Abstract
Defects in FAM83H on human chromosome 8q24.3 cause autosomal-dominant hypocalcified amelogenesis imperfecta (ADHCAI). FAM83H does not encode a recognizable signal peptide, so we predicted that the Fam83h protein functions within the cell. We tested this hypothesis by constitutively expressing mouse Fam83h with green fluorescent protein (GFP) fused to its C-terminus in HEK293 and HeLa cell lines. Green fluorescent signal from the Fam83h-GFP fusion protein was associated with perinuclear vesicles, usually in the vicinity of the Golgi apparatus. No signal was observed within the nucleus. In addition, we identified FAM83H nonsense mutations in Hispanic (C1330C>T; p.Q444X) and Caucasian (c.1192C>T; p.Q398X) families with ADHCAI. We conclude that Fam83h localizes in the intracellular environment, is associated with vesicles, and plays an important role in dental enamel formation. FAM83H is the first gene involved in the etiology of amelogenesis imperfecta (AI) that does not encode a secreted protein.
Keywords: ADHCAI, Golgi, enamel, tooth, amelogenesis imperfecta
Introduction
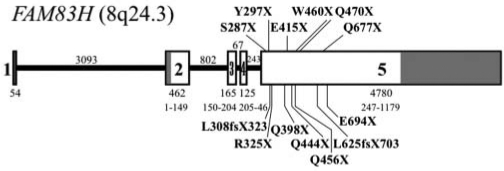
Amelogenesis imperfecta (AI) is the collective designation for a diverse group of inherited diseases exhibiting defects in dental enamel in the absence of significant non-dental symptoms (Witkop and Sauk, 1976). Because the phenotype is primarily restricted to enamel, significant research efforts have been devoted to identifying the suite of extracellular matrix proteins that control enamel biomineralization, and to determining if the genes encoding them contribute to the etiology of AI. The major proteins expressed in secretory stage enamel are amelogenin (Snead et al., 1983, 1985), enamelin (Hu et al., 1997b), ameloblastin (Cerny et al., 1996; Krebsbach et al., 1996; Hu et al., 1997a), and enamelysin (MMP-20) (Bartlett et al., 1996). In addition, kallikrein 4 (KLK4) (Simmer et al., 1998), amelotin (Iwasaki et al., 2005; Moffatt et al., 2006), and apin (Moffatt et al., 2008) are expressed during the maturation stage. AI-causing mutations in AMELX, ENAM, MMP20, and KLK4 have been identified (Hu et al., 2007; Wright et al., 2009a); however, mutational analyses of AI kindreds with these candidate genes successfully identified the causative mutation in only 25% of cases (Kim et al., 2006). So when a genome-wide scan mapped autosomal-dominant hypocalcified amelogenesis imperfecta (ADHCAI) to a 2.1-Mb region of chromosome 8q24.3 (Mendoza et al., 2007) that contained no AI candidate genes, it started an informal race to identify the disease-causing gene. A gene-by-gene search determined that defects in FAM83H cause ADHCAI (OMIM #130900) (Kim et al., 2008), and subsequent reports have brought to 14 the number of novel disease-causing mutations in FAM83H (Lee et al., 2008; Hart et al., 2009; Wright et al., 2009b). Remarkably, all 14 of the FAM83H mutations are in the last exon (exon 5), and cause premature termination of the Fam83h protein (Fig. 1).
Figure 1.
FAM83H gene diagram showing mutations identified in families with ADHCAI. The 5 exons are indicated by numbered boxes. The non-coding regions of exons 1, 2, and 5 are shaded. The number of nucleotides in each exon is shown immediately under the box. The range of amino acids in the Fam83h protein encoded by each exon is below the number of nucleotides. Introns are represented by a solid line, with the number of nucleotides in the intron shown above. The 14 disease-causing mutations in FAM83H are labeled in bold above and below the 5’ part of exon 5.
FAM83H (family with sequence similarity 83 member H on chromosome 8q24.3) is a gene that was originally discovered during computer analyses of the human genomic sequence. Expressed sequence tags (ESTs) indicate that Fam8h3 is expressed in many tissues. How could defects in a gene that is broadly expressed cause a phenotype that is restricted to enamel? The pattern of FAM83H mutations that cause ADHCAI may provide a clue. Premature translation termination codons in the last coding exon generally escape non-sense-mediated decay (Shyu et al., 2008), suggesting that truncated proteins are translated, and that the phenotype is not due to haploinsufficiency (a simple 50% reduction in the amount of Fam83h protein). Human Fam83h has 1179 amino acids. All of the reported FAM83H mutations shorten the protein so that as little as 286 or as many as 693 amino acids of the normal protein are synthesized. Apparently, half the normal amount of Fam83h does not cause ADHCAI (since no mutations predicted to cause haploinsufficiency have been observed), but synthesis of reduced amounts of wild-type protein, along with truncated Fam83h, does. ADHCAI may be caused by the dominant-negative effects of expressing truncated Fam83h. Truncated Fam83h might interact with wild-type Fam83h (expressed from the unmutated allele) or with another protein, forming a non-functional dimer or multimer.
Knowing where Fam83h normally functions is an important early step in understanding its functional role in dental enamel formation. In this study, we report FAM83H mutations in two families with ADHCAI and test the hypothesis that Fam83h functions within the cell by expressing Fam83h fused to green fluorescent protein (GFR) in cell lines and then localizing it by fluorescent microscopy.
Materials & Methods
Protocol Approval
The study protocol and participant consent forms were reviewed and approved by the Institutional Review Board at the University of Michigan.
Recruitment and Examination of Participants
All participants were given oral and radiographic examinations to characterize their dental phenotype and affection status. The proband of family 1 was a Hispanic boy, who was the only person recruited from this family. The medical history revealed no health concerns or allergies. The proband of family 2 was a Caucasian girl. The five-person nuclear family was recruited.
Mutation Analysis
Based upon the clinical diagnosis of ADHCAI, we limited our mutation analysis to exon 5 of the FAM83H gene. Genomic DNA was isolated from 2 mL of peripheral whole blood by means of the QIAamp® DNA Blood Maxi Kit (Qiagen, Valencia, CA, USA). PCR amplifications used Platinum® DNA polymerase (Invitrogen, Carlsbad, CA, USA). Five primer pairs were used to amplify overlapping segments of exon 5: Set 1 (748 bp, 57°C annealing T), ACCTCACATCCCTGCGTCCTC, CGGGTCCCACTGGTA CTGCT; Set 2 (725 bp, 57°C annealing T), GTGTCGCGGCAGACGTTCCTC: GAGCGGAATGAGTCCTGCTTGG; Set 3 (610 bp, 60°C annealing T), AGCAGGACTCATTCCGCTC, GACTCATTATGGAGCACCTGG; Set 4 (726 bp, 64°C annealing T), GCCTGCCTTCCCGCTTCC, CTAGCTCG GGGCTGTTGTGGG; and Set 5 (709 bp, 64°C annealing T), GCCAACGCCTTGTACAGCAGC, TGCTGAGGGGAGAA AGGCACTG. The PCR amplifications were performed in 50-µL reaction mixes containing 21.5 µL sterile water, 1 µL each of 10 pmol of sense and antisense primers, 1 µL 0.5 µg/µL template, 0.5 µL FailSafe PCR Enzyme Mix, and 25 µL FailSafe PCR 2x Premix J (Epicenter Biotechnologies, Madison, WI, USA). The reactions had a five-minute denaturation at 94°C, followed by 40 cycles at 94°C for 30 sec, primer annealing at 57-64°C for 30 sec, and product extension at 72°C for 60 sec. In the final cycle, the 72°C extension was for 7 min. PCR amplification products were run in a 1.2% agarose gel, excised with a razor blade, purified via the QIAquick Gel Extraction Kit (Qiagen), and characterized by direct DNA sequencing. The same oligonucleotide primers used to generate the PCR amplification products were used to prime the sequencing reactions. The sequencing results were checked against the Homo sapiens chromosome 8 genomic contig reference assembly AC105219 and cDNA reference sequence NM_198488.3.
Fabrication of the Fam83h-GFP Expression Construct
A full-length mouse Fam83h cDNA clone (BY722193) in pBluescriptIIKS+ (Stratagene, La Jolla, CA, USA) was obtained from K.K. DNAFORM (Yokohama, Japan). Restriction sites were introduced before the Fam83h translation initiation codon (BglII) and replacement of the translation termination codon (SalI) by PCR, which generated a 3651-bp amplicon (primer set: aagatcTGGCCCCAACATGGCCCGTC, agtcgacTTTTTTGCTTTTGAATGTGCCC). The PCR conditions included an initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 sec, annealing at 65°C for 30 sec, and extension at 72°C for 4 min. The final extension was carried out at 72°C for 7 min. The products were resolved by electrophoresis on a 1.0% agarose gel buffered with 1×TAE and visualized by ethidium bromide. The amplification product was extracted from the gel and ligated into pCR2.1-TOPO (Invitrogen). The Fam83h coding region was excised by double digestion with BglII and SalI and ligated into phrGFP-C (Stratagene), which had been restricted with BamHI and SalI. Proper construction of the recombinant expression plasmid was verified by DNA sequencing.
Intracellular Localization of Fam83h
HEK293 or HeLa cells were cultured with DMEM with 10% FBS, 1% 100 mM sodium pyruvate, and 1% GluTA MAX-1 in a Lab-Tek chamber slide with cover (Nalge Nunc International, Rochester, NY, USA) and used to seed glass coverslips to a density of 2 × 105 cells/mm2, one day prior to transfection. The phrGFP-Fam83h plasmid in Lipofectamine2000 (Invitrogen) was diluted with DMEM without serum to a final concentration of 1.2 µg/µL, and incubated for 20 min at room temperature. The phrGFP-Fam83h/Lipofectamine2000 complexes were then added to the seeded coverslips. After 24 hrs, the cells were fixed with 4% paraformaldehyde for 10 min at room temperature. Golgi apparatus was stained with 1 µM BODIPY TR complexed to BSA (Invitrogen), mounted with ProLong Gold anti-fade reagent with DAPI (Invitrogen) for staining of the nucleus, and photographed by means of a Leica DM 5000B fluorescence microscope.
Results
Evaluation of Participants
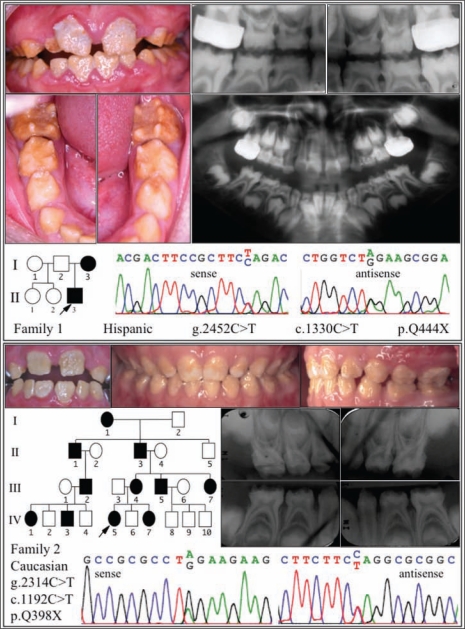
The proband of family 1 was a Hispanic boy (9 yrs, 5 mos) with age-appropriate dental development, but his dentition exhibited rough hypoplastic enamel, having a brownish discoloration (Fig. 2, top). Both the primary and permanent dentitions were affected. The surface roughness and sensitivity hindered effective oral hygiene, and the proband presented with heavy plaque and calculus accumulation and generalized gingivitis. The enamel layer was thin (contributing to spacing of the teeth) and did not contrast with dentin on radiographs. The thinness of the enamel layer appeared to be the result of rapid attrition following eruption of the teeth. The proband’s teeth were highly sensitive to thermal changes. Removal of the surface enamel on the permanent maxillary central incisors exposed a thin layer of whiter enamel, which bound composite restorative materials adequately (data not shown). The biological mother of the proband was not available for examination, but reportedly has similar problems with her teeth, indicating a dominant pattern of inheritance. The stepmother reported that the proband was otherwise healthy. Mutation analysis of genomic DNA from the proband identified a FAM83H nonsense mutation (g.2452C>T; c.1330C>T; p.Q444X), which was previously shown to cause ADHCAI in a Turkish family (Hart et al., 2009).
Figure 2.
Phenotypic features and mutation identification. (top) Oral photographs and radiographs of the proband of family 1, who was in the mixed-dentition stage at age 9 yrs and 5 mos. Note the plaque accumulation and associated gingivitis in the anterior region. Both the primary and secondary dentitions were affected. The enamel was generally brown and rough, and cannot be distinguished from dentin on radiographs. DNA sequencing chromatograms reveal a C-to-T transition in one of the two FAM83H alleles, changing codon 444 from a glutamine (Q) to a stop (X) codon. (bottom) Oral photographs and radiographs of the proband of family 2. The permanent maxillary central incisors (left) at 8 yrs and 10 mos were rough and stained. The primary dentition (center and right), shown at 5 yrs of age, was also affected. The enamel and dentin are not distinguishable on radiographs taken at 4 yrs and 10 mos. DNA sequencing chromatograms show an A-to-G transition in one of the two FAM83H alleles, changing codon 398 from a glutamine (Q) to a stop (X) codon.
The dental phenotype of the proband in family 2 was similar to that of family 1 (Fig. 2, bottom). The proband was a Caucasian girl (8 yrs, 10 mos) with no known systemic abnormalities. The individual had dental hypersensitivity. The enamel layer was yellowish brown, thin, and did not contrast with dentin on radiographs. The enamel was rough, covered with plaque and calculus, and associated with generalized gingivitis. Wear facets on the proband’s primary teeth suggested more than average enamel attrition. The primary dentition displayed islands of normal-looking enamel, particularly near the incisal edges or cusp tips. The enamel phenotype in this family can be traced back for over four generations, with virtually all affected individuals having extensive dental histories. Participant I1, at age 88 yrs, was reported to be healthy, other than having AI. Mutation analysis of genomic DNA from the proband identified a FAM83H nonsense mutation (g.2314C>T; c.1192C>T; p.Q398X), which was previously identified as a spontaneous nonsense mutation in a Korean family with ADHCAI (Kim et al., 2008) and also in a Turkish family (Hart et al., 2009). The highly varied ethnic backgrounds of the three AI families with this FAM83H mutation indicate that they arose independently.
Intracellular Localization of Fam83h
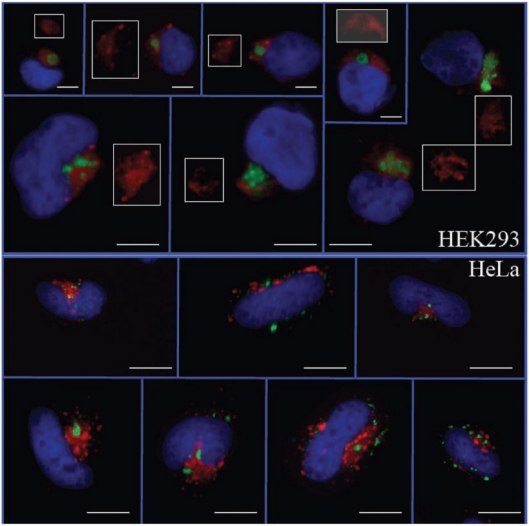
The full-length mouse Fam83h protein fused to green fluorescent protein, expressed in a human kidney epithelial cell line (HEK293), suggests that Fam83h is associated with peri-nuclear vesicles in the vicinity of the Golgi apparatus, perhaps in the trans-Golgi network or in the late endosomes (Fig. 3, top). The localization pattern varies with cell type: The association with the Golgi is less strict in HeLa cells (Fig. 3, bottom). A more definitive localization requires the development of Fam83h-specific antibodies to immuno-localize Fam83h in amelo-blasts at the ultrastructural level.
Figure 3.
HEK293 (top) and HeLa (bottom) cells transfected with a construct expressing Fam83h-GFP. In HEK293 cells, Fam83h (green) is always associated with the Golgi (red) and never appears inside the nucleus (blue). Typically, the Golgi stain is weaker or negative where the GFP localizes (white-outlined insets). In HeLa cells, the association between Fam83H-GFP and the Golgi is not as strict. The green fluorescence is always associated with small vesicle-like structures that only sometimes overlap with the Golgi. Bars = 4 µm.
Discussion
The human FAM83 family of genes is comprised of 8 members, designated FAM-83A through FAM83H. Fam83h (1179 amino acids) is the largest; Fam83a (434 amino acids) is the smallest. Alignment of the human Fam83 proteins shows that homology common to the entire group begins near the N-terminus and ends at Ser281 (Appendix). This conserved N-terminal region also corresponds closely to the part of Fam83h (Arg4 to Leu284) that is included in the conserved domain database (CDD) for the phospholipase D (PLD) superfamily (Marchler-Bauer et al., 2007). The homology with PLD, however, is trace, and may be indicative of a conserved three-dimensional fold. The N-terminal domain that shows trace similarity to PLD bears a much greater resemblance to the other members of the FAM83 family and is the shared element that gives the group its identity. The 14 FAM83H mutations that cause ADHCAI delete the sequence on the C-terminal side of the PLD-like domain, with the closest truncation terminating at Ser287.
A BLASTP search of GenBank identified FAM83H homologues going back to fish. Alignment of selected vertebrate sequences (human NM_198488; mouse AK035531; cattle XM603315; opossum XM_001370407; bird XM_423955; frog BC078004; and zebrafish NM_001045090) showed homology throughout the entire protein, although the homology for the C-terminal segment starting with Ser287 was lower than the homology for the PDL-like region (Appendix). Thus, the Fam83h gene might have arisen early in vertebrate evolution by duplication of a gene in the Fam83 family. It appears that ameloblasts are dependent upon the Fam83h protein for proper dental enamel formation.
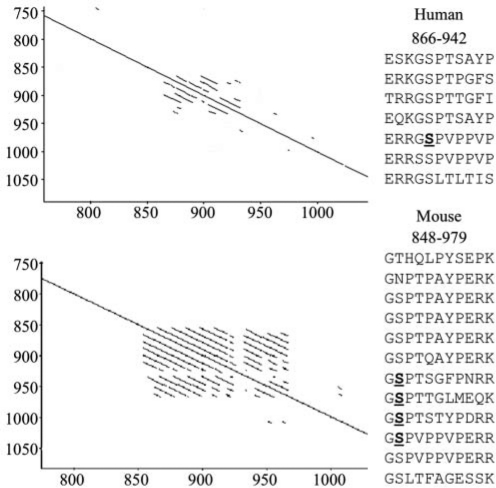
While the protein sequence of Fam83h on the C-terminal side of the PLD-like domain is not homologous with other proteins, it does have some notable features. This part of Fam83h contains a tandem repeat region that varies in length between species. The repeated segment is 11 amino acids in length, with at least 7 copies in humans (E866 to S942) and 12 copies in the mouse (G848 to K979) (Fig. 4). Tandem repeat sequences of variable length are also observed in amelogenin (Takagi et al., 1984; Hu et al., 1996) and enamelin (Hu et al., 1998). Large-scale characterization of phosphorylated sequences in the mouse liver and in a human (HeLa) cell line identified multiple serine residues that are sometimes phosphorylated in Fam83h ([mouse] Ser5, Ser7, Ser8, Ser9, Ser512, Ser513, Ser515, Ser522, Ser832, Ser834, Ser841, Ser915, Ser926, Ser937, Ser948, and Ser1041; and [human] Ser523, Ser731, Ser914, and Ser1003). Most of the phosphorylation sites are in the area deleted by FAM83H truncation mutations in ADHCAI kindreds, and some are in the tandem repeat region. The finding that Fam83h is phosphorylated suggests that it is regulated by kinases.
Figure 4.
Tandem repeats in human (top) and mouse (bottom) Fam83h. Self-diagonal plots highlight repeated sequences in the Fam83h protein. In this type of analysis, a protein is aligned against itself. The solid line along the diagonal indicates the protein’s identity with itself. Amino acid identities to upstream or downstream sequences are marked by lines above or below the diagonal. Inspection of the sequences corresponding to lines off the diagonal showed the repeats are 11 amino acids in length. The human Fam83h sequence between E866 and S942 and the mouse Fam83h sequence between Gly848 and Lys979 are shown at the right. Phosphorylated serines in the repeat region are underlined and in bold.
Phospholipase D (PLD) is a key facilitator of membrane vesicle trafficking. There are two human PLD genes: PLD1 (3q26) and PLD2 (17p13.1). Our fluorescent images suggest that Fam83h is associated with intracellular vesicles in the perinuclear region, usually closely associated with the Golgi complex. This localization pattern is similar to that of PLD1, but not PLD2. GFP-tagged PLD1 localizes mainly to the perinuclear region, including the trans-Golgi apparatus, multivesicular endosomes, and late endosomes, but not to other organelles, while PLD2 localizes to the plasma membrane (Du et al., 2004; Hiroyama and Exton, 2005). Phospholipase D catalyzes the hydrolysis of phosphatidylcholine (a membrane phospholipid) to phosphatidic acid and choline. Because the homology with PLD is low, it seems unlikely that Fam83h could catalyze this reaction, but this cannot be ruled out. At present we speculate that Fam83h might be a peripheral membrane protein that plays a role in intracellular trafficking or cytoskeletal re-organization.
Supplementary Material
Footnotes
This investigation was supported by USPHS Research Grants DE015846 and DE019622 (NIDCR/NIH) and by the Korea Science and Engineering Foundation (KOSEF) grant funded by the Korean government (MEST) [No. R01-2008-000-10174-0 (2008)].
All authors declare that there are no conflicting interests.
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
References
- Bartlett JD, Simmer JP, Xue J, Margolis HC, Moreno EC. (1996). Molecular cloning and mRNA tissue distribution of a novel matrix metalloproteinase isolated from porcine enamel organ. Gene 183:123-128 [DOI] [PubMed] [Google Scholar]
- Cerny R, Slaby I, Hammarström L, Wurtz T. (1996). A novel gene expressed in rat ameloblasts codes for proteins with cell binding domains. J Bone Min Res 11:883-891 [DOI] [PubMed] [Google Scholar]
- Du G, Huang P, Liang BT, Frohman MA. (2004). Phospholipase D2 localizes to the plasma membrane and regulates angiotensin II receptor endocytosis. Mol Biol Cell 15:1024-1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart PS, Becerik S, Cogulu D, Emingil G, Ozdemir-Ozenen D, Han ST, et al. (2009). Novel FAM83H mutations in Turkish families with autosomal dominant hypocalcified amelogenesis imperfecta. Clin Genet 75:401-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroyama M, Exton J. (2005). Localization and regulation of phospholipase D2 by ARF6. J Cell Biochem 95:149-164 [DOI] [PubMed] [Google Scholar]
- Hu CC, Zhang C, Qian Q, Ryu OH, Moradian-Oldak J, Fincham AG, et al. (1996). Cloning, DNA sequence, and alternative splicing of opossum amelogenin mRNAs. J Dent Res 75:1728-1734 [DOI] [PubMed] [Google Scholar]
- Hu CC, Fukae M, Uchida T, Qian Q, Zhang CH, Ryu OH, et al. (1997a). Sheathlin: cloning, cDNA/polypeptide sequences, and immunolocalization of porcine enamel proteins concentrated in the sheath space. J Dent Res 76:648-657 [DOI] [PubMed] [Google Scholar]
- Hu CC, Fukae M, Uchida T, Qian Q, Zhang CH, Ryu OH, et al. (1997b). Cloning and characterization of porcine enamelin mRNAs. J Dent Res 76:1720-1729 [DOI] [PubMed] [Google Scholar]
- Hu CC, Simmer JP, Bartlett JD, Qian Q, Zhang C, Ryu OH, et al. (1998). Murine enamelin: cDNA and derived protein sequences. Connect Tissue Res 39:47-61 [DOI] [PubMed] [Google Scholar]
- Hu JC, Chun YH, Al Hazzazzi T, Simmer JP. (2007). Enamel formation and amelogenesis imperfecta. Cells Tissues Organs 186:78-85 [DOI] [PubMed] [Google Scholar]
- Iwasaki K, Bajenova E, Somogyi-Ganss E, Miller M, Nguyen V, Nourkeyhani H, et al. (2005). Amelotin—a novel secreted, ameloblast-specific protein. J Dent Res 84:1127-1132 [DOI] [PubMed] [Google Scholar]
- Kim JW, Simmer JP, Lin BP, Seymen F, Bartlett JD, Hu JC. (2006). Mutational analysis of candidate genes in 24 amelogenesis imperfecta families. Eur J Oral Sci 114(Suppl 1):3-12 [DOI] [PubMed] [Google Scholar]
- Kim JW, Lee SK, Lee ZH, Park JC, Lee KE, Lee MH, et al. (2008). FAM83H mutations in families with autosomal-dominant hypocalcified amelogenesis imperfecta. Am J Hum Genet 82:489-494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebsbach PH, Lee SK, Matsuki Y, Kozak CA, Yamada K, Yamada Y. (1996). Full-length sequence, localization, and chromosomal mapping of ameloblastin: a novel tooth-specific gene. J Biol Chem 271:4431-4435 [DOI] [PubMed] [Google Scholar]
- Lee SK, Hu JC, Bartlett JD, Lee KE, Lin BP, Simmer JP, et al. (2008). Mutational spectrum of FAM83H: the C-terminal portion is required for tooth enamel calcification. Hum Mutat 29:E95-E99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson J, Derbyshire M, DeWeese-Scott C, Gonzales N, Gwadz M, et al. (2007). CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res 35:D237-D240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza G, Pemberton TJ, Lee K, Scarel-Caminaga R, Mehrian-Shai R, Gonzalez-Quevedo C, et al. (2007). A new locus for autosomal dominant amelogenesis imperfecta on chromosome 8q24.3. Hum Genet 120:653-662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt P, Smith CE, St-Arnaud R, Simmons D, Wright JT, Nanci A. (2006). Cloning of rat amelotin and localization of the protein to the basal lamina of maturation stage ameloblasts and junctional epithelium. Biochem J 399:37-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt P, Smith CE, St-Arnaud R, Nanci A. (2008). Characterization of Apin, a secreted protein highly expressed in tooth-associated epithelia. J Cell Biochem 103:941-956 [DOI] [PubMed] [Google Scholar]
- Shyu AB, Wilkinson MF, van Hoof A. (2008). Messenger RNA regulation: to translate or to degrade. EMBO J 27:471-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer JP, Fukae M, Tanabe T, Yamakoshi Y, Uchida T, Xue J, et al. (1998). Purification, characterization, and cloning of enamel matrix serine proteinase 1. J Dent Res 77:377-386 [DOI] [PubMed] [Google Scholar]
- Snead ML, Zeichner-David M, Chandra T, Robson KJ, Woo SL, Slavkin HC. (1983). Construction and identification of mouse amelogenin cDNA clones. Proc Natl Acad Sci USA 80:7254-7258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead ML, Lau EC, Zeichner-David M, Fincham AG, Woo SL, Slavkin HC. (1985). DNA sequence for cloned cDNA for murine amelogenin reveal the amino acid sequence for enamel-specific protein. Biochem Biophys Res Commun 129:812-818 [DOI] [PubMed] [Google Scholar]
- Takagi T, Suzuki M, Baba T, Minegishi K, Sasaki S. (1984). Complete amino acid sequence of amelogenin in developing bovine enamel. Biochem Biophys Res Commun 121:592-597 [DOI] [PubMed] [Google Scholar]
- Witkop CJ, Jr, Sauk JJ., Jr (1976). Heritable defects of enamel. In: Oral facial genetics. Stewart RE, Prescott GH., Jr editors. St. Louis: C.V. Mosby Co., pp. 151-226 [Google Scholar]
- Wright JT, Hart TC, Hart PS, Simmons D, Suggs C, Daley B, et al. (2009a). Human and mouse enamel phenotypes resulting from mutation or altered expression of AMEL, ENAM, MMP20 and KLK4. Cells Tissues Organs 189:224-229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JT, Frazier-Bowers S, Simmons D, Alexander K, Crawford P, Han ST, et al. (2009b). Phenotypic variation in FAM83H-associated amelogenesis imperfecta. J Dent Res 88:356-360 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.