Abstract
Mutagenesis screens to isolate a variety of alleles leading to null and non-null phenotypes represent an important approach for the characterization of gene function. Genetic schemes that use visible markers permit the efficient recovery of chemically induced mutations. We have developed a universal reporter system to visibly mark chromosomes for genetic screens in the mouse. The dualcolor reporter is based on a single vector that drives the ubiquitous coexpression of the enhanced GFP (EGFP) spectral variants yellow and cyan. We show that widespread expression of the dual-color reporter is readily detected in embryonic stem cells, mice, and throughout developmental stages. CRE-loxP- and FLPe-FRT-mediated deletion of each color cassette demonstrates the modular design of the marker system. Random integration followed by plasmid rescue and sequence-based mapping was used to introduce the marker to a defined genomic location. Thus, single-step placement will simplify the construction of a genomewide bank of marked chromosomes. The dual-color nature of the marker permits complete identification of genetic classes of progeny as embryos or mice in classic regionally directed screens. The design also allows for more efficient and novel schemes, such as marked suppressor screens, in the mouse. The result is a versatile reporter that can be used independently or in combination with the growing sets of deletion and inversion resources to enhance the design and application of a wide variety of genetic schemes for the functional dissection of the mammalian genome.
With complete genomic sequence data, the genetic make-up of a number of organisms is available for comparison and study and the majority of the protein coding genes in several species have been identified. Nevertheless, the challenge remains to classify gene activities and identify interactions that form the basis of protein assemblies and biological networks. Mutagenesis screens are a powerful approach toward this goal. They can be used to generate a set of related reagents that reveal gene function and define genetic relationships. This information is useful for ordering components of a pathway or identifying associations critical for the formation of protein complexes. Based on this principle, a variety of screens have been used to build collections of mutations valuable for the functional genetic characterization of biological processes in model organisms from bacteria to mice.
One of the most extensively developed model organisms for genetic analysis is Drosophila melanogaster (www.flybase.org). A key to the success of Drosophila is the availability of genetic reagents that increase the ease and efficiency of performing mutagenesis screens. Chromosomal rearrangements together with visible markers based on variations in traits such as eye color or wing morphology are used to isolate, map, and maintain mutations for loci within the marked genomic interval. In combination with the point mutagen ethylmethanesulfonate (EMS) these genetic reagents have been used in screens to recover allelic series for sets of genes involved in a wide variety of biological processes (1).
The practical benefits afforded by the collection of visibly marked chromosomal reagents in Drosophila also offer attractive advantages for genetic analyses in the mouse. Early mutagenesis screens, such as the specific locus test (SLT), used classic coat color loci as visible markers to recover chemical- and radiation-induced chromosomal rearrangements at defined locations in the genome (2). The SLT screen was also used to establish the chemical N-ethyl-N-nitrosourea (ENU) as a highly effective point mutagen in the mouse (3). The coat color markers and their corresponding chromosomal deletion complexes have permitted the functional dissection of discrete portions of the mouse genome (4), from mapping phenotypes associated with megabase (Mb)-scale chromosomal deficiencies to the generation of allelic series of ENU-induced mutations, revealing a range of functions for individual genes (5, 6).
The existing sets of visible markers and chromosomal deletions cover only a fraction of the mouse genome. Genomewide sets of genetic reagents in the mouse are needed for the global application of phenotype-based approaches that have been invaluable for functional studies in Drosophila. Recently, two strategies have been developed for generating large-scale chromosomal rearrangements at any genomic location by using mouse embryonic stem (ES) cells. One approach involves irradiation to remove a selection cassette permitting the isolation of overlapping deletions (7, 8). Another strategy uses CRE-loxP-mediated chromosome engineering to generate Mb-scale deletions, nested deletion complexes or marked chromosomal inversions (9, 10). Traditionally, the markers used in genetic screens have taken advantage of mutations in endogenous loci that alter coat color or morphology. The K14-Agouti transgene (11) has been used to extend genetic marker applications beyond endogenous loci and linked to CRE-loxP-engineered inversions to produce marked balancer chromosomes (12). However, many of the chromosomal reagents are not tagged and a single marker does not offer a complete range of options in the design of mutagenesis screens. Thus, visible markers are needed to match with these chromosomal resources to develop a more complete set of tools for genetic studies in the mouse.
In this study we have developed a stand-alone genetic marker that can be used in conjunction with the various chromosomal aberration resources being assembled across the mouse genome. The marker is based on a single transgene, pCX-YNC, that drives the ubiquitous coexpression of the enhanced GFP (EGFP) color variants cyan and yellow in embryos and mice. Thus, genotypic classes of offspring can be readily identified in newborn and adult mice, as well as throughout development. Each color cassette can be removed independently by using the CRE or FLPe site-specific recombinases. The result is a modular, dominant marker that can be used at any location in the genome to increase the efficiency and flexibility in the design of genetic screens. The development of complementary sets of chromosomal rearrangements and visible markers will enhance the application of phenotype-based strategies for genetic characterization of the mouse.
Materials and Methods
Plasmid Construction. The pCX-YNC vector was created by cloning the EGFP expression cassette (cytomegalovirus immediate early enhancer with chicken β-actin promoter and first intron driving EGFP) from pCX-EGFP (13, 14) into pPNTlox2, a modified derivative of pPNT (15). The HSV-tk cassette of pPNTlox2 was removed to generate the vector pCX-EGFPneolox, where the loxP sites flank the neomycin (neo) resistance cassette. The EGFP coding sequence in the vector pCX-EGFPneolox was replaced with the enhanced yellow fluorescent protein (EYFP)-coding region (from pEYFP-C1, Clontech) to generate pCX-EYFPneolox. The EYFP and neo expression cassettes from pCX-EYFPneolox were cloned into pPNTlox2, again with the HSV-tk cassette removed, and the pCX-EGFP origin of replication and ampicillin resistance gene were added to generate pCX-EYFPneolox2, where the tandem EYFP and neo expression cassettes were flanked by loxP sites. Next, the enhanced cyan fluorescent protein (ECFP)-coding region (from pECFP-1, Clontech) was used to replace the EGFP coding region in pCX-EGFP and the ECFP expression cassette was cloned between the FRT sites in pFRT2 (16). The FRT flanked ECFP expression cassette was cloned into pCX-EYFPneolox2 to generate pCX-YNC.
Cell Culture and Mice. CJ7 and v6.4 (129S4/SvJae × C57BL/6J)F1 ES cells were grown by using standard culture conditions. A total of 1 × 107 cells were electroporated with 15 μg of SalI linearized pCX-EGFPneolox or pCX-YNC and grown in media with 200 μg/ml G418 (GIBCO) for 7–9 days to select stable integrants. Individual colonies were picked onto 96-well plates and scored for fluorescence. Selected ES cell lines were expanded and frozen with a subset of these used for injection into C57BL/6J blastocysts for production of mice. Chimeric mice were scored for fluorescence and 129-derived agouti coat color and mated to establish germ-line competence and transmission of the fluorescent transgene. S1C2/EGFP mice produced by stable integration of pCX-EGFPneolox into 129-derivative CJ7 ES cells have been maintained on a 129S3/SvImJ background. YNC mice, such as E10/YNC, produced by integration of pCX-YNC into v6.4 ES cells have been maintained on a C57BL/6J background (currently N4). CRE and FLPe recombinase-derived single-color variants were generated by mating E10/YNC mice with EIIa-CRE transgenic mice (17) or FLPeR transgenic mice (18).
Molecular Characterization. Genomic DNA was extracted from either ES cells, liver, or tail tip by using standard techniques. Single-copy integration of the pCX-YNC vector was determined by Southern blot analysis of BamHI-digested genomic DNA by using a 950-bp 32P-labeled SnaBI–SgrAI fragment from the cytomegalovirus enhancer region of the vector. This assay identifies multicopy inserts showing an orientation-dependent, vector-specific unit length BamH1 band and single-copy inserts showing only two unique bands derived from plasmid arms flanked by genomic DNA. PCR analysis was also used to screen for single-copy integrants by using primers C1.F (CAGGGTTATTGTCTCATGAGC) and C10.R (CCTCGACCCATGGTAATAGC) that amplified orientation-specific products to identify multicopy integrants. PCR analysis was used for molecular confirmation of CRE-loxP and FLPe-FRT-mediated rearrangements using primer pairs EYFP.F (GCCACCATGGTGAGCAAGG), EYFP.R (CATGCCGAGAGTGATCCCG) and ECFP.F (CTGACCCTGAAGTTCATCTGC), ECFP.R (TCGTTGGGGTCTTTGCTCAGG) to monitor the loss of the EYFP and ECFP cassettes and the primer pair CRE.F (GACGTTAACATGCATATAACTTC), CRE.R (GCAACATATGCCCATATGCTG) that amplifies a 469-bp CRE-mediated, recombined junction fragment and primer pair FLP.F (CTTCGTATAATGTATGCTATACG), FLP.R (CTTTCCTGCGTTATCCCCTG) that amplifies a 395-bp FLPe-mediated, recombined junction fragment. In addition to visualization, transmission of the fluorescent transgenes was monitored by PCR analysis using the EYFP.F/EYFP.R, ECFP.F/ECFP.R and EGFP.F (GTAAACGGCCACAAGTTCAGC), EGFP.R (CATGCCGAGAGTGATCCCG) primer pairs.
Plasmid Rescue and Mapping. DNA flanking the pCX-YNC integration site was cloned from BamHI-digested E10/YNC genomic DNA by using plasmid rescue as described (19). Genomic DNA captured in the rescued plasmid was sequenced by using the pCX-YNC vector-based primer (TTGTCTCATGAGCGGATAC). Genomic sequence was used to search the Ensembl database (www.ensembl.org) to determine a unique chromosomal position for pCX-YNC integration. The integration site was confirmed by PCR analysis using the primer pair E10–5.F (CTGCT TCTGTCTGT T TGCT TG), E10–5.R (GAAGCTTATAAACATACAGCAAG) flanking the integration site to produce a 664-bp genome-specific product in wild-type or E10/YNC hemizygous mice and the primer pair C1.F, E10–5.R that amplifies a 393-bp junction-specific fragment in E10/YNC hemizygous or homozygous mice.
Fluorescent Imaging. Multispectral fluorescence of ES cells and preimplantation embryos was detected by using a Nikon Diaphot 300 inverted fluorescent microscope fitted with filter sets for excitation [green (HQ470/40X), yellow (HQ500/20X), and cyan (D436/20X)] and emission [green (HQ525/50M), yellow (HQ535/30M), and cyan (D480/40M)]. Postimplantation embryos were viewed by using a Nikon SMZ-U fluorescent stereoscope fitted with filter sets for excitation [green (D470/40X), yellow (Q515LP), and cyan (D460DCLP)] and emission [green (OG515), yellow (HQ535/30M), and cyan (D470/30M)]. Photomicrographs were taken by using a Spot RT cooled color digital camera. Newborn and adult mice were viewed by using a gooseneck fiber-optic light source (100-W lamp) fitted with filter sets for excitation [green (D470/20X), yellow (HQ495/30X), and cyan (D436/20X)] and photographed with a digital camera using filter sets for emission [green (OG515), yellow (OG530), and cyan (D480/40M)]. The same emission filters were also used for routine viewing of animals (filter sets from Chroma Technology, Brattleboro, VT).
Results
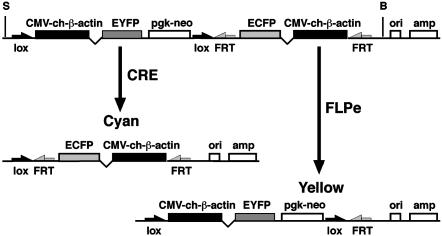
An EGFP-Based Reporter for Enhancing Genetic Screens in the Mouse. The EGFP derived from the bioluminescent jellyfish, Aequorea victoria, has emerged as a widely used reporter in several organisms, including the mouse (20). Detection of EGFP expression is noninvasive, offering the major advantage of in vivo imaging. As a result, EGFP is increasingly used to monitor gene expression and study protein localization and trafficking. Initial studies in transgenic mice used a cytomegalovirus immediate early enhancer and the chicken β-actin promoter to deliver widespread and readily detectable expression of EGFP (13, 21), fulfilling basic criteria for a reporter to be used in genetic screens. We modified the pCX-EGFP vector used in these initial studies to provide additional features that increase its utility as a marker tool. The strategy was to design a reporter construct that would offer the greatest degree of flexibility, but only require a single integration event to tag a defined location in the mouse genome (Fig. 1).
Fig. 1.
A single vector for dual-color marking of chromosomes. View of the linearized pCX-YNC vector showing the three independent expression cassettes for EYFP, neo, and ECFP. The EYFP and neo cassettes are flanked by loxP sites, and the ECFP cassette is flanked by FRT sites. CRE- and FLPe-mediated recombination generates single-color ECFP or EYFP expression as shown. The bacterial origin of replication (ori) and ampicillin resistance gene (amp) can be used for plasmid rescue of adjacent mouse genomic DNA. (S, SalI; B, BamHI).
Generation of Dual-Color ES Cell Lines and Mice. Marked ES cell lines were produced by electroporation of the pCX-YNC vector followed by selection for G418-resistant colonies. Individual clones were randomly picked and evaluated for EYFP and ECFP coexpression. In pilot studies we used the pCX-EGFPneolox single-color vector to establish that a high percentage (58%) of randomly picked colonies surviving selection expressed detectable levels of EGFP. Only ES cell lines exhibiting robust and ubiquitous expression were selected for production of mice. Of the pCX-EGFPneolox ES cell lines, 33% showed strong, uniform fluorescence. In our studies using the pCX-YNC vector, 45% of the stable integrant ES cell lines showed readily detectable, ubiquitous coexpression of EYFP and ECFP. Thus, having the three independent expression cassettes (EYFP, neo, and ECFP) together on a single construct did not appear to significantly interfere with their coexpression. Application of the genetic marker depends on CRE- and FLPe-mediated manipulation. Therefore, pCX-YNC integrants need to be single-copy and produce detectable levels of the fluorescent reporters. We performed Southern blot and PCR analysis to determine that 58% of the dual-color ES cell lines (22 of 38) that were selected for readily detectable, uniform expression contained a single-copy of the pCX-YNC reporter (data not shown).
We used one of the single-color pCX-EGFPneolox ES cell lines, S1C2, to generate mice for our initial studies to evaluate the EGFP-based reporter. The S1C2/EGFP transgenic mouse line showed ubiquitous expression resulting in bright green fluorescence in developing embryos as well as in newborn and adult mice (Fig. 2A). We have maintained the S1C2/EGFP mouse line for seven generations as a hemizygous colony. PCR analysis was used to determine that the transgene was transmitted at the expected ≈50% ratio with all carriers exhibiting green fluorescence, and we have not observed an appreciable reduction in EGFP expression levels over successive generations.
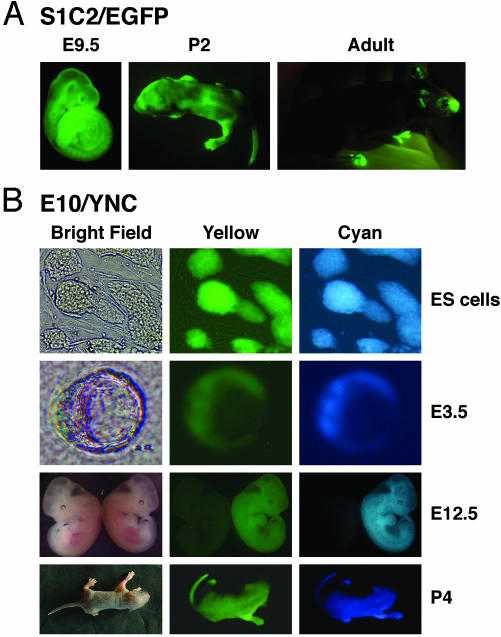
Fig. 2.
Bright dual-color fluorescence in pCX-YNC embryos and mice. (A) The S1C2/EGFP transgenic line shows strong, widespread expression of EGFP in embryonic day 9.5 (E9.5) embryos, 2-day-old pups (P2), and adult mice. (B) The E10/YNC transgenic line illustrates strong, widespread coexpression of EYFP and ECFP in ES cells, blastocysts (E3.5), E12.5 embryos, and 4-day-old pups (P4). Selected stages are shown, both lines showed ubiquitous expression throughout development and as mice.
Six of the brightest dual-color fluorescent pCX-YNC ES cell lines were used for blastocyst injection to generate mice. Five of the lines produced high ES cell contribution chimeras, determined either by viewing yellow and cyan fluorescence at birth or 129-derived agouti coat color. Germ-line competent chimeras for each of the five lines were used to establish colonies of mice. We examined the E10/YNC line to determine the extent of EYFP and ECFP expression. Dual-color fluorescence was easily visualized throughout development, including pre- and postimplantation embryonic stages, as well as in newborn pups and adult mice (Fig. 2B and data not shown). In addition, all adult organs and tissues examined, e.g., brain, heart, lung, skin, and skeletal muscle, exhibited widespread yellow and cyan fluorescence (data not shown). As seen in the E10/YNC line, each of the four other pCX-YNC lines of mice exhibited easily visualized, uniform yellow and cyan fluorescence in developing embryos and in newborn and adult mice (data not shown). Thus, a single-copy of the pCX-YNC reporter construct is able to drive the widespread coexpression of the EYFP and ECFP reporters, permitting the visual selection and efficient production of multiple lines of marked, dual-color ES cells and mice.
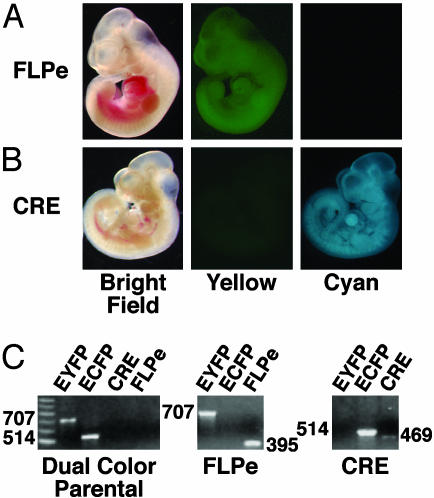
Evaluating Marker Performance in Mice. One of the requirements for executing a variety of marked genetic screens is that segregating chromosomes can be independently tagged and used to distinguish specific classes of offspring. Therefore, we designed the pCX-YNC reporter to be modular by making it possible to remove each color cassette by using CRE-loxP and FLPe-FRT site-specific recombination. The CRE and FLPe recombinases efficiently delete DNA flanked by their specific recognition sequences (22). To demonstrate the modular nature of the reporter, mice from the E10/YNC line were mated to FLPeR mice to excise the FRT flanked ECFP cassette (18). A number of F1 progeny showed a mosaic pattern of cyan fluorescence while retaining ubiquitous yellow fluorescence. These mice were selected as likely germ-line mosaics for production of F2 progeny. As predicted, a significant subset of F2 progeny inherited the recombined pCX-YNC transgene and exhibited only single-color yellow fluorescence (Fig. 3A). PCR analysis was used to confirm that FLPe-mediated recombination had deleted the ECFP cassette (Fig. 3C). The E10/YNC mice were also mated with EIIa-CRE mice to excise the loxP flanked EYFP cassette (17). In this instance, F1 progeny showing a loss of yellow fluorescence were used to generate F2 progeny carrying the recombined pCX-YNC transgene. These mice exhibited only single-color cyan fluorescence (Fig. 3B). Again, PCR analysis was used to confirm that CRE-mediated recombination had deleted the EYFP cassette (Fig. 3C). Thus, one or both color cassettes can be removed, permitting sets of ES cells or mice to be generated that are dual-color, single-color, or no-color depending on the engineering of the integrated vector.
Fig. 3.
CRE- and FLPe-mediated manipulation of the pCX-YNC reporter. (A) E10/YNC midgestation (embryonic day 10.5) embryo showing exclusive yellow fluorescence after FLPe-FRT-based removal of the ECFP cassette. (B) E10/YNC midgestation embryo showing exclusive cyan fluorescence after CRE-loxP-based removal of the EYFP cassette. (C) Molecular confirmation of specific rearrangements. (Left) PCR analysis of the dual-color E10/YNC parental line demonstrates the presence of both the EYFP (707 bp) and ECFP (514 bp) cassettes and the absence of the CRE-loxP- and FLPe-FRT-specific recombination products. (Center) The retention of the EYFP cassette, loss of the ECFP cassette, and the FLPe-FRT vector-specific rearrangement product (395 bp) after FLPe-mediated recombination. (Right) The loss of the EYFP cassette, retention of the ECFP cassette, and the CRE-loxP vector-specific rearrangement product (469 bp) after CRE-mediated recombination.
In various marked genetic schemes, mice are often homozygous for the pCX-YNC transgene or carry the transgene opposite a chromosomal deletion (Fig. 4). Therefore, integration of the pCX-YNC reporter cannot disrupt an essential gene leading to lethality or infertility. We intercrossed E10/YNC hemizygous mice to produce homozygous E10/YNC mice. E10/YNC homozygous mice were viable and fertile with no overtly deleterious phenotypes associated with the insertion event or expression of the fluorescent reporters. All of the offspring of these mice expressed both reporters and displayed consistent dual-color fluorescence. As with E10/YNC, of the two additional pCX-YNC lines tested thus far both have produced normal, viable homozygous mice.
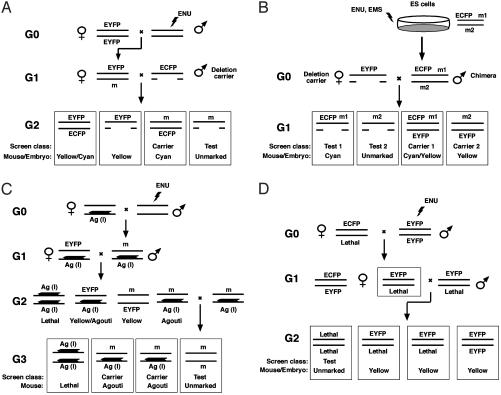
Fig. 4.
Marking directed mutagenesis screens using the pCX-YNC reporter. (A) A traditional hemizygosity screen using visibly marked chromosomes and deletions. In this example an ENU mutagenized male (G0) is mated with an EYFP homozygous female to generate an EYFP marked G1 (G1) female heterozygous for an ENU-induced mutation (m). Mutations in the region are uncovered in a test cross with an ECFP marked deletion carrier male. The G2 (G2) test class progeny are color-coded to visually distinguish the test class (unmarked), mutation carrier (cyan), and two discard classes (yellow/cyan and yellow). (B) A hemizygosity screen combining visibly marked chromosomes, deletions, and ES cell mutagenesis. ENU or EMS mutagenized ES cell clones are used to produce germ-line competent chimeric mice. Mutations in the region produced in the diploid ES cells residing on the ECFP marked chromosome (m1) and on the unmarked chromosome (m2) are uncovered in a single-generation test cross with an EYFP marked deletion carrier female. The G1 test class progeny are color-coded to visually distinguish the m1 test class (cyan), the m1 carrier (cyan/yellow), the m2 test class (unmarked), and the m2 carrier (yellow). An EGFP-marked C57BL/6J host blastocyst can be used to ensure that offspring are ES cell-derived. Host-derived embryos and mice are green and can be distinguished from the cyan and unmarked ES cell-derived test class. Although yellow and green fluorescence are not readily distinguished, the m2 carrier can be identified based on 129-derived ES cell agouti coat color. (C) A regional homozygosity screen using a visibly marked, recessive lethal inversion. An ENU-treated G0 male is mated with a marked inversion-heterozygote female. CRE-loxP mediated chromosome engineering has been used to produce recessive lethal inversion chromosomes dominantly marked with the K14-Agouti transgene (Ag). The mutation (m) carrying marked inversion-heterozygote G1 offpsring are mated with a marked inversion-heterozygote that is also EYFP- (or ECFP)-marked to produce G2 progeny. The mutation carrying marked inversion-heterozygous G2 mice are selected based on the absence of yellow fluorescence. These G2 offspring can be intercrossed to produce the G3 test class progeny that are marked to distinguish the homozygous mutation test class (unmarked) and carrier (e.g., agouti). Homozygosity for the marked inversion is lethal (l), typically owing to the disruption of an essential locus at an inversion breakpoint. (D) A genomewide dominant suppressor screen using visibly marked chromosomes. In this example an ENU-treated EYFP homozygous G0 male is mated with an ECFP marked female heterozygous for the sensitizing recessive mutation (e.g., embryonic lethality). In this two-generation scheme, G1 females heterozygous for the lethal mutation that are also heterozygous for a genomewide complement of ENU-induced mutations are selected based on yellow fluorescence and mated with an EYFP marked male that is heterozygous for the sensitizing lethal mutation to produce the G2 test cross progeny. Typically, the unmarked class homozygous for the sensitizing recessive lethal mutation will be absent in the G2 litter. The recovery of unmarked G2 progeny reveals an ENU-induced mutation that acts as a dominant suppressor of the lethal phenotype.
Assigning Genomic Location. The pCX-YNC transgene was randomly integrated to generate marked lines of ES cells that can be characterized and selected to produce mice. This offers a rapid approach for creating an initial bank of ES cells harboring the pCX-YNC reporter distributed across the genome. Once marked ES cells are evaluated for dual-color expression and transgene copy number, a chromosomal position needs to be determined for the reporter. To this end, the pCX-YNC construct contains plasmid sequences for isolating adjacent genomic DNA. Plasmid rescue permits the efficient recovery of flanking DNA to map the insertion site of randomly integrated plasmids (9). We used plasmid rescue to clone a genomic fragment at the junction of the E10/YNC integration site (19). Sequence from this fragment was used to search the genome for a specific address. The sequence showed a unique match mapping the location of the E10/YNC integration site at ≈40.3 Mb on mouse chromosome 5 (Ensembl mgsc v15.30.1). Complete genomic sequence data were used to design PCR primers on opposite sides of the integration site to confirm the map position. PCR analysis using DNA from wild-type or E10/YNC hemizygous mice amplified the appropriate 664-bp endogenous segment, whereas this product was not amplified by using DNA from E10/YNC homozygous mice (data not shown).
Discussion
Mutations induced by ENU or EMS are an important reagent for studying gene function. As point mutagens, these agents produce a variety of alleles leading to null, hypomorphic, and hypermorphic phenotypes. Such an allelic series is useful for revealing a complete range of gene activity, as well as identifying critical aspects of protein structure and function by unveiling amino acids essential for domain interaction and activity (23). A number of screening strategies are used to isolate ENU- and EMS-induced mutations. Genomewide screens represent a valuable survey to recover sets of mutations related to a specific biological process. Focused screens to identify modifiers or directed at a defined chromosomal interval extend mutagenesis approaches. Such directed screens are most powerful when they incorporate selection schemes or visible genetic markers. To this end, we have developed an EGFP-based reporter, pCX-YNC, that serves as a modular, dominant marker to efficiently recover mutations in mice by using various phenotype-based mutagenesis screening strategies.
Marked Regional Screens Using a Chromosomal Deletion. An unmarked, genomewide screen requires a three generation breeding scheme to recover recessive mutations. Therefore, considerations such as expensive animal husbandry and a ≈10-week generation time limit the number of genomes that can be screened. In addition, all of the offspring need to be phenotyped to identify a mutant. Thus, it is labor intensive to screen for embryonic lethality and often impractical to screen for late-onset diseases. In contrast, a traditional hemizygosity screen using a chromosomal deletion uncovers recessive mutations in a two-generation breeding scheme (4, 24, 25). By incorporating the pCX-YNC reporter all genetic classes of offspring are visually marked (Fig. 4A). As a result, embryonic lethal phenotypes can be simply identified as a missing class or appropriate offspring can be selected at developmental as well as adult stages for high investment phenotyping. The carrier class mouse that is heterozygous for the mutation is also easily identified for recovery of the ENU-induced allele.
Recently, ENU and EMS have been used to mutagenize mouse ES cells that can then be used to produce mice (26, 27). A high mutation load, different classes of mutagen, and shortened breeding schemes are advantages afforded by chemical mutagenesis of ES cells (28). When ES cell mutagenesis is used, a regional hemizygosity screen can be performed by using a single-cross breeding scheme. Incorporating the pCX-YNC reporter identifies two test classes of mice for phenotype assessment and marks each corresponding heterozygous carrier of the induced mutation (Fig. 4B). With their shortened breeding schemes and visual genotyping of progeny, marked regional screens allow a greater number of genomes to be surveyed and reduce the effort associated with phenotyping. Given these benefits, regional screens are well established as a practical approach for the functional dissection of a defined chromosomal region. In addition, genomewide transcript maps and QTL analyses are revealing clusters of genes that are coexpressed or perform related biological activities (29, 30). Thus, tools that facilitate a more comprehensive analysis of a defined chromosomal interval are also valuable for understanding relationships between genome organization and function.
Marked Regional Screens Using a Chromosomal Inversion. A regional screen using a marked chromosomal inversion offers similar advantages to a deletion-based screen for uncovering lethal mutations as well as highlighting test and carrier class offspring for phenotyping and stock maintenance, respectively. A traditional inversion-based screen uses a three-generation breeding scheme to isolate recessive mutations (4). However, inversions have unique advantages in that they suppress recombination and can span a larger interval (e.g., 20–40 centimorgans) without the risk of presenting haploinsufficient phenotypes that restrict the use of Mb-scale deletions. Thus, marked inversions can be generated across the genome. Still, these reagents require a second dominant visible marker for use in a three-generation regional homozygosity screen. The pCX-YNC reporter can serve as a second region-specific marker to be used with inversions for performing regional mutagenesis screens (Fig. 4C).
Marking Sensitized Screens to Uncover Suppressor Mutations. Focused screens can also be directed at understanding protein assemblies or genetic pathways. An existing mutation is used in these sensitized screens to uncover mutations that enhance or suppress the original phenotype. Modifier screens have been extensively used in Drosophila, for example, to identify components of the receptor tyrosine kinase signaling pathway (31). Although not widely used in mice, modifier screens have the potential to reveal important genetic interactions. Tools that improve their efficiency or permit screens that would otherwise be impossible are needed to increase the application of modifier screens in the mouse. When the pCX-YNC reporter is used, it is possible to design a two-generation, genomewide screen to reveal dominant mutations that suppress a phenotype such as embryonic lethality (Fig. 4D). A number of key developmental regulators exhibit dosage sensitive genetic interactions. Mutations that suppress a phenotype, for example, by modifying negative feedback and permitting reduced signaling or alternative pathways to elicit a normal cellular response would provide important insights into the intricate thresholds and interplay between pathway components that establish signaling networks.
Building a Visible Marker Resource to Extend Genetic Studies in the Mouse. There is increasing emphasis on understanding the functional composition of the mammalian genome to learn more about the genetic basis of development and disease. Numerous annotation and comparative analysis tools are being developed to study genomic sequence data. In addition, there is a growing list of methods, both gene- and phenotype-driven, to generate mutations for investigating gene activity. Given the goal of genomewide functional annotation and the scope of genetic complexity, it is critical that reagents that enhance approaches for increasing the mouse mutant resource continue to be developed (32). The pCX-YNC reporter offers a flexible reagent that adds to the growing list of resources for performing focused mutagenesis screens at an increasing number of genomic locations. CRE- and FLPe-mediated recombination can be performed in ES cells or mice to readily generate various color combinations. Using the color combinations, it is possible to design a number of diverse screens, breeding schemes (e.g., stock maintenance), and reporter assays (e.g., lineage analysis) in addition to the examples provided in Fig. 4. Thus, we envision that the pCX-YNC marker could be used in a wide variety of genetic applications.
We have demonstrated that a significant percentage of the pCX-YNC ES cell lines (26% of the total number of colonies picked) show dual-color fluorescence as a single-copy integrant. In addition, all of the mouse lines produced from the dual-color ES cells exhibited widespread fluorescence suitable as a universal genetic marker. Visual monitoring, from expression in ES cells to CRE- and FLPe-mediated manipulations, is an added advantage for easy development and application of the resource. Finally, targeting or random integration followed by plasmid rescue can be used to place the vector in a defined chromosomal location. Taken together, these results indicate that the pCX-YNC reporter is well suited for the production of a bank of ES cells that can be used as a genetic marker tool in mice. Ideally, for genomewide application, the marker density and distribution would provide tight linkage with target intervals to minimize labor associated with verifying recombination events that would “misidentify” marked offspring. In model organisms, such as Drosophila and zebrafish, where extensive sets of mutations already exist, a variety of inventive screens continue to enrich the collections of mutants for genetic studies (1, 33). Genomewide sets of visible markers and chromosomal aberrations offer important reagents for building collections of mutant mice for genetic characterization of the mammalian genome.
Acknowledgments
We thank M. Okabe (Genome Information Research Center, Osaka University, Osaka) and S. Dymecki (Department of Genetics, Harvard Medical School, Boston) for generously providing the pCX-EGFP and pFRT2 vectors, respectively. We thank T. Gridley and J. Schimenti for ES cells, and G. Cox and S. John for helpful discussions and comments on the manuscript. This work was supported by National Institutes of Health Grant GM62928 (to T.P.O.), the shared service facilities of The Jackson Laboratory (TJL) Cancer Center (CORE Grant CA34196), and TJL Institutional Mutagenesis Program Awards NS41215 and HL66611.
Abbreviations: ECFP, enhanced cyan fluorescent protein; EGFP, enhanced GFP; EYFP, enhanced yellow fluorescent protein; EMS, ethylmthanesulfonate; ENU, N-ethyl-N-nitrosourea; ES, embryonic stem; Mb, megabase.
References
- 1.St. Johnston, D. (2002) Nat. Rev. Genet. 3, 176–188. [DOI] [PubMed] [Google Scholar]
- 2.Davis, A. P. & Justice, M. J. (1998) Genetics 148, 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russell, W. L., Kelly, E. M., Hunsicker, P. R., Bangham, J. W., Maddux, S. C. & Phipps, E. L. (1979) Proc. Natl. Acad. Sci. USA 76, 5818–5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinchik, E. M. (2000) Mamm. Genome 11, 489–499. [DOI] [PubMed] [Google Scholar]
- 5.Roix, J. J., Hagge-Greenberg, A., Bissonnette, D. M., Rodick, S., Russell, L. B. & O'Brien, T. P. (2001) Genetics 157, 803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinchik, E. M. & Carpenter, D. A. (1999) Genetics 152, 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.You, Y., Bergstrom, R., Klemm, M., Lederman, B., Nelson, H., Ticknor, C., Jaenisch, R. & Schimenti, J. (1997) Nat. Genet. 15, 285–288. [DOI] [PubMed] [Google Scholar]
- 8.Thomas, J. W., LaMantia, C. & Magnuson, T. (1998) Proc. Natl. Acad. Sci. USA 95, 1114–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LePage, D. F., Church, D. M., Millie, E., Hassold, T. J. & Conlon, R. A. (2000) Proc. Natl. Acad. Sci. USA 97, 10471–10476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mills, A. A. & Bradley, A. (2001) Trends Genet. 17, 331–339. [DOI] [PubMed] [Google Scholar]
- 11.Kucera, G. T., Bortner, D. M. & Rosenberg, M. P. (1996) Dev. Biol. 173, 162–173. [DOI] [PubMed] [Google Scholar]
- 12.Zheng, B., Sage, M., Cai, W. W., Thompson, D. M., Tavsanli, B. C., Cheah, Y. C. & Bradley, A. (1999) Nat. Genet. 22, 375–378. [DOI] [PubMed] [Google Scholar]
- 13.Okabe, M., Ikawa, M., Kominami, K., Nakanishi, T. & Nishimune, Y. (1997) FEBS Lett. 407, 313–319. [DOI] [PubMed] [Google Scholar]
- 14.Niwa, H., Yamamura, K. & Miyazaki, J. (1991) Gene 108, 193–199. [DOI] [PubMed] [Google Scholar]
- 15.Tybulewicz, V. L., Crawford, C. E., Jackson, P. K., Bronson, R. T. & Mulligan, R. C. (1991) Cell 65, 1153–1163. [DOI] [PubMed] [Google Scholar]
- 16.Dymecki, S. M. (1996) Gene 171, 197–201. [DOI] [PubMed] [Google Scholar]
- 17.Lakso, M., Pichel, J. G., Gorman, J. R., Sauer, B., Okamoto, Y., Lee, E., Alt, F. W. & Westphal, H. (1996) Proc. Natl. Acad. Sci. USA 93, 5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farley, F. W., Soriano, P., Steffen, L. S. & Dymecki, S. M. (2000) Genesis 28, 106–110. [PubMed] [Google Scholar]
- 19.Hicks, G. G., Shi, E. G., Li, X. M., Li, C. H., Pawlak, M. & Ruley, H. E. (1997) Nat. Genet. 16, 338–344. [DOI] [PubMed] [Google Scholar]
- 20.Hadjantonakis, A. K. & Nagy, A. (2001) Histochem. Cell Biol. 115, 49–58. [DOI] [PubMed] [Google Scholar]
- 21.Hadjantonakis, A. K., Gertsenstein, M., Ikawa, M., Okabe, M. & Nagy, A. (1998) Mech. Dev. 76, 79–90. [DOI] [PubMed] [Google Scholar]
- 22.Kilby, N. J., Snaith, M. R. & Murray, J. A. (1993) Trends Genet. 9, 413–421. [DOI] [PubMed] [Google Scholar]
- 23.Justice, M. J., Noveroske, J. K., Weber, J. S., Zheng, B. & Bradley, A. (1999) Hum. Mol. Genet. 8, 1955–1963. [DOI] [PubMed] [Google Scholar]
- 24.Hagge-Greenberg, A., Snow, P. & O'Brien, T. P. (2001) Mamm. Genome 12, 938–941. [DOI] [PubMed] [Google Scholar]
- 25.Justice, M. J., Zheng, B., Woychik, R. P. & Bradley, A. (1997) Methods 13, 423–436. [DOI] [PubMed] [Google Scholar]
- 26.Munroe, R. J., Bergstrom, R. A., Zheng, Q. Y., Libby, B., Smith, R., John, S. W., Schimenti, K. J., Browning, V. L. & Schimenti, J. C. (2000) Nat. Genet. 24, 318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen, Y., Yee, D., Dains, K., Chatterjee, A., Cavalcoli, J., Schneider, E., Om, J., Woychik, R. P. & Magnuson, T. (2000) Nat. Genet. 24, 314–317. [DOI] [PubMed] [Google Scholar]
- 28.Chen, Y., Schimenti, J. & Magnuson, T. (2000) Mamm. Genome 11, 598–602. [DOI] [PubMed] [Google Scholar]
- 29.de Haan, G., Bystrykh, L. V., Weersing, E., Dontje, B., Geiger, H., Ivanova, N., Lemischka, I. R., Vellenga, E. & Van Zant, G. (2002) Blood 100, 2056–2062. [DOI] [PubMed] [Google Scholar]
- 30.Caron, H., van Schaik, B., van der Mee, M., Baas, F., Riggins, G., van Sluis, P., Hermus, M. C., van Asperen, R., Boon, K., Voute, P. A., et al. (2001) Science 291, 1289–1292. [DOI] [PubMed] [Google Scholar]
- 31.Therrien, M., Morrison, D. K., Wong, A. M. & Rubin, G. M. (2000) Genetics 156, 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nadeau, J. H., Balling, R., Barsh, G., Beier, D., Brown, S. D., Bucan, M., Camper, S., Carlson, G., Copeland, N., Eppig, J., et al. (2001) Science 291, 1251–1255. [DOI] [PubMed] [Google Scholar]
- 33.Patton, E. E. & Zon, L. I. (2001) Nat. Rev. Genet. 2, 956–966. [DOI] [PubMed] [Google Scholar]