Abstract
The ability to image and quantify multiple biomarkers in disease necessitates the development of split reporter fragment platforms. We have divided the β-galactosidase enzyme into unique, independent polypeptides that are able to re-assemble and complement enzymatic activity in bacteria and in mammalian cells. We created two sets of complementing pairs that individually have no enzymatic activity. However, when brought into close geometric proximity, the complementing pairs associated resulting in detectable enzymatic activity. We then constructed a stable ligand complex comprised of reporter fragment, linker, and targeting moiety. The targeting moiety, in this case a ligand, allowed cell surface receptor targeting in vitro. Further, we were able to simultaneously visualize two cell surface receptors implicated in cancer development, epidermal growth factor receptor and transferrin receptor, using complementing pairs of the ligand-reporter fragment complex.
Keywords: β-galactosidase, complementation, enzyme fragments, targeted-reporter complex
INTRODUCTION
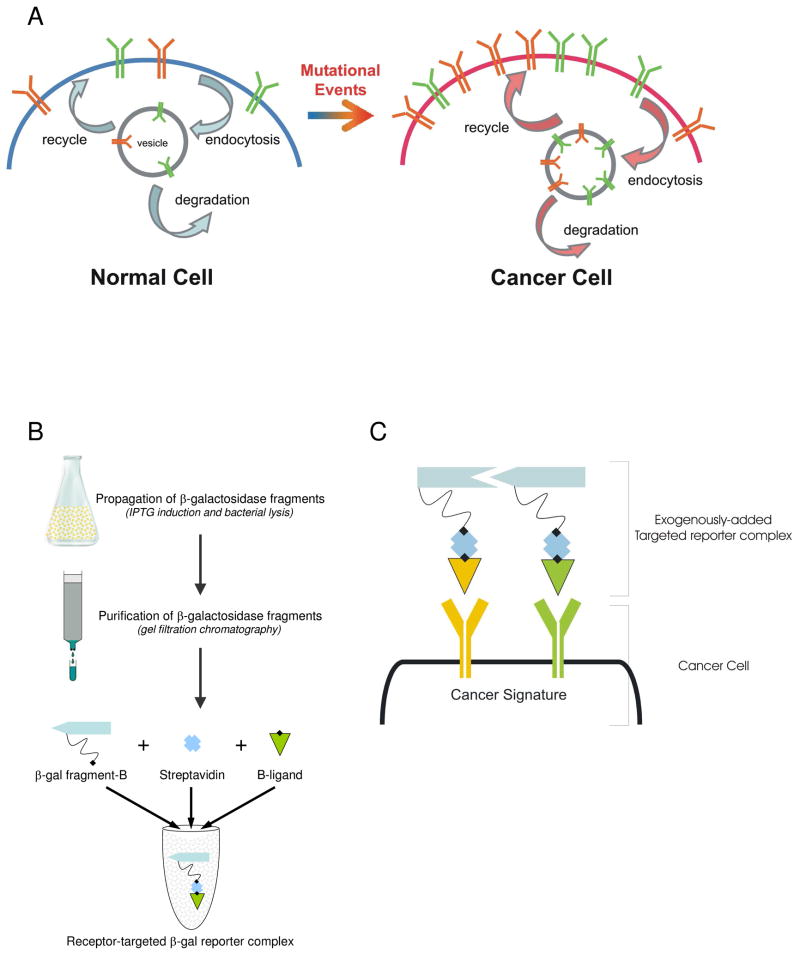
Targeted-reporter imaging agent platforms answer a critical unmet need and have real application for imaging the multi-step progression of cancer growth.1 The development of these platforms to investigate molecular signatures associated with disease creates the next frontier in in vivo imaging. Mutational events that drive a normal cell to become a cancer cell require the coordinated overexpression of multiple biomarkers, e.g., cell surface receptors (Figure 1A). For instance, the most common gain of function mutation observed in invasive phenotypes associated with breast, ovarian, skin, brain, and lung cancers is the amplification and overexpression of the epidermal growth factor receptor (EGFR).2–6 Overexpression of the transferrin receptor (TfR) has also been implicated by our laboratory as well as others in the malignant phenotype.7–9 Tumors that overexpress EGFR and TfR have increased activity that leads to uncontrolled cell growth accompanied by decreased apoptosis and increased angiogenesis. The overexpression of these receptors also leads to activation of other genes that promote cancer growth through such means as invasion and metastasis, as well as resistance to chemotherapy and radiotherapy.10,11 These expression patterns of multiple biomarkers can thus be indicative of the type, stage, or severity of the disease.12
Figure 1.
Targeted-reporter fragment complementation in identifying the cancer signature. A Cancer progression. B Targeted-reporter complex formation. C Receptor targeted-reporter complementation schematic.
A growing body of evidence asserts that several upregulated biomarkers contribute to tumor behavior.13,14 This is an especially intriguing development since most disease-associated assays rely on single biomarker identification and few of the hundreds of single markers evaluated to date have demonstrated significant clinical or diagnostic utility. Given that disease is recognized both by its complexity and progression, single biomarker utilities are self-delimiting, but by surveying many biomarkers at once with the use of microarray-based gene expression profiling or proteomic technologies, it is now possible to read the molecular signature of an individual patient’s tumor. In fact, determining the status of various cell surface receptors has become routine in the care of cancer patients and has proven useful in guiding standard of care treatment.15–17 By developing multi-marker imaging, we will provide an imaging tool to exploit the accruing molecular understanding of cancers allowing eventual imaging of combinatorial biomarkers that will uniquely identify cancers and predict prognosis non-invasively.
Advances in the identification of suitable cancer biomarkers have been the hallmark of the genomic and proteomic revolutions and allow researchers to develop imaging tools that are both more specific and sensitive for detection of disease. The desire to label multiple biomarkers has lead to high-throughput serial (HTS) identification schemes that take a parallel approach to compound analysis by incorporating diverse assay types to correlate protein expression to specific types or stages of cancer. These assays use tumor biopsy samples and therefore can only sample a limited portion of the entire tumor, yielding only partial information regarding tumor biomarker expression. The results of such studies are often limited by technical variability between assays, lack of appropriate controls, and a paucity of direct interactions among the biomarkers examined.18 Most of these techniques are not amenable to translation into non-invasive in vivo imaging paradigms.
Further investigations have allowed for imaging of direct protein-protein interactions.19–21 The most promising methodology is referred to as the Protein fragment Complementation Assay (PCA).22 PCAs are based on protein-protein interaction strategies that dissect a reporter protein into two fragments and fuse each fragment to one of two known interacting proteins of interest. Reassembly and activity of the reporter protein fragments occurs via oligomerization-assisted interaction of the proteins of interest. Most PCAs include small, monomeric split reporter proteins with well-characterized domains and “bait and prey” proteins of interest. The split reporter proteins are necessarily inactive as fragments and do not spontaneously interact. Instead, reporter protein assembly is driven by the high affinity protein interactions of the fused proteins of interest. It is important to highlight the key consequence of using most PCAs - genetic manipulation of either bacteria or mammalian cells. As a result, this decreases the translation of these assays into clinical settings and instead limits them to in vitro cell imaging or in vivo small animal imaging applications. Other key obstacles that are not necessarily addressed by direct protein-protein imaging include identification of suitable cancer biomarkers, exploitation of these biomarkers in detection protocols, and development of technologies that improve selectivity, sensitivity, and specificity of biomarker-targeted delivery of imaging agents or therapeutics.
To develop imaging tools that take advantage of the diagnostic molecular signature but do not require protein-protein interactions or genetic manipulation of the target cells, new technologies must employ a contrast agent or signal-amplifying material conjugated to a molecular targeting agent. By linking a reporter enzyme to a targeting moiety, signal-amplification at the molecular level can be achieved. To expand this approach, we have engineered enzyme fragments that in themselves have no activity, but will complement in trans to provide robust activation at the cell surface.
One of the most popular and widely used reporter enzymes is Escherichia coli β-galactosidase (β-gal) encoded by the lacZ gene. β-gal can hydrolyze disaccharides such as β-galactosides, including lactose, into monosaccharides. The protein product is extremely stable and resistant to proteolytic degradation. Many substrate detection reagents to measure β-gal’s catalytic activity are commercially available for use in calorimetric, fluorescent, and chemiluminescent assays.23–25
β-gal was one of the first enzymes to be broken into spontaneously re-combinable fragments.26 First sequenced in 1970, β-gal is a homotetrameric protein comprised of four polypeptide chains, each 1023 amino acids long.27 β-gal monomers are further subdivided into five domains with much of the active site formed at the carboxy terminal end of domain 3.27,28 β-gal, like most other enzymes, is constitutively active as long as it is properly folded. The ability to restore enzymatic activity from cleaved fragments is the fundamental basis of α-complementation, a staple of blue/white clonal screening.29,30 Based on subtractive deletion mutants, β-gal was originally split into a small amino terminal fragment (residues 3-41, alpha-donor) and the large remaining subunit (alpha-acceptor). The two fragments are inactive separately. But when an alpha-donor subunit links two alpha-acceptor dimers together, intracistronic complementation (dimer-dimer interaction) occurs and restores the active quaternary conformation of β-gal.31–33
Historically, molecular biology reporter assays were developed to monitor gene expression by integrating reporters that were either intrinsically fluorescent or enzymatic. Enzymes, such as β-galactosidase, firefly luciferase, β-lactamase, and alkaline phosphatase, are particularly effective reporters because they are specific, sensitive, and stable in diverse applications.20,34–36 In addition, most enzyme reporters have little to no endogenous expression in mammalian cells, effectively reducing background complications. Enzyme reporters when compared to their fluorescent counterparts are also favored because they can be amplified by prolonged incubation with substrate, thereby increasing reporter sensitivity.
To date, enzyme reporters have been successfully linked to innumerable targets: genes, antibodies, and peptide proteins. Investigators have envisioned using these reporter fragments to reveal protein-protein interactions, intracellular localization and translocation, and non-invasive imaging using activity-based reporters.19,37,38 Their biological information on cellular status, however, is limited for these non-invasive imaging approaches, which until recently have sought to link an enzyme’s single catalytic activity to changes in expression of a single biomarker. Reporter fragmentation expands the functionality of the reporter assay.20,34,39
To create a targeted-reporter complex (Figure 1B), the biotinylated β-gal fragment is first induced in bacteria. Next, the protein fragments are fractionated by gel filtration column chromatography and fractions containing the fragment of interest are combined, dialyzed, and concentrated. Finally, the biotinylated β-gal fragment is combined with streptavidin and biotinylated receptor ligand and incubated for 1 hr. at room temperature; the result is referred to as the receptor-targeted β-gal reporter complex. Once the complex is created, it can be exogenously added in complementing pairs to live cell cultures (Figure 1C). Therefore, investigators are only constrained by the number of fragments with which the reporter enzyme can be broken. Thus, by expanding the number of biomarkers available to diagnostic and therapeutic purview, we envision a novel and innovative platform-based approach to disease identification, staging, and treatment.
Here, we have engineered completely unique β-gal fragments from those previously identified by truncation mutants. These studies show that the β-gal activity can be obtained by biomarker-facilitated trans-complementation of the individual subunits. Individually, the monomer fragments are small, stable, and enzymatically inactive. When the correct combination of β-gal fragments are incorporated in bacteria or correctly oriented in mammalian cells, trans-complementation occurs and enzymatic activity is restored.31–33 Further, we demonstrate the utility of these fragments for targeted-complementation in live cell assays and demonstrate the robustness of the system with an eye on utilizing these fragments to investigate molecular signatures associated with disease.
EXPERIMENTAL SECTION
Plasmids and bacterial strains
pSV-β-gal was kindly provided by Dr. Antonio Choicca. pHAT10 vector was purchased from Clontech (Mountain View, CA); pAN4 vector was purchased from Avidity, Inc. (Aurora, CO). E. coli K12 ER1793 bacteria were purchased from New England BioLabs (Ipswich, MA). Rat glioma C6 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat inactivated fetal calf serum (FCS).
β-galactosidase constructs creation
Full-length β-gal from the pSV-β-gal plasmid (Promega; Madison, WI) was used as the starting genetic template. DNA encoding the split-β-gal fragment(s) is amplified by PCR using full length β-gal template and primers that introduce flanking restriction enzyme sequences. The PCR product was ligated into a pHAT10 vector (Clontech; Mountain View, CA) containing a suitable antibiotic selectable marker for bacterial propagation. The resulting coding sequence, consisting of the His-tag and split-β-gal fragment, was excised from the vector and inserted into the pAN4 vector (Avidity; Aurora, CO). The pAN4 vector was used to express a single N-terminal biotin-protein fusion.
Propagation of β-galactosidase extracts
Luria-Bertani (LB) broth (5 mL) containing ampicillin (50 μg/μl) was inoculated with a bacterial scrape (~25 μL) containing one of the discrete β-gal plasmids and allowed to grow overnight (18–20 hr.) in an incubator/orbital shaker at 37 °C. Following the growth period, LB broth (1 L) with ampicillin (50 μg/μl) was inoculated with the 5 mL overnight growth and placed in the incubator/orbital shaker at 37 °C until an absorbance of 0.4 was observed at 600 nm (5.5 hr.). The bacterial culture was then induced with 1 mM IPTG. For subunits α-4, 1-ω, and3 ω, as well as full length β-gal, the IPTG-induced culture was grown in an incubator/orbital shaker at 37 °C until a reading between 0.8 and 1.0 was observed at absorbance 600 nm (6 hr.). For subunits α-1 and 1–4, the IPTG-induced bacterial culture was grown on a rocker at room temperature for 19 hr. Upon completion, the cells were pelleted and the supernatant discarded. The cell pellet was lysed with lysozyme at room temperature for 20 min. and then stored at −80°C until purification.
Purification of β-galactosidase extracts
β-gal subunits were purified from contaminating bacterial cell degradation products and other particles using affinity chromatography. The whole cell lysate was thawed at 37 °C, sonicated, and centrifuged. The lysate was passed over a nickel Talon affinity column (Clontech; Mountain View, CA) using gravity flow. The column was washed twice with extraction buffer [50 mM NaHPO4, pH 7; 300 mM NaCl in ddH2O]. The His-tagged protein was eluted off the column in 0.5mL fractions with elution buffer [0.15 M imidazole in extraction buffer]. Each fraction was analyzed for the presence and concentration of purified protein using standard protein analysis (Bio Rad DC Protein Assay kit; Bio Rad; Hercules, CA) and immunoblot analysis with anti-His (Upstate, Billerica, MA) and HRP-conjugated streptavidin (Chemicon; Temecula, CA). Fractions containing the desired protein were combined, dialyzed against PBS, and stored at 4 °C.
β-galactosidase activity assay
For the bacteria-based activity assay, E. coli K12 ER1793, deficient in β-galactosidase activity, were transfected with individual β-galactosidase fragment plasmids or combinations of plasmids that would produce a complementing full length enzyme. Bacteria were electroporated with plasmid (1 ng) for 5.2 msec at 2.5 kV. After 1 hr incubation in LB broth, bacteria were streaked on LB agar plates containing ampicillin (0.1 g/L) and X-gal (50 μg/ml). Plates were incubated at 37 °C overnight. Colonies containing active β-galactosidase stained blue.
For the solution-based activity assay, β-gal fragments were added, either individually or in complementing pairs, to uncoated 96-well microtiter assay plates in equal molar amounts and incubated at room temperature on an orbital rocker for 1 hr. Full-length β-gal (1 mg/ml) was serially diluted to create a standardized concentration curve. At time zero, the assay was initiated by using a 12-channel pipettor to add 20 μl of ONPG (4 mg/ml) to each well of the microplate. In the endpoint assay, the microplates were incubated at room temperature for the appropriate length of time, e.g., 30 min, before the reaction was terminated by the addition of 50 μl of 1 M Na2CO3. Then, the absorbance (420 nm) was read in a Tecan Infinite 200 plate reader (Tecan; San Jose, CA). The absorbance data was transferred to a Microsoft Excel spreadsheet and the amount of ONPG substrate hydrolyzed was calculated.
Western blot analysis
Samples were lysed in 200 μL cell lysis buffer (Cell Signaling Technology, Inc.; Danvers, MA) on ice for 10 min., sonicated, and centrifuged at 4°C, 3 min., 13,200 rpm. Protein concentrations were determined by modified Bradford assays performed on supernatants using the DC Bio-Rad Protein Assay Dye kit (Bio-Rad; Hercules, CA). Absorbance was measured at 750 nm using the Tecan Infinite 200 (Tecan; San Jose, CA). Equal amounts (100 μg) of the protein samples were boiled for 5 minutes in 1X final concentration reducing sample buffer. The samples were run in 10% bis-acrylamide SDS-PAGE running gels for 60 min. at 150V using the mini-Protean 3 electrophoresis system (Bio-Rad; Hercules, CA), then transferred to nitrocellulose membrane using a mini-Protean Transblotter system (Bio-Rad; Hercules, CA) for 60 minutes at 200V. For immunoblotting, the membranes were blocked with 5% condensed milk solution in Tris Buffered Saline-Tween 20 (TBST, 1h, RT). Cells stably overexpressing the human receptors EGFR and TfR were stained with either monoclonal anti-EGFR (1:500 dilution; DAKO; M7298) or monoclonal anti-TfR (1:500 dilution; US Biologicals; T8199) for 1 hr. at room temperature. Blots were then incubated with HRP-conjugated secondary antibody (1:1000 dilution) for mammalian whole cell extracts in TBST for 1 hr. at room temperature. Immunoblots containing electrophoresed bacterial lysates were incubated with HRP-conjugated streptavidin (1:1000 dilution) to recognize the biotinylated β-galactosidase fragments. After three TBST washes (5 min. each), the blots were incubated with ECL chemiluminescence reagent (Immobilon Western Kit; Millipore; Billerica, MA) for 1 min. and exposed to KODAK BioMax Light Film (Kodak; Rochester, NY).
Receptor uptake immunofluorescence
Cells overexpressing both human EGFR and human TfR were simultaneously incubated over time (10–60 min.) with Alexa488-EGF (5 μg/ml) and Alexa594-Tf (50 μg/ml) at 37 °C. Cells were rinsed briefly, fixed, counterstained with DAPI to visualize the nuclei, and mounted for observation.
Histochemical assay for β-galactosidase activity
The assay was performed according to a protocol described previously.31,33 After transfection with the β-gal constructs for 48 h in a 12-well plate, cells were fixed in 4% paraformaldehyde in phosphate buffered saline (PBS) at 25°C for 5 min. and rinsed twice with PBS for 5 min. X-gal (5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside; Sigma; St Louis, MO, USA) was diluted to a final concentration of 1 mg/mL in 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 2 mM MgCl2 in PBS, applied to cells, and incubated at 37 °C overnight. Cells were rinsed twice with PBS for 5 min. Images were captured by a Retiga EXi camera connected to a Leica DM4000 B upright microscope (Leica Microsystems; Wetzlar, Germany).
Targeted-reporter complex assay for live cells
Biotinylated ligands for the epidermal growth factor receptor (EGFR) or transferrin receptor (TfR) (B-EGF and B-Tf, Invitrogen; Carlsbad, CA) were linked to biotinylated split-β-gal fragments using streptavidin. Ligand, linker, and reporter fragment were mixed in a molar ratio 1:1:3 at room temperature for 1 hr. Excess D-biotin was added to block any remaining unbound streptavidin sites. In the case of control assays, untargeted reporter complex was prepared with D-biotin, in place of the ligand moiety, mixed with linker and reporter fragment in a molar ration 1:1:3. The ligand-complex was then diluted to 500 μl with cell feeding media (DMEM, 10% fetal bovine serum, 1% penicillin-streptomycin) and added directly to coverslips seeded with cells overexpressing both human EGFR and TfR. Cells were pre-incubated with EGF ligand-complex (0.66 μM based on targeting moiety) for 15 min and rinsed prior to the addition of Tf ligand-complex (0.66 μM based on targeting moiety) for an additional 10 min at 37 °C. The cells were then fixed with 4% paraformaldehyde, rinsed with X-gal wash buffer, and stained overnight at 37 °C with 1 mg/ml X-gal as described previously. Images were captured by a Retiga EXi camera connected to a Leica DM4000 B upright microscope (Leica Microsystems; Wetzlar, Germany).
RESULTS
Enzyme candidates for the split-protein fragment approach are readily accessible in nature. We utilized selection criteria that took into account basic enzyme characteristics such as high substrate specificity, lack of (or low level) endogenous expression in mammalian cells, activity at physiologic pH, defined molecular structure including sub-domain functional activities, and low toxicity when the enzyme is subsequently introduced into eukaryotic cells. β-gal was a potentially robust enzyme that fit the criteria for enzyme fragmentation and imaging. Further, β-gal has defined domains that make it amenable to fragment design.
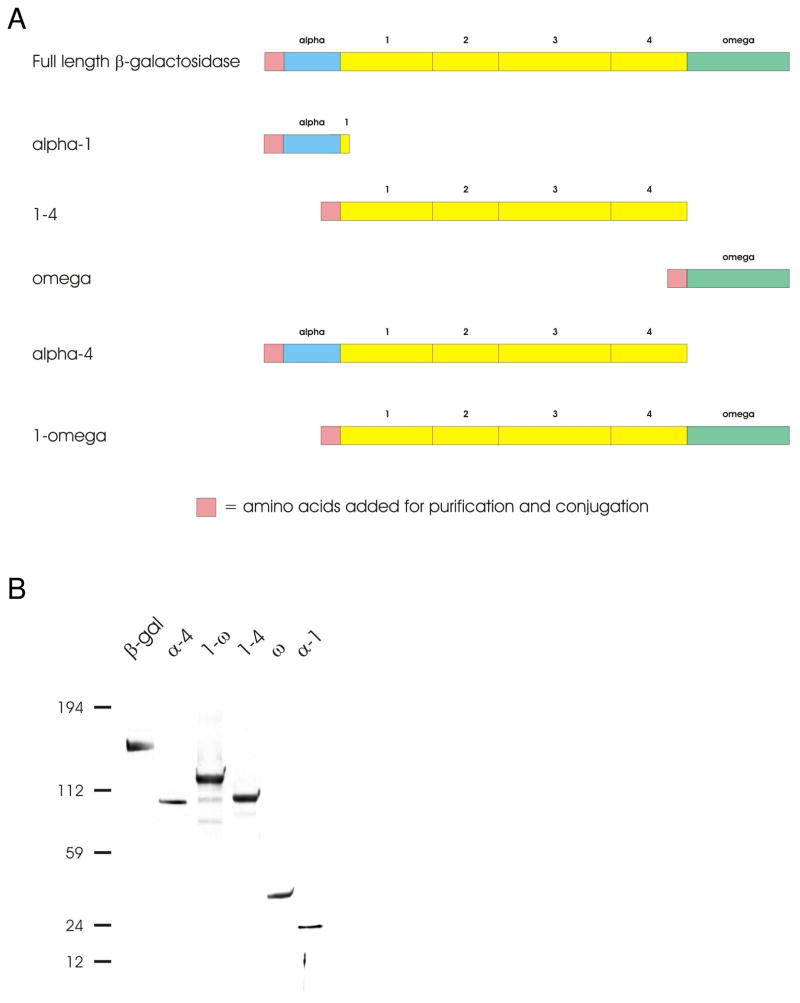
To achieve this, β-gal was first split into multiple polypeptides (Figure 2A). The polypeptides were engineered based on the five discrete domains identified by x-ray crystallography28. We used the β-gal gene encoding amino acids 10 through 1023 from the plasmid pSV-β-gal (Promega; Madison, WI) as the starting genetic code. The split-β-gal fragments were designed to create individual bi-complementing pairs: alpha-1 domain (α-1) and the 1-omega domain (1-ω), alpha-4 domain (α-4) and the omega domain (ω). A third domain, 1–4, was also constructed to recreate tri-complementation between the alpha domain and the omega domain.
Figure 2.
β-galactosidase constructs. A Schematic of β-galactosidase constructs. Full-length β-galactosidase cDNA was cut into varying lengths by restriction enzyme digestion. Each fragment incorporates a necessary domain for β-galactosidase complementation. B Purification of biotinylated β-galactosidase. cDNA fragments were ligated in frame into the bacterial expression plasmid pAN, which adds a biotin to the amino terminus of each β-galactosidase fragment. Each plasmid was expressed in bacteria, IPTG-induced, and purified over a Talon resin column. The expression and purity of the proteins was verified by Western blot analysis. An equal amount of whole cell lysate (100 μg) was electrophoresed and transblotted onto nitrocellulose paper. The nitrocellulose was immunoblotted with streptavidin-HRP (1:1000 dilution). Lane 1: β-galactosidase, 135 kDa; lane 2: α-4, 100 kDa; lane 3: 1-ω, 116 kDa; lane 4: 1-4, 80 kDa; lane 5: ω, 35 kDa; lane 6:α-1, 20 kDa.
To yield acceptable levels of β-gal fragments from bacteria, the exact location of the translational start and stop sites for each polypeptide was empirically determined. The resulting biotin-tagged, His-tagged split-β-gal fragments were then propagated, expressed, and purified from E. coli (Figure 2B). Data demonstrate the success of the initial step and show the purity of several β-gal fragments after single step purification over a cobalt affinity column. The yield for each of the fragments was approximately 1 mg per liter bacteria.
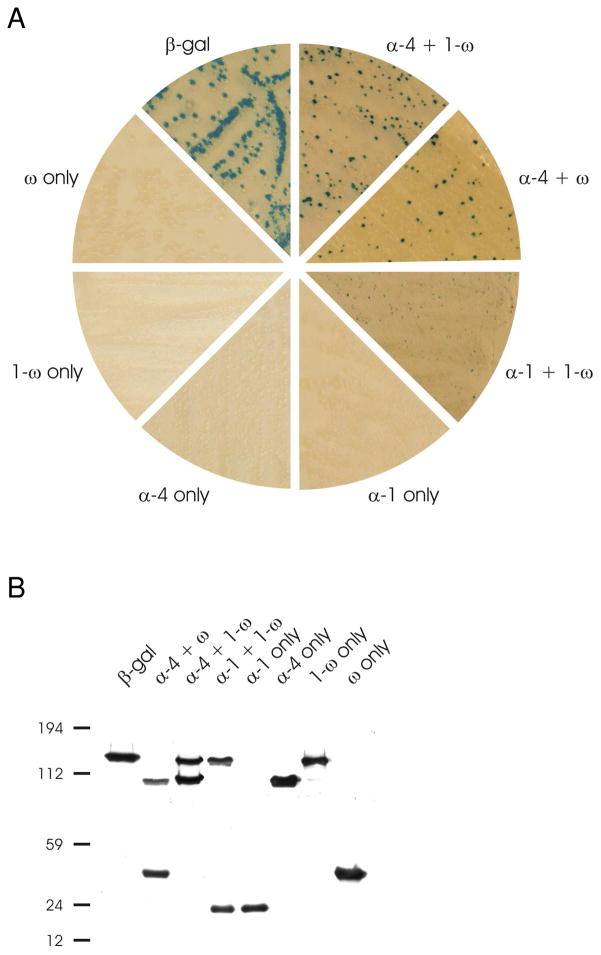
β-gal activity can be detected in vivo in E. coli in the presence of X-gal (β-gal substrate) and isopropyl β-D-1-thiogalactopyranoside (IPTG), a feature that can be used to screen for bacterial colonies that express β-gal. In a systematic screen for trans-complementation-competent β-gal fragments in bacteria that does not express endogenous β-gal, strain ER1793, we found several fragment pairs that complement to recapitulate enzyme activity (Figure 3A). Different combinations of the constructs were used to transform ER1793 bacteria (a bacterium devoid of any β-gal protein). Only combinations of fragments that contain the full complement of the five β-gal domains, α, 1–4, and ω,3 yielded activity via trans-complementation. The complementing pairs, α-4 + ω and α-4 + 1-ω, stained very intensely when incubated with X-gal substrate. Of the three complementing pairs, α-1 + 1-ω stained less intensely. None of the individual fragments restore activity to the β-gal deletion strain ER1793. Also, incomplete combinations of fragments are devoid of β-gal activity (not shown). Western blot analysis of whole bacterial extracts from these clones confirmed the expression of split-β-gal fragment(s) in the bacteria (Figure 3B). Although the proteins were expressed in the bacteria at similar levels, some complementing pairs, i.e., α-4 and 1-ω and α-4 and ω, were better transactivating partners when compared to α-1 and 1-ω.
Figure 3.
Bacterial bi-molecular complementation. A Expression of active β-galactosidase assayed by X-gal staining. E. coli K12 ER1793 were transfected with individual β-galactosidase fragment plasmids or combinations of plasmids that would produce a complementing full length enzyme. Colonies containing active β-galactosidase stained blue. B Western blot analysis of bacteria expressing β-galactosidase fragments. E. coli K12 ER1793 transformed with complementing pairs of β-galactosidase plasmids express both β-galactosidase fragments. Lane 1: β-galactosidase, 135 kDa; lane 2: α-4 + ω, 100 kDa and 35 kDa; lane 3: α-4 + 1-ω, 100 kDa and 116 kDa; lane 4: α-1 + 1-ω, 20 kDa and 116 kDa; lane 5: α-1, 20 kDa; lane 6: α-4, 100 kDa; lane 7: 1-ω, 116 kDa; lane 8:ω, 35 kDa.
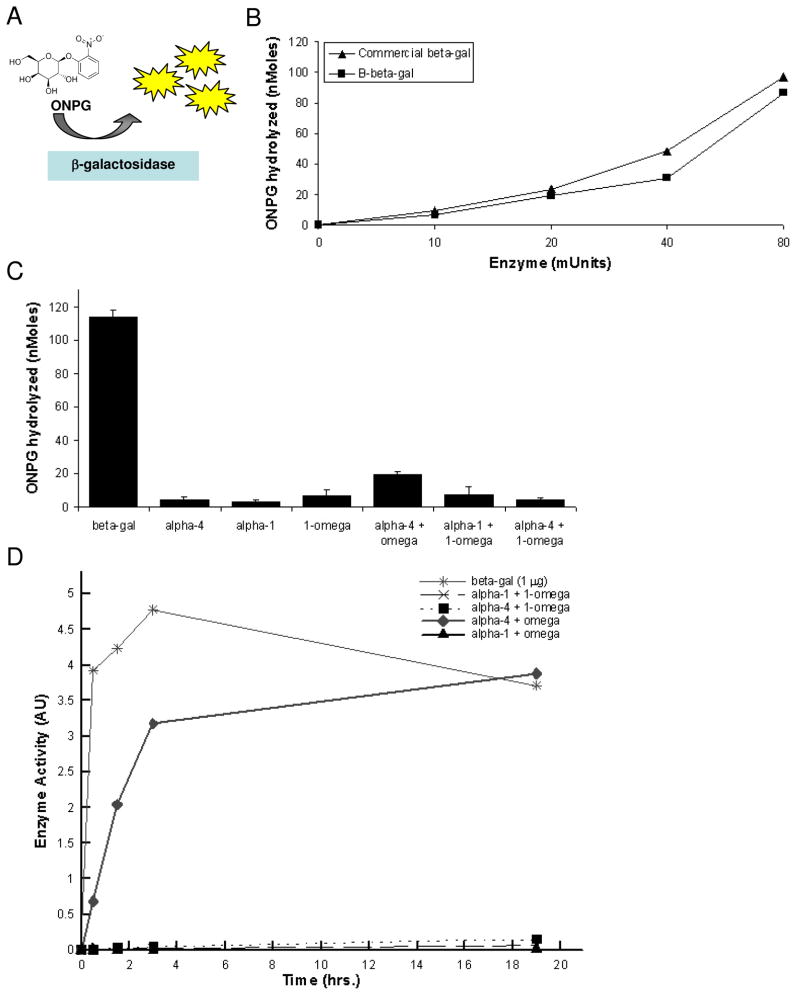
To test the ability of the split-fragments to spontaneously aggregate into tetramers in solution, complementing pairs of β-gal fragments were combined and assessed in a solution-based activity assay. Levels of active β-gal expression were measured by its catalytic hydrolysis of ortho-nitrophenyl-β-D-galactopyranoside (ONPG), a synthetic chromogenic substrate, to orthonitrophenol (ONP), a yellow product (λmax = 420 nm). Enzyme activity was measured by the rate of appearance of the yellow color using a spectrophotometer (Fig. 4A). First, purified full length biotinylated β-gal activity was measured against commercially available, lyophilized β-gal (Figure 4B). Increasing amounts of enzyme were assayed in the presence of ONPG. Enzyme activity was virtually identical for purified recombinant biotinylated-β-gal prepared in the laboratory versus purchased β-gal.
Figure 4.
β-galactosidase subunit association in solution. A Schematic of β-galactosidase activity assay. B Comparison of β-galactosidase activity of full length enzyme prepared in the laboratory versus commercially available standard. C Enzyme activity of β-galactosidase protein fragments. β-gal fragments were added, either individually or in complementing pairs, to uncoated 96-well microtiter assay plates in equal molar amounts and incubated at room temperature on an orbital rocker for 30 min. in the presence of β-galactosidase fluorescent substrate ONPG. Minimal to no enzyme activity was observed for individual or complementing fragments in solution. D Enzyme activity of β-galactosidase protein fragments over time. β-galactosidase protein fragments were added individually or in complementing pairs in the presence of ONPG over a 19 hr incubation period. Full length β-galactosidase was optimally active at 30 min. All fragment combinations were inactive in solution over a 3 hr. time period except the combination α-4 and ω which gradually increased after 30 min.
The folding mechanism of β-gal is known to occur in three stages: formation of secondary dimer structures from monomeric primary structure (fast), structural rearrangement of dimers (slow), and association of dimers into tetramers (fast)40. Assuming that the slowest tetrameric folding proteins require many minutes or hours to fold under simulated conditions, we combined β-gal fragments in equal molar amounts and allowed them to interact for 1 hr at room temperature (Figure 4C). We then incubated the combinations with ONPG substrate for 30 min. and measured absorbance at 420 nm. Most complementing pairs of β-gal fragments did not reconstitute full β-gal activity in solution. This is clearly a desired result since we did not want indiscriminate complementation of the β-gal fragments in solution. To check our selected refolding/association time, we assayed β-gal enzymatic activity over a range of time points (Figure 4D) and measured enzyme activity. Full-length β-gal was used as the standard control. Complementing pairs α-1, 1-ω and α-4, 1-ω did not produce any perceivable enzyme activity. Non-complementing pairs that do not reconstruct the full-length β-gal also did not have enzymatic activity. Only the combination of α-4 and ω resulted in the spontaneous reformation of β-gal after a 0.5 hr. association time.
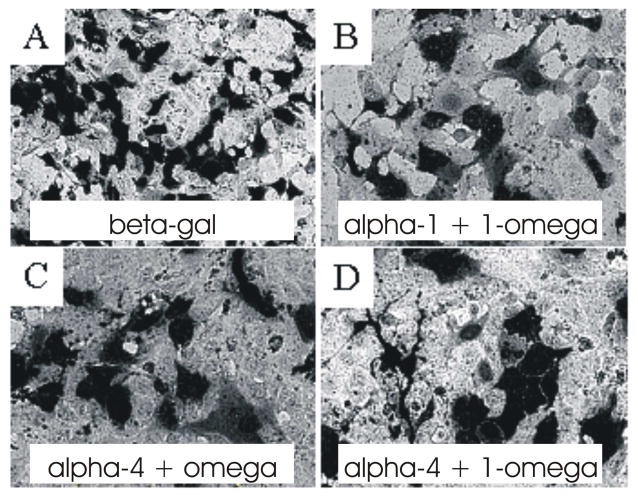
Next, we examined whether split-β-gal fragments could recombine when transfected into mammalian cells in vivo. Rat C6 glioma cells were transiently transfected with each of the constructs, either singly or in combination. Only in cells co-transfected with combinations of complementing fragments could any β-gal activity be detected (Figure 5). In control transfections, in which C6 cells did not receive a complete complement of all the subunits, no β-gal activity was detectable (not shown).
Figure 5.
Bi-molecular complementation of β-galactosidase subunits in mammalian cells. Rat glioma C6 cells were transiently transfected using Lipofectamine 2000 as per manufacturers instructions with pcDNA3.1 plasmids containing either full length β-galactosidase cDNA (1 μg) or individual β-galactosidase fragment cDNA (1 μg each). Forty-eight hours after transfection, the cells were rinsed, fixed in paraformaldehyde, and incubated overnight with X-gal staining buffer. Phase contrast images are shown. A C6 cells expressing full length β-galactosidase. B Cells transfected with β-galactosidase fragments α-1 and 1-ω, expressing enzyme activity. C Cells transfected with β-galactosidase fragments α-4 and ω, expressing enzyme activity. D C6 cells transfected with β-galactosidase fragments α-4 and 1-ω, expressing enzyme activity.
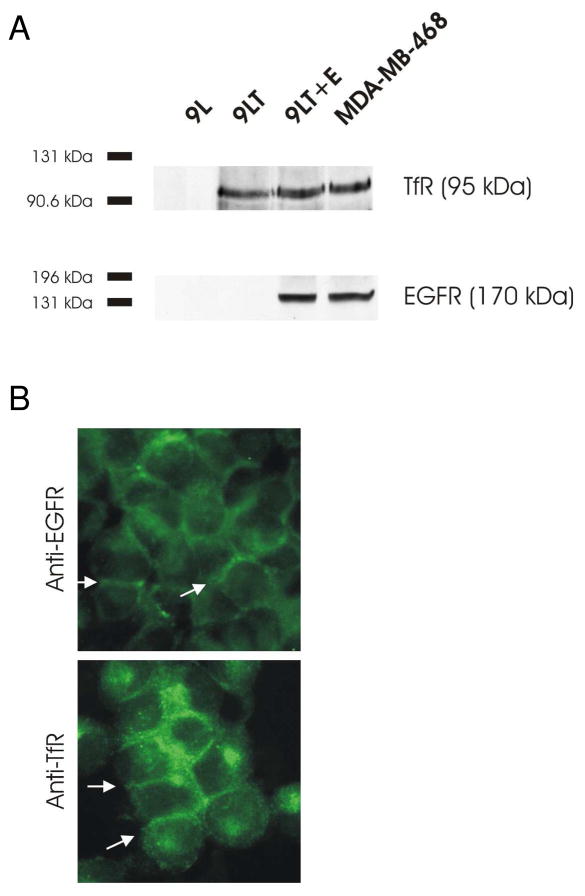
To demonstrate the utility of our reporter fragments in identifying multiple biomarkers, we created a complex as outlined in Figure 1B that consisted of a reporter fragment, a linker, and a targeting moiety, in this case a ligand. We designed two experiments to determine whether the individual targeted ligand-complexes retained their ability to bind cell surface receptors and whether the individual β-gal fragment ligand-complexes were able to re-establish full β-gal activity when oriented on the cell surface. First, we created a rat 9L gliosarcoma cell line stably over-expressing two human cell surface receptors, TfR and EGFR. We assayed the expression level of the human receptors by immunoblot analysis (Figure 6A). As a control to monitor receptor integrity MDA-MB-468 cells were also included in this assay. MDA-MB-468 cells express high levels of EGFR and TfR. In addition, we demonstrated the localization of the human receptors at the cell membrane and to a lesser extent within the cell’s interior (Figure 6B). The level of both receptors in the engineered 9L cells were similar to those measured in a panel of different human tumor cell lines (Supplementary Figure 1).
Figure 6.
Receptor overexpression in mammalian cells. A The rat gliosarcoma cell line, 9L, was stably transfected with none, one, or two human receptors. Western-blot analysis using equal amounts of total protein from the corresponding lysates was used to determine human EGFR expression. The same blot was stripped and re-probed to determine total human TfR protein levels. B Immunolocalization of human EGFR and TfR in cells overexpressing both EGFR and TfR. Cell surface staining is indicated by white arrows.
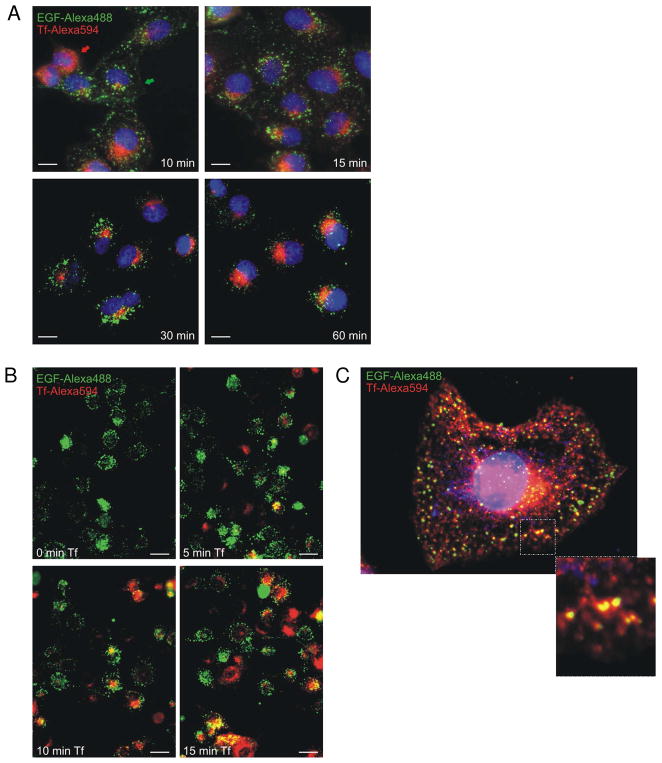
To show that the human receptors were functionally active, cells overexpressing both receptors were incubated with a cocktail of Alexa488-conjugated EGF and Alexa594-conjugated Tf (Molecular Probes; Eugene, OR) and observed using fluorescence microscopy. Activated EGFR and TfR rapidly internalized their respective ligands into clearly delineated endocytotic vesicles (Figure 7A). The majority of human EGFR and TfR cycled independently, however, when cells were stimulated simultaneously by their respective ligands. To shift the receptors to the same temporal location within the endocytotic vesicles, we examined receptor co-localization by first pre-loading cells overexpressing both receptors with Alexa488-EGF for 15 min. and then incubating them with Alexa594-Tf at increasing time points (Figure 7B). Minimal receptor co-localization was detected after a 5 min. exposure to Alexa594-Tf. Increased co-localization was observed as early as 10 min. and persisted for 15 min. Figure 7C shows a representative cell at high magnification in which a sub-population of vesicles contained both human receptors after a 15 min. pre-incubation with Alexa488-EGF and 10 min. incubation with Alexa594-Tf.
Figure 7.
Receptor co-localization in mammalian cells. A Double labeling of EGFR and TfR with fluorophore-conjugated ligand. Green arrow indicates EGFR localized at the cell surface. Red arrow indicates rapidly internalized TfR. Photographs were captured at 40X magnification. Scale bar indicates 10 μm. B Serial addition of fluorophore-conjugated ligands EGF and Tf increases internal receptor co-localization. Cells overexpressing both EGFR and TfR were first pre-loaded with Alexa488-EGF (5 μg/ml) for 15 min. and then incubated with Alexa594-Tf (50 μg/ml) over increasing time at 37 °C. Cells were washed, fixed, and mounted for fluorescent observation. Photographs were captured at 40X magnification. Scale bar: 20 μm. C Representative cell with receptor co-localization of EGFR and TfR. Photograph was taken at 100× magnification. Inset emphasizes receptor co-localization.
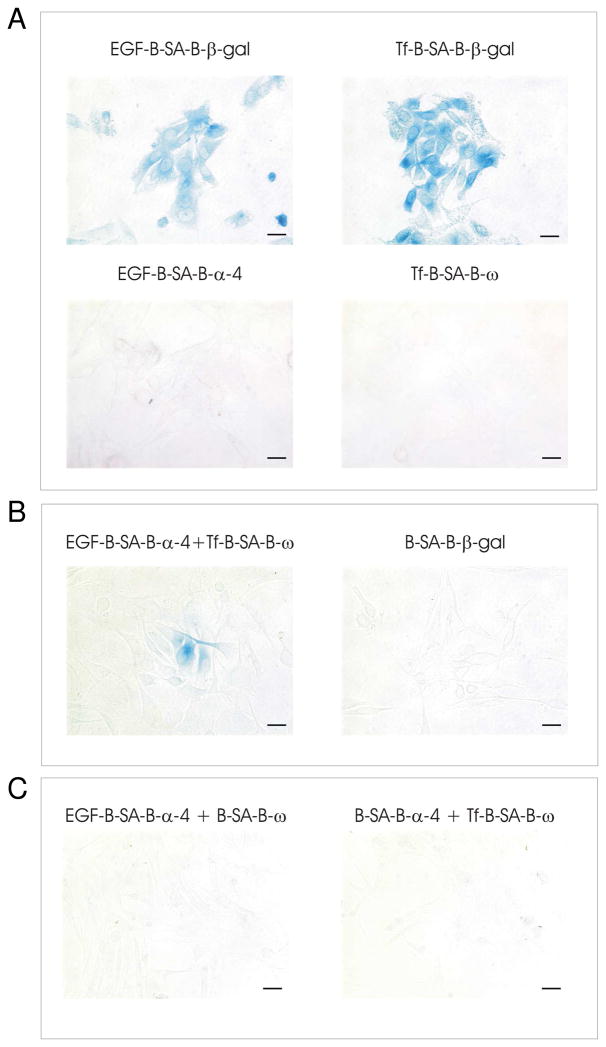
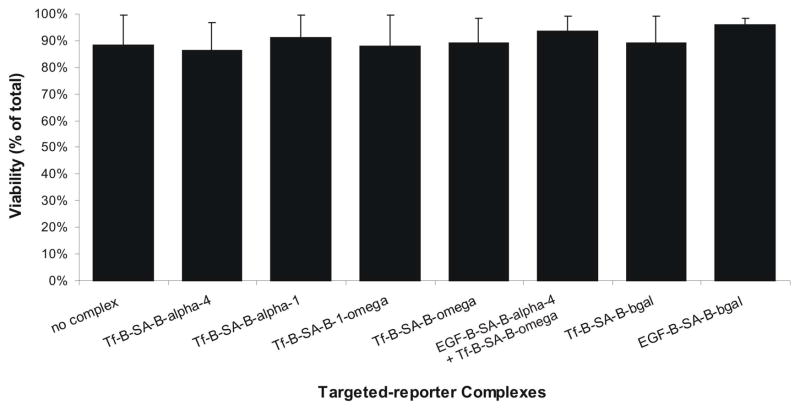
Next, ligand-complexes were formed with the full-length β-gal enzyme reporter. To create the ligand-complexes, a biotinylated-ligand is linked via streptavidin to a biotinylated-β-gal enzyme reporter. Ligand, linker, and reporter fragment were mixed in a molar ratio 1:1:3, respectively, and incubated at room temperature for 1 hr. Excess D-biotin was added after 1 hr. to block any remaining unbound streptavidin sites. The resulting ligand-complex was diluted in cell culture media. When the cells were incubated at 37 °C with either the EGF ligand-complex or the Tf ligand-complex, for 15 min or 10 min, respectively, enzymatic activity was successfully visualized after X-gal staining (Figure 8A, top panel). More than 75% of all the cells overexpressing the two human receptors were labeled with either EGF-B-SA-B-β-gal or Tf-B-SA-B-β-gal. In an attempt to capture the sub-population of receptors cycling together in vesicles illustrated in Figure 7B, individual ligand-complexes were generated with a β-gal reporter fragment instead of the full length enzyme. Biotinylated-EGF was linked to biotinylated-α-4 and biotinylated-Tf was linked to biotinylated ω. Live cells expressing both human receptors were serially incubated with first one ligand-complex, washed, and then the other complementing ligand-complex. The cells were extensively washed and fixed with 4% paraformaldehyde. Enzymatic activity was visualized by overnight X-gal staining (Figure 8B). Cells incubated with individual ligand-reporter fragment complexes, EGF-B-SA-B-α-4 or Tf-B-SA-B-ω, did not stain positive for enzymatic activity (Figure 8A, bottom panel). As a control to demonstrate that reassembly of the β-gal enzyme was driven by receptor proximity (targeting) and not by spontaneous reassembly of the reporter fragments, live cells expressing both human receptors were incubated for 25 min. simultaneously with complementing pairs of β-gal fragments, one of each pair was targeted using ligand moiety while the other was untargeted using biotin in place of the ligand: complementing pairs [EGF-B-SA-B-α-4 and B-SA-B-ω] or [B-SA-B-α-4 and Tf-B-SA-B-ω] (Fig. 8C). After overnight incubation with X-gal stain, no enzymatic activity was observed. In other studies, sequential addition of the same complementing control pairs of targeted and untargeted complexes also resulted in no enzymatic activity (data not shown). Ligand-reporter fragment complexes did not adversely affect cell viability after incubation or growth rate of cells after complex removal as demonstrated by trypan blue cell staining (Figure 9).
Figure 8.
Bi-molecular complementation of receptor-targeted complexes in mammalian cells. A Receptor-targeted β-galactosidase complementation. Cells overexpressing human receptors, EGFR and TfR, were incubated with full length β-galactosidase reporter (0.1 nMole) targeted to either the EGFR or TfR. X-gal staining reveals uptake of the ligand-targeted reporter complex (top panels). B EGF was linked to the α-4 β-galactosidase fragment and Tf was linked to the ω β-galactosidase fragment. When serially incubated on live cells, enzymatic complementation was observed after X-gal staining. Images were taken at 20X magnification. Scale bar represents 10 μm. C Cells overexpressing human receptors, EGFR and TfR, were incubated with EGF linked to the α-4 β-galactosidase fragment and biotin linked to the ω β-galactosidase fragment (left panel). Cells overexpressing human receptors, EGFR and TfR, were incubated with biotin linked to the α-4 β-galactosidase fragment and Tf linked to the ω β-galactosidase fragment (right panel). When simultaneously incubated on live cells, no enzymatic complementation was observed after X-gal staining. Images were taken at 20× magnification. Scale bar represents 10 μm.
Figure 9.
Cell viability after incubation with ligand-targeted reporter complexes. β-galactosidase reporter fragment complexes were incubated on live cells for 1 hr. at 37 °C. The cells were washed, stained with trypan blue, and counted. Cell counts were graphed as a percentage of total cells.
DISCUSSION
Reporter fragmentation is a fundamental complement to genomic applications and especially intriguing as an intrinsic monitor of cancer development. Monitoring cancer could easily incorporate imaging cellular and molecular processes as well as tracking drug delivery. As a platform, for instance, split reporter fragments can be used in therapeutic delivery vehicles where one fragment is targeted to the therapeutic candidate cell and the other fragment delivered by the delivery vehicle; only in the presence of both complementing pairs would a positive response be observed, indicating successful delivery of the therapeutic payload.
Split reporters, both non-enzymatic and enzymatic, have been widely used to examine protein-protein interactions. Non-enzymatic proteins, such as ubiquitin and green fluorescent protein (GFP), have been effectively utilized in split fragment reporter assays.41–44 However, the utility of ubiquitin as an endogenous enzyme substrate and GFP as an intrinsic fluorescent reporter do not reflect the ultimate goal of our proposed platform, to create an exogenously targeted self-amplifying reporter complex. Split ubiquitin fragments act together as a single-domain protein substrate which is rapidly cleaved by endogenous ubiquitin-specific proteases. Since it is not an activity-based reporter, it does not have amplification built into the assay design. Once the substrate has been acted upon, the imageable signal is lost. GFP split reporter fragments, on the other hand, do not possess any enzyme activity and are inherently incapable of amplification.
Enzymes like β-lactamase, dihydrofolate reductase (DHFR), and firefly luciferase have all been successfully spliced into smaller fragments and are more similar to our design approach.20,22,45,46 The majority of these enzymes have been used in a PCA designed-format. The basic concept is simple: only when two enzyme fragments are brought together as a result of interacting proteins is there enzymatic activity. The extent of the protein–protein interaction is then evaluated by measuring the magnitude of enzyme activity. These assays are very different paradigms and differ significantly from the one suggested in this study as outlined in Table 1. The prevailing PCA system utilizes genetically engineered fusion constructs. The site of fusion between the reporter fragment and protein of interest is crucial. For instance, β-lactamase does not have any clearly defined domains and requires experimentation to select the proper fusion site.47 The reporter is classically a monomeric enzyme with inactive, non-spontaneously complementing subunits; the fused proteins of interest are high affinity interacting proteins. The resulting fusion chimera are then expressed in genetically transformed mammalian cells and potential interactions are observed. Predictions of protein interactions must, in some way, be exercised prior to evaluating the interactions within the cell. Even more restrictive is the fact that these reporter constructs require genetic manipulation of the host cells, excluding them from functional translation into human subjects. Their utility is subsequently relegated to biochemical in vitro assays or genetically engineered small animal in vivo imaging.
Table 1.
Comparison of our Targeted-Reporter Complex System to the Protein-fragment Complementation Assay (PCA).
| Targeted-Reporter Complex Assay | Protein fragment Complementation Assay (PCA) | |
|---|---|---|
| Genetic Manipulation | None | Engineered cell lines Chimeric fusion proteins |
| Complementation Driven By | Close proximity - (i.e. receptor clustering or vesicle internalization) | Direct protein-protein interactions of fusion proteins |
| Spontaneous Complementation | Low | None |
| Sensitivity | Reporter (enzyme) amplification | Most reporters – no amplification (i.e. GFP, ubiquitin) Exceptions β-lactamase Renilla luciferase |
| Specificity | Ligand-driven | Known protein-protein interactions |
| Detection Modality | Optical MRI Nuclear |
Optical |
In contrast, our targeted-reporter complex can evaluate endogenous changes in multiple biomarker expression by labeling external cell surface receptors using exogenously-added targeted-split reporter fragments. Several potential enzymes undergo spontaneous reassembly to form active enzyme, however, formation of protein aggregates is a major drawback of the spontaneous reassembly of the protein fragments.48 The β-gal fragments engineered for these studies do not have sufficient affinity to drive enzyme activity unless the targeted receptors are in close proximity either at the surface or in internalized vesicles (Fig. 8B). The further development and eventual application of these techniques in vivo will allow surveying of cancer biomarkers over the entire tumor surface (accessible via vascular delivery of the components) rather than the limited sampling currently obtained using biopsies and ex vivo analysis.
In our study, we capitalize on the common endocytotic internalization of the two receptors, EGFR and TfR. To demonstrate that these receptors are good representatives of the cancer signature, we first qualitatively measured the endogenous expression level of these receptors in a variety of different human cancer cell lines using immunofluorescence techniques and show that they are co-expressed at high levels in many of the cell lines (Supplementary Fig. 1). Representative human cancer cell lines are shown that demonstrate unique, observable expression patterns for the two receptors. Next, we examined the expression of each receptor in the cell lines and graphed the densitometric values. This underscores the validity that our cell model does, in fact, represent cancer-associated endogenous changes in receptor expression.
PCA techniques that rely on protein chimera to interrogate cytosolic protein-protein interactions and receptor ligand-induced dimerization have been useful for non-invasive imaging of genetically modified cells where the components are functionally related.49–51 In this study, we introduce the technology to image dynamic alterations in cancer that are not necessarily related functionally, but are related molecularly for diagnostic purposes. Here, we show that it is possible to visualize non-interacting receptors. To image non-interacting biomarkers identified as diagnostically important by genomic technologies, we exploit receptor proximity or common internalization pathways to assay changes in expression that may not be related functionally, but are implicated in the malignant phenotype. Therefore, any number of potential cell surface markers, whether interacting or not, are amenable for imaging using our technology.
The two receptors observed, EGFR and TfR, are overexpressed in many cancers, but receptor-mediated internalization appears to diverge along the endocytotic pathway.7 Studies have suggested that some receptors, like EGFR, are diffusely distributed on the cell surface prior to ligand binding and cluster within clathrin-coated pits after ligand binding and receptor dimerization. TfR also binds ligand rapidly and internalizes through clathrin-coated vesicles. Ultimately, TfR is returned to the cell surface intact, while EGFR proceeds to lysosomes where it is degraded.52–55 It remains unclear, however, how much of a common pathway these two receptors share. As shown in Fig. 7, the co-localization of the two chosen receptors occurs in only 10–20% of the total cells as evidenced by yellow overlay of independent fluorescent channel image acquisition using fluorophore-conjugated ligands. This quantitative approximation is mimicked by our visual observations shown in Fig. 8; only 10–20% of the cells containing both EGFR and TfR in the same location allow for β-gal fragment complementation. Recent findings by Leonard et al corroborate our findings in that although TfR and EGFR follow distinct routes of internalization under physiological conditions, for a short period of time (6–20 min. of simultaneous incubation with EGF and Tf) the two receptors intersect at a common point along the endocytic pathway representing 20–30% co-localization in vesicles 300 nm between from the cell membrane.56
The current study describes novel β-gal protein fragments that allow directed (or targeted) complementation. By creating a receptor-targeted reporter complex, we have increased the latitude and functionality of the enzymatic complementation assay. As described above, previous assays required direct protein-protein interactions to maximize the utility of the enzyme reporter fragments, but are limited in use since they require genetic manipulations of target tissues. Our approach utilizes cancer biomarker proximity and clustering to re-assemble active β-gal from exogenously-added components. This approach enables visualization of proteins (receptors) near each other, but not necessarily interacting, without genetically altering the target tissues.
We designed the ligand-complex to deliver either EGF or Tf with a β-gal fragment to the cell surface receptors to maximize enzyme self-assembly at the cell surface or within endocytotic vesicles. Biotin-β-gal fragments, streptavidin, and biotin-ligands were combined in molar ratios (3:1:1) to take advantage of streptavidin’s four biotin binding sites. This strategy allows us to amplify the probability of enzyme reformation since the majority of streptavidin was bound by biotin-β-gal fragments. Ultimately, it was critical to show that the creation of the complex did not compromise bioactivity or receptor functionality.
Our platform-based approach adds an element of interchangeability that is hampered by reporter fusion constructs. Others, such as Gillies and colleagues, have also recognized the importance of binding multiple targets simultaneously, but have undertaken alternative methods. For example, the Gillies’ laboratory has designed a single multimeric backbone capable of targeting multiple receptors.1,57,58 To target a signature of three markers, they exploit the specificity of low affinity ligands to recognized only cells expressing three surface markers. Steric hindrance associated with biomarker order along the backbone, however, is a major constraint for this schema.
By designing components and assembling them in a modular fashion, our platform multi-functionalizes the ligand-complexes adoptable for imaging and therapeutics. These complexes satisfy two main requirements: interchangeability with new constituents as more specific cancer biomarkers are discovered and independent reconfiguration or refinement to meet evolving imaging needs. Our findings are not the only biomarker combinations available. Using unrelated receptor family members from the receptor tyrosine kinase family (EGFR) and the transferrin receptor family (TfR), we are able to prove the feasibility of our imaging constructs. Other biomarkers will allow us to use these targeted-reporter complexes across several different cancer types in a diagnostic manner in much the same way high throughput serial (HTS) assays have revolutionized a more personalized approach to epigenetic factors.
Supplementary Material
Supplementary Figure 1 Relative overexpression of the cell surface membrane receptors, EGFR and TfR, in various human cancer cell lines. A Immunofluorescence and Western blot data correlate well with receptor expression levels. B The graph demonstrates the quantitation of the endogenous levels of each receptor from the Western autoradiograph. C The table assigns values to the expression levels to aid in the choice of cancer cell lines for future in vivo studies.
Acknowledgments
The authors would like to thank Ms. Kari Lavik, Mr. Alex Liggett, and Mr. John Smetona for technical assistance.
This work was supported by an ongoing center grant from the National Foundation for Cancer Research (to JPB) and a fellowship award (to AMB). The project described was also supported by Grant Number K01EB006910 (to AMB) from the National Institute of Biomedical Imaging and Bioengineering. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Biomedical Imaging and Bioengineering or the National Institutes of Health.
Footnotes
SUPPORTING INFORMATION
Supplementary Figure 1 depicting representative cancer cell line overexpression of two cancer biomarkers, EGFR and TfR, as assayed by fluorescence and Western blot analysis of whole cell lysate. Table evaluating relative receptor overexpression. Electronic Supporting Information files are available without a subscription to ACS Web Editions. This information is available free of charge via the Internet at http://pubs.acs.org/. All files are copyrighted by the American Chemical Society. Files may be downloaded for personal use; users are not permitted to reproduce, republish, redistribute, or resell any Supporting Information, either in whole or in part, in either machine-readable form or any other form. For permission to reproduce this material, contact the ACS Copyright Office by copyright@acs.org or by fax at 202-776-8112.
REFERENCE LIST
- 1.Stephen RM, Gillies RJ. Promise and progress for functional and molecular imaging of response to targeted therapies. Pharm Res. 2007;24(6):1172–1185. doi: 10.1007/s11095-007-9250-3. [DOI] [PubMed] [Google Scholar]
- 2.Alper O, Bergmann-Leitner ES, Bennett TA, Hacker NF, Stromberg K, Stetler-Stevenson WG. Epidermal growth factor receptor signaling and the invasive phenotype of ovarian carcinoma cells. J Natl Cancer Inst. 2001;93(18):1375–1384. doi: 10.1093/jnci/93.18.1375. [DOI] [PubMed] [Google Scholar]
- 3.Magkou C, Nakopoulou L, Zoubouli C, Karali K, Theohari I, Bakarakos P, Giannopoulou I. Expression of the epidermal growth factor receptor (EGFR) and the phosphorylated EGFR in invasive breast carcinomas. Breast Cancer Res. 2008;10(3):R49. doi: 10.1186/bcr2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoelzinger DB, Mariani L, Weis J, Woyke T, Berens TJ, McDonough WS, Sloan A, Coons SW, Berens ME. Gene expression profile of glioblastoma multiforme invasive phenotype points to new therapeutic targets. Neoplasia. 2005;7(1):7–16. doi: 10.1593/neo.04535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakada M, Nakada S, Demuth T, Tran NL, Hoelzinger DB, Berens ME. Molecular targets of glioma invasion. Cell Mol Life Sci. 2007;64(4):458–478. doi: 10.1007/s00018-007-6342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krahn G, Leiter U, Kaskel P, Udart M, Utikal J, Bezold G, Peter RU. Coexpression patterns of EGFR, HER2, HER3 and HER4 in non-melanoma skin cancer. Eur J Cancer. 2001;37(2):251–259. doi: 10.1016/s0959-8049(00)00364-6. [DOI] [PubMed] [Google Scholar]
- 7.Hogemann-Savellano D, Bos E, Blondet C, Sato F, Abe T, Josephson L, Weissleder R, Gaudet J, Sgroi D, Peters PJ, Basilion JP. The transferrin receptor: a potential molecular imaging marker for human cancer. Neoplasia. 2003;5(6):495–506. doi: 10.1016/s1476-5586(03)80034-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Habashy HO, Powe DG, Staka CM, Rakha EA, Ball G, Green AR, Aleskandarany M, Paish EC, Douglas MR, Nicholson RI, Ellis IO, Gee JM. Transferrin receptor (CD71) is a marker of poor prognosis in breast cancer and can predict response to tamoxifen. Breast Cancer Res Treat. 2009 doi: 10.1007/s10549-009-0345-x. [DOI] [PubMed] [Google Scholar]
- 9.Liu TF, Tatter SB, Willingham MC, Yang M, Hu JJ, Frankel AE. Growth factor receptor expression varies among high-grade gliomas and normal brain: epidermal growth factor receptor has excellent properties for interstitial fusion protein therapy. Mol Cancer Ther. 2003;2(8):783–787. [PubMed] [Google Scholar]
- 10.Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28(Suppl 1):S24–S31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki M, Shigematsu H, Hiroshima K, Iizasa T, Nakatani Y, Minna JD, Gazdar AF, Fujisawa T. Epidermal growth factor receptor expression status in lung cancer correlates with its mutation. Hum Pathol. 2005;36(10):1127–1134. doi: 10.1016/j.humpath.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Gillies RJ, Hoffman JM, Lam KS, Menkens AE, Piwnica-Worms DR, Sullivan DC, Weissleder R. Meeting report: high-throughput technologies for in vivo imaging agents. Mol Imaging. 2005;4(2):98–103. doi: 10.1162/15353500200505115. [DOI] [PubMed] [Google Scholar]
- 13.Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: modifications, screening, and therapy. Annu Rev Med. 2008;59:267–280. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- 14.Sharma S, Kelly TK, Jones PA. Epigenetics in Cancer. Carcinogenesis. 2009 doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen B, O’ Connell CD, Boone DJ, Amos JA, Beck JC, Chan MM, Farkas DH, Lebo RV, Richards CS, Roa BB, Silverman LM, Barton DE, Bejjani BA, Belloni DR, Bernacki SH, Caggana M, Charache P, Dequeker E, Ferreira-Gonzalez A, Friedman KJ, Greene CL, Grody WW, Highsmith WE, Jr, Hinkel CS, Kalman LV, Lubin IM, Lyon E, Payne DA, Pratt VM, Rohlfs E, Rundell CA, Schneider E, Willey AM, Williams LO, Willey JC, Winn-Deen ES, Wolff DJ. Developing a sustainable process to provide quality control materials for genetic testing. Genet Med. 2005;7(8):534–549. doi: 10.1097/01.gim.0000183043.94406.81. [DOI] [PubMed] [Google Scholar]
- 16.Gregg JP, Grody WW. Molecular pathology and cancer genetic screening. West J Med. 1995;162(5):449–450. [PMC free article] [PubMed] [Google Scholar]
- 17.Grody WW. Molecular genetic risk screening. Annu Rev Med. 2003;54:473–490. doi: 10.1146/annurev.med.54.101601.152127. [DOI] [PubMed] [Google Scholar]
- 18.Maddox CB, Rasmussen L, White EL. Adapting Cell-Based Assays to the High Throughput Screening Platform: Problems Encountered and Lessons Learned. JALA Charlottesv Va. 2008;13(3):168–173. doi: 10.1016/j.jala.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borloo J, De SL, Vergauwen B, Van Beeumen JJ, Devreese B. A beta-galactosidase-based bacterial two-hybrid system to assess protein-protein interactions in the correct cellular environment. J Proteome Res. 2007;6(7):2587–2595. doi: 10.1021/pr070037j. [DOI] [PubMed] [Google Scholar]
- 20.Galarneau A, Primeau M, Trudeau LE, Michnick SW. Beta-lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein protein interactions. Nat Biotechnol. 2002;20(6):619–622. doi: 10.1038/nbt0602-619. [DOI] [PubMed] [Google Scholar]
- 21.Luker KE, Piwnica-Worms D. Optimizing luciferase protein fragment complementation for bioluminescent imaging of protein-protein interactions in live cells and animals. Methods Enzymol. 2004;385:349–360. doi: 10.1016/S0076-6879(04)85019-5. [DOI] [PubMed] [Google Scholar]
- 22.Remy I, Michnick SW. Clonal selection and in vivo quantitation of protein interactions with protein-fragment complementation assays. Proc Natl Acad Sci U S A. 1999;96(10):5394–5399. doi: 10.1073/pnas.96.10.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong H, Zhang B, Little G, Kovar J, Chen H, Xie W, Schutz-Geschwender A, Olive DM. beta-Galactosidase activity assay using far-red-shifted fluorescent substrate DDAOG. Anal Biochem. 2009;386(1):59–64. doi: 10.1016/j.ab.2008.11.031. [DOI] [PubMed] [Google Scholar]
- 24.Ho NH, Weissleder R, Tung CH. A self-immolative reporter for beta-galactosidase sensing. Chembiochem. 2007;8(5):560–566. doi: 10.1002/cbic.200600386. [DOI] [PubMed] [Google Scholar]
- 25.Tung CH, Zeng Q, Shah K, Kim DE, Schellingerhout D, Weissleder R. In vivo imaging of beta-galactosidase activity using far red fluorescent switch. Cancer Res. 2004;64(5):1579–1583. doi: 10.1158/0008-5472.can-03-3226. [DOI] [PubMed] [Google Scholar]
- 26.Ullmann A, JACOB F, MONOD J. Characterization by in vitro complementation of a peptide corresponding to an operator-proximal segment of the beta-galactosidase structural gene of Escherichia coli. J Mol Biol. 1967;24(2):339–343. doi: 10.1016/0022-2836(67)90341-5. [DOI] [PubMed] [Google Scholar]
- 27.Jacobson RH, Zhang XJ, DuBose RF, Matthews BW. Three-dimensional structure of beta-galactosidase from E. coli. Nature. 1994;369(6483):761–766. doi: 10.1038/369761a0. [DOI] [PubMed] [Google Scholar]
- 28.Jacobson RH, Matthews BW. Crystallization of beta-galactosidase from Escherichia coli. J Mol Biol. 1992;223(4):1177–1182. doi: 10.1016/0022-2836(92)90269-p. [DOI] [PubMed] [Google Scholar]
- 29.JACOB F, ULLMAN A, MONOD J. THE PROMOTOR, GENETIC ELEMENT NECESSARY TO THE EXPRESSION OF AN OPERON. C R Hebd Seances Acad Sci. 1964;258:3125–3128. [PubMed] [Google Scholar]
- 30.Villarejo MR, Zabin I. Beta-galactosidase from termination and deletion mutant strains. J Bacteriol. 1974;120(1):466–474. doi: 10.1128/jb.120.1.466-474.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossi F, Charlton CA, Blau HM. Monitoring protein-protein interactions in intact eukaryotic cells by beta-galactosidase complementation. Proc Natl Acad Sci U S A. 1997;94(16):8405–8410. doi: 10.1073/pnas.94.16.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langley KE, Fowler AV, Zabin I. Amino acid sequence of beta-galactosidase. IV. Sequence of an alpha-complementing cyanogen bromide peptide, residues 3 to 92. J Biol Chem. 1975;250(7):2587–2592. [PubMed] [Google Scholar]
- 33.Mohler WA, Blau HM. Gene expression and cell fusion analyzed by lacZ complementation in mammalian cells. Proc Natl Acad Sci U S A. 1996;93(22):12423–12427. doi: 10.1073/pnas.93.22.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wehrman T, Kleaveland B, Her JH, Balint RF, Blau HM. Protein-protein interactions monitored in mammalian cells via complementation of beta -lactamase enzyme fragments. Proc Natl Acad Sci U S A. 2002;99(6):3469–3474. doi: 10.1073/pnas.062043699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulmurugan R, Gambhir SS. Monitoring protein-protein interactions using split synthetic renilla luciferase protein-fragment-assisted complementation. Anal Chem. 2003;75(7):1584–1589. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paulmurugan R, Gambhir SS. Firefly luciferase enzyme fragment complementation for imaging in cells and living animals. Anal Chem. 2005;77(5):1295–1302. doi: 10.1021/ac0484777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fung P, Peng K, Kobel P, Dotimas H, Kauffman L, Olson K, Eglen RM. A homogeneous cell-based assay to measure nuclear translocation using beta-galactosidase enzyme fragment complementation. Assay Drug Dev Technol. 2006;4(3):263–272. doi: 10.1089/adt.2006.4.263. [DOI] [PubMed] [Google Scholar]
- 38.Hammer MM, Wehrman TS, Blau HM. A novel enzyme complementation-based assay for monitoring G-protein-coupled receptor internalization. FASEB J. 2007;21(14):3827–3834. doi: 10.1096/fj.07-8777com. [DOI] [PubMed] [Google Scholar]
- 39.Villalobos V, Naik S, Piwnica-Worms D. Detection of protein-protein interactions in live cells and animals with split firefly luciferase protein fragment complementation. Methods Mol Biol. 2008;439:339–352. doi: 10.1007/978-1-59745-188-8_23. [DOI] [PubMed] [Google Scholar]
- 40.Nichtl A, Buchner J, Jaenicke R, Rudolph R, Scheibel T. Folding and association of beta-galactosidase. Journal of Molecular Biology. 1998;282(5):1083–1091. doi: 10.1006/jmbi.1998.2075. [DOI] [PubMed] [Google Scholar]
- 41.Ozawa T, Takeuchi TM, Kaihara A, Sato M, Umezawa Y. Protein splicing-based reconstitution of split green fluorescent protein for monitoring protein-protein interactions in bacteria: improved sensitivity and reduced screening time. Anal Chem. 2001;73(24):5866–5874. doi: 10.1021/ac010717k. [DOI] [PubMed] [Google Scholar]
- 42.Lehming N. Analysis of protein-protein proximities using the split-ubiquitin system. Brief Funct Genomic Proteomic. 2002;1(3):230–238. doi: 10.1093/bfgp/1.3.230. [DOI] [PubMed] [Google Scholar]
- 43.Reichel C, Johnsson N. The split-ubiquitin sensor: measuring interactions and conformational alterations of proteins in vivo. Methods Enzymol. 2005;399:757–776. doi: 10.1016/S0076-6879(05)99050-2. [DOI] [PubMed] [Google Scholar]
- 44.Muller J, Johnsson N. Split-ubiquitin and the split-protein sensors: chessman for the endgame. Chembiochem. 2008;9(13):2029–2038. doi: 10.1002/cbic.200800190. [DOI] [PubMed] [Google Scholar]
- 45.Piwnica-Worms D, Luker KE. Imaging protein-protein interactions in whole cells and living animals. Ernst Schering Res Found Workshop. 2005;(49):35–41. doi: 10.1007/3-540-26809-x_2. [DOI] [PubMed] [Google Scholar]
- 46.Luker KE, Smith MC, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms D. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc Natl Acad Sci U S A. 2004;101(33):12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michnick SW, Remy I, Campbell-Valois FX, Vallee-Belisle A, Pelletier JN. Detection of protein-protein interactions by protein fragment complementation strategies. Methods Enzymol. 2000;328:208–230. doi: 10.1016/s0076-6879(00)28399-7. [DOI] [PubMed] [Google Scholar]
- 48.Shiba K, Schimmel P. Functional assembly of a randomly cleaved protein. Proc Natl Acad Sci U S A. 1992;89(5):1880–1884. doi: 10.1073/pnas.89.5.1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dibya D, Sander S, Smith EA. Identifying cytoplasmic proteins that affect receptor clustering using fluorescence resonance energy transfer and RNA interference. Anal Bioanal Chem. 2009 doi: 10.1007/s00216-009-3146-5. [DOI] [PubMed] [Google Scholar]
- 50.Guo C, Levine H. A thermodynamic model for receptor clustering. Biophys J. 1999;77(5):2358–2365. doi: 10.1016/S0006-3495(99)77073-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eglen RM, Singh R. Beta galactosidase enzyme fragment complementation as a novel technology for high throughput screening. Comb Chem High Throughput Screen. 2003;6(4):381–387. doi: 10.2174/138620703106298473. [DOI] [PubMed] [Google Scholar]
- 52.Stahl PD, Barbieri MA. Multivesicular bodies and multivesicular endosomes: the “ins and outs” of endosomal traffic. Sci STKE 2002. 2002(141):e32. doi: 10.1126/stke.2002.141.pe32. [DOI] [PubMed] [Google Scholar]
- 53.Pastan I, Hanover J, Willingham M. The cellular entry of EGF and transferrin: a problem in intracellular sorting. Curr Top Cell Regul. 1985;26:17–25. doi: 10.1016/b978-0-12-152826-3.50009-7. [DOI] [PubMed] [Google Scholar]
- 54.Dargemont C, Le BA, Rothenberger S, Iacopetta B, Kuhn LC. The internalization signal and the phosphorylation site of transferrin receptor are distinct from the main basolateral sorting information. EMBO J. 1993;12(4):1713–1721. doi: 10.1002/j.1460-2075.1993.tb05816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roberts RL, Fine RE, Sandra A. Receptor-mediated endocytosis of transferrin at the blood-brain barrier. J Cell Sci. 1993;104(Pt 2):521–532. doi: 10.1242/jcs.104.2.521. [DOI] [PubMed] [Google Scholar]
- 56.Leonard D, Hayakawa A, Lawe D, Lambright D, Bellve KD, Standley C, Lifshitz LM, Fogarty KE, Corvera S. Sorting of EGF and transferrin at the plasma membrane and by cargo-specific signaling to EEA1-enriched endosomes. J Cell Sci. 2008;121(Pt 20):3445–3458. doi: 10.1242/jcs.031484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Handl HL, Vagner J, Han H, Mash E, Hruby VJ, Gillies RJ. Hitting multiple targets with multimeric ligands. Expert Opin Ther Targets. 2004;8(6):565–586. doi: 10.1517/14728222.8.6.565. [DOI] [PubMed] [Google Scholar]
- 58.Xu L, Vagner J, Josan J, Lynch RM, Morse DL, Baggett B, Han H, Mash EA, Hruby VJ, Gillies RJ. Enhanced targeting with heterobivalent ligands. Mol Cancer Ther. 2009;8(8):2356–2365. doi: 10.1158/1535-7163.MCT-08-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Relative overexpression of the cell surface membrane receptors, EGFR and TfR, in various human cancer cell lines. A Immunofluorescence and Western blot data correlate well with receptor expression levels. B The graph demonstrates the quantitation of the endogenous levels of each receptor from the Western autoradiograph. C The table assigns values to the expression levels to aid in the choice of cancer cell lines for future in vivo studies.