Abstract
A general relationship between fluctuation and response in a biological system is presented. The fluctuation is given by the variance of some quantity, whereas the response is given as the average change of that quantity for a given parameter change. We propose a relationship where the two are proportional, in a similar way to the fluctuation–dissipation theorem in physics. By studying an evolution experiment where fluorescence of protein in bacteria increases, we confirm our relation by observing a positive correlation between the speed of fluorescence evolution and the phenotypic fluctuation of the fluorescence over clone bacteria. The generality of the relationship as well as its relevance to evolution is discussed.
Living organisms are composed of many different types of molecules, such as proteins, RNA molecules, phospholipid molecules, and so on. Because these molecules are synthesized and decomposed by chemical processes occurring at finite temperatures, the molecules are inevitably affected by thermal fluctuations, as a consequence of the laws of physics and chemistry, no matter how well the mechanism of the organism is designed. Even when experimental conditions are controlled as carefully as possible, most cellular variables, such as the quantities of any types of molecules, will vary from cell to cell to some extent, i.e., there will inevitably be fluctuations. Therefore, to obtain experimentally reproducible data, one needs to measure the entire distribution of the variable in question.
Indeed, fluctuations in both quantity and behavior are inevitable for living organisms. The relevance of fluctuations has been previously investigated with regard to the enzymatic function of protein (1), a molecular machine (2), and also a macromolecular system (3), whereas fluctuations in gene expression in cells have been extensively investigated (4–6).
Organisms, however, respond to changes in their surroundings. For example, bacteria tumble with a frequency that depends on the temperature of the external medium as well as on the concentrations of the various chemicals in the medium (7). If the external medium changes, the bacterial tumbling frequency changes. In fact, the average numbers of any given molecules, such as proteins, in a cell will depend on the surrounding conditions and will change in response to changes in the surroundings, such as changes in temperature, pH value, and so on (8). This change gives a basis for a cell to respond. Indeed, understanding how organisms and cells respond to environmental changes is of great importance for the understanding of all biological functions and the processes of adaptation. Also, of course a great many biological experiments have been performed to study and measure such effects.
In the present paper, we propose a relationship between fluctuation and response that should hold in a broad class of systems, discuss its relevance to biological systems, and give an explicit experimental demonstration of the relationship in an experiment on the evolution of functional protein in a cell. In the evolution experiment we report here, fluctuation is defined to be the variance of the fluorescence of a bacterial protein in genetically identical clone bacteria. The fluorescence fluctuates due to phenotypic fluctuations. The response is defined to be the average change in this fluorescence per genetic mutation. We compare our proposed theoretical relationship with the experimental results and find good agreement. We then discuss the relevance of phenotypic fluctuations to evolution, in the framework of our fluctuation–response relationship.
In the analysis of both theory and experiment, we adopt the following terminology. We refer to a measurable quantity (e.g., the concentration of a protein) in a biological system (a cell or an organism) as a “variable” of the system. We adopt the term “parameter” for a quantity that specifies a condition of the system, which influences the system's variables and can be controlled externally in each experiment. According to this definition, the DNA sequence of a gene in a cell is to be regarded as one of the system's parameters, in the artificial evolution experiment that we will discuss later. Last, the terms “average” and “variance” apply to the distribution of the variable over biological systems, such as over cells or organisms.
To begin with, we will state our theoretical proposition as follows: When we change the value of a parameter a slightly so that a → a + Δa, the change in the average value of a variable x will be proportional to its variance at the initial parameter value a, i.e.,
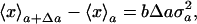 |
[1] |
where the coefficient b is a constant independent of the parameter a, and 〈x〉a and 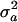 are the average and variance of the variable x at the initial parameter value a, respectively. They are explicitly defined as 〈x〉a = ∫Ω xP(a, x)dx and
are the average and variance of the variable x at the initial parameter value a, respectively. They are explicitly defined as 〈x〉a = ∫Ω xP(a, x)dx and 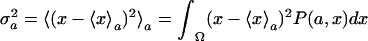 , where P(a, x) is the normalized distribution function of x at the parameter a, and the symbol Ω under the integral symbol means that the integral is taken over the whole range of x. In this paper, the symbol 〈 〉a denotes the average of a given function of x between the brackets with respect to the distribution function P(a, x), i.e., for any function g(x), 〈g(x)〉a:= ∫Ω g(x)P(a, x)dx. We assume that the parameter a and the variable x are both scalars.
, where P(a, x) is the normalized distribution function of x at the parameter a, and the symbol Ω under the integral symbol means that the integral is taken over the whole range of x. In this paper, the symbol 〈 〉a denotes the average of a given function of x between the brackets with respect to the distribution function P(a, x), i.e., for any function g(x), 〈g(x)〉a:= ∫Ω g(x)P(a, x)dx. We assume that the parameter a and the variable x are both scalars.
In the next section, we present the mathematical assumptions underlying the derivation of Eq. 1 and explain the general conditions the distribution P(a, x) must satisfy for our formula to apply to biological systems. In Experiment, we examine the validity of our formula by studying an experiment on the evolution of GFP in bacteria. As explained, in this experiment, the variable is taken to be the magnitude of fluorescence per cell, whereas the parameter and its change correspond to the DNA sequence and its change by mutation, respectively. In Summary and Discussion, we discuss the relevance of our results to a few biological issues.
Formal Derivation of Formula 1
Before we formally derive formula 1, we first derive a general relation holding for distributions having the following property: The distribution can be approximately written in Gaussian form even when the control parameter a is changed, i.e., the distribution function is approximated by the following form for any value of a:
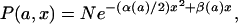 |
where N is a normalization constant so that ∫Ω P(a, x)dx = 1, and α and β are functions of the parameter a [α(a) must be positive for all a]. In other words, this property can be expressed as follows: If the logarithm of the distribution P is expanded in powers of x, the terms with powers of x higher than x2 are always much smaller than those with x and x2. We label such a distribution a “Gaussian-like” distribution in this paper and restrict our theory to this distribution. Such distributions are observed quite often in biological experiments, thus the relation derived below does not hold for special cases only but holds for most practical cases prevalent in biology.
Assuming this distribution holds, we now consider the difference between the average values of x evaluated at the parameter values a and a + Δa, where Δa is supposed to be such a small quantity that both the average value and the variance of x do not much vary. We first write the distribution function at the parameter a + Δa in terms of that at the parameter a as follows:
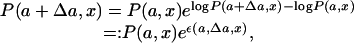 |
where we have introduced, for convenience of notation, the quantity ε(a, Δa, x):= log P(a + Δa, x) – log P(a,x). By expanding this term in powers of (x – 〈x〉a), with 〈x〉a as the average of x at the parameter value a, we obtain
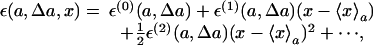 |
where ε(n)(a, Δa):= (∂nε(a, Δa, x)/∂xn)|x=〈x〉a with n = 0, 1, 2,.... Because of our assumption that the distribution be a Gaussian-like one, the terms of powers of x higher than x2 can be neglected. Using this expression, we obtain
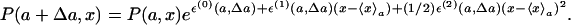 |
[2] |
Because the distribution must satisfy the normalization condition, it must be written as
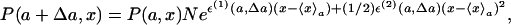 |
[3] |
where N = 1/〈exp[ε(1)(a, Δa)(x–〈x〉a)+ (1/2)ε(2)(a, Δa)(x–〈x〉a)2]〉a. Note that this rewriting is always correct, i.e., the factor eε(0)(a, Δa) is almost equal to N, as long as the distribution is a Gaussian-like one. Using expression 3 valid to the first order in Δa, we obtain
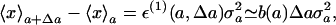 |
[4] |
where b(a) is the first-order derivative of ε(1)(a, Δa) with respect to Δa, i.e., b(a) = ∂ε(1)(a, Δa)/∂Δa|Δa=0. Eq. 4 is a general relation, which always holds for a Gaussian-like distribution up to the order of Δa. This expression shows that the difference between the average values of x evaluated at the parameters a and a + Δa is proportional to its variance at a with the coefficient of proportionality ε(1). As long as we set up an experiment in which the parameter a changes slightly, this assumption at the linear relationship with Δa is guaranteed.
So far, the coefficient b(a) can depend on a. Now, if we assume its dependence is negligible, we obtain the relationship
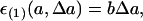 |
[5] |
where b is a constant independent of the parameter a. This leads to the first relationship (Eq. 1). For example, if parameter a can vary only across a limited range, it may be possible to neglect the a dependence.
Now we reconsider the assumptions underlying this derivation.∥ First, note that the above form is quite similar to the fluctuation–dissipation relation in statistical physics (9). In thermodynamics, the term ε(1) in relation 4 is called a generalized force that produces a deviation of the variable x from an equilibrium point. The size of the deviation is of the order of Δa.
In this case, however, because the fluctuation–dissipation relation in statistical physics is restricted to linear nonequilibrium thermodynamics, the relation holds only near the equilibrium point x = 0, where Δa is small, and therefore the dependence on a need not be considered.
For example, consider a bead attached to a spring placed in a fluid with shear flow, where the magnitude of shear flow, Δa, is controlled by the speed of a wall of the container enclosing the fluid. The force acting on the bead is proportional to the relative velocity of the fluid around the bead, and the deviation of the bead from the equilibrium position x – 〈x〉 is proportional to the force and the fluctuation σ2 (10).
Although formula 1 is formally similar to the fluctuation–dissipation theorem, the system in question is not near thermal equilibrium, and hence neglecting the a dependence is an assumption here. The validity of this assumption and the plausibility of the correlation between the fluctuation and response should be examined experimentally, as will be discussed later. Finally, we remark on the choice of parameter a at the beginning of the above derivation. If we choose the parameter arbitrarily, however, relationship 1 is expected to hold only in some rare cases. Indeed, often the distribution of a given variable x is modified only slightly by a change of some parameter value. In the above argument, it is assumed that parameter a and variable x are “closely associated,” in the sense that change in parameter a strongly influences the change in the distribution of x. Although our term “closely associated” is a little ambiguous, it is generally possible to determine from one's experience which parameter is closely associated with any variable in question. For example, consider the measurement of the growth rate of Escherichia coli in a medium with some source of nutrition. The growth rate is influenced by the concentration of the nutrient. Hence, the concentration, controlled externally, is the parameter most closely associated with the growth rate. As another example, consider the evolution of some characteristic of a cell that is under the control of a particular gene. If the gene's DNA sequence changes through a mutation, its influence on the characteristic will also change. In this case, the parameter most closely associated with the characteristic is the degree of substitution in the gene's DNA sequence. And here the rate of change of the parameter will be given by the gene mutation rate. In fact, in general, there is usually little difficulty in choosing the associated parameter.
From the above argument, we propose that for a certain class of biological systems, relationship 1 holds for variables with Gaussian-like distributions if the controllable parameter is closely associated with the variable. Of course, this proposal must be carefully verified and the relationship validated by several experiments. As a first step in this direction we now discuss our experiment on the evolution of a functional protein.
Experiment
To check whether the proposed relationship holds true in biological systems, we analyzed data from an evolution experiment. In our experiment, we artificially selected GFP mutants with higher fluorescence and evolved them for several generations (11, 12).
Previously, one of us (T.Y.) and colleagues (13, 14) succeeded in synthesizing a protein with a random sequence. By genetically attaching an arbitrarily chosen random sequence to the N terminus of a wild-type GFP gene and transforming E. coli with the gene, a GFP mutant with low fluorescence was formed as initial material for the evolution experiment. By applying random mutagenesis to only the attached fragment before GFP, a mutant pool with a diversity of 106 was prepared. One of the transformed E. coli cells possessing the different mutant genes was selected based on its level of fluorescence intensity to be the parent clone of the next generation. Successive generations were then generated so that the (n + 1)th generation was generated from a parent clone selected from the nth generation, while always keeping diversity size and selection pressure the same as in the first generation.
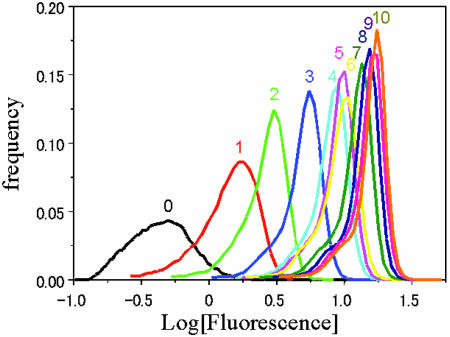
To observe the diversity of green fluorescence shown by the single gene under unvarying experimental conditions, the clone selected at each generation was cultured in a liquid medium, and the grown cells were applied to flow cytometry to measure the distribution of fluorescence intensity per cell (11). In this experiment, the fluorescence magnitude is the natural choice for the variable x. The fluorescence distributions obtained in each generation are shown in Fig. 1. It should be noted first that each distribution has a finite width, although the E. coli cells are “clones” generated of the same genetic information from the same gene under the same conditions. The distribution here originates from the phenotypic variation (i.e., the fluorescence variation) of the clones and does not relate to the gene variation (4, 5).
Fig. 1.
Histogram of the logarithm of fluorescence intensity for each generation, in the experiment described in the text. The number above the peak of each distribution indicates its generation number. We evaluated the fluorescence intensity of each E. coli in each generation by dividing its measured fluorescence intensity value (FL) by its forward scatter (FS) value measured with the cytometry, because the FS value roughly indicates the size of E. coli, whereas the FL value is usually proportional to the FS value.
However, genetic mutation at each generation changes the protein structure, leading to a change in the (average) fluorescence, with the average fluorescence increasing through the selection process. Hence the mutation, i.e., amino acid substitution of the polypeptide sequence attached to the GFP, corresponds to a scalar parameter a that controls the variable x, the fluorescence intensity. If the mutation rate is sufficient (but not too high) in the experiment, the average fluorescence intensity produced by the clone selected at each generation will increase further, which shows that the parameter and the variable are closely associated with each other. Note also that in artificial evolution experiments, the number of mutations away from the original clone is often adopted as a scalar parameter representing the fitness landscape describing the property in concern.
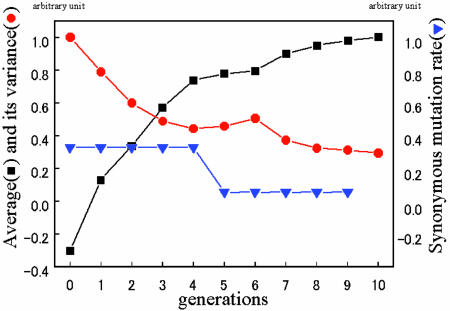
Shown in Fig. 2 is the average value of the distribution and its variance plotted against generation number. Note that both the change in the average value between two successive generations and the variance at each generation decrease generation by generation. Thus this result shows a positive correlation between the response and the fluctuation roughly consistent with our result (Eq. 1), which predicts that the change of average value should be proportional to its variance.
Fig. 2.
The average (black squares) and variance (red circles) of fluorescence over clone cells selected at each generation, plotted vs. the generation number. The average values and variances are computed from the Gaussian-like distributions as their peak position and half-width in Fig. 1. The synonymous mutation rate for each generation, which was determined by DNA sequencing, is also plotted as blue triangles.
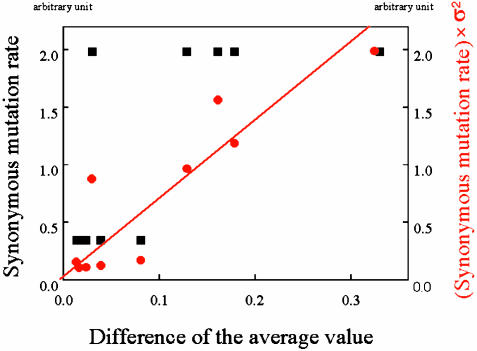
To check the validity of relationship 1 more quantitatively from the experimental results, we plot in Fig. 3 the change in the average value per generation against the variance multiplied by the parameter change, where the parameter change is given by the synonymous mutation rate, also shown there. If the proportionality in Eq. 1 holds perfectly, all these points should lie on a straight line that passes through the origin. Also in Fig. 3, to serve as a comparison, we plot the change in the average value per generation directly against the parameter, that is, without multiplying by the variance. The correlation shown by the results where we multiply by the variance is clearly much better than that shown by the results when we do not. Indeed, the correlation coefficient of the variance-multiplied results is 0.79, whereas the unmultiplied is only 0.21. From this result, it is likely that the change of fluorescence is more highly correlated with the mutation rate multiplied by the variance than with only the mutation rate. It should also be noted that this correlation appears irrespective of the sudden change in the mutation rate that occurred in this experiment at the fifth generation, as can be seen in Fig. 2. This observed correlation is not a trivial result because the variance is about one-third decreased in later generations compared to the first generation. In other words, explicit a dependence of b(a) in Eq. 5 is not observed.
Fig. 3.
σ2 (variance of fluorescence intensity) multiplied by Δa (synonymous mutation rate) is plotted vs. the change of average fluorescence intensity as red circles by using the data from Fig. 2. The red line is a linear fit to the data, which turn out to pass through the origin. For reference, the synonymous mutation rate Δa vs. the change of average fluorescence intensity value is also plotted as black squares. The correlation coefficient for the linear fit is 0.79, whereas that for the synonymous mutation rate (black squares) is 0.21.
The present relationship may remind us of the fundamental theorem of natural selection established by Fisher (15, 16), which asserts that evolution speed and genetic variance are proportional. Here, we should note again that in contrast to this celebrated theorem, our relationship holds for the phenotypic variance of clones, having the same genes, i.e., with null genetic variance.
To close this section, we note that the variable of concern here is not the fluorescence intensity itself but its logarithm. Indeed, the distribution functions in Fig. 1 are plotted by using the logarithm of fluorescence intensity, and the average in Fig. 2 is also computed from the logarithm of the fluorescence. In other words, we adopted the logarithm of the fluorescence as the variable x of our theory. The reason for this choice is as follows. First, the distribution is close to Gaussian in the logarithmic scale. If we directly use the intensity itself, the distribution is highly asymmetric about the peak position and has a larger tail on the higher value side. In fact, we have recently confirmed both in theory and experiments that the distribution of abundances of most chemicals (e.g., proteins) in a cell shows log-normal distributions, i.e., the logarithms of the abundances obey the Gaussian distribution (ref. 17; C. Furusawa and A. Kashiwagi, private communication). Because our theory is based on Gaussian-like distributions, it is natural to use the logarithm of the abundances as the variable x. Indeed, the observed linear relationship between the response and variance is true only with this choice of variable. Finally, it should also be noted that most chemical characteristics depend on the logarithm of chemical abundances (recall that in chemical thermodynamics equations, the logarithm of the concentration generally appears as a relevant term; an example is seen in the definition of pH).
Summary and Discussion
We proposed relation 1 between fluctuation (variance) and response (change of variable with change of parameter) in biological systems and examined the derivation of relation 1, to reveal the underlying assumptions for it. The validity of the relation was examined experimentally by an experiment on the evolution of GFP in E. coli, and a positive correlation between the fluctuation and response was confirmed.
As mentioned above, our relation 1 is analogous to the fluctuation–dissipation theorem in statistical physics (9). Of course, that theorem is directly applicable to the fluctuation and response of biomolecules near thermodynamic equilibrium. For example, by measuring the average and variance of the DNA radius and then by measuring the change under a force acting on two sides of the molecule, one could straightforwardly confirm relation 1. This application, however, is not surprising at all, because the condition for the fluctuation–dissipation theorem in physics is satisfied, and the DNA molecule is regarded as a circular polymer in an aqueous solution at equilibrium.
On the other hand, the main issue of the present paper is the application of relation 1 to a more general biological system, where the fluctuation–dissipation theorem in physics is no longer applicable. Indeed, in the experiment we adopted, the variable and the parameter are not thermodynamic quantities, and the fluctuation–dissipation theorem in physics is not applicable at all. However, the experimental result suggests the relevance of relationship 1 to such a system.
The argument for deriving relation 1 is rather general, as long as the system in concern is stable and the distribution of variables in concern is Gaussian-like. For example, by measuring the abundances of some proteins in a cell with cytometry, the distribution is obtained. Then, by measuring the change of distribution against the change of the medium condition such as the pH value, concentration of nutrition chemical, and so forth, we can test the validity of the relation.
By closely analyzing the derivation of the relation, we have shown that the following three conditions are necessary for relation 1 to hold: (i) that the distribution of the variable (the measurable quantity) is Gaussian-like; (ii) that the parameter we control is closely associated with the variable and finely influences the distribution of the variable; and (iii) that the parameter dependence on the proportionality coefficient is negligible.
Requirement i is necessary for general relation 4 to hold, whereas condition ii is necessary for relation 5 to hold and iii, for relation 1. Following these considerations, we propose that relation 1 will be observed, by suitably choosing a variable that shows a Gaussian-like distribution and a corresponding parameter. Even though assumption iii and accordingly the proportionality in the relationship may not be completely satisfied, a positive correlation between the fluctuation and response itself should be generally observed, and the relation, once verified in several biological systems, may introduce novel directions in the study of biology.
Based on relationship 1, we are able to introduce the notion of “softness” into a biological system: When the variance is large, the system is said to be soft with respect to the variable, because the system shows a large response to the change in the parameter associated with the variable.
In biology, we often use the term “plasticity” to represent the impression that a system is easily changeable in response to external change. In this case, the left-hand side of Eq. 1 is large. Relation 1 implies that a system having significant softness is plastic in this sense. Fluctuations can also be a way to measure and characterize the adaptability of a system.
As our experiment shows, application of the relation to evolution will have biological significance. In this case, a change of phenotype by genetic mutation gives a response (the left-hand side of Eq. 1) that is proportional to the fluctuation of phenotypes for the clone organisms of the same gene, hence the greater the phenotypic fluctuation, the higher the evolution speed. Organisms with larger phenotypic diversity may have higher evolution speed. Possible relationships of phenotypic variability with evolution have often been discussed, but so far no quantitative means to examine the validity of the discussion are available. Our relation 1 may provide one such means.
As mentioned, the fundamental theorem of natural selection by Fisher (15, 16) proves that evolution speed and genetic variance are proportional. As a mathematical expression, our relationship takes a similar form with regard to the proportionality between the phenotypic fluctuation and the speed of evolution. In the case of the theorem by Fisher (15, 16), the formula for the genetic variance is established by genetics, and the proportionality derived is exact. In contrast, the relationship we have proposed here is concerned with the phenotypic fluctuations of clones having identical genes. In this case, the detailed mechanism producing the phenotypic fluctuation has not yet been elucidated, and no established formula is available concerning the degree of phenotypic fluctuation. Still, we have obtained the approximate formula that states that the variance of the phenotype and the evolution speed are proportional. This formula is valid under a certain condition, and its plausibility has been experimentally verified. In this sense, our findings provide insight into the relationship between phenotypic fluctuation and evolution.
We finally comment on ε(2) appearing in Eq. 3. Although the term ε(2) does not appear in relations 1 and 4, and thus it is not involved in the main subject of this paper, it is worth discussing this ε(2) coefficient a little from a biological point of view. As we have seen, ε(1) is the coefficient relating the change of average value of the variable to its variance. On the other hand, ε(2) is the coefficient connecting the change of the variance with the variance. Indeed, calculating the change of the variance in a way similar to the derivation of Eq. 4, we get the relation
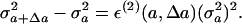 |
[6] |
It is thus possible to use the coefficient ε(2) to explain the change of variance. For example, in our experiment, the variance shrinks as the generation progresses. This decrease can be interpreted by a negative value of the coefficient ε(2). It should be noted that the positive sign of Δa is defined here to correspond to the direction of optimization in the evolution. Hence, the variance of phenotype, i.e., the softness of a certain property, decreases as optimization progresses in evolution. This general statement, of course, needs to be confirmed through further experiments.
The negative value of ε(2) might also provide an understanding for the empirical rule that the more optimized a certain gene, the less frequently mutants with higher fitness will appear. The rule is confirmed by many evolution experiments [see for example, Matsuura et al. (18)]. In our evolution experiment, the change in fluorescence intensity at each generation decreases as fluorescence intensity increases and as optimization progresses. This occurs because around a parent clone with a high fluorescence, the probability that mutants that bring about much higher fluorescence will appear is lower. Although this is generally just because the gene gets closer to a local maximum in the fitness landscape, it does not explain why there is a local maximum. On the other hand, if ε(2) is negative, the cell system in question will lose its softness as generations progress. When this happens, the system, according to Eq. 1, will lose its plasticity, and the rate of evolutionary optimization will decrease. In other words, the decrease in softness due to the negative ε(2) brings the system to a local maximum in the fitness landscape.
Basically, the two equations, Eqs. 1 and 6, predict the evolution rate of the property in question and its deceleration, in terms of the variance of the phenotype and of the variance. In the study of evolution, where many researchers try to trace back to biological events in the past, the two relationships might offer a base for experimental studies with quantitative predictions.
Acknowledgments
We acknowledge the helpful support of K. Ohnuma, C. Furusawa, and A. Kashiwagi, and we thank A. Ponzi for proofreading. This research was supported in part by Grants-in-Aid (nos. 11CE2006, 15207020, and 15013235) and the 21st Century Center of Excellence Program of the Ministry of Education, Culture, Sports, Science, and Technology, Japan, and by a grant from Takeda Science Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Footnotes
The derivation could be a little simplified, if we assume the Gaussian distribution completely and take the Δa → 0 limit in the beginning, as is often adopted in standard statistical physics. We adopted the present derivation to see under what conditions the linearity approximation holds and also to conveniently discuss the decrease of variance through evolution observed in the experiment (to be shown later). Further elaboration on our framework and the linearity condition should be pursued in the future.
References
- 1.Tang, K. E. S. & Dill, K. A. (1998) J. Biomol. Struct. Dyn. 16, 397–411. [DOI] [PubMed] [Google Scholar]
- 2.Ishijima, A., Kojima, H., Funatsu, T., Tokunaga, M., Higuchi, H., Tanaka, H. & Yanagida, T. (1998) Cell 92, 161–171. [DOI] [PubMed] [Google Scholar]
- 3.Osawa, F. (1975) J. Theor. Biol. 52, 175–186. [DOI] [PubMed] [Google Scholar]
- 4.Elowitz, M. B., Levine, A. J., Siggia, E. D. & Swain, P. S. (2002) Science 297, 1183–1186. [DOI] [PubMed] [Google Scholar]
- 5.Swain, P. S., Elowitz, M. B. & Siggia, E. D. (2002) Proc. Natl. Acad. Sci. USA 99, 12795–12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hasty, J., Pradines, J., Dolnik, M. & Collins, J. J. (2000) Proc. Natl. Acad. Sci. USA 97, 2075–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spudich, J. & Koshland, D. (1976) Nature 262, 467–471. [DOI] [PubMed] [Google Scholar]
- 8.Ueda, M., Sako, Y., Tanaka, T., Devreotes, P. & Yanagida, T. (2001) Science 294, 864–867. [DOI] [PubMed] [Google Scholar]
- 9.Landau, L. D. & Lifshitz, E. M. (1980) Statistical Physics (Pergamon, Oxford), pp. 384–396.
- 10.Reichl, L. E. (1980) A Modern Course in Statistical Physics (Univ. of Texas Press, Austin) pp. 557–560.
- 11.Ito, Y., Kawama, T., Urabe, I. & Yomo, T. J. (2003) Mol. Evol., in press. [DOI] [PubMed]
- 12.Waldo, G. S., Standish, B. M., Berendzen, J. & Terwilliger, T. C. (1999) Nat. Biotechnol. 17, 691–695. [DOI] [PubMed] [Google Scholar]
- 13.Prijambada, I. D., Yomo, T., Tanaka, F., Kawama, T., Yamamoto, K., Hasegawa, A., Shima, Y., Negoro, S. & Urabe, I. (1996) FEBS Lett. 382, 21–25. [DOI] [PubMed] [Google Scholar]
- 14.Yamauchi, A., Yomo, T., Tanaka, F., Prijambada, I. D., Ohhashi, S., Yamamoto, K., Shima, Y., Ogasahara, K., Yutani, K., Kataoka, M., et al. (1998) FEBS Lett. 421, 147–151. [DOI] [PubMed] [Google Scholar]
- 15.Fisher, R. A. (1999) in The Genetical Theory of Natural Selection (Oxford Univ. Press, Oxford) pp. 22–47.
- 16.Edwards, A. W. F. (2000) in Foundations of Mathematical Genetics (Cambridge Univ. Press, Cambridge, U.K.), pp. 8–20.
- 17.Kaneko, K. (2003) Phys. Rev. E 68, 031909-1–031909-5. [Google Scholar]
- 18.Matsuura, T., Yomo, T., Trakulnaleamsai, S., Ohashi, Y., Yamamoto, K. & Urabe, I. (1998) Protein Eng. 11, 789–795. [DOI] [PubMed] [Google Scholar]