylor-Papadimitriou J, Duhig T, Peat N, Burchell J, Pemberton L, Lalani EN, Wilson D. Molecular cloning and expression of human tumor-associated polymorphic epithelial mucin. J. Biol. Chem. 1990;265:15286–15293. [PubMed] [Google Scholar]
- 16.Moniaux N, Nollet S, Porchet N, Degand P, Laine A, Aubert JP. Complete sequence of the human mucin MUC4: a putative cell membrane-associated mucin. Biochem. J. 1999;338:325–333. [PMC free article] [PubMed] [Google Scholar]
- 17.Asker N, Axelsson MA, Olofsson SO, Hansson GC. Dimerization of the human MUC2 mucin in the endoplasmic reticulum is followed by a N-glycosylation-dependent transfer of the mono- and dimers to the Golgi apparatus. J. Biol. Chem. 1998;273:18857–18863. doi: 10.1074/jbc.273.30.18857. [DOI] [PubMed] [Google Scholar]
- 18.Godl K, Johansson ME, Lidell ME, Morgelin M, Karlsson H, Olson FJ, Gum JR, Jr., Kim YS, Hansson GC. The N terminus of the MUC2 mucin forms trimers that are held together within a trypsin-resistant core fragment. J. Biol. Chem. 2002;277:47248–47256. doi: 10.1074/jbc.M208483200. [DOI] [PubMed] [Google Scholar]
- 19.Bobek LA, Tsai H, Biesbrock AR, Levine MJ. Molecular cloning, sequence, and specificity of expression of the gene encoding the low molecular weight human salivary mucin (MUC7) J. Biol. Chem. 1993;268:20563–20569. [PubMed] [Google Scholar]
- 20.Moniaux N, Andrianifahanana M, Brand RE, Batra SK. Multiple roles of mucins in pancreatic cancer, a lethal and challenging malignancy. Br. J. Cancer. 2004;91:1633–1638. doi: 10.1038/sj.bjc.6602163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khorrami AM, Choudhury A, Andrianifahanana M, Varshney GC, Bhattacharyya SN, Hollingsworth MA, Kaufman B, Batra SK. Purification and characterization of a human pancreatic adenocarcinoma mucin. J. Biochem. (Tokyo) 2002;131:21–29. doi: 10.1093/oxfordjournals.jbchem.a003073. [DOI] [PubMed] [Google Scholar]
- 22.Porchet N, Nguyen VC, Dufosse J, Audie JP, Guyonnet-Duperat V, Gross MS, Denis C, Degand P, Bernheim A, Aubert JP. Molecular cloning and chromosomal localization of a novel human tracheo-bronchial mucin cDNA containing tandemly repeated sequences of 48 base pairs. Biochem. Biophys. Res. Commun. 1991;175:414–422. doi: 10.1016/0006-291x(91)91580-6. [DOI] [PubMed] [Google Scholar]
- 23.Gross MS, Guyonnet-Duperat V, Porchet N, Bernheim A, Aubert JP, Nguyen VC. Mucin 4 (MUC4) gene: regional assignment (3q29) and RFLP analysis. Ann. Genet. 1992;35:21–26. [PubMed] [Google Scholar]
- 24.Escande F, Lemaitre L, Moniaux N, Batra SK, Aubert JP, Buisine MP. Genomic organization of MUC4 mucin gene. Towards the characterization of splice variants. Eur. J. Biochem. 2002;269:3637–3644. doi: 10.1046/j.1432-1033.2002.03032.x. [DOI] [PubMed] [Google Scholar]
- 25.Moniaux N, Escande F, Batra SK, Porchet N, Laine A, Aubert JP. Alternative splicing generates a family of putative secreted and membrane-associated MUC4 mucins. Eur. J. Biochem. 2000;267:4536–4544. doi: 10.1046/j.1432-1327.2000.01504.x. [DOI] [PubMed] [Google Scholar]
- 26.Singh AP, Moniaux N, Chauhan SC, Meza JL, Batra SK. Inhibition of MUC4 expression suppresses pancreatic tumor cell growth and metastasis. Cancer Res. 2004;64:622–630. doi: 10.1158/0008-5472.can-03-2636. [DOI] [PubMed] [Google Scholar]
- 27.Timpl R. Structure and biological activity of basement membrane proteins. Eur. J. Biochem. 1989;180:487–502. doi: 10.1111/j.1432-1033.1989.tb14673.x. [DOI] [PubMed] [Google Scholar]
- 28.Mayer U, Kohfeldt E, Timpl R. Structural and genetic analysis of laminin-nidogen interaction. Ann. N. Y. Acad. Sci. 1998;857:130–142. doi: 10.1111/j.1749-6632.1998.tb10113.x. [DOI] [PubMed] [Google Scholar]
- 29.Ciccarelli FD, Doerks T, Bork P. AMOP, a protein module alternatively spliced in cancer cells. Trends Biochem. Sci. 2002;27:113–115. doi: 10.1016/s0968-0004(01)02049-7. [DOI] [PubMed] [Google Scholar]
- 30.Colombatti A, Bonaldo P, Doliana R. Type A modules: interacting domains found in several non-fibrillar collagens and in other extracellular matrix proteins. Matrix. 1993;13:297–306. doi: 10.1016/s0934-8832(11)80025-9. [DOI] [PubMed] [Google Scholar]
- 31.Ruggeri ZM, Ware J. von Willebrand factor. FASEB J. 1993;7:308–316. doi: 10.1096/fasebj.7.2.8440408. [DOI] [PubMed] [Google Scholar]
- 32.Aksoy N, Akinci OF. Mucin macromolecules in normal, adenomatous, and carcinomatous colon: evidence for the neotransformation. Macromol. Biosci. 2004;4:483–496. doi: 10.1002/mabi.200300099. [DOI] [PubMed] [Google Scholar]
- 33.Hsuan JJ, Panayotou G, Waterfield MD. Structural basis for epidermal growth factor receptor function. Prog. Growth Factor Res. 1989;1:23–32. doi: 10.1016/0955-2235(89)90039-2. [DOI] [PubMed] [Google Scholar]
- 34.Van Zoelen EJ, Stortelers C, Lenferink AE, Van de Poll ML. The EGF domain: requirements for binding to receptors of the ErbB family. Vitam. Horm. 2000;59:99–131. doi: 10.1016/s0083-6729(00)59005-0. [DOI] [PubMed] [Google Scholar]
- 35.McInnes C, Sykes BD. Growth factor receptors: structure, mechanism, and drug discovery. Biopolymers. 1997;43:339–366. doi: 10.1002/(SICI)1097-0282(1997)43:5<339::AID-BIP2>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 36.Choudhury A, Moniaux N, Winpenny JP, Hollingsworth MA, Aubert JP, Batra SK. Human MUC4 mucin cDNA and its variants in pancreatic carcinoma. J. Biochem. (Tokyo) 2000;128:233–243. doi: 10.1093/oxfordjournals.jbchem.a022746. [DOI] [PubMed] [Google Scholar]
- 37.Choudhury A, Moniaux N, Ringel J, King J, Moore E, Aubert JP, Batra SK. Alternate splicing at the 3′-end of the human pancreatic tumor-associated mucin MUC4 cDNA. Teratog. Carcinog. Mutagen. 2001;21:83–96. doi: 10.1002/1520-6866(2001)21:1<83::aid-tcm8>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 38.Komatsu M, Arango ME, Carraway KL. Synthesis and secretion of Muc4/sialomucin complex: implication of intracellular proteolysis. Biochem. J. 2002;368:41–48. doi: 10.1042/BJ20020862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vinall LE, Hill AS, Pigny P, Pratt WS, Toribara N, Gum JR, Kim YS, Porchet N, Aubert JP, Swallow DM. Variable number tandem repeat polymorphism of the mucin genes located in the complex on 11p15.5. Hum. Genet. 1998;102:357–366. doi: 10.1007/s004390050705. [DOI] [PubMed] [Google Scholar]
- 40.Duraisamy S, Ramasamy S, Kharbanda S, Kufe D. Distinct evolution of the human carcinoma-associated transmembrane mucins, MUC1, MUC4 AND MUC16. Gene. 2006;373:28–34. doi: 10.1016/j.gene.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 41.Buisine MP, Devisme L, Degand P, Dieu MC, Gosselin B, Copin MC, Aubert JP, Porchet N. Developmental mucin gene expression in the gastroduodenal tract and accessory digestive glands. II. Duodenum and liver, gallbladder, and pancreas. J. Histochem. Cytochem. 2000;48:1667–1676. doi: 10.1177/002215540004801210. [DOI] [PubMed] [Google Scholar]
- 42.Gipson IK. Mucins of the human endocervix. Front. Biosci. 2001;6:D1245–1255. doi: 10.2741/gipson. [DOI] [PubMed] [Google Scholar]
- 43.Gipson IK. Distribution of mucins at the ocular surface. Exp. Eye Res. 2004;78:379–388. doi: 10.1016/s0014-4835(03)00204-5. [DOI] [PubMed] [Google Scholar]
- 44.Reid CJ, Gould S, Harris A. Developmental expression of mucin genes in the human respiratory tract. Am. J. Respir. Cell Mol. Biol. 1997;17:592–598. doi: 10.1165/ajrcmb.17.5.2798. [DOI] [PubMed] [Google Scholar]
- 45.Liu B, Offner GD, Nunes DP, Oppenheim FG, Troxler RF. MUC4 is a major component of salivary mucin MG1 secreted by the human submandibular gland. Biochem. Biophys. Res. Commun. 1998;250:757–761. doi: 10.1006/bbrc.1998.9390. [DOI] [PubMed] [Google Scholar]
- 46.Juusola J, Ballantyne J. Multiplex mRNA profiling for the identification of body fluids. Forensic Sci. Int. 2005;152:1–12. doi: 10.1016/j.forsciint.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 47.Buisine MP, Devisme L, Copin MC, Durand-Reville M, Gosselin B, Aubert JP, Porchet N. Developmental mucin gene expression in the human respiratory tract. Am. J. Respir. Cell Mol. Biol. 1999;20:209–218. doi: 10.1165/ajrcmb.20.2.3259. [DOI] [PubMed] [Google Scholar]
- 48.Lopez-Ferrer A, Curull V, Barranco C, Garrido M, Lloreta J, Real FX, de Bolos C. Mucins as differentiation markers in bronchial epithelium. Squamous cell carcinoma and adenocarcinoma display similar expression patterns. Am. J. Respir. Cell Mol. Biol. 2001;24:22–29. doi: 10.1165/ajrcmb.24.1.4294. [DOI] [PubMed] [Google Scholar]
- 49.Buisine MP, Devisme L, Savidge TC, Gespach C, Gosselin B, Porchet N, Aubert JP. Mucin gene expression in human embryonic and fetal intestine. Gut. 1998;43:519–524. doi: 10.1136/gut.43.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guillem P, Billeret V, Buisine MP, Flejou JF, Lecomte-Houcke M, Degand P, Aubert JP, Triboulet JP, Porchet N. Mucin gene expression and cell differentiation in human normal, premalignant and malignant esophagus. Int. J. Cancer. 2000;88:856–861. doi: 10.1002/1097-0215(20001215)88:6<856::aid-ijc3>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 51.Buisine MP, Devisme L, Maunoury V, Deschodt E, Gosselin B, Copin MC, Aubert JP, Porchet N. Developmental mucin gene expression in the gastroduodenal tract and accessory digestive glands. I. Stomach. A relationship to gastric carcinoma. J. Histochem. Cytochem. 2000;48:1657–1666. doi: 10.1177/002215540004801209. [DOI] [PubMed] [Google Scholar]
- 52.Liu B, Lague JR, Nunes DP, Toselli P, Oppenheim FG, Soares RV, Troxler RF, Offner GD. Expression of membrane-associated mucins MUC1 and MUC4 in major human salivary glands. J. Histochem. Cytochem. 2002;50:811–820. doi: 10.1177/002215540205000607. [DOI] [PubMed] [Google Scholar]
- 53.Patton S, Gendler SJ, Spicer AP. The epithelial mucin, MUC1, of milk, mammary gland and other tissues. Biochim. Biophys. Acta. 1995;1241:407–423. doi: 10.1016/0304-4157(95)00014-3. [DOI] [PubMed] [Google Scholar]
- 54.Ruvoen-Clouet N, Mas E, Marionneau S, Guillon P, Lombardo D, Le Pendu J. Bile-salt-stimulated lipase and mucins from milk of “secretor” mothers inhibit the binding of Norwalk virus capsids to their carbohydrate ligands. Biochem. J. 2006;393:627–634. doi: 10.1042/BJ20050898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J, Perez A, Yasin M, Soto P, Rong M, Theodoropoulos G, Carothers Carraway CA, Carraway KL. Presence of MUC4 in human milk and at the luminal surfaces of blood vessels. J. Cell. Physiol. 2005;204:166–177. doi: 10.1002/jcp.20277. [DOI] [PubMed] [Google Scholar]
- 56.Price-Schiavi SA, Andrechek E, Idris N, Li P, Rong M, Zhang J, Carothers Carraway CA, Muller WJ, Carraway KL. Expression, location, and interactions of ErbB2 and its intramembrane ligand Muc4 (sialomucin complex) in rat mammary gland during pregnancy. J. Cell. Physiol. 2005;203:44–53. doi: 10.1002/jcp.20200. [DOI] [PubMed] [Google Scholar]
- 57.Yolken RH, Peterson JA, Vonderfecht SL, Fouts ET, Midthun K, Newburg DS. Human milk mucin inhibits rotavirus replication and prevents experimental gastroenteritis. J. Clin. Invest. 1992;90:1984–1991. doi: 10.1172/JCI116078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schroten H, Hanisch FG, Plogmann R, Hacker J, Uhlenbruck G, Nobis-Bosch R, Wahn V. Inhibition of adhesion of S-fimbriated Escherichia coli to buccal epithelial cells by human milk fat globule membrane components: a novel aspect of the protective function of mucins in the nonimmunoglobulin fraction. Infect. Immun. 1992;60:2893–2899. doi: 10.1128/iai.60.7.2893-2899.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Habte HH, Kotwal GJ, Lotz ZE, Tyler MG, Abrahams M, Rodriques J, Kahn D, Mall AS. Antiviral activity of purified human breast milk mucin. Neonatology. 2007;92:96–104. doi: 10.1159/000100808. [DOI] [PubMed] [Google Scholar]
- 60.Alos L, Lujan B, Castillo M, Nadal A, Carreras M, Caballero M, de Bolos C, Cardesa A. Expression of membrane-bound mucins (MUC1 and MUC4) and secreted mucins (MUC2, MUC5AC, MUC5B, MUC6 and MUC7) in mucoepidermoid carcinomas of salivary glands. Am. J. Surg. Pathol. 2005;29:806–813. doi: 10.1097/01.pas.0000155856.84553.c9. [DOI] [PubMed] [Google Scholar]
- 61.Gipson IK, Ho SB, Spurr-Michaud SJ, Tisdale AS, Zhan Q, Torlakovic E, Pudney J, Anderson DJ, Toribara NW, Hill JA., 3rd Mucin genes expressed by human female reproductive tract epithelia. Biol. Reprod. 1997;56:999–1011. doi: 10.1095/biolreprod56.4.999. [DOI] [PubMed] [Google Scholar]
- 62.Watanabe H. Significance of mucin on the ocular surface. Cornea. 2002;21:S17–22. doi: 10.1097/00003226-200203001-00005. [DOI] [PubMed] [Google Scholar]
- 63.Corrales RM, Calonge M, Herreras JM, Saez V, Mayo A, Chaves FJ. Levels of mucin gene expression in normal human conjunctival epithelium in vivo. Curr. Eye Res. 2003;27:323–328. doi: 10.1076/ceyr.27.5.323.17221. [DOI] [PubMed] [Google Scholar]
- 64.Idris N, Carraway KL. Sialomucin complex (Muc4) expression in the rat female reproductive tract. Biol. Reprod. 1999;61:1431–1438. doi: 10.1095/biolreprod61.6.1431. [DOI] [PubMed] [Google Scholar]
- 65.Price-Schiavi SA, Carraway CA, Fregien N, Carraway KL. Post-transcriptional regulation of a milk membrane protein, the sialomucin complex [ascites sialoglycoprotein (ASGP)-1/ASGP-2, rat muc4], by transforming growth factor beta. J. Biol. Chem. 1998;273:35228–35237. doi: 10.1074/jbc.273.52.35228. [DOI] [PubMed] [Google Scholar]
- 66.Carraway KL, Idris N. Regulation of sialomucin complex/Muc4 in the female rat reproductive tract. Biochem. Soc. Trans. 2001;29:162–166. doi: 10.1042/0300-5127:0290162. [DOI] [PubMed] [Google Scholar]
- 67.Koscinski I, Viville S, Porchet N, Bernigaud A, Escande F, Defossez A, Buisine MP. MUC4 gene polymorphism and expression in women with implantation failure. Hum. Reprod. 2006;21:2238–2245. doi: 10.1093/humrep/del189. [DOI] [PubMed] [Google Scholar]
- 68.Arul GS, Moorghen M, Myerscough N, Alderson DA, Spicer RD, Corfield AP. Mucin gene expression in Barrett’s oesophagus: an in situ hybridisation and immunohistochemical study. Gut. 2000;47:753–761. doi: 10.1136/gut.47.6.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buisine MP, Desreumaux P, Debailleul V, Gambiez L, Geboes K, Ectors N, Delescaut MP, Degand P, Aubert JP, Colombel JF, Porchet N. Abnormalities in mucin gene expression in Crohn’s disease. Inflamm. Bowel Dis. 1999;5:24–32. doi: 10.1097/00054725-199902000-00004. [DOI] [PubMed] [Google Scholar]
- 70.Buisine MP, Desreumaux P, Leteurtre E, Copin MC, Colombel JF, Porchet N, Aubert JP. Mucin gene expression in intestinal epithelial cells in Crohn’s disease. Gut. 2001;49:544–551. doi: 10.1136/gut.49.4.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Corfield AP, Carroll D, Myerscough N, Probert CS. Mucins in the gastrointestinal tract in health and disease. Front. Biosci. 2001;6:D1321–1357. doi: 10.2741/corfield. [DOI] [PubMed] [Google Scholar]
- 72.Handra-Luca A, Lamas G, Bertrand JC, Fouret P. MUC1, MUC2, MUC4, and MUC5AC expression in salivary gland mucoepidermoid carcinoma: diagnostic and prognostic implications. Am. J. Surg. Pathol. 2005;29:881–889. doi: 10.1097/01.pas.0000159103.95360.e8. [DOI] [PubMed] [Google Scholar]
- 73.Weed DT, Gomez-Fernandez C, Yasin M, Hamilton-Nelson K, Rodriguez M, Zhang J, Carraway KL. MUC4 and ErbB2 expression in squamous cell carcinoma of the upper aerodigestive tract: correlation with clinical outcomes. Laryngoscope. 2004;114:1–32. doi: 10.1097/00005537-200408001-00001. [DOI] [PubMed] [Google Scholar]
- 74.Weed DT, Gomez-Fernandez C, Pacheco J, Ruiz J, Hamilton-Nelson K, Arnold DJ, Civantos FJ, Zhang J, Yasin M, Goodwin WJ, Carraway KL. MUC4 and ERBB2 expression in major and minor salivary gland mucoepidermoid carcinoma. Head Neck. 2004;26:353–364. doi: 10.1002/hed.10387. [DOI] [PubMed] [Google Scholar]
- 75.Llinares K, Escande F, Aubert S, Buisine MP, de Bolos C, Batra SK, Gosselin B, Aubert JP, Porchet N, Copin MC. Diagnostic value of MUC4 immunostaining in distinguishing epithelial mesothelioma and lung adenocarcinoma. Mod. Pathol. 2004;17:150–157. doi: 10.1038/modpathol.3800027. [DOI] [PubMed] [Google Scholar]
- 76.Tsutsumida H, Goto M, Kitajima S, Kubota I, Hirotsu Y, Wakimoto J, Batra SK, Imai K, Yonezawa S. MUC4 expression correlates with poor prognosis in small-sized lung adenocarcinoma. Lung Cancer. 2007;55:195–203. doi: 10.1016/j.lungcan.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 77.Andrianifahanana M, Moniaux N, Schmied BM, Ringel J, Friess H, Hollingsworth MA, Buchler MW, Aubert JP, Batra SK. Mucin (MUC) gene expression in human pancreatic adenocarcinoma and chronic pancreatitis: a potential role of MUC4 as a tumor marker of diagnostic significance. Clin. Cancer Res. 2001;7:4033–4040. [PubMed] [Google Scholar]
- 78.Swartz MJ, Batra SK, Varshney GC, Hollingsworth MA, Yeo CJ, Cameron JL, Wilentz RE, Hruban RH, Argani P. MUC4 expression increases progres-sively in pancreatic intraepithelial neoplasia. Am. J. Clin. Pathol. 2002;117:791–796. doi: 10.1309/7Y7N-M1WM-R0YK-M2VA. [DOI] [PubMed] [Google Scholar]
- 79.Hruban RH, Goggins M, Parsons J, Kern SE. Progression model for pancreatic cancer. Clin. Cancer Res. 2000;6:2969–2972. [PubMed] [Google Scholar]
- 80.Leikauf GD, Borchers MT, Prows DR, Simpson LG. Mucin apoprotein expression in COPD. Chest. 2002;121:166S–182S. doi: 10.1378/chest.121.5_suppl.166s. [DOI] [PubMed] [Google Scholar]
- 81.Singh AP, Chauhan SC, Andrianifahanana M, Moniaux N, Meza JL, Copin MC, van Seuningen I, Hollingsworth MA, Aubert JP, Batra SK. MUC4 expression is regulated by cystic fibrosis transmembrane conductance regulator in pancreatic adenocarcinoma cells via transcriptional and post-translational mechanisms. Oncogene. 2006;26:30–41. doi: 10.1038/sj.onc.1209764. [DOI] [PubMed] [Google Scholar]
- 82.Bhattacharyya SN, Dubick MA, Yantis LD, Enriquez JI, Buchanan KC, Batra SK, Smiley RA. In vivo effect of wood smoke on the expression of two mucin genes in rat airways. Inflammation. 2004;28:67–76. doi: 10.1023/b:ifla.0000033022.66289.04. [DOI] [PubMed] [Google Scholar]
- 83.Chauhan SC, Singh AP, Ruiz F, Johansson SL, Jain M, Smith LM, Moniaux N, Batra SK. Aberrant expression of MUC4 in ovarian carcinoma: diagnostic significance alone and in combination with MUC1 and MUC16 (CA125) Mod. Pathol. 2006;19:1386–1394. doi: 10.1038/modpathol.3800646. [DOI] [PubMed] [Google Scholar]
- 84.Singh AP, Chauhan SC, Bafna S, Johansson SL, Smith LM, Moniaux N, Lin MF, Batra SK. Aberrant expression of transmembrane mucins, MUC1 and MUC4, in human prostate carcinomas. Prostate. 2006;66:421–429. doi: 10.1002/pros.20372. [DOI] [PubMed] [Google Scholar]
- 85.Singh AP, Chaturvedi P, Batra SK. Emerging roles of MUC4 in cancer: a novel target for diagnosis and therapy. Cancer Res. 2007;67:433–436. doi: 10.1158/0008-5472.CAN-06-3114. [DOI] [PubMed] [Google Scholar]
- 86.Moniaux N, Chaturvedi P, Varshney GC, Meza JL, Rodriguez-Sierra JF, Aubert JP, Batra SK. Human MUC4 mucin induces ultra-structural changes and tumorigenicity in pancreatic cancer cells. Br. J. Cancer. 2007;97:345–357. doi: 10.1038/sj.bjc.6603868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carraway KL, Ramsauer VP, Haq B, Carothers Carraway CA. Cell signaling through membrane mucins. Bioessays. 2003;25:66–71. doi: 10.1002/bies.10201. [DOI] [PubMed] [Google Scholar]
- 88.Chaturvedi P, Singh AP, Moniaux N, Senapati S, Chakraborty S, Meza JL, Batra SK. MUC4 mucin potentiates pancreatic tumor cell proliferation, survival, and invasive properties and interferes with its interaction to extracellular matrix proteins. Mol. Cancer Res. 2007;5:309–320. doi: 10.1158/1541-7786.MCR-06-0353. [DOI] [PubMed] [Google Scholar]
- 89.Warren CM, Landgraf R. Signaling through ERBB receptors: multiple layers of diversity and control. Cell. Signal. 2006;18:923–933. doi: 10.1016/j.cellsig.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 90.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 91.Klos KS, Wyszomierski SL, Sun M, Tan M, Zhou X, Li P, Yang W, Yin G, Hittelman WN, Yu D. ErbB2 increases vascular endothelial growth factor protein synthesis via activation of mammalian target of rapamycin/p70S6K leading to increased angiogenesis and spontaneous metastasis of human breast cancer cells. Cancer Res. 2006;66:2028–2037. doi: 10.1158/0008-5472.CAN-04-4559. [DOI] [PubMed] [Google Scholar]
- 92.Holbro T, Hynes NE. ErbB receptors: directing key signaling networks throughout life. Annu. Rev. Pharmacol. Toxicol. 2004;44:195–217. doi: 10.1146/annurev.pharmtox.44.101802.121440. [DOI] [PubMed] [Google Scholar]
- 93.Firth LC, Baker NE. EGF receptor signaling: a prickly proposition. Curr. Biol. 2003;13:R773–774. doi: 10.1016/j.cub.2003.09.020. [DOI] [PubMed] [Google Scholar]
- 94.Sweeney C, Carraway KL., 3rd Ligand discrimination by ErbB receptors: differential signaling through differential phosphorylation site usage. Oncogene. 2000;19:5568–5573. doi: 10.1038/sj.onc.1203913. [DOI] [PubMed] [Google Scholar]
- 95.Carraway KL, 3rd, Rossi EA, Komatsu M, Price-Schiavi SA, Huang D, Guy PM, Carvajal ME, Fregien N, Carraway CA, Carraway KL. An intramembrane modulator of the ErbB2 receptor tyrosine kinase that potentiates neuregulin signaling. J. Biol. Chem. 1999;274:5263–5266. doi: 10.1074/jbc.274.9.5263. [DOI] [PubMed] [Google Scholar]
- 96.Jepson S, Komatsu M, Haq B, Arango ME, Huang D, Carraway CA, Carraway KL. Muc4/sialomucin complex, the intramembrane ErbB2 ligand, induces specific phosphorylation of ErbB2 and enhances expression of p27(kip), but does not activate mitogen-activated kinase or protein kinaseB/Akt pathways. Oncogene. 2002;21:7524–7532. doi: 10.1038/sj.onc.1205970. [DOI] [PubMed] [Google Scholar]
- 97.Ramsauer VP, Pino V, Farooq A, Carothers Carraway CA, Salas PJ, Carraway KL. Muc4-ErbB2 complex formation and signaling in polarized CACO-2 epithelial cells indicate that Muc4 acts as an unorthodox ligand for ErbB2. Mol. Biol. Cell. 2006;17:2931–2941. doi: 10.1091/mbc.E05-09-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pochampalli MR, el Bejjani RM, Schroeder JA. MUC1 is a novel regulator of ErbB1 receptor trafficking. Oncogene. 2007;26:1693–1701. doi: 10.1038/sj.onc.1209976. [DOI] [PubMed] [Google Scholar]
- 99.Pino V, Ramsauer VP, Salas P, Carothers Carraway CA, Carraway KL. Membrane mucin Muc4 induces density-dependent changes in ERK activation in mammary epithelial and tumor cells: role in reversal of contact inhibition. J. Biol. Chem. 2006;281:29411–29420. doi: 10.1074/jbc.M604858200. [DOI] [PubMed] [Google Scholar]
- 100.Ramsauer VP, Carraway CA, Salas PJ, Carraway KL. Muc4/sialomucin complex, the intramembrane ErbB2 ligand, translocates ErbB2 to the apical surface in polarized epithelial cells. J. Biol. Chem. 2003;278:30142–30147. doi: 10.1074/jbc.M303220200. [DOI] [PubMed] [Google Scholar]
- 101.Funes M, Miller JK, Lai C, Carraway KL, 3rd, Sweeney C. The mucin Muc4 potentiates neuregulin signaling by increasing the cell-surface populations of ErbB2 and ErbB3. J. Biol. Chem. 2006;281:19310–19319. doi: 10.1074/jbc.M603225200. [DOI] [PubMed] [Google Scholar]
- 102.Komatsu M, Yee L, Carraway KL. Overexpression of sialomucin complex, a rat homologue of MUC4, inhibits tumor killing by lymphokine-activated killer cells. Cancer Res. 1999;59:2229–2236. [PubMed] [Google Scholar]
- 103.Komatsu M, Tatum L, Altman NH, Carothers Carraway CA, Carraway KL. Potentiation of metastasis by cell surface sialomucin complex (rat MUC4), a multifunctional anti-adhesive glycoprotein. Int. J. Cancer. 2000;87:480–486. doi: 10.1002/1097-0215(20000815)87:4<480::aid-ijc4>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 104.Otvos L, Jr., Cudic M. Conformation of glycopeptides. Mini Rev. Med. Chem. 2003;3:703–711. doi: 10.2174/1389557033487809. [DOI] [PubMed] [Google Scholar]
- 105.Komatsu M, Carraway CA, Fregien NL, Carraway KL. Reversible disruption of cell-matrix and cell-cell interactions by overexpression of sialomucin complex. J. Biol. Chem. 1997;272:33245–33254. doi: 10.1074/jbc.272.52.33245. [DOI] [PubMed] [Google Scholar]
- 106.Dumic J, Dabelic S, Flogel M. Galectin-3: an open-ended story. Biochim. Biophys. Acta. 2006;1760:616–635. doi: 10.1016/j.bbagen.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 107.Komatsu M, Jepson S, Arango ME, Carothers Carraway CA, Carraway KL. Muc4/sialomucin complex, an intramembrane modulator of ErbB2/HER2/Neu, potentiates primary tumor growth and suppresses apoptosis in a xenotransplanted tumor. Oncogene. 2001;20:461–470. doi: 10.1038/sj.onc.1204106. [DOI] [PubMed] [Google Scholar]
- 108.Bax DA, Haringsma J, Einerhand AW, van Dekken H, Blok P, Siersema PD, Kuipers EJ, Kusters JG. MUC4 is increased in high grade intraepithelial neoplasia in Barrett’s oesophagus and is associated with a proapoptotic Bax to Bcl-2 ratio. J. Clin. Pathol. 2004;57:1267–1272. doi: 10.1136/jcp.2004.017020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dunn GA, Ireland GW. New evidence that growth in 3T3 cell cultures is a diffusion-limited process. Nature. 1984;312:63–65. doi: 10.1038/312063a0. [DOI] [PubMed] [Google Scholar]
- 110.Holley RW, Kiernan JA. “Contact inhibition” of cell division in 3T3 cells. Proc. Natl. Acad. Sci. U. S. A. 1968;60:300–304. doi: 10.1073/pnas.60.1.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stoker MG. Role of diffusion boundary layer in contact inhibition of growth. Nature. 1973;246:200–203. doi: 10.1038/246200a0. [DOI] [PubMed] [Google Scholar]
- 112.Vinals F, Pouyssegur J. Confluence of vascular endothelial cells induces cell cycle exit by inhibiting p42/p44 mitogen-activated protein kinase activity. Mol. Cell. Biol. 1999;19:2763–2772. doi: 10.1128/mcb.19.4.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wille JJ, Jr., Maercklein PB, Scott RE. Neoplastic transformation and defective control of cell proliferation and differentiation. Cancer Res. 1982;42:5139–5146. [PubMed] [Google Scholar]
- 114.Harrington KJ, Syrigos KN. The role of E-cadherin-catenin complex: more than an intercellular glue? Ann. Surg. Oncol. 2000;7:783–788. doi: 10.1007/s10434-000-0783-5. [DOI] [PubMed] [Google Scholar]
- 115.Agrawal B, Krantz MJ, Parker J, Longenecker BM. Expression of MUC1 mucin on activated human T cells: implications for a role of MUC1 in normal immune regulation. Cancer Res. 1998;58:4079–4081. [PubMed] [Google Scholar]
- 116.Taylor-Papadimitriou J, Burchell J, Miles DW, Dalziel M. MUC1 and cancer. Biochim. Biophys. Acta. 1999;1455:301–313. doi: 10.1016/s0925-4439(99)00055-1. [DOI] [PubMed] [Google Scholar]
- 117.Taylor-Papadimitriou J, Burchell JM, Plunkett T, Graham R, Correa I, Miles D, Smith M. MUC1 and the immunobiology of Cancer. J. Mammary Gland Biol. Neoplasia. 2002;7:209–221. doi: 10.1023/a:1020360121451. [DOI] [PubMed] [Google Scholar]
- 118.Perrais M, Pigny P, Ducourouble MP, Petitprez D, Porchet N, Aubert JP, Van Seuningen I. Characterization of human mucin gene MUC4 promoter: importance of growth factors and proinflammatory cytokines for its regulation in pancreatic cancer cells. J. Biol. Chem. 2001;276:30923–30933. doi: 10.1074/jbc.M104204200. [DOI] [PubMed] [Google Scholar]
- 119.Choudhury A, Moniaux N, Ulrich AB, Schmied BM, Standop J, Pour PM, Gendler SJ, Hollingsworth MA, Aubert JP, Batra SK. MUC4 mucin expression in human pancreatic tumours is affected by organ environment: the possible role of TGFbeta2. Br. J. Cancer. 2004;90:657–664. doi: 10.1038/sj.bjc.6601604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Andrianifahanana M, Agrawal A, Singh AP, Moniaux N, van Seuningen I, Aubert JP, Meza J, Batra SK. Synergistic induction of the MUC4 mucin gene by interferon-gamma and retinoic acid in human pancreatic tumour cells involves a reprogramming of signalling pathways. Oncogene. 2005;24:6143–6154. doi: 10.1038/sj.onc.1208756. [DOI] [PubMed] [Google Scholar]
- 121.Mariette C, Perrais M, Leteurtre E, Jonckheere N, Hemon B, Pigny P, Batra S, Aubert JP, Triboulet JP, Van Seuningen I. Transcriptional regulation of human mucin MUC4 by bile acids in oesophageal cancer cells is promoter-dependent and involves activation of the phosphatidylinositol 3-kinase signalling pathway. Biochem. J. 2004;377:701–708. doi: 10.1042/BJ20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Choudhury A, Singh RK, Moniaux N, El-Metwally TH, Aubert JP, Batra SK. Retinoic acid-dependent transforming growth factor-beta 2-mediated induction of MUC4 mucin expression in human pancreatic tumor cells follows retinoic acid receptor-alpha signaling pathway. J. Biol. Chem. 2000;275:33929–33936. doi: 10.1074/jbc.M005115200. [DOI] [PubMed] [Google Scholar]
- 123.Jonckheere N, Perrais M, Mariette C, Batra SK, Aubert JP, Pigny P, Van Seuningen I. A role for human MUC4 mucin gene, the ErbB2 ligand, as a target of TGF-beta in pancreatic carcinogenesis. Oncogene. 2004;23:5729–5738. doi: 10.1038/sj.onc.1207769. [DOI] [PubMed] [Google Scholar]
- 124.Piessen G, Jonckheere N, Vincent A, Hemon B, Ducourouble MP, Copin MC, Mariette C, Van Seuningen I. Regulation of the human mucin MUC4 by taurodeoxycholic and taurochenodeoxycholic bile acids in oesophageal cancer cells is mediated by hepatocyte nuclear factor 1alpha. Biochem. J. 2006;402:81–91. doi: 10.1042/BJ20061461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Damera G, Xia B, Ancha HR, Sachdev GP. IL-9 modulated MUC4 gene and glycoprotein expression in airway epithelial cells. Biosci. Rep. 2006;26:55–67. doi: 10.1007/s10540-006-9000-5. [DOI] [PubMed] [Google Scholar]
- 126.Damera G, Xia B, Sachdev GP. IL-4 induced MUC4 enhancement in respiratory epithelial cells in vitro is mediated through JAK-3 selective signaling. Respir. Res. 2006;7:39. doi: 10.1186/1465-9921-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]