Abstract
Most of the phenotypes in nature are complex and are determined by many quantitative trait loci (QTLs). In this study we identify gene sets that contribute to one important complex trait: the ability of yeast cells to survive under alkali stress. We carried out an in-lab evolution (ILE) experiment, in which we grew yeast populations under increasing alkali stress to enrich for beneficial mutations. The populations acquired different sets of affecting alleles, showing that evolution can provide alternative solutions to the same challenge. We measured the contribution of each allele to the phenotype. The sum of the effects of the QTLs was larger than the difference between the ancestor phenotype and the evolved strains, suggesting epistatic interactions between the QTLs. In parallel, a clinical isolated strain was used to map natural QTLs affecting growth at high pH. In all, 17 candidate regions were found. Using a predictive algorithm based on the distances in protein-interaction networks, candidate genes were defined and validated by gene disruption. Many of the QTLs found by both methods are not directly implied in pH homeostasis but have more general, and often regulatory, roles.
Keywords: congenic lines, growth on alkali, in-lab evolution, QTL mapping, Saccharomyces cerevisiae
Introduction
Already at the beginning of the twentieth century, Mendel's theory was attacked on the ground that the simple rules he had set did not apply to the variation typically observed in nature (East, 1909, 1916; Castle, 1914). Indeed, most of the phenotypes in nature are characterized by a complex pattern of heredity, and are shaped by multiple, often intricately interacting genetic and environmental factors (Lander and Schork, 1994; Darvasi and Pisante-Shalom, 2002; Abiola et al, 2003), and are also affected by many genes (usually called quantitative trait loci (QTLs)). Variation in the phenotype of individuals carrying the same Mendelian allele is also a common observation. This natural variability is often explained by the presence of ‘genetic modifiers’ or variations in the ‘genetic background’, that is, by the effects that particular combinations of alleles have on the expression of the gene in question (Lander and Botstein, 1989; Lander and Schork, 1994; Lander and Kruglyak, 1995; Glazier et al, 2002). Dissecting complex networks, identifying their genetic components and studying the highly intricate interactions among them are some of the major challenges facing modern biology. Polygenic traits include economically and medically important determinants, including agricultural crop yields and propensity to diseases such as cancer, schizophrenia or myocardial arrest in humans. Nevertheless, so far, only very few affecting genes have been identified (Glazier et al, 2002; Maher, 2008). Moreover, usually the QTLs identified by large mapping efforts account for only a very small fraction of the variability observed in nature (Maher, 2008). Part of the reason for this failure resides in the fact that each QTL contributes only marginally to the studied phenotype, and each phenotype can result from different allelic combinations. Moreover, as we show in this study, several independent genetic networks can shape the same complex phenotype.
In this study we focused on the ability of the budding yeast Saccharomyces cerevisiae to grow at high pH, identifying gene sets responsible for this complex trait. Environmental pH has dramatic effects on the physiology of the cell, and affects nutrient availability and protein activity. Furthermore, environmental pH serves as a potent inducer of differentiation and development. In the opportunistic fungal pathogen Candida albicans, for example, high pH induces hyphal growth and pathogenesis (Davis, 2003). Moreover, appropriate responses to high pH govern fungal virulence in plants, insects and animals (Penalva et al, 2008). Yeast cells grow best at acidic pH that is maintained by the activity of the plasma membrane H+-ATPase, which actively extrudes protons and maintains a proton gradient crucial for the uptake of nutrients (van der Rest et al, 1995). Adaptation of S. cerevisiae to alkaline pH involves a change in its expression profile (Causton et al, 2001; Lamb et al, 2001) and mutations in more than 100 genes reduce growth at alkaline conditions (Serrano et al, 2004), suggesting that a large number of genes may affect the ability of yeast cells to grow at high pH. In this paper, we use two different strategies, in-lab evolution (ILE) and backcrossing (congenic lines), to map QTLs in yeast. Different sets of QTLs were found in independently evolved selection lines. Almost all candidate QTLs uncovered affected regulatory genes, such as ubiquitin ligases, proteins involved in GPI anchoring as well as copper/iron sensing and chromatin remodelers, and not structural components of the pH homeostasis machinery (such as proton pumps) (van der Rest et al, 1995; Forgac, 1998; MacLeod et al, 1998). Our results provide insights on the architecture of complex trait networks and their evolution.
Results
The ability to thrive under alkali stress varies among different yeast isolates and behaves as a complex trait. We examined the capacity of 37 clinically isolated and laboratory S. cerevisiae strains (Supplementary Information 3.1) to grow on media of varying pH, and determined the maximal pH (MP) at which each strain was able to grow (Supplementary Information 1.1 and 1.2). The MP phenotypes of this population showed a normal distribution, ranging between 7.4 and 8.6, with a mean of approximately pH 8.0 (Supplementary Figure 1A). The standard laboratory strain BY4741 (Brachmann et al, 1998) was chosen as a representative of yeast strains with low MP. As expected, in crosses between low MP and high MP strains (Supplementary Information 3.2), the ability to thrive on alkaline media also shows a similar normal distribution (Supplementary Information and Supplementary Figures 1A and 1B). We used a combination of two independent methods (Figure 1) to dissect the architecture of the QTLs underlying the trait. Our combined strategy enables the identification of different sets of genes that evolved in laboratory and clinically isolated strains.
Figure 1.
General strategy for mapping quantitative trait loci (QTLs) affecting genetic variability. See text for details. (A) In-lab evolution. A strain with poor ability to grow under alkali stress was serially transferred to media of increasing pH. (B) Construction of congenic lines up to the eighth generation. (C) Serially transferred cultures accumulate beneficial mutations. After long-term selection at increasing pH levels, cells were isolated from low pH media and individual colonies were grown and tested for their ability to grow at high pH. The ancestor (BY4741) and colonies derived from individual selection lines are shown. (D) Map of QTLs affecting the MP phenotype. Schematic representation of the 16 chromosomes of S. cerevisiae (black circles represent centromeres). Regions that were inherited from the high MP parent in the congenic lines and were identified by hybridization to oligonucleotide arrays are marked in red. QTLs showing mutations in the in-lab-evolved strains are marked green, yellow and orange.
ILE and QTL identification
The ILE experiment was carried out by a serial transfer method. The low MP laboratory strain BY4741 was subjected to a stepwise selection procedure during 120–150 generations, enabling enrichment for beneficial mutations that allow growth at increasing pH. We started cultures from single colonies grown at pH 7.4, and successively transferred samples of the population to rich medium at higher and higher pH levels up to pH 8.6 (Figure 1A). Five independent lines went through this procedure in parallel to create five separate populations.
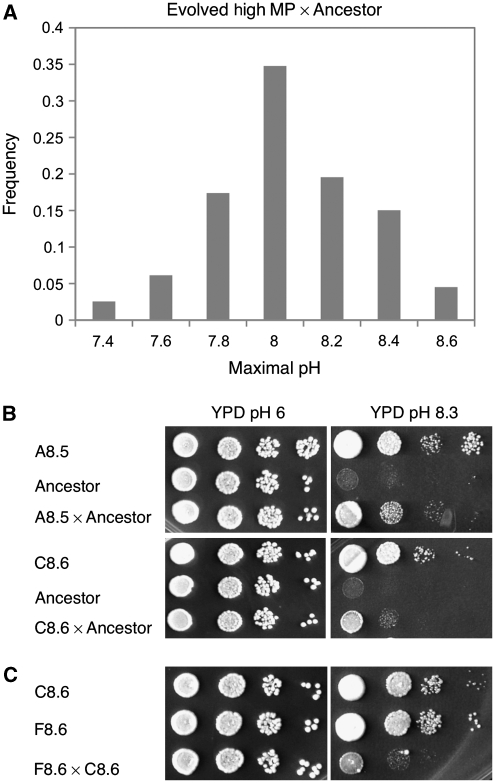
At the end of this long-term selection, individual clones were isolated from each population and their ability to grow at high pH was examined after nonselective growth; all the cells tested retained their high MP phenotype (Figure 1C). We crossed yeast cells able to grow at pH 8.6 to the low MP ancestor and examined the ability of the haploid progeny to grow on media of various pH levels. As expected, a normal distribution of phenotypes was observed in each case (Figure 2A). To see whether different mutations had evolved in different lines, we carried out crosses between clones from different ILE lines and the low MP ancestor. On the basis of the phenotype of each hybrid we conclude that the mutations selected included both recessive and dominant (or at least co-dominant) alleles (examples are shown in Figure 2B). We also carried out crosses between clones from different lines. The MP phenotypes observed in the hybrids were different from those of the selected parents, indicating that different sets of alleles had evolved in each clone (Figure 2C). To identify the mutations responsible for the ability to grow at high pH, we used tiling arrays to genotype the low MP ancestor and three evolved strains: two clones from two independent lines that had acquired the ability to grow at pH 8.5 and pH 8.6 (A8.5 and C8.6) and one clone from an intermediate stage of line C (C8.0). The identity and nature of the mutated alleles was further confirmed using direct DNA sequencing. Table I summarizes all the mutations uncovered by the ILE analysis.
Figure 2.
Several genetic networks were selected during the in-lab evolution. (A) Phenotype distribution among 210 spores derived from a cross between a selected line (A8.5) and its low MP ancestor. (B) Clones from populations A8.5 and C8.6 were crossed to the low MP ancestor. The 10-fold dilutions of diploid yeast cells were plated on YPD at pH 6 and high pH solid media. In each case, the hybrid had a phenotype different from that of each parent. The hybrids also show different MP phenotypes. (C) The hybrid between two selected lines shows a low MP phenotype, indicating that most of the mutations that occurred in these lines are recessive and affect different QTLs.
Table 1. Location and nature of the mutations acquired during selection for the ability to grow at high pH in the ILE procedure.
| Clone | Gene | Mutation | Function | Fixation stagea | Validationb |
|---|---|---|---|---|---|
| RHA, reciprocal hemizygosity assay; AS, allele swapping assay. | |||||
| aStage at the ILE procedure at which the given allele is observed as the sole allele in the cell population. | |||||
| bAssay that demonstrated a biological effect on pH homeostasis for the gene analyzed. | |||||
| cYFR057w is a telomeric ORF. All attempts to delete or replace it failed. | |||||
| dApproximately 80% of population in B8.6 has the mutation in MAC1. | |||||
| C8.0, C8.6 | GTT2/MMP1 | Converging 3′ UTRs | Glutathione S-transferase/ S-methylmethionine permease | pH 7.7 | No significant effect found |
| C8.0, C8.6 | YFR057w | Converging 3′ UTRs | Unknown | pH 7.8 | Could not be testedc |
| C8.6 | ECM21 | Nonsense at codon 193 | Ubiquitin-ligase adaptor | No fixation | Deletion, RHA, AS |
| C8.6 | NMD4/YLR363w-a | Base substitution at NMD4 promoter (−262 bp) | Nonsense-mediated mRNA decay/unknown | pH 8.4 | Deletion, RHA, AS |
| C8.6 | GPI17 | S63 to L | GPI-anchor transamidase | No fixation | Essential gene, not tested |
| C8.6 | YHR140w/SPS100 | Base substitution at SPS100 promoter (−384 bp) | Unknown/spore wall maturation | No fixation | Deletion, RHA, AS |
| C8.6 | MAC1 | C271 to W | Copper-sensing transcription factor | pH 8.4 | Deletion, RHA, AS |
| A8.5 | C271 to S, P384 Silent | pH 7.6 | Deletion, RHA, AS | ||
| G8.6 | C271 to Y | pH 8.6 | Deletion, RHA, AS | ||
| B8.6 | C271 to Y | pH 8.6d | Deletion, RHA, AS | ||
| A8.5 | CDC23 | M71 to I | Ubiquitin ligase (APC/C) | pH 8.4 | RHA |
| A8.5 | CTF3 | R697 Silent | Outer kinetochore protein | No fixation | Deletion (mild effect) |
| C8.0 | OAC1 | I48 to F | Oxaloacetate carrier | No fixation | No significant effect found |
| C8.0 | RRI2 | A138 to P | Ubiquitin ligase regulation | No fixation | No significant effect found |
| C8.0 | GPH1 | 3′ UTR | Glycogen phosphorylase | No fixation | Deletion, AS |
| C8.0 | IES2 | Base substitution at promoter (−65 bp) | Chromatin remodeling | No fixation | Deletion, AS |
| C8.0 | SGT2 | Start codon M1 to I | Glutamine-rich chaperone | No fixation | AS |
| C8.0 | HRD1 | L19 to F | Ubiquitin ligase | No fixation | AS |
In total, mutations were found in 15 genes (Table I) in the three clones that we had genotyped. In each clone, different sets of genes were mutated during the ILE (except MAC1, which acquired mutations in both line A (C271S) and line C (C271W)). The mutations observed in strain C8.0, which represents an intermediate stage in the evolution of line C (to which C8.6 also belongs), only partially overlap those of clone C8.6. Thus, some of the mutations that appeared in the population selected for growth at pH 8.0 did not reach fixation in the population and therefore do not appear at later stages of selection (pH 8.6). To pinpoint the stage at which each mutation attained fixation within the population and to test whether other lines had acquired the same mutations, we sequenced all the genes in Table I, including upstream and downstream sequences, in populations from intermediate stages of the evolution in all lines. We found that mutations appeared and reached fixation at different stages in different lines (Table I and Supplementary Information 6). Some of the mutations found in the particular clones subjected to DNA sequencing were still polymorphic within the selected population, showing the existence of multiple clonal lineages within each cell population. Interestingly, populations B8.6 and G8.6 (but not F8.6) showed mutations in the same codon (cysteine 271) of the MAC1 gene mutated in lines A and C. Although affecting the same codon, the actual changes were different in the various cell lines. None of the other genes in Table I were affected in lines B, F and G, despite the ability of those strains to grow at high pH. We therefore conclude that different mutations were selected in each independent selection line, resulting in different QTL combinations that yield the same high MP phenotype.
Validation of the gene set identified in the ILE experiment
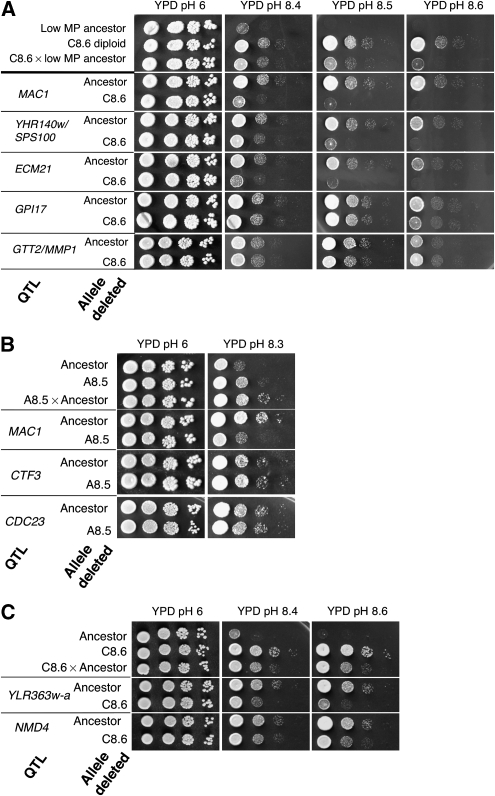
As a first step to determine whether all 15 genes mutated during ILE contribute to the ability to thrive at high pH, we deleted the individual genes (except for GPI17 and CDC23, which are essential) in the low MP ancestor (Supplementary Figure 2A) and in each of the evolved clones (Supplementary Figure 2B and C) and tested their ability to grow at high pH. In principle, it is not to be expected that a full deletion of the gene should show the same phenotype as the point mutations found during the ILE procedure. Yet, some of the deleted strains, such as Δmac1 and Δgph1, showed a reduction in their ability to grow at high pH, whereas Δecm21 showed improved fitness under these conditions (Supplementary Figure 2). This is consistent with the fact that a nonsense mutation in ECM21 was acquired by clone C8.6 (Table I). In agreement, deletion of ECM21 from the low MP background increased the fitness at high pH, whereas the same deletion did not improve the phenotype of the C8.6 strain (Supplementary Figure 2A and B).
Reciprocal hemizygotes
Reciprocal hemizygosity is a tool for analyzing the contribution of each allele in the same (hybrid) genetic background (Steinmetz et al, 2002). Isogenic pairs of strains were constructed, all with the same hybrid genetic background (low MP × selected high MP), and differing by which of the two alleles of each gene was deleted (Figure 3A and B). We tested the differences in the ability to grow at high pH between pairs of reciprocal hemizygotes of nine alleles found in A8.5 and C8.6. The majority of alleles tested had a visible effect, with the exception of GTT2 (which has a mutation in the 3′ UTR region), CTF3 (R697, silent mutation), CDC23 (M71I) and GPI17 (S63L) (Figure 3A–C). This analysis confirms the contribution to the MP phenotype of five out of the nine strains tested.
Figure 3.
Validation of the effect of each mutation found in evolved lines. (A) Drop assay for reciprocal hemizygote pairs to test the relative effect of each allele. In each pair of strains one of the alleles (from the ancestor or evolved origin) was deleted. (B) Results of a drop assay for reciprocal hemizygote pairs derived from line A8.5. (C) A single SNP affects two genes that contribute to the ability to grow at high pH. Drop assay for reciprocal hemizygote pairs of NMD4 and its adjacent, divergently transcribed ORF YLR363w-A (of unknown function). Both genes contribute to the ability to grow under alkali stress.
Some SNPs affect the phenotype through two genes
One of the mutations identified in C8.6 occurs in chromosome XII, at the shared regulatory region of two divergently transcribed genes, NMD4 and YLR363W-A. Whereas NMD4 is a well-characterized gene that affects nonsense-mediated decay and mRNA stability (He and Jacobson, 1995), YLR363W-A encodes a nuclear protein of unknown function. To determine which of the two genes is the one contributing to the high MP phenotype, we constructed two pairs of reciprocal hemizygotes: one pair with reciprocal deletions of the NMD4 ORF, and the other pair with reciprocal deletions of YLR363W-A. The ability of the two pairs to grow on media of different pH was tested. Figure 3C shows that deletion of any of the ORFs in the C8.6 background leads to reduced MP, indicating that both genes contribute to the phenotype. Thus, a single mutation in the promoter of these divergent genes was selected in C8.6, apparently affecting two neighboring genes that contribute to survival under alkali stress.
Allele swapping
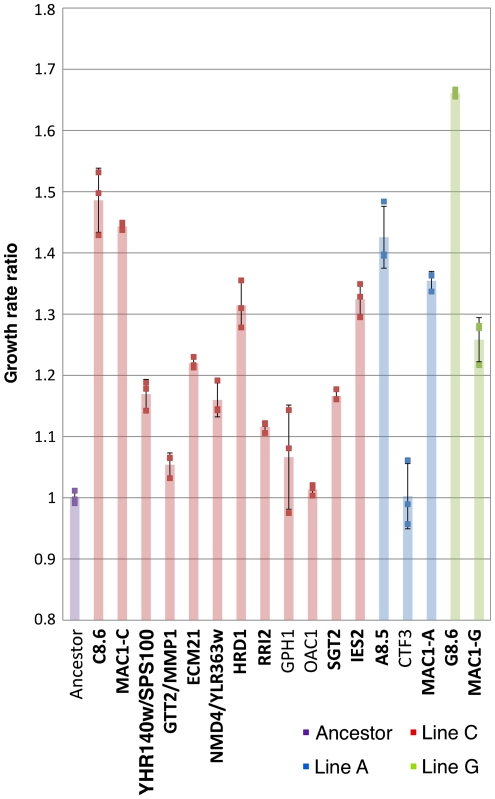
Finally, we wanted to estimate the contribution of each mutation to the phenotype. To do so, we created a set of 14 strains from the low MP haploid background, in which one allele was swapped by the allele from the evolved strain (11 different QTLs and 3 independent alleles of MAC1). We measured the fitness of these strains (as well as that of the evolved strains and the low MP wild-type control) in triplicate at pH 6 and at pH 8 (Figure 4). Out of the 14 mutations, 11 showed a significant effect on the phenotype (Figure 4). Among them, the three different alleles of MAC1 (C271Y, C271W and C271S) had the highest effect. Under this genetic configuration (single allele swapping), most of the ability to grow at high pH can be attributed to MAC1, whereas other QTLs have more modest contributions (Figure 4).
Figure 4.
Quantification of the effect of each mutation using allele swapping. In all, 14 ‘Allele swap’ strains were created by introducing, in the low MP ancestor background, a single mutated allele from a high MP evolved strain. Growth curves were obtained for three different cultures of each of these strains and the controls at pH 6.0 and pH 8.0. Each column represents the ratio between the growth rate of a given strain and the average growth rate of the ancestor strain at high pH (8.0). The three dots on each column represent the calculated ratios for each strain. The colors represent alleles identified in different parent lines. The pale bars in the background provide the ratio based on the average of the three repeats. The strains that improve significantly growth at high pH (P<0.05, FDR corrected) are bolded. Source data is available for this figure at www.nature.com/msb.
Construction of congenic lines
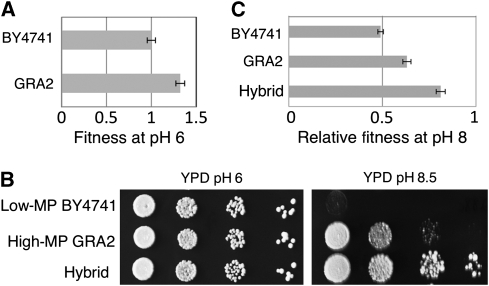
The experiments described above were able to uncover several new sets of genes that acquired mutations during ILE. To explore additional variability that may have developed in nature (under different environmental conditions), we also analyzed a naturally occurring yeast isolate that is able to grow at high pH. Strain GRA2 is a haploid spore derived from a clinically isolated diploid strain (MKF12) (Supplementary Information 3.2.). In comparison with the low MP strain BY4741, GRA2 shows a shorter doubling time under standard growth conditions (Figure 5A) and higher relative fitness at high pH (Figure 5B and C). Hereafter, we refer to GRA2 as the High MP strain. A cross between the strains yielded a diploid hybrid whose relative fitness at high pH media was slightly higher than that of diploid strains isogenic to either parent, a phenomenon known in quantitative genetics as heterosis (Figure 5B and C). The mild phenotypic superiority of the hybrid implies that both parents carry alleles contributing to the phenotype (Lippman and Zamir, 2007). The hybrid strain was subjected to meiosis and the ability of 258 haploid segregants to grow at various pH levels was monitored. As expected, a normal distribution of MP was observed (Supplementary Figure 1B). We could thus estimate the heritability of the MP phenotype, calculated as the ratio between the genetic and the phenotypic variance (see Materials and methods; Brem et al, 2002). We estimate that 88% of the phenotypic variance is of genetic, rather than environmental, origin.
Figure 5.
Genetic variability in the ability to grow at high pH. (A) Fitness measurement of strains GRA2 and BY4741. The doubling time (DT) of each strain was measured while growing on YPD pH 6 or pH 7.9 during logarithmic phase. (B) A cross between the low MP parent (BY4741) and the high MP parent (GRA2) results in hybrid vigor (heterosis). Serial 10-fold dilutions of diploid strains were plated on regular and high-pH media. (C) Relative fitness of the three strains.
To dissect the network of genes contributing to the high MP phenotype of this strain, we created congenic lines up to the eighth generation (Figure 1B). Starting with the high MP and the low MP parents, a series of backcrosses was carried out. At each generation, segregants that are able to grow at pH 8.6 were selected for, and backcrossed, to the low MP parent. Four independent congenic lines were thus created in parallel. After eight rounds of such backcrosses, all segregants with high MP should have inherited from the high MP parent a set of genes that allows growth at high pH, whereas the rest of the genome predominantly reflects that of the low MP parent. At each generation, random recombination events are expected to reduce the size of the chromosomal intervals inherited from the high MP parent. However, the reduction in size decreases with each successive generation. Simulations showed that after eight generations additional backcrosses would not significantly reduce interval size (data not shown).
To analyze whether all congenic lines carried the same set of genes affecting the growth at high pH, or whether each one acquired a different set of genes allowing growth at high pH, we carried out crosses between individuals from the four different congenic lines at the fourth generation. If the gene sets of two lines are identical, we expect all the progeny to be able to grow at high pH. If, however, each parent inherited from the high MP strain a different sub-network, then we should encounter, among the progeny, spores with poor alkali resistance. The proportion of high MP segregants was 0.85–0.9 for all inter-line crosses. This frequency is similar to the estimated heritability (0.88), indicating that the same gene set was inherited in all four lines.
We took advantage of the naturally occurring variation in sequence between GRA2 and BY4741 to identify the genomic regions potentially carrying the high MP genetic network. Four strains from two independent congenic lines were genotyped by tiling arrays. Two of the strains (N8HA and N8HB) were obtained independently after eight successive backcrosses and two additional strains represented intermediate stages (N4HA and N6HB—fourth and sixth backcrosses, respectively). As expected, the strains from the eighth generation shared most of their genomes with the low MP parent, whereas <10% of their genomes was inherited from the high MP parent. The genetic information potentially containing the high MP alleles was present in chromosomal intervals that were larger in the fourth and in the sixth generation backcross than in the strains that underwent eight backcrosses, illustrating the size reduction of genomic regions originating from the high MP parent with each backcross. Once the regions were identified by the tiling arrays, we confirmed them using sequencing. We also sequenced the same regions in two additional independent congenic lines to see whether they had also inherited the same regions from the high MP parent. Almost all the regions identified were conserved in at least two independently obtained congenic lines (Supplementary Table 1).
Figure 1D and Table II present the 17 regions derived from the high MP parent, which are candidates to contain QTLs. On average, each region is 42 kb long and contains 17.6 genes (Supplementary Table 2). MAC1 was the only gene identified by the ILE procedure that was also present in one of the congenic regions. To systematically predict the gene in each genomic region identified, we took advantage of the large amount of information available about the function of the yeast genome. We applied a predictive algorithm (see Materials and methods) that takes into consideration the network proximity of each gene within the defined intervals to the genes identified in the ILE procedure, and their similarity in GO annotation, generating ranked probabilities of being a relevant gene. This method allowed us to predict, for each interval, the genes most likely to affect the MP phenotype in the high MP strain (Table II). Out of the predicted genes, five (MAC1, CTR1, FEN1, KEM1 and PMR1) have been previously identified in a systematic genome-wide screen as required for growth at high pH (Serrano et al, 2004), and we have confirmed this phenotype (Supplementary Figure 3). From each region we picked the candidates with the highest and the lowest scores and tested the ability of their deletion to affect growth at high pH. Out of the 29 deletions with the best scores tested, 13 decreased the ability to grow at high pH (Table II, Supplementary Figure 3) whereas only 4 out of the 28 lowest-ranking candidates affected the phenotype (P-value=0.015).
Table 2. Regions inherited from the high MP parent (present in all the congenic lines), and best candidate genes in each region.
| Ch. | Position | N | Candidate | Function |
|---|---|---|---|---|
| aNot tested (essential gene). | ||||
| Deletion of genes in bold resulted in a decreased MP relative to the wild type. | ||||
| N represents the number of genes in each region. | ||||
| III | 168966–200800 | 13 | FEN1 | Fatty acid elongase that affects cell wall synthesis. Deletion confers sensitivity to high pH (Serrano et al, 2004) |
| V | 1–29961 | 7 | SIT1 | Ferrioxamine B transporter, induced during iron deprivation |
| VI | 30356–51606 | 8 | ALR2 | Probable Mg(2+) transporter |
| VII | 143903–200324 | 26 | PMR1 | High-affinity Ca2+/Mn2+ P-type ATPase. Deletion confers sensitivity to high pH (Serrano et al, 2004) |
| KEM1 | Exonuclease involved in mRNA stability. Deletion confers sensitivity to high pH (Serrano et al, 2004) | |||
| VII | 325256–396816 | 31 | HNM1 | Choline transporter |
| VIII | 478122–517418 | 20 | GPI16 a | Subunit of GPI transamidase complex |
| RPN10 | Subunit of the 19S regulatory particle (RP) of the 26S proteasome | |||
| IX | 424859–431838 | 1 | YPS6 | GPI-anchored aspartic protease |
| X | 633129–683951 | 20 | ILM1 | Unknown function |
| XI | 533219–574982 | 18 | TIF1 | Translation initiation factor eIF4A |
| XII | 92342–114724 | 7 | KNS1 | Nonessential putative protein kinase |
| XII | 262946–280320 | 8 | RPL22A | Protein component of the large (60S) ribosomal subunit |
| XII | 345291–374049 | 7 | MDN1 a | Peroxyredoxin, protects against oxidative damage |
| XII | 606720–775305 | 67 | MMS22 | Protein involved in resistance to ionizing radiation |
| XII | 937245–949420 | 2 | CTR3 | High-affinity Copper transporter |
| XIII | 307000–355700 | 24 | MAC1 | Transcriptional activator of Cu transporters. Deletion confers sensitivity to high pH (Serrano et al, 2004) |
| XIV | 453937–509171 | 28 | RPL16B | N-terminally acetylated protein component of the large (60S) ribosomal subunit |
| XVI | 785313–804496 | 12 | CTR1 | High-affinity Copper transporter. Deletion confers sensitivity to high pH (Serrano et al, 2004) |
One of the smallest identified regions carried a single gene, CTR3, encoding a high-affinity copper transporter of the plasma membrane, which is regulated by Mac1 (Pena et al, 2000). We sequenced the CTR3 ORF and promoter region of the high MP and the low MP parents and found no differences (with the exception of a transition at position 95 of the ORF, which led to a conservative change between acidic amino acids (aspartic to glutamic acid)). Notably, however, the low MP strain carries a Ty element (a yeast retrotransposon) at ∼100 bp upstream of the CTR3 ORF, which is missing from the high MP strain. Analysis of several clones from the ILE lines C and A showed that they lack the Ty element (originally present in the low MP ancestor), which was excised by an LTR–LTR recombination event (Kupiec and Petes, 1988) during the selection procedure.
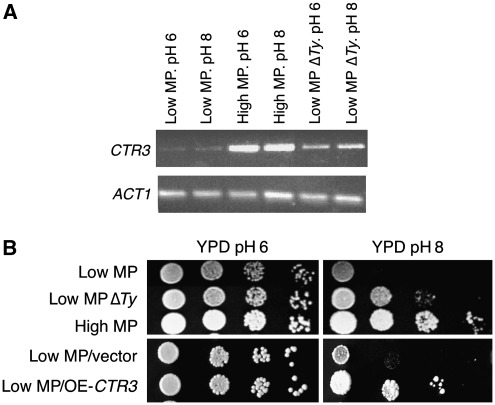
We used RT–PCR to measure the transcription level of CTR3 in both parents and found that the mRNA level of CTR3 is 5 times higher in the high MP strain than in the low MP parental strain (Figure 6A). Deletion of the Ty in the low MP strain led to a 2.5-fold increase in CTR3 transcription, but it is still lower than the transcription level observed in the high MP strain (Figure 6A). These results indicate that in addition to factors acting in cis, such as the presence or absence of the Ty element, additional trans-acting factors have a role in the regulation of CTR3. A good candidate for such a regulator is MAC1, which is identified in several of the ILE lines (Table I) and also found in one of the congenic regions (Table II). Deleting the Ty also led to a noticeable improvement in its ability to grow at high pH, but, as expected, not to a complete high MP phenotype (Figure 6B). In agreement, over-expression of CTR3 from a high-copy-number plasmid also improved the ability to grow at high pH (Figure 6B).
Figure 6.
The CTR3 gene contributes to the MP phenotype. (A) RT–PCR showing the differences in transcription levels of CTR3 in different strains. ACT1 served as a control. (B) The ability of the various strains to grow at regular or high pH media (10-fold serial dilutions). Overexpression of CTR3 allows growth at higher MP.
Discussion
In this paper we present a whole-genome strategy for QTL identification. We were able to map four independent gene sets affecting the ability to grow under alkali stress. pH affects almost all intracellular processes, and governs fungal virulence in plants and animals (Davis, 2003; Penalva et al, 2008). Using the ILE procedure we identified 15 QTLs. By analyzing strains deleted for the candidate genes, as well as by reciprocal hemizygosity and allele swapping assays, we have verified that at least nine of them affect the ability of yeast cells to thrive at high pH. We have also carried out an estimation of their effect (Figure 4). In addition, our congenic lines identified 12 additional candidate QTLs affecting the same trait. MAC1 was identified by both approaches. The fact that only a low degree of overlap was detected between the two experimental approaches may indicate that different selective forces exert an effect during selection procedures in the laboratory and evolution in the wild. Understanding the architecture of quantitative variation has become the new frontier of genetic research, and many studies are focusing on whole-genome strategies (Deutschbauer and Davis, 2005; Ben-Ari et al, 2006; Ellegren and Sheldon, 2008; Gerke et al, 2009). Although these studies are improving our understanding of the genetic basis of many complex traits, each study can identify only a small fraction of the variants causing a certain phenotype. Moreover, groups analyzing the same phenotype usually find little overlap between QTLs (Gudbjartsson et al, 2008; Lettre et al, 2008; Maher, 2008; Weedon et al, 2008). The small overlap suggests that the variance observed in nature may be due to many alternative allelic combinations with similar effects. Indeed, our study revealed that even under a uniform selection regime, different combinations of mutations that lead to the same phenotype can appear (Figure 2B and C).
Inspection of the QTLs affecting resistance to high pH reveals a striking absence of structural genes directly in charge of the cellular machinery responsible for pH homeostasis, such as proton pumps (Forgac, 1998). Instead, the majority of the mutations identified affect global regulators, such as ubiquitin ligases, chromatin remodelers, proteins involved in cell wall maintenance and copper sensing factors.
Copper exerts an effect as a cofactor for many enzymes in yeast, and its intracellular levels are strictly regulated. Copper and iron ions were shown to affect tolerance to high pH (Serrano et al, 2004). Four of five separate selection lines independently acquired different mutations at the same position (Cysteine 271) of the copper-sensing transcription factor, MAC1. Mac1p is a transcriptional activator of copper-responding genes (such as CTR1 and CTR3, which were also found by the congenic lines strategy; Jungmann et al, 1993). Copper binds the trans-activating Cysteine-rich domain (residues 264–279) of Mac1p and inhibits its activity (Jensen and Winge, 1998). Mutations at Cys271 are therefore likely to abrogate copper binding, thus elevating transcription in the Mac1 gene cohort. We have shown that changes in the expression level of CTR3 can alter fitness at high pH. In the wild-type strain GRA2 (and in several of the ILE lines), the high expression of CTR3 was caused by excision of a transposon (Knight et al, 1996), underscoring the importance of mobile elements in evolution (Dunham et al, 2002; Gresham et al, 2008) and gene expression, as suggested by McClintock (McClintock, 1984). A second group of regulatory genes affected cell wall maintenance. Glycosylphosphatidylinositol (GPI) anchoring is essential for the correct localization of many proteins that exert an effect in the assembly and remodeling of fungal cell wall as a response to environmental pH (Ohishi et al, 2001; Serrano et al, 2006). GPI17 (identified in the ILE lines), as well as FEN2, YPS6 and GPI16, (found in the congenic lines) have important roles in this process. Moreover, both ECM21 and YHR140w show physical and genetic interactions with members of the GPI addition pathway (Tong et al, 2004; Miller et al, 2005), underscoring the central role of GPI anchoring in pH homeostasis.
A third group of QTL genes was related to ubiquitin metabolism. They either encode ubiquitin ligases (HRD1 and CDC23; Scrimale et al, 2009), or adaptors/regulators of ubiquitin ligases (ECM21 and RRI2; Lin et al, 2008).
Mutations in genes with wide regulatory functions seem therefore to be preferred to mutations with narrow functions affecting solely the trait selected (e.g., proton pumps). This fact reflects the inherent architecture of the genomes, which places genes with related functions under common regulatory circuits. Mutations in key regulators can thus affect several genes at once, conferring an immediate effect on fitness. Such an arrangement is advantageous in the long run, despite the possibility of mutations that may inactivate the whole network. Mutations simultaneously affecting two adjacent genes were also observed (Table I).
Many models (Fisher, 1930; Orr, 2003) and experiments (Brem and Kruglyak, 2005) have been designed to answer evolutionary questions regarding the accumulation of QTLs, the effect of each QTL and the interactions among them (St Onge et al, 2007; Shachar et al, 2008). Using allele swapping we could isolate and estimate the contribution of each QTL in the low MP background. The alleles analyzed showed different effects on the MP phenotype, and no correlation could be observed between their effect and the time the mutation appeared in the selected population. Some genes such as MAC1 and YHR140w have a major effect. MAC1 alone, for example, can explain approximately 80% of the difference between the low MP and the high MP strains. Those results show that a simple additive model alone is insufficient to explain the phenotype, as the sum of the individual effects measured is greater than the effect observed in the selected high MP strain. Our results thus suggest that epistatic effects among the various alleles dampen the individual contribution measured as a single allele. Similarly, several of the mutations tested that failed to show a significant effect on the MP phenotype may do so in the appropriate genetic background, that is, when combined with the appropriate QTLs. A combinatorial approach is required to study the way in which the identified genes interact. Future plans include the creation of a combinatorial collection of mutants to analyze the hierarchies of regulation and the genetic relations between the mutations found. With the availability of new methodologies to map and identify QTLs, such as the one presented in this study, many issues can be addressed. One important lesson from the explosion of whole-genome studies during the past years is that ‘simple’ monogenic Mendelian inheritance is often not so simple and the traditional distinction between Mendelian and quantitative traits begins to blur. What was considered in the past a gene with a Mendelian inheritance pattern can be regarded today as a QTL with a strong phenotypic effect. Mutational or genotypic heterogeneity can explain some of the phenotypic variation observed in monogenic traits, but usually not all of it (Botstein and Risch, 2003). The residual variation is probably due to modifier genes, genetic background and environmental contributors. Identifying such modifiers is an important challenge for the future. Our work provides new insights and novel tools for handling this challenge and sheds light on the genetic basis of fitness variation.
Materials and methods
Yeast growth
Yeast cells were grown in yeast extract-peptone-dextrose medium (YPD) at pH 6 at 30 °C (standard growth conditions). The pH was adjusted and kept constant using 100 mM Tris buffer.
Enrichment of beneficial mutations by ILE
Strain BY4741 (MATa Δura3, Δleu2, Δhis3 and Δmet15) grows extremely slowly at pH 7.4. Five independent lines were established from a single colony. When cultures reached saturation, aliquots (5 μl) were transferred to fresh medium (5 ml) at a slightly higher pH (0.1 pH unit steps) and were similarly incubated. This procedure was repeated until populations that were able to grow at pH 8.5 or 8.6 were obtained. Samples were taken at each passage and the genotype of the strain tested.
At the end of this procedure cells were streaked on YPD at pH 6, and individual colonies were tested for their ability to grow at high pH using a drop assay.
Congenic lines
A cross between BY4741 and GRA2 was performed, and after meiosis, haploid spores were plated on medium at pH 8.4. Spores that could grow were then re-tested in a drop assay (as described above) and the best-growing segregants backcrossed to the BY4741 strain. This procedure was repeated for eight generations, establishing four independent congenic lines.
Phenotypic determination
(A) Serial 10fold dilutions were spotted on YPD plates at various pH. For each strain, four independent replicas were made. (B) Fitness measurements: Strains were inoculated into 2 ml of YPD at pH 6 and incubated until exponential growth (∼107 cells/ml) was reached. Optical density at 600 nm (OD600) of cultures was measured and all strains were diluted to OD 0.1 in 2 ml YPD at either pH 6 or pH 8. Cultures were grown to OD 1.5. The growth rate of each culture was monitored by measuring the OD600 every 30 min, and the growth rate was calculated by fitting an exponential curve to the measurements. The relative fitness of a strain was defined as the ratio of the growth rate between the mutant and the ancestor (see details in Supplementary Information).
Heritability and QTL estimations
Heritability was calculated using the formula:
in which Var(seg) and Var(p) stand for the variance in MP phenotype of the segregants and the parents, respectively (Brem et al, 2002).
Genotyping
Yeast strains were grown under standard conditions. Genomic DNA was extracted and hybridized to 25-mer oligonucleotide microarrays (Affymetrix GeneChip® S. cerevisiae tiling 1.0R) that provide complete and redundant coverage of the ∼12 Mb S. cerevisiae genome. This design provides for multiple measurements of each nucleotide's contribution to hybridization efficiency and therefore has the ability to detect the presence and location of single-nucleotide polymorphisms and deletion events throughout the entire yeast genome with near nucleotide precision (Gresham et al, 2006; Schacherer et al, 2007). Potential SNPs were detected by: (1) applying a genome-wide SNP detection algorithm on each DNA microarray separately, (2) statistically analyzing and integrating detections across replicates and, (3) (for the congenic strains) mapping parental regions (for details see Supplementary Information). This process was followed by sequencing of some SNPs in the genotyped congenic lines and in other congenic lines to verify the borders of the regions. The cell files for the DNA microarray experiments are available at ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/) under accession numbers: a) in Lab Evolution: E-MEXP-2520; b) Congenics: E-MEXP-2521.
Allele swapping
To swap each allele we used the ‘delitto perfetto’ method (Storici and Resnick, 2003). In brief, we used the low MP strain (BY4741) as genetic background. For each candidate QTL, we deleted the region containing the mutation by insertion of a fragment carrying the URA3 gene and the gene I-SceI with its restriction site. We then induced a double-strand break at the I-SceI site to enhance homologous recombination and replaced the URA3-I-SceI fragment by a PCR fragment containing the mutation and plated on 5FOA to select for candidates. Candidates were then sequenced to verify that the allele was replaced and no other mutations occurred in the region during the process. Essential genes (GPI17 and CDC23) and the telomeric gene YFR057w were not tested using allele swapping because of technical difficulties.
Estimating the effect of each QTL
To evaluate the contribution of each QTL, we used the information from the allele swapping experiment. The effect of each mutation was defined as the growth rate of each strain compared with the ancestor, all measured at high pH (Supplementary Information 7). Figure 4 presents the contribution of each gene. The total contribution of all genes is greater than the difference between the fitness of the evolved and the wild-type parent, suggesting that the contribution model is not additive and the mutant alleles interact, each masking part of the effects of the others (epistasis).
Systematic prediction of QTLs in each genomic region identified by the congenic lines
We constructed a joint protein–protein and protein–DNA interaction network, by combining protein–protein interaction data from BioGrid (Breitkreutz et al, 2008), KEGG (Kanehisa and Goto, 2000), Reactome (Joshi-Tope et al, 2005) and (Yu et al, 2008) with protein–DNA interactions from three large-scale studies (Simon et al, 2001; Harbison et al, 2004; Workman et al, 2006) (Supplementary Table 2). An edge was added to the network if the pair of genes was reported as interacting in any of these data sets. We then calculated, for each gene in the identified intervals, its minimal network distance (the length of the shortest path in the graph) from any gene uncovered using ILE (ILE gene set) and picked within each interval that gene for which the distance was minimal. In case several such genes were found, we picked the gene that had a path of minimal length to more ILE genes. Finally, in case of ties, we picked the gene that was most similar to some ILE gene based on GO semantic similarity (Lord et al, 2003) (taking into account only ‘biological process’ annotations). Validations were carried out with the two higher-and lower-ranking deletion strains available (essential genes were not tested).
Supplementary Material
Supplementary Methods, Supplementary Figures S1–3, Supplementary Table S1
Supplementary Table
Legend for Supplementary Table S2
Acknowledgments
We thank Lior David, Adam Deutschbauer, Caroline Uhlik, Pat Brown, Daniel Melamed, Rachel Brem and Yoav Arava for discussions and technical help. We also thank Benny Yakir for his input on statistical matters during the initial stages of this project. This work was supported by grants from the Israeli Science Foundation, the Israeli Ministry of Science and Technology and the US-Israel Bi-national Fund (BSF) to Martin Kupiec and partly from the Wolfson Foundation to Ron Shamir. We thank all members of the Kupiec lab for help and encouragement. Gal Hagit Romano was supported by an Eshkol scholarship from the Israeli Ministry of Science and Technology. Igor Ulitsky and Gal H Romano were supported in part by a fellowship from the Edmond J Safra Bioinformatics program at Tel-Aviv University.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abiola O, Angel JM, Avner P, Bachmanov AA, Belknap JK, Bennett B, Blankenhorn EP, Blizard DA, Bolivar V, Brockmann GA, Buck KJ, Bureau JF, Casley WL, Chesler EJ, Cheverud JM, Churchill GA, Cook M, Crabbe JC, Crusio WE, Darvasi A et al. (2003) The nature and identification of quantitative trait loci: a community's view. Nat Rev Genet 4: 911–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari G, Zenvirth D, Sherman A, David L, Klutstein M, Lavi U, Hillel J, Simchen G (2006) Four linked genes participate in controlling sporulation efficiency in budding yeast. PLoS Genet 2: e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Risch N (2003) Discovering genotypes underlying human phenotypes: past successes for Mendelian disease, future approaches for complex disease. Nat Genet 33(Suppl): 228–237 [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 [DOI] [PubMed] [Google Scholar]
- Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, Dolinski K, Tyers M (2008) The BioGRID interaction database: 2008 update. Nucleic Acids Res 36: D637–D640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem RB, Kruglyak L (2005) The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc Natl Acad Sci USA 102: 1572–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L (2002) Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755 [DOI] [PubMed] [Google Scholar]
- Castle WE (1914) Multiple factors in heredity. Science 39: 686–689 [DOI] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA (2001) Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12: 323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvasi A, Pisante-Shalom A (2002) Complexities in the genetic dissection of quantitative trait loci. Trends Genet 18: 489–491 [DOI] [PubMed] [Google Scholar]
- Davis D (2003) Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr Genet 44: 1–7 [DOI] [PubMed] [Google Scholar]
- Deutschbauer AM, Davis RW (2005) Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat Genet 37: 1333–1340 [DOI] [PubMed] [Google Scholar]
- Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F, Botstein D (2002) Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 99: 16144–16149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- East EM (1909) A note concerning inheritance in sweet corn. Science 29: 465–467 [DOI] [PubMed] [Google Scholar]
- East EM (1916) Studies on size inheritance in nicotiana. Genetics 1: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, Sheldon BC (2008) Genetic basis of fitness differences in natural populations. Nature 452: 169–175 [DOI] [PubMed] [Google Scholar]
- Fisher RA (1930) The Genetical Theory of Natural Selection. Oxford: Oxford University Press [Google Scholar]
- Forgac M (1998) Structure, function and regulation of the vacuolar (H+)-ATPases. FEBS Lett 440: 258–263 [DOI] [PubMed] [Google Scholar]
- Gerke J, Lorenz K, Cohen B (2009) Genetic interactions between transcription factors cause natural variation in yeast. Science 323: 498–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazier AM, Nadeau JH, Aitman TJ (2002) Finding genes that underlie complex traits. Science 298: 2345–2349 [DOI] [PubMed] [Google Scholar]
- Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, DeSevo CG, Botstein D, Dunham MJ (2008) The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet 4: e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham D, Ruderfer DM, Pratt SC, Schacherer J, Dunham MJ, Botstein D, Kruglyak L (2006) Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science 311: 1932–1936 [DOI] [PubMed] [Google Scholar]
- Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, Sulem P, Thorlacius S, Gylfason A, Steinberg S, Helgadottir A, Ingason A, Steinthorsdottir V, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T et al. (2008) Many sequence variants affecting diversity of adult human height. Nat Genet 40: 609–615 [DOI] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA (2004) Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Jacobson A (1995) Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev 9: 437–454 [DOI] [PubMed] [Google Scholar]
- Jensen LT, Winge DR (1998) Identification of a copper-induced intramolecular interaction in the transcription factor Mac1 from Saccharomyces cerevisiae. EMBO J 17: 5400–5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi-Tope G, Gillespie M, Vastrik I, D’Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, Matthews L, Lewis S, Birney E, Stein L (2005) Reactome: a knowledgebase of biological pathways. Nucleic Acids Res 33: D428–D432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J, Reins HA, Lee J, Romeo A, Hassett R, Kosman D, Jentsch S (1993) MAC1, a nuclear regulatory protein related to Cu-dependent transcription factors is involved in Cu/Fe utilization and stress resistance in yeast. EMBO J 12: 5051–5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SA, Labbe S, Kwon LF, Kosman DJ, Thiele DJ (1996) A widespread transposable element masks expression of a yeast copper transport gene. Genes Dev 10: 1917–1929 [DOI] [PubMed] [Google Scholar]
- Kupiec M, Petes TD (1988) Allelic and ectopic recombination between Ty elements in yeast. Genetics 119: 549–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb TM, Xu W, Diamond A, Mitchell AP (2001) Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J Biol Chem 276: 1850–1856 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11: 241–247 [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D (1989) Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Schork NJ (1994) Genetic dissection of complex traits. Science 265: 2037–2048 [DOI] [PubMed] [Google Scholar]
- Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, Eyheramendy S, Voight BF, Butler JL, Guiducci C, Illig T, Hackett R, Heid IM, Jacobs KB, Lyssenko V, Uda M, Boehnke M, Chanock SJ, Groop LC, Hu FB et al. (2008) Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet 40: 584–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD (2008) Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135: 714–725 [DOI] [PubMed] [Google Scholar]
- Lippman ZB, Zamir D (2007) Heterosis: revisiting the magic. Trends Genet 23: 60–66 [DOI] [PubMed] [Google Scholar]
- Lord PW, Stevens RD, Brass A, Goble CA (2003) Investigating semantic similarity measures across the gene ontology: the relationship between sequence and annotation. Bioinformatics 19: 1275–1283 [DOI] [PubMed] [Google Scholar]
- MacLeod KJ, Vasilyeva E, Baleja JD, Forgac M (1998) Mutational analysis of the nucleotide binding sites of the yeast vacuolar proton-translocating ATPase. J Biol Chem 273: 150–156 [DOI] [PubMed] [Google Scholar]
- Maher B (2008) Personal genomes: the case of the missing heritability. Nature 456: 18–21 [DOI] [PubMed] [Google Scholar]
- McClintock B (1984) The significance of responses of the genome to challenge. Science 226: 792–801 [DOI] [PubMed] [Google Scholar]
- Miller JP, Lo RS, Ben-Hur A, Desmarais C, Stagljar I, Noble WS, Fields S (2005) Large-scale identification of yeast integral membrane protein interactions. Proc Natl Acad Sci USA 102: 12123–12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi K, Inoue N, Kinoshita T (2001) PIG-S and PIG-T, essential for GPI anchor attachment to proteins, form a complex with GAA1 and GPI8. EMBO J 20: 4088–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA (2003) The distribution of fitness effects among beneficial mutations. Genetics 163: 1519–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena MM, Puig S, Thiele DJ (2000) Characterization of the Saccharomyces cerevisiae high affinity copper transporter Ctr3. J Biol Chem 275: 33244–33251 [DOI] [PubMed] [Google Scholar]
- Penalva MA, Tilburn J, Bignell E, Arst HN Jr (2008) Ambient pH gene regulation in fungi: making connections. Trends Microbiol 16: 291–300 [DOI] [PubMed] [Google Scholar]
- Schacherer J, Ruderfer DM, Gresham D, Dolinski K, Botstein D, Kruglyak L (2007) Genome-wide analysis of nucleotide-level variation in commonly used Saccharomyces cerevisiae strains. PLoS ONE 2: e322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrimale T, Didone L, de Mesy Bentley KL, Krysan DJ (2009) The unfolded protein response is induced by the cell wall integrity mitogen-activated protein kinase signaling cascade and is required for cell wall integrity in Saccharomyces cerevisiae. Mol Biol Cell 20: 164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano R, Bernal D, Simon E, Arino J (2004) Copper and iron are the limiting factors for growth of the yeast Saccharomyces cerevisiae in an alkaline environment. J Biol Chem 279: 19698–19704 [DOI] [PubMed] [Google Scholar]
- Serrano R, Martin H, Casamayor A, Arino J (2006) Signaling alkaline pH stress in the yeast Saccharomyces cerevisiae through the Wsc1 cell surface sensor and the Slt2 MAPK pathway. J Biol Chem 281: 39785–39795 [DOI] [PubMed] [Google Scholar]
- Shachar R, Ungar L, Kupiec M, Ruppin E, Sharan R (2008) A systems-level approach to mapping the telomere length maintenance gene circuitry. Mol Syst Biol 4: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Zeitlinger J, Gifford DK, Jaakkola TS, Young RA (2001) Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106: 697–708 [DOI] [PubMed] [Google Scholar]
- St Onge RP, Mani R, Oh J, Proctor M, Fung E, Davis RW, Nislow C, Roth FP, Giaever G (2007) Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat Genet 39: 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz LM, Sinha H, Richards DR, Spiegelman JI, Oefner PJ, McCusker JH, Davis RW (2002) Dissecting the architecture of a quantitative trait locus in yeast. Nature 416: 326–330 [DOI] [PubMed] [Google Scholar]
- Storici F, Resnick MA (2003) Delitto perfetto targeted mutagenesis in yeast with oligonucleotides. Genet Eng (NY) 25: 189–207 [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L et al. (2004) Global mapping of the yeast genetic interaction network. Science 303: 808–813 [DOI] [PubMed] [Google Scholar]
- van der Rest ME, Kamminga AH, Nakano A, Anraku Y, Poolman B, Konings WN (1995) The plasma membrane of Saccharomyces cerevisiae: structure, function, and biogenesis. Microbiol Rev 59: 304–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, Freathy RM, Perry JR, Stevens S, Hall AS, Samani NJ, Shields B, Prokopenko I, Farrall M, Dominiczak A, Johnson T, Bergmann S, Beckmann JS, Vollenweider P, Waterworth DM et al. (2008) Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet 40: 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman CT, Mak HC, McCuine S, Tagne JB, Agarwal M, Ozier O, Begley TJ, Samson LD, Ideker T (2006) A systems approach to mapping DNA damage response pathways. Science 312: 1054–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual JF, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C et al. (2008) High-quality binary protein interaction map of the yeast interactome network. Science 322: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods, Supplementary Figures S1–3, Supplementary Table S1
Supplementary Table
Legend for Supplementary Table S2