Abstract
The autophagy pathway is the major degradation pathway of the cell for long-lived proteins and organelles. Dysfunction of autophagy has been linked to several neurodegenerative disorders that are associated with an accumulation of misfolded protein aggregates. Alzheimer’s disease, the most common neurodegenerative disorder, is characterized by 2 aggregate forms, tau tangles and amyloid-β plaques. Autophagy has been linked to Alzheimer’s disease pathogenesis through its merger with the endosomal-lysosomal system, which has been shown to play a role in the formation of the latter amyloid-β plaques. However, the precise role of autophagy in Alzheimer’s disease pathogenesis is still under contention. One hypothesis is that aberrant autophagy induction results in an accumulation of autophagic vacuoles containing amyloid-β and the components necessary for its generation, whereas other evidence points to impaired autophagic clearance or even an overall reduction in autophagic activity playing a role in Alzheimer’s disease pathogenesis. In this review, we discuss the current evidence linking autophagy to Alzheimer’s disease as well as the uncertainty over the exact role and level of autophagic regulation in the pathogenic mechanism of Alzheimer’s disease.
Keywords: Alzheimer’s disease, amyloidogenesis, autophagy, dysfunction, endosomal-lysosomal pathway
The cellular process of autophagy, called the waste management or housekeeping system of the cell,1 eliminates cytoplasmic “waste” material and aids in the recycling of some of its components. The autophagic pathway is responsible for the delivery of cellular debris such as damaged organelles and obsolete intracellular macromolecules to lysosomes for degradation.2,3 The word autophagy is derived from the Greek words auto (meaning “self”) and phage (meaning “eat”), which precisely define the intent of autophagy to clear cellular contents.4 There are 3 subtypes of autophagy distinguishable by their mechanisms and cellular functions: macroautophagy, microautophagy, and chaperone-mediated autophagy.5– 8
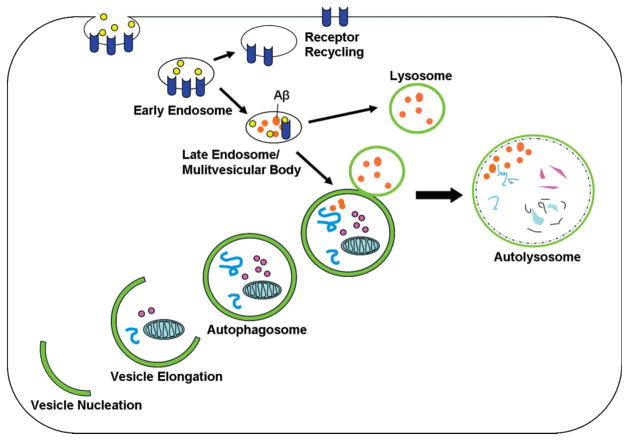
The most prevalent form of autophagy is macroautophagy, which is discussed in this review and hereafter will be referred to as autophagy. Autophagy is the main mechanism used in eukaryotes for the catabolism of organelles and long-lived proteins. Such cytoplasmic debris is brought to lysosomes through double-membraned vacuoles known as autophagosomes. The cellular process of autophagy involves a series of ordered steps (Figure 1). Morphologically, the first step is the nucleation process, by which an isolation membrane, also known as a phagophore, is formed. The membrane then elongates, and the edges of the membrane fuse; this results in the formation of enclosed vacuoles termed autophagosomes, which hold cytoplasmic contents. Lastly, the autophagosome fuses with a lysosome, which forms an autolysosome in which the engulfed material and the inner membrane are degraded.8– 10
Fig 1.
The endosomal-lysosomal pathway merges with the autophagic pathway. The autophagic pathway is used for organelle and protein turnover. Upon the induction of autophagy, a double-membraned structure, which is called the isolation membrane, forms by vesicle nucleation around cytoplasmic contents and becomes an autophagosome. Cytoplasmic substrates are then degraded after fusion of the autophagosome with the lysosome, and this results in the autolysosome. The endosomal pathway, which is important for the uptake and recycling of nutrients and transmembrane receptors, merges with the autophagic pathway. Aβ is generated in multivesicular bodies of the endosomal pathway and may also be generated in autophagosomes. Abbreviation: Aβ, amyloid-β.
Autophagy has roles in both normal cellular homeostasis and disease states. The most studied form of autophagy is its induced response to a variety of stress conditions including nutrient starvation. The activation of autophagy results in enhanced protein degradation accompanied by an increase in the amino acid pool, which provides an energy source and allows for necessary protein synthesis.11 In addition to its response to nutrient availability, a growing body of evidence indicates that autophagy is involved in an array of normal physiological processes as well as pathological conditions, such as development, cell death, aging, antigen presentation, bacterial degradation, and tumor suppression.7,12 Most, if not all, mammalian cells also maintain a basal level of autophagy that is responsible for the normal intracellular turnover of proteins and organelles. This constitutively active autophagy (basal level) is believed to be especially critical for cells that are postmitotic, such as neurons, possibly because of their inability to dilute aberrant components through cell division.13– 15
Despite the rapid growth of autophagy study, the knowledge of the autophagic process in neurons is extremely limited. Recent studies, demonstrating that loss of basal autophagy in the central nervous system causes an accumulation of ubiquitinated protein inclusions and neurodegeneration,13,15 suggest an essential role for autophagy in the continuous removal of proteins destined for degradation and neuroprotection. Emerging evidence has indeed implicated autophagy in several major neurodegenerative disorders, such as frontotemporal dementia, Alzheimer’s disease (AD), Parkinson’s disease, and Huntington’s disease.16– 19 Although many studies have suggested a neuroprotective role, the exact way in which autophagy is connected to these neuropathological conditions is unclear. Consistent with a beneficial, cytoprotective function of autophagy, studies have shown that inhibition of autophagy results in an accumulation of α-synuclein in Parkinson’s disease20– 22 and increased formation of huntingtin aggregates in Huntington’s disease,23 whereas increases in autophagic activity can aid in the clearance of these toxic aggregate species.24 The involvement of autophagy in the clearance of protein aggregates associated with neurodegenerative disease is of great interest and has stimulated studies that seek the potential therapeutic value of autophagy in the treatment of neurodegenerative diseases associated with protein aggregates. However, some reports, questioning the universality of neuroprotective autophagy, have suggested that inappropriate autophagic induction contributes to cytotoxicity in AD.25
Of particular interest in the link of autophagy to disease pathogenesis is a series of studies that suggest dysfunctional autophagy in AD.25– 28 We review the current evidence that links autophagy to AD. In addition, we also discuss the uncertainty over the precise role that autophagy may play in the context of AD pathogenesis.
BEGINNINGS OF AN ALZHEIMER’S DISEASE–AUTOPHAGY CONNECTION: ENDOSOMAL-LYSOSOMAL PATHWAY
AD is the most common neurodegenerative disorder;29 its symptomatic hallmarks include slow progression of memory impairment and loss of cognitive function. The AD-affected brain is characterized by 2 forms of aggregates: the intracellular tau tangle and the extracellular amyloid-β (Aβ) peptide neuritic plaque. The former is composed of tau, a microtubule-associated protein that is aberrantly phosphorylated in AD and aggregates into paired helical filaments,30 whereas the latter is derived from proteolysis of amyloid precursor protein (APP).31 Mutations in APP32 and presenilin,33,34 a protein involved in APP-to-Aβ proteolysis, cause familial Alzheimer’s disease (FAD), which is rare and autosomal-dominant. Sporadic AD, which is more prevalent, shows many of the pathological characteristics of FAD, and this suggests that factors affecting the APP-to-Aβ pathway and subsequent Aβ-amyloidogenesis play a significant role in sporadic forms of the disease.35
The investigation of the role of autophagy in AD began because of the initial findings of the involvement of the endosomal-lysosomal system in the pathogenesis of AD and specifically Aβ-amyloidogenesis. Several studies have recognized the endosomal-lysosomal pathway as an important regulator of the processing of APP.36– 38 The endocytic pathway enables a cell to sample its surrounding environment and plays a critical role in the uptake of extracellular nutrients and regulation of cell-surface receptor expression. Sorting of internalized molecules occurs in the early endosome, which directs the material back to the plasma membrane for recycling, to the trans-Golgi network for further processing, or to late endosomes and lysosomes for degradation.39 APP potentially undergoes processing at each of these locations. Early endosomes produce Aβ from APP in normal cells and mediate the uptake of Aβ and soluble APP. In the AD brain, activation of the endocytic pathway is the earliest noted intracellular manifestation of the disease, and neurons in susceptible brain areas exhibit progressive abnormalities in the endocytic pathway that include increases in the size and volume of early endosomes.40 Aβ has also been detected in these enlarged endosomes that are immunopositive for the early endosomal marker rab5.41
Abnormalities in the lysosomal pathway also occur early in AD pathogenesis before the appearance of neurofibrillary tangles or senile plaques. An up-regulation in the lysosmal system occurs in vulnerable cell populations and results in increased numbers of lysosomes with elevated expression of lysosomal hydrolases.42 These hydrolases include cathepsins, which have been shown to be both directly43 and indirectly39,44 involved in Aβ formation. As AD pathogenesis progresses, lysosomal dysfunction appears to occur with the buildup of vacuolar structures and the accumulation of Aβ1– 42. The degeneration of the compromised neurons leads to the release of these structures into the extracellular space, where they associate with deposits of Aβ.45
Although endocytic pathway induction and dysfunction can lead to lysosomal abnormalities, the autophagic pathway also converges on the lysosome (Figure 1). Additionally, the endocytic pathway in neurons exhibits a significant degree of merger with the autophagic pathway with the fusion of late endosomes with autophagosomes.46 However, only in the last 10 years with advances in knowledge of the autophagic process has AD research begun to focus on the pathological effects of possible irregularities in the autophagic pathway.39,47
AMYLOID-β AMYLOIDOGENESIS IN AUTOPHAGOSOMES
A long-recognized characteristic feature of the AD brain is the presence of granulovacuolar structures in dystrophic (swollen) neurites.48 As previously mentioned, the presence of these structures has been associated with an up-regulation/dysfunction of the endosomal-lysosomal pathway, as these structures tend to be lysosome-dense bodies that are acid phosphatase–positive and abundant in cathepsins, lysosomal proteases. More recent studies have identified many of these accumulated bodies as autophagosomes and autophagolysosomes, or early and late autophagic vacuoles (AVs), both intermediate structures of autophagy.28 What could be of particular importance is the claim that these numerous AVs contain the components for Aβ generation, APP, and its processing enzymes.25,49 APP has 3 sites of proteolytic cleavage (α, β, and γ), and Aβ is generated from a β-cleaved C-terminal APP fragment that subsequently undergoes γ-cleavage to create 40- to 42-mer Aβ peptides. The γ-secretase protein complex that includes presenilin mediates this γ-cleavage and has been shown to be highly active in accumulated AVs.
ABERRANT INDUCTION AND A DYSFUNCTIONAL AUTOPHAGIC PATHWAY
Presenilin 1 Studies
One theory linking autophagy and AD is that irregular autophagy stimulation results in increased Aβ production through the accumulation of AVs containing the γ-secretase machinery.25 As a component of the γ-secretase complex, the transmembrane protein presenilin 1 (PS1) plays a significant role in the production of Aβ.50 Specifically, PS1 has been found to regulate the processing of APP C-terminal fragments in the endoplasmic reticulum and Golgi.51 In FAD, the most common mutations are found in PS1 and PS2. These mutations cause increased production of Aβ peptide leading to the accumulation of this protein and the formation of Aβ plaques.52,53
There is evidence suggesting a possible link between PS1 and AD through autophagy. The recent studies of PS1 mutated cell lines and mouse models have elucidated this potential link. In the PS1/APP mouse model of β-amyloidogenesis, there was an accumulation of AVs containing APP, PS1, and other proteins.25 Similarly, in PS1-null neurons, there was a buildup of degradative vacuoles accompanied by both an abnormal presence of α-synuclein and β-synuclein proteins in these vacuoles and an increased level of both proteins. Interestingly, in control neurons, stimulation of autophagy caused a similar mislocalization of these synucleins. The effects seen in the PS1-deficient neurons were proposed to be due not to abnormal γ-secretase activity but possibly to abnormal regulation of calcium channels.54
Telencephalin (TLN), a neural intercellular adhesion molecule with which PS1 interacts,55 has also been shown to accumulate in autophagic-like vacuoles in PS1-null hippocampal neurons. However, the prolonged half-life and enrichment of TLN were not a result of abnormal γ-secretase cleavage because TLN is not a substrate of γ-secretase. Rather, the authors hypothesized that the deficiency in PS1 caused a failure of autophagy to clear TLN by possibly disallowing proper vacuole and lysosome fusion.56 All of the aforementioned evidence supports the proposition that impaired presenilin function translates into possible autophagic dysfunction either causing or exacerbating protein accumulation and, therefore, perhaps playing a significant role in the pathogenesis of neurodegenerative diseases such as AD.
Oxidative Stress
Oxidative damage to neurons has been reported to be an early occurrence in the pathogenesis of AD.57 Zheng et al.58,59 showed that the exposure of differentiated neuroblastoma cells to oxidative stress resulted in the accumulation of Aβ-enriched lysosomes. This Aβ accumulation was suppressed by 3-methyladenine (3-MA), an inhibitor of autophagosome formation, and this suggested the involvement of autophagy. The authors of the aforementioned study postulated that this increase in intralysosomal Aβ was due to oxidative stress inducing reparative autophagy, which resulted in the delivery of additional Aβ to autophagolysosomes. Other suggested mechanisms have included oxidative stress–induced enhancement of APP processing and decreased Aβ degradation due to either a decrease in lysosomal enzyme activity or a structural modification of Aβ that would make it more resistant to hydrolysis. Oxidative stress and damage in AD have also been proposed to lead to autophagy of mitochondria with an accumulation of mitochondrial degradation products also in the lysosomal pathway.60– 62
Okadaic Acid Model
Okadaic acid (OA), a protein phosphatase-2A inhibitor, has been shown to be a useful tool for mimicking several pathological hallmarks of AD, including enhanced tau phosphorylation, Aβ deposition, and neuronal death. Use of OA is not sufficient for characterizing the mechanisms of the development of AD pathology, but it has been used to examine the role of tau phosphorylation in AD neuronal death.63 A recent study demonstrated that treatment of rat cortical neurons with OA also resulted in the accumulation of AVs and elevated levels of the autophagosome marker microtubule-associated protein 1 light chain 3 II (LC3-II) as well as increases in the lysosomal system. OA both activated mammalian target of rapamycin, an inhibitor of autophagy, and up-regulated expression of the autophagy protein beclin 1, and this led the authors to conclude that the observed autophagic induction was due to the beclin 1–dependent autophagic pathway in their system. Blocking autophagy with 3-MA ameliorated OA-induced neurotoxicity.
BLOCKING AUTOPHAGY DEGRADATION
Impaired Clearance
Although several studies have implicated an induction of autophagy with the accumulation of AVs in AD models, all of these studies have also conceded that dysfunction in the autophagic pathway most likely plays a role. More specifically, the enrichment of AVs in AD suggests impairment of autophagic clearance.27
The role and level of autophagy in the normal brain are often disputed. Because of the relative dearth of AV intermediates in comparison to AD brain tissue, many contend that basal autophagic activity is low in the healthy brain. A recent study by Boland et al.,26 however, has argued that autophagy in healthy neurons is extremely efficient, with autophagosomes being rapidly cleared by fusion with lysosomes. Whether basal activity is low or “efficiently” high, 2 separate studies have shown that autophagy is critical for neuronal survival. The deletion of 2 specific autophagy genes (autophagy-related 5 and autophagy-related 7) resulted in the blockage of constitutive autophagy and thereby promoted the formation of ubiquitinated protein aggregates, neurodegeneration, behavioral abnormalities, and early mortality.13,15 The importance of a functional autophagy pathway was also affirmed by the aforementioned study by Boland and colleagues. In the study, rat cortical neurons were treated with rapamycin, lysosomal inhibitors, or vinblastine, which both induces autophagy and blocks AV clearance by lysosomes. Neurons treated with lysosomal inhibitors or vinblastine exhibited an accumulation of AVs with morphologies similar to those in the AD brain and the PS1/APP mouse model. The authors, therefore, concluded that autophagy dysfunction due to impaired AV clearance, and not autophagy induction alone, resulted in pathological patterns similar to AD.
Tau and Autophagic Dysfunction
Like Aβ, altered processing and impaired degradation by the lysosomal system have also been implicated in the buildup of tau neurofibrillary tangles in AD. At least 2 different tau transgenic mouse lines have exhibited autophagic-lysosomal abnormalities upon ultrastructural analysis.64,65 The first study focused on transgenic mice expressing human tau with 4 tubulin-binding repeats [increased by frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17) splice donor mutations] and 3 FTDP-17 missense mutations: G272V, P301L, and R406W. An examination of these mice revealed an increase in lysosomal neuronal complexes and an up-regulation of acid phosphatases in regions that had accumulated filamentous tau aggregates.65 In a second study, transgenic mice expressing mutant human (P301L) tau demonstrated axonal spheroids that were enriched with tau filaments as well as autophagosomes, with some spheroids appearing similar to those in the characteristic dystrophic neurites of AD.64
Overexpression of tau in other model systems has also demonstrated a possible link to autophagy dysfunction. In a study using an inducible tau expression system in cultured cells, the acidotropic agent chloroquine was used to disrupt the lysosomal system after tau induction, and this resulted in a delayed clearance of tau. Chloroquine treatment before and during tau induction led to an accumulation of tau with the formation of aggregates. This aggregation was not due to changes in tau phosphorylation, but an altered functioning of specific lysosomal proteases was found to be likely. Tau accumulation and aggregation were also induced when autophagy was inhibited in the cells with 3-MA treatment.66 A second more recent study used an inducible mouse neuroblastoma N2a cell model featuring the repeat domain of tau with the FTDP-17 mutation and found that autophagy could degrade soluble mutant tau and its associated aggregates. Additionally, autophagy inhibition led to an increase in mutant tau aggregation and toxicity.67 Similarly, induction of autophagy by rapamycin in Drosophila overexpressing wild-type and mutant tau resulted in increased tau clearance.68 Like other aggregate-prone proteins, such as α-synuclein and huntingtin mutants, tau (or phosphorylated tau) can also be a substrate of autophagy; however, the mechanism underlying the degradation of specific disease-related proteins (likely in soluble form prior to the aggregates) is unclear.
Although never investigated, possible interference of dysfunctional tau with autophagosome clearance may contribute to AD neuropathology. Tau has been shown to regulate the stability of microtubules; therefore, modification of tau by phosphorylation or a change in the amount of tau can result in destabilization of neuronal microtubules. This can affect the placement and function of mitochondria, lysosomes, and other cellular structures and potentially put the cell in great danger.69 Specifically, autophagosome clearance by lysosomes is a microtubule-dependent process.70– 72 Therefore, abnormal function of tau may disrupt neuronal function including axonal transport, which also involves trafficking of autophagic cargos,73 and lead to the buildup of autophagosomes in AD dystrophic neurites or axons.28
ROLE FOR REDUCTION OF AUTOPHAGY
Beclin 1–Deficient Mouse Model
In what appears to be in opposition to some of the previously mentioned studies, recently there has also been evidence suggesting that decreased autophagic action may play a role in the pathogenic mechanisms of AD. Pickford et al.74 reported that beclin 1 levels are decreased in the affected gray matter of patients with early stages of AD. Beclin 1 is part of a lipid kinase complex that is essential for autophagy and has been to shown to be involved in tumor suppression, neuroprotection, and cell death.75– 77 In contrast to the reduced cortical beclin 1 levels in AD patients, AD pathology in APP transgenic mice was not sufficient to cause a deficiency in beclin 1. However, APP transgenic mice heterozygous for beclin 1 demonstrated an increased accumulation of Aβ peptide, synaptic degeneration, and subsequent neuronal death. A reduction of autophagy in beclin 1–deficient mice was indicated by reduced levels of LC3-II in comparison with those of the control mice. Autophagic reduction in these mice was accompanied by an accumulation of disrupted lysosomal structures. A change in the microglial response to Aβ peptide suggested an increase in brain injury. Overexpression of beclin 1, perhaps by the stimulation of autophagy, resulted in a reduction of both extracellular and intracellular Aβ peptide.74 In this set of experiments, reduction of autophagy appeared to potentiate AD pathology, and this suggests that autophagy may also be neuroprotective in AD. This would indicate autophagy stimulation as a potential therapeutic route for AD.
Herpes Simplex Virus Type 1
Herpes simplex virus type 1 (HSV1) was initially proposed as an AD causal agent because of the parallels between the brain regions affected by the virus in herpes simplex encephalitis and those affected in AD.78 Nine years later, Jamieson et al.79 reported that latent HSV1 DNA is present in a large proportion of AD patients and in normal elderly populations. Subsequent studies have further strengthened the idea of a possible connection by linking apolipoprotein E with the severity of HSV1 infection. Wozniak’s group80 has also claimed to have demonstrated that HSV1 infection can result in Aβ deposition and phosphorylation of tau.
HSV1 can disrupt autophagy by counteracting the protein kinase R (PKR) defense mechanism, which is activated by the presence of double-stranded RNA. Activated PKR would normally enhance autophagy by phosphorylating elongation initiation factor 2α (eIF2α), and this would result in the termination of protein synthesis and the initiation of apoptosis. HSV1 interferes with the rescue action of PKR by dephosphorylating eIF2α.81 Specifically, a protein encoded by HSV1, infected cell polypeptide 34.5, has been reported to bind beclin 1 and inhibit its autophagy function.82 Autophagic interference by HSV1 has been proposed to prevent the degradation of Aβ and neurofibrillary tangles that accumulate in AD,81 and this is consistent with the hypothesis that impaired autophagy in neurons in part contributes to the neuropathology of AD.
LATE BREAKING STUDIES AND FUTURE PERSPECTIVES
Much recent work in the field of Alzheimer’s disease has provided molecular and cellular evidence that links autophagy to the pathogenesis of AD.26,63,66,74,81 These studies undoubtedly strengthen the mechanistic view that centralizes on an axis of endocytosis, autophagy, and lysosomal degradation in the cellular pathway of AD, especially given the emerging evidence showing the convergence of endosomes and autophagosomes in the lysosomal degradation route. Although these studies implicate aberrant autophagic activity in the process of Aβ synthesis or deposits, they seem to suggest 2 competing hypotheses with respect to the levels of autophagosomes or autophagic activity in the pathological process of AD27,74 (Figure 2).
Fig 2.
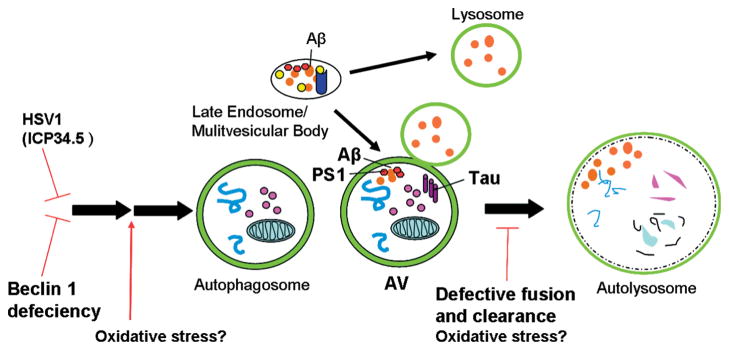
Possible alterations of the autophagic pathway lead to the accumulation of AVs in AD. Changes in the autophagic pathway (especially the accumulation of AVs in dystrophic neurites) have been linked to AD. The specific involvement of autophagy is unclear, but 2 competing hypotheses have emerged. One hypothesis links autophagic inhibition to AD pathogenesis. Autophagy could be inhibited in the AD brain by a reduction in beclin 1 or by the HSV1 protein ICP34.5, which binds and inhibits beclin 1. A second hypothesis focuses on aberrant autophagy induction and/or defective lysosomal fusion and clearance. Oxidative stress has been proposed to cause increases in Aβ by up-regulating autophagy or by affecting its capacity for degradation and clearance. The mechanism for defective lysosomal fusion and subsequent clearance is not known, but the accumulation of intracellular Aβ or tau may play a role. Abbreviations: Aβ, amyloid-β; AD, Alzheimer’s disease; AV, autophagic vacuole; HSV1, herpes simplex virus type 1; ICP34.5, infected cell polypeptide 34.5; PS1, presenilin 1.
Two studies that were published during the preparation of this review emphasize the polarized views of autophagy’s role in Aβ-induced neurotoxicity.83,84 Using Drosophila as a model system, Ling et al.84 reported that flies expressing Aβ1– 42 demonstrated an age-dependent increase in autophagic dysfunction. Moreover, down-regulating autophagy in flies in which specific autophagy genes were either reduced in copy or silenced in selective neurons resulted in a protective effect on the Aβ1– 42 toxicity. In the opposite manner, Aβ1– 42 flies treated with rapamycin to induce autophagy experienced increased toxicity and a shortened lifespan. A second recent article, however, demonstrated protective effects of autophagy on Aβ1– 42 toxicity in a neuroblastoma cell model. Cells were treated with extracellular Aβ, which induced an autophagic process and resulted in the accumulation of reactive oxygen species. Cells given a cotreatment of rapamycin exhibited a decrease in reactive oxygen species accumulation and increased survival, whereas those treated with the autophagic inhibitor wortmannin did not display the same protective effects.83
Many factors, such as different animal models, cellular models, and experimental paradigms, can contribute to the differences in conclusions. Also, the different model systems are likely representative of different particular stages of the AD pathogenic process. Therefore, properly functioning autophagy as a protective mechanism in one study may not necessarily contradict the finding that enhanced dysfunctional autophagy is neurotoxic in a different study. However, the major impediment to the revelation of the exact mechanism is undoubtedly the lack of knowledge of the neuronal autophagic process. Future work will be needed not only to further investigate the details of altered autophagy in AD neurons but also to characterize basal autophagy function under normal conditions.
Footnotes
DISCLOSURES
Potential conflict of interest: Nothing to report.
References
- 1.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunn WA., Jr Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol. 1994;4:139–143. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 3.Mortimore GE, Poso AR, Lardeux BR. Mechanism and regulation of protein degradation in liver. Diabetes Metab Rev. 1989;5:49–70. doi: 10.1002/dmr.5610050105. [DOI] [PubMed] [Google Scholar]
- 4.de Duve D. The peroxisome: a new cytoplasmic organelle. Proc R Soc Lond B Biol Sci. 1969;173:71–83. [PubMed] [Google Scholar]
- 5.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaushik S, Cuervo AM. Chaperone-mediated autophagy. Methods Mol Biol. 2008;445:227–244. doi: 10.1007/978-1-59745-157-4_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizushima N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 2005;12(suppl 2):1535–1541. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- 8.Klionsky DJ, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 9.Dunn WA., Jr Studies on the mechanisms of autophagy: maturation of the autophagic vacuole. J Cell Biol. 1990;110:1935–1945. doi: 10.1083/jcb.110.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn WA., Jr Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol. 1990;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 12.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 13.Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 14.Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 16.Martinez-Vicente M, Cuervo AM. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol. 2007;6:352–361. doi: 10.1016/S1474-4422(07)70076-5. [DOI] [PubMed] [Google Scholar]
- 17.Rusten TE, Simonsen A. ESCRT functions in autophagy and associated disease. Cell Cycle. 2008;7:1166–1172. doi: 10.4161/cc.7.9.5784. [DOI] [PubMed] [Google Scholar]
- 18.Williams A, Jahreiss L, Sarkar S, et al. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr Top Dev Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]
- 19.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 20.Vogiatzi T, Xilouri M, Vekrellis K, Stefanis L. Wild type alpha-synuclein is degraded by chaperone-mediated autophagy and macroautophagy in neuronal cells. J Biol Chem. 2008;283:23542–23556. doi: 10.1074/jbc.M801992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Webb JL, Ravikumar B, Atkins J, et al. Alpha-synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 22.Paxinou E, Chen Q, Weisse M, et al. Induction of alpha-synuclein aggregation by intracellular nitrative insult. J Neurosci. 2001;21:8053–8061. doi: 10.1523/JNEUROSCI.21-20-08053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006;172:719–731. doi: 10.1083/jcb.200510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu WH, Cuervo AM, Kumar A, et al. Macroautophagy–a novel beta-amyloid peptide-generating pathway activated in Alzheimer’s disease. J Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boland B, Kumar A, Lee S, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nixon RA. Autophagy, amyloidogenesis and Alzheimer disease. J Cell Sci. 2007;120(pt 23):4081–4091. doi: 10.1242/jcs.019265. [DOI] [PubMed] [Google Scholar]
- 28.Nixon RA, Wegiel J, Kumar A, et al. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 29.Ferri CP, Prince M, Brayne C, et al. Global prevalence of dementia: a Delphi consensus study. Lancet. 2005;366:2112–2117. doi: 10.1016/S0140-6736(05)67889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goedert M. Tau protein and the neurofibrillary pathology of Alzheimer’s disease. Trends Neurosci. 1993;16:460–465. doi: 10.1016/0166-2236(93)90078-z. [DOI] [PubMed] [Google Scholar]
- 31.Glenner GG, Wong CW. Alzheimer’s disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 32.Goate A, Chartier-Harlin MC, Mullan M, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 33.Levy-Lahad E, Wasco W, Poorkaj P, et al. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 34.Sherrington R, Rogaev EI, Liang Y, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 35.Hardy J. Amyloid, the presenilins and Alzheimer’s disease. Trends Neurosci. 1997;20:154–159. doi: 10.1016/s0166-2236(96)01030-2. [DOI] [PubMed] [Google Scholar]
- 36.Grbovic OM, Mathews PM, Jiang Y, et al. Rab5-stimulated up-regulation of the endocytic pathway increases intracellular beta-cleaved amyloid precursor protein carboxyl-terminal fragment levels and Abeta production. J Biol Chem. 2003;278:31261–31268. doi: 10.1074/jbc.M304122200. [DOI] [PubMed] [Google Scholar]
- 37.Koo EH, Squazzo SL. Evidence that production and release of amyloid beta-protein involves the endocytic pathway. J Biol Chem. 1994;269:17386–17389. [PubMed] [Google Scholar]
- 38.Pasternak SH, Bagshaw RD, Guiral M, et al. Presenilin-1, nicastrin, amyloid precursor protein, and gamma-secretase activity are co-localized in the lysosomal membrane. J Biol Chem. 2003;278:26687–26694. doi: 10.1074/jbc.m304009200. [DOI] [PubMed] [Google Scholar]
- 39.Nixon RA, Cataldo AM, Mathews PM. The endosomal-lysosomal system of neurons in Alzheimer’s disease pathogenesis: a review. Neurochem Res. 2000;25:1161–1172. doi: 10.1023/a:1007675508413. [DOI] [PubMed] [Google Scholar]
- 40.Cataldo AM, Barnett JL, Pieroni C, Nixon RA. Increased neuronal endocytosis and protease delivery to early endosomes in sporadic Alzheimer’s disease: neuropathologic evidence for a mechanism of increased beta-amyloidogenesis. J Neurosci. 1997;17:6142–6151. doi: 10.1523/JNEUROSCI.17-16-06142.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cataldo AM, Petanceska S, Terio NB, et al. Abeta localization in abnormal endosomes: association with earliest Abeta elevations in AD and Down syndrome. Neurobiol Aging. 2004;25:1263–1272. doi: 10.1016/j.neurobiolaging.2004.02.027. [DOI] [PubMed] [Google Scholar]
- 42.Cataldo AM, Barnett JL, Berman SA, et al. Gene expression and cellular content of cathepsin D in Alzheimer’s disease brain: evidence for early up-regulation of the endosomal-lysosomal system. Neuron. 1995;14:671–680. doi: 10.1016/0896-6273(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 43.Chevallier N, Vizzavona J, Marambaud P, et al. Cathepsin D displays in vitro beta-secretase-like specificity. Brain Res. 1997;750:11–19. doi: 10.1016/s0006-8993(96)01330-3. [DOI] [PubMed] [Google Scholar]
- 44.Beyreuther K, Pollwein P, Multhaup G, et al. Regulation and expression of the Alzheimer’s beta/A4 amyloid protein precursor in health, disease, and Down’s syndrome. Ann N Y Acad Sci. 1993;695:91–102. doi: 10.1111/j.1749-6632.1993.tb23035.x. [DOI] [PubMed] [Google Scholar]
- 45.Cataldo AM, Hamilton DJ, Nixon RA. Lysosomal abnormalities in degenerating neurons link neuronal compromise to senile plaque development in Alzheimer disease. Brain Res. 1994;640:68–80. doi: 10.1016/0006-8993(94)91858-9. [DOI] [PubMed] [Google Scholar]
- 46.Larsen KE, Sulzer D. Autophagy in neurons: a review. Histol Histopathol. 2002;17:897–908. doi: 10.14670/HH-17.897. [DOI] [PubMed] [Google Scholar]
- 47.Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct. 2002;27:421–429. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki K, Terry RD. Fine structural localization of acid phosphatase in senile plaques in Alzheimer’s presenile dementia. Acta Neuropathol. 1967;8:276–284. doi: 10.1007/BF00688828. [DOI] [PubMed] [Google Scholar]
- 49.Yu WH, Kumar A, Peterhoff C, et al. Autophagic vacuoles are enriched in amyloid precursor protein-secretase activities: implications for beta-amyloid peptide over-production and localization in Alzheimer’s disease. Int J Biochem Cell Biol. 2004;36:2531–2540. doi: 10.1016/j.biocel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 50.Xia W, Ostaszewski BL, Kimberly WT, et al. FAD mutations in presenilin-1 or amyloid precursor protein decrease the efficacy of a gamma-secretase inhibitor: evidence for direct involvement of PS1 in the gamma-secretase cleavage complex. Neurobiol Dis. 2000;7(pt B):673–681. doi: 10.1006/nbdi.2000.0322. [DOI] [PubMed] [Google Scholar]
- 51.Xia W, Zhang J, Ostaszewski BL, et al. Presenilin 1 regulates the processing of beta-amyloid precursor protein C-terminal fragments and the generation of amyloid beta-protein in endoplasmic reticulum and Golgi. Biochemistry. 1998;37:16465–16471. doi: 10.1021/bi9816195. [DOI] [PubMed] [Google Scholar]
- 52.Xia W. Role of presenilin in gamma-secretase cleavage of amyloid precursor protein. Exp Gerontol. 2000;35:453–460. doi: 10.1016/s0531-5565(00)00111-x. [DOI] [PubMed] [Google Scholar]
- 53.Gandy S. The role of cerebral amyloid beta accumulation in common forms of Alzheimer disease. J Clin Invest. 2005;115:1121–1129. doi: 10.1172/JCI25100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson CA, Murphy DD, Giasson BI, et al. Degradative organelles containing mislocalized alpha-and beta-synuclein proliferate in presenilin-1 null neurons. J Cell Biol. 2004;165:335–346. doi: 10.1083/jcb.200403061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gahmberg CG. Leukocyte adhesion: CD11/CD18 integrins and intercellular adhesion molecules. Curr Opin Cell Biol. 1997;9:643–650. doi: 10.1016/s0955-0674(97)80117-2. [DOI] [PubMed] [Google Scholar]
- 56.Esselens C, Oorschot V, Baert V, et al. Presenilin 1 mediates the turnover of telencephalin in hippocampal neurons via an autophagic degradative pathway. J Cell Biol. 2004;166:1041–1054. doi: 10.1083/jcb.200406060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu X, Raina AK, Lee HG, et al. Oxidative stress signalling in Alzheimer’s disease. Brain Res. 2004;1000:32–39. doi: 10.1016/j.brainres.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 58.Zheng L, Roberg K, Jerhammar F, et al. Oxidative stress induces intralysosomal accumulation of Alzheimer amyloid beta-protein in cultured neuroblastoma cells. Ann N Y Acad Sci. 2006;1067:248–251. doi: 10.1196/annals.1354.032. [DOI] [PubMed] [Google Scholar]
- 59.Zheng L, Marcusson J, Terman A. Oxidative stress and Alzheimer disease: the autophagy connection? Autophagy. 2006;2:143–145. doi: 10.4161/auto.2.2.2444. [DOI] [PubMed] [Google Scholar]
- 60.Arendt T, Holzer M, Fruth R, et al. Phosphorylation of tau, Abeta-formation, and apoptosis after in vivo inhibition of PP-1 and PP-2A. Neurobiol Aging. 1998;19:3–13. doi: 10.1016/s0197-4580(98)00003-7. [DOI] [PubMed] [Google Scholar]
- 61.Tanaka T, Zhong J, Iqbal K, et al. The regulation of phosphorylation of tau in SY5Y neuroblastoma cells: the role of protein phosphatases. FEBS Lett. 1998;426:248–254. doi: 10.1016/s0014-5793(98)00346-9. [DOI] [PubMed] [Google Scholar]
- 62.Yoon S, Choi J, Yoon J, et al. Okadaic acid induces JNK activation, bim overexpression and mitochondrial dysfunction in cultured rat cortical neurons. Neurosci Lett. 2006;394:190–195. doi: 10.1016/j.neulet.2005.10.034. [DOI] [PubMed] [Google Scholar]
- 63.Yoon SY, Choi JE, Kweon HS, et al. Okadaic acid increases autophagosomes in rat neurons: implications for Alzheimer’s disease. J Neurosci Res. 2008;86:3230–3239. doi: 10.1002/jnr.21760. [DOI] [PubMed] [Google Scholar]
- 64.Lin WL, Lewis J, Yen SH, et al. Ultrastructural neuronal pathology in transgenic mice expressing mutant (P301L) human tau. J Neurocytol. 2003;32:1091–1105. doi: 10.1023/B:NEUR.0000021904.61387.95. [DOI] [PubMed] [Google Scholar]
- 65.Lim F, Hernandez F, Lucas JJ, et al. FTDP-17 mutations in tau transgenic mice provoke lysosomal abnormalities and tau filaments in forebrain. Mol Cell Neurosci. 2001;18:702–714. doi: 10.1006/mcne.2001.1051. [DOI] [PubMed] [Google Scholar]
- 66.Hamano T, Gendron TF, Causevic E, et al. Autophagic-lysosomal perturbation enhances tau aggregation in transfectants with induced wild-type tau expression. Eur J Neurosci. 2008;27:1119–1130. doi: 10.1111/j.1460-9568.2008.06084.x. [DOI] [PubMed] [Google Scholar]
- 67.Wang Y, Martinez-Vicente M, Kruger U, et al. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum Mol Genet. 2009;18:4153–4170. doi: 10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berger Z, Ravikumar B, Menzies FM, et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 69.Avila J, Lucas JJ, Perez M, Hernandez F. Role of tau protein in both physiological and pathological conditions. Physiol Rev. 2004;84:361–384. doi: 10.1152/physrev.00024.2003. [DOI] [PubMed] [Google Scholar]
- 70.Matteoni R, Kreis TE. Translocation and clustering of endosomes and lysosomes depends on microtubules. J Cell Biol. 1987;105:1253–1265. doi: 10.1083/jcb.105.3.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jahreiss L, Menzies FM, Rubinsztein DC. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic. 2008;9:574–587. doi: 10.1111/j.1600-0854.2008.00701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kovacs AL, Seglen PO. Inhibition of hepatocytic protein degradation by inducers of autophagosome accumulation. Acta Biol Med Ger. 1982;41:125–130. [PubMed] [Google Scholar]
- 73.Yue Z. Regulation of neuronal autophagy in axon: implication of autophagy in axonal function and dysfunction/degeneration. Autophagy. 2007;3:139–141. doi: 10.4161/auto.3602. [DOI] [PubMed] [Google Scholar]
- 74.Pickford F, Masliah E, Britschgi M, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yue Z, Jin S, Yang C, et al. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cao Y, Klionsky DJ. Physiological functions of Atg6/beclin 1: a unique autophagy-related protein. Cell Res. 2007;17:839–849. doi: 10.1038/cr.2007.78. [DOI] [PubMed] [Google Scholar]
- 77.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 78.Ball MJ. Limbic predilection in Alzheimer dementia: is reactivated herpesvirus involved? Can J Neurol Sci. 1982;9:303–306. doi: 10.1017/s0317167100044115. [DOI] [PubMed] [Google Scholar]
- 79.Jamieson GA, Maitland NJ, Wilcock GK, et al. Latent herpes simplex virus type 1 in normal and Alzheimer’s disease brains. J Med Virol. 1991;33:224–227. doi: 10.1002/jmv.1890330403. [DOI] [PubMed] [Google Scholar]
- 80.Wozniak MA, Itzhaki RF, Shipley SJ, Dobson CB. Herpes simplex virus infection causes cellular beta-amyloid accumulation and secretase upregulation. Neurosci Lett. 2007;429:95–100. doi: 10.1016/j.neulet.2007.09.077. [DOI] [PubMed] [Google Scholar]
- 81.Itzhaki RF, Cosby SL, Wozniak MA. Herpes simplex virus type 1 and Alzheimer’s disease: the autophagy connection. J Neurovirol. 2008;14:1–4. doi: 10.1080/13550280701802543. [DOI] [PubMed] [Google Scholar]
- 82.Orvedahl A, Alexander D, Talloczy Z, et al. HSV-1 ICP34. 5 confers neurovirulence by targeting the beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 83.Hung SY, Huang WP, Liou HC, Fu WM. Autophagy protects neuron from Abeta-induced cytotoxicity. Autophagy. 2009;5:502–510. doi: 10.4161/auto.5.4.8096. [DOI] [PubMed] [Google Scholar]
- 84.Ling D, Song HJ, Garza D, et al. Abeta42-induced neurodegeneration via an age-dependent autophagic-lysosomal injury in Drosophila. PLoS One. 2009;4:e4201. doi: 10.1371/journal.pone.0004201. [DOI] [PMC free article] [PubMed] [Google Scholar]