Abstract
Background
The Gram-negative bacterium Yersinia pestis is the causative agent of the bubonic plague. Efficient iron acquisition systems are critical to the ability of Y. pestis to infect, spread and grow in mammalian hosts, because iron is sequestered and is considered part of the innate host immune defence against invading pathogens. We used a proteomic approach to determine expression changes of iron uptake systems and intracellular consequences of iron deficiency in the Y. pestis strain KIM6+ at two physiologically relevant temperatures (26°C and 37°C).
Results
Differential protein display was performed for three Y. pestis subcellular fractions. Five characterized Y. pestis iron/siderophore acquisition systems (Ybt, Yfe, Yfu, Yiu and Hmu) and a putative iron/chelate outer membrane receptor (Y0850) were increased in abundance in iron-starved cells. The iron-sulfur (Fe-S) cluster assembly system Suf, adapted to oxidative stress and iron starvation in E. coli, was also more abundant, suggesting functional activity of Suf in Y. pestis under iron-limiting conditions. Metabolic and reactive oxygen-deactivating enzymes dependent on Fe-S clusters or other iron cofactors were decreased in abundance in iron-depleted cells. This data was consistent with lower activities of aconitase and catalase in iron-starved vs. iron-rich cells. In contrast, pyruvate oxidase B which metabolizes pyruvate via electron transfer to ubiquinone-8 for direct utilization in the respiratory chain was strongly increased in abundance and activity in iron-depleted cells.
Conclusions
Many protein abundance differences were indicative of the important regulatory role of the ferric uptake regulator Fur. Iron deficiency seems to result in a coordinated shift from iron-utilizing to iron-independent biochemical pathways in the cytoplasm of Y. pestis. With growth temperature as an additional variable in proteomic comparisons of the Y. pestis fractions (26°C and 37°C), there was little evidence for temperature-specific adaptation processes to iron starvation.
Background
Yersinia pestis, a Gram-negative bacterium, is the causative agent of the bubonic and pneumonic plague. The pathogenic lifestyle of this microbe involves two distinct life stages, one in the flea vector, the other in mammalian hosts, primarily rodents [1]. Genome sequencing and analyses have been completed for four major Y. pestis biovars, including the chromosome [2] and three virulence/transmission-associated plasmids [3,4] of the KIM strain, which belongs to the biovar mediaevalis. In addition to plasmid-encoded virulence factors, the genetically unstable chromosomal 102-kb pgm locus is also important for full virulence of Y. pestis in mammals and for its transmission via blocked fleas [5,6]. This locus encodes the yersiniabactin-dependent iron transport (Ybt) system and the hemin storage (Hms)-dependent biofilm system. Biofilm formation allows colonization of the flea proventriculus causing blockage which in turn induces active feeding behavior [7,8].
Efficient iron acquisition systems are critical to the ability of Yersinia pestis to infect, spread and grow in mammalian hosts, because iron is sequestered and is considered part of the innate host immune defence against invading pathogens [9]. The Ybt system includes a series of enzymes responsible for the siderophore's biosynthesis. Following secretion and iron chelation, the iron/yersiniabactin complex is bound by the outer membrane (OM) receptor Psn and transferred into the periplasm via TonB-dependent energy transmission. Binding of the complex to the periplasmic surface of the inner membrane-localized ATP-binding cassette (ABC) transporter YbtP/YbtQ, which contains two permease and two ATP-binding domains, initiates iron import into the cytoplasm. A functional Ybt transporter is required for bacterial infection by subcutaneous routes and important for iron acquisition in early stages of the bubonic plague in mice [10-12]. The manganese- and iron-specific ABC transporter Yfe is also important for full Y. pestis virulence according to data from a bubonic plague mouse model [13]. Other ABC transporters for iron (Yfu and Yiu) and hemin (Hmu) were functionally characterized, but were not found to be required for virulence in the mouse model [14-16]. The transporters Yfe and Feo serve somewhat redundant roles in ferrous iron uptake under microaerophilic growth conditions [17]. Genomic analysis suggests the existence of other transporters and OM receptors for iron/siderophores but have not been functionally characterized to date [2,18].
The ferric uptake regulator Fur is a dominant transcription factor controlling iron assimilation in many bacterial species [19]. Iron transporters, iron storage proteins and some proteins requiring iron cofactors for function feature conserved binding sites for Fur upstream of their genes. These sites are termed Fur-boxes [20]. Under iron-rich conditions, Fur binds Fe2+, assumes a conformation resulting in tight binding to the Fur-box and repression of gene transcription [21]. Low iron levels result in the loss of this metal ion and allosteric conformational changes in Fur that alleviate transcriptional repression. Positive regulation by Fur in Gram-negative bacteria seems to be primarily indirect via negative transcriptional control of small RNAs [22-24]. The Fur-dependent E. coli small RNA is termed RyhB, and two RyhB orthologs were discovered in the Y. pestis CO92 genome [22]. E. coli RyhB controls the expression of genes whose products store iron or contain iron cofactors such as heme and iron-sulfur (Fe-S) clusters [25,26]. The Fe-S cluster proteins FNR, IscR and SoxR are important global regulators [27]. Some enzymes with functions in diverse branches of cellular energy metabolism [28-30] also contain Fe-S clusters. Thus, widespread changes in the proteome and metabolome of bacteria occur due to iron starvation. In E. coli, the Fur regulon was reported to overlap functionally with the regulons of the catabolite repressor protein [31] and the oxidative stress regulator OxyR [32]. These overlaps suggest intriguing networks of metabolic inter-connectivity, allowing bacterial survival and growth under iron-deficient conditions. Iron homeostasis has not been thoroughly investigated to date in Y. pestis. Human plasma is an iron-limiting environment, and growth condition-dependent comparisons of Y. pestis transcriptional patterns have included growth in human plasma [33]. Many genes involved in iron acquisition and storage and the response to oxidative stress were found to be differentially expressed [33-35]. There was reasonably good agreement between the aforementioned studies and DNA microarray data comparing a Δfur mutant with its Fur+ parent strain [20].
Our objective was to assess iron acquisition and intracellular consequences of iron deficiency in the Y. pestis strain KIM6+ at two physiologically relevant temperatures (26°C and 37°C). Bacterial cultures weregrown in the absence and presence of 10 μM FeCl3. Cell lysis was followed by fractionation into periplasm, cytoplasm and mixed membranes. Upon pooling of two biological replicate samples for each growth condition, proteins were analysed by differential 2D gel display. Considering the high number of distinct experimental groups (fractions) and at least three required technical 2D gel replicates per experiment for meaningful statistical analyses, the rationale for sample pooling was to keep 2D gel runs at a manageable level. Sample pooling has the disadvantage that information on quantitative variability of proteins comparing biological replicates is not obtained. Proteome analysis was performed for two equivalent growth time points (13-15 h), which represented the stationary phase for iron-replete conditions (OD600 ~2.0) and growth arrest at the OD600 of ~0.8 for iron-deficient conditions.
Methods
Bacterial strains and culture conditions
The Y. pestis strain KIM6+ used in this study is an avirulent derivative of the fully virulent KIM strain, which was cured of the pCD1 plasmid but retained the chromosomal pgm locus and the plasmids pMT1 and pPCP1 [36]. We used strain maintenance and cell growth procedures and verified the presence of the pgm locus on Congo Red agar as described previously [37]. Bacterial colonies were grown on tryptose blood agar at 30°C, harvested after 48 h and stored at -80°C. Aliquots of these cell stocks were used to grow 5-10 mL cultures in chemically defined PMH2 medium [14] supplemented with 10 μM FeCl3, followed by dilution to an OD600 of ~0.05 with 0.3-1 L of PMH2. PMH2 was deferrated by incubation with Chelex-100 resin overnight at 4°C [14]. Two passages of cell stocks in 10-30 mL of this medium were followed by dilution to an OD600 of ~0.05 with 0.3-1 L of deferrated PMH2. Overnight cell cultures (13-15 h) reached OD600s of ca. 1.8-2.5 and 0.6-0.9 for iron-rich and iron-deficient cells, respectively. Chelex-100 treatment was previously shown to reduce contaminating iron levels to 0.2-0.3 μM, and replenishment of this medium with 10 μM FeCl3 resulted in full recovery of the normal Y. pestis growth rate and yield. Chelex-100 treatment likely removes some other metal ions as well. However, in contrast to iron, addition of Mn, Zn and Cu did not enhance the observed growth rate or yield. Cell pellets were harvested by centrifugation at 8,000 × g for 15 min at 4°C and washed with ca. 30 volumes of 33 mM K2HPO4 (pH 7.5).
Subcellular fractionation of Y. pestis cells
K2HPO4-washed Y. pestis cells were subjected to a lysozyme/EDTA spheroplasting method, followed by lysis of spheroplasts via sonication in a hypotonic buffer as previously described [38,39]. Soluble periplasmic and cytoplasmic fractions were exchanged into buffer A (25 mM NH4HCO3, 1 mM Na-EDTA and 1 mM benzamidine) and concentrated to 2-5 mg/mL protein at 3,000 × g using membrane filtration units (NMWL ~10,000). Protein concentrations were measured with the bicinchoninic acid assay, unless stated otherwise. Mixed membrane pellets were isolated from spheroplast lysates by centrifugation at 50,000 × g for 1 h at 4°C. These pellets were homogenized in 0.25 M sucrose, 150 mM NaCl, 10 mM Tris-OAc, pH 7.8, 5 mM Na-EDTA, 0.2 mM DTT, 10 μg/ml Leupeptin, 5 μg/ml Pepstatin, 10 μg/ml Nα-p-Tosyl-L-arginine methyl ester and 2 mM PMSF (ca. 10 mL/g pellet weight), and washed to remove most soluble protein contaminants. Sodium bromide (2.5 M final concentration) was added to the suspended membrane pellet, stirred for 1 h at 20°C and centrifuged at 50,000 × g for 1 h at 4°C. Insoluble pellets were then extracted with an ice-cold solution of 0.18 M Na2CO3, pH 11.3, 50 mM DTT, 1 mM CaCl2, 1 mM MgCl2 and 1 mM MnCl, stirred for 1 h and spun at 50,000 × g for 1 h at 4°C. The supernatants were not processed further. The membrane protein-enriched pellets were solubilised with 8 M urea, 2 M thiourea, 1% (w/v) amidosulfobetaine 14, 2 mM tributylphosphine and 0.5% Bio-Lyte pH 3-10 carrier ampholytes for analysis in 2D gels. Following incubation for 30 min at 20°C and centrifugation at 16,200 × g for 15 min, soluble aliquots of the extract, termed urea/amidosulfobetaine 14-extracted membrane (usb-MBR) fraction, were run in SDS-PAGE gels. Protein amounts were estimated from Coomassie Brilliant Blue G-250 (CBB)-stained band intensities. Integral OM proteins were more enriched than lipoproteins and integral IM proteins. The latter proteins tend to resist solubilisation or re-precipitate during the IEF separation step.
Enzyme assays
Spectrophotometric enzyme assay were performed in 96-well microtiter plates using soluble fractions of Y. pestis cell lysates. Cells were harvested during the mid-exponential phase (OD600 ~0.5-0.7) and stationary phase (OD600 ~1.8-2.1) time points from iron-replete conditions in PMH2 medium at 26°C. Cells from two equivalent time points (OD600 ~0.4-0.6 and OD600 ~0.7-0.9, respectively) were harvested when growth occurred in iron-free media at 26°C. In a 100 mM NaH2PO4 buffer (pH 6.5) with 75 μg/mL lysozyme, 1 mM Na-EDTA, 1 mM PMSF and 0.1% Triton X-100, cells were subjected to pressure cycling (12 cycles of 35 kPsi for 5 sec and 0 Psi for 20 sec). After the addition of 5 mM MgCl, 10 μg/mL DNAse I and 10 μg/mL RNAse cell lysates were incubated for 45 min at 20°C and centrifuged at 16,200 × g for 30 min at 4°C. The supernatants were frozen at -80°C in the presence of 15% glycerol until used for enzyme assays.
Pyruvate oxidase activities were determined using sodium pyruvate and Na3Fe(CN)6 as substrates and monitoring the rate of absorbance decrease of Na3Fe(CN)6 at A450 (E450 = 0.218 cm-1 mM-1) while incubating at 30°C. Cell lysates were adjusted to ~0.4 mg/mL protein and assayed at pH 6.0 in 120 mM NaH2PO4 as previously reported [40], with one modification: 1% Nonidet-P40 was added to the assay buffer, because this detergent increased the activity of PoxB. Aconitase activities were determined using a coupled enzyme assay converting citrate to isocitrate and, via activity of supplemented isocitric dehydrogenase (IcdA), isocitrate to α-ketoglutarate as previously described [41] (assay kit from Cayman Chemicals, Ann Arbor, MI). The rate of absorbance increases at A340 (E340 = 0.00622 cm-1 μM-1) due to formation of the IcdA product NADPH was monitored while incubating at 37°C. To increase the pH and stabilize aconitase, crude extracts were exchanged into 50 mM Tris-HCl (pH 7.5), 0.6 mM MnCl2 and 2 mM sodium citrate, and adjusted to ~0.5 mg/mL protein. To distinguish the aconitase/IcdA activity from other NADP+-dependent oxidoreductive enzymes, the aconitase inhibitor oxalomalate was added at a 18.7 mM concentration to the assay mixture. Catalase activities were determined assessing the peroxidatic function with methanol as a substrate and formaldehyde as a product in the presence of optimal concentrations of H2O2 at 22°C. Crude extracts were diluted to ~0.15 mg/mL protein, and formaldehyde was measured colorimetrically at A540 in an endpoint assay via addition of the chromogen 4-amino-3-hydrozino-5-mercapto-1,2,4-triazole, as previously described [42] (assay kit from Cayman Chemicals). Superoxide dismutase (SOD) activities were determined using a coupled enzyme assay measuring the dismutation of the superoxide radical formed by xanthine oxidase at 22°C. The reaction was coupled to the conversion of a tetrazolium salt to formazan whose absorbance was measured at A450 in an endpoint assay as described [43] (assay kit from Cayman Chemicals). Cell lysates were diluted to ~1.1 μg/mL protein to measure SOD activities.
Protein separation and differential display in 2D gels
Equal protein amounts from two biological replicates of periplasmic and cytoplasmic fractions were combined and diluted in a 1:5 to 1:10 ratio with RB buffer, which contained 8 M urea, 2 M thiourea, 4% (w/v) CHAPS, 18 mM DTT and 0.5% (v/v) Bio-Lyte pH 3-10 carrier ampholytes. Equal protein amounts from solubilised biological replicates of mixed membrane fractions were also combined. The rationale for sample pooling is described at the end of the 'Background' section. Circa 75 μg protein for Sypro Ruby®-stained gels and 130 μg for Coomassie Brilliant Blue G250 (CBB)-stained gels were loaded via rehydration loading onto 24 cm IPG gel strips (pH ranges 4-7 and 3-10) and separated in the 1st dimension as previously described [39]. Established methods were also used for 2nd dimension slab gel electrophoresis (25 × 19.5 × 0.15 cm), gel staining with CBB, scanning and gel image import into the analysis software Proteomweaver v.4.0 [44]. The scope of differential 2D display analysis was extensive, with three subcellular fractions and four growth conditions (fourteen experimental groups for seven group-to-group comparisons, among them two analyses for the periplasmic fraction with 2D gels in the pH ranges 4-7 and 6.5-10). Software-assisted gel image analysis included spot matching, pre-match and post-match spot normalization and spot intensity averaging. The analysis mode did not require internal standards for spot normalization. The Mann-Whitney Test was used for statistical significance analysis of spot abundance changes. It is a non-parametric two sample distribution-free t-test and assesses whether two independent samples of observations come from the same distribution: 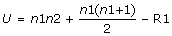 , where n1 and n2 are numbers of observations in the samples and R1 is the sum of the ranks of the observations in sample 1. P-values determined by this test are based on 3 ≤ n ≤ 5 observations, which reflect 2D spot intensity data from an equal number of replicate gels. Provided that spot abundance ratios were ≥1.5, p-values < 0.02 were considered statistically significant. Protein spots observed in only one the two sample groups required spot matches among at least three technical replicates, but p-values could not be determined.
, where n1 and n2 are numbers of observations in the samples and R1 is the sum of the ranks of the observations in sample 1. P-values determined by this test are based on 3 ≤ n ≤ 5 observations, which reflect 2D spot intensity data from an equal number of replicate gels. Provided that spot abundance ratios were ≥1.5, p-values < 0.02 were considered statistically significant. Protein spots observed in only one the two sample groups required spot matches among at least three technical replicates, but p-values could not be determined.
Mass spectrometry and bioinformatic protein analysis
Nearly all spots derived from 2D gels of the three Y. pestis subcellular fractions were analyzed by mass spectrometry (MS) two or more times. This was necessary in order to link each spot abundance change unambiguously to identification of a distinct protein; limitation of spot resolution in 2D gels is a known problem when the analyzed samples are highly complex. Prerequisites for confident spot identification were known protein identities of surrounding spots with equal or higher abundance and the comparison of Mascot scores in those spots. Methods for spot cutting and protein digestion with trypsin were reported previously [45]. Peptide digests were analyzed using a MALDI-TOFTOF mass spectrometer (4700 Proteomics Analyzer, Applied Biosystems) and a nano-electrospray LC-MS/MS system (LTQ ion trap mass spectrometer, Thermo-Finnigan, San Jose, CA) equipped with an Agilent 1100 series solvent delivery system (Agilent, Palo Alto, CA). Reversed phase peptide separations for LC-MS/MS analysis were performed at nanoflow rates (350 nL/min). Technical details of MS and MS2 analysis methods have been described [45]. The data were searched against the latest release of the Y. pestis KIM strain subset of the NCBInr database, using the Mascot searching engine v.2.1 (Matrix Science, London, UK). Carbamidomethyl was invariably selected as a fixed modification and one missed tryptic cleavage was allowed. MALDI search parameters (+1 ions) included mass error tolerances of ± 100 ppm for peptide precursor ions and ± 0.2 Da for fragment ions. LTQ ion trap search parameters (+1, +2 and +3 ions) included mass error tolerances of ± 1.4 Da for peptide precursor ions and ± 0.5 Da for fragment ions. Protein identifications were accepted as significant when Mascot protein scores >75 were obtained. Using a randomized decoy database, setting a default significance threshold of 0.05 in the Mascot algorithm and requiring two peptide e-values < 0.1 per protein identification, the false positive rate of proteins by LC-MS/MS was estimated to be <0.5%. Bioinformatic predictions of Y. pestis KIM iron transporters and binding proteins, of transmembrane domains, of protein export signal motifs and of β-barrel OM protein motifs were derived from the algorithms utilized in TransportDB http://www.membranetransport.org, TMHMM and SignalP http://www.cbs.dtu.dk/services and PRED-TMBB [46], respectively.
Results
Using subcellular fractionation and differential 2D gel display to assess the response of Y. pestis to iron starvation
Three subcellular fractions of the Y. pestis strain KIM6+, a periplasmic, a cytoplasmic and a membrane fraction enriched in integral OM proteins, were isolated from cells cultured at two growth temperatures (26°C and 37°C), without FeCl3 or supplemented with 10 μM FeCl3. Data published on the periplasmic and membrane proteomes of this strain facilitated the study [39,44,47]. Extensive protein identification efforts from soluble cytoplasmic fractions were performed for this study. We estimated the coverage of subcellular proteomes by comparing predicted localizations of experimentally identified proteins with those in silico assigned to the ORFs annotated in the Y. pestis KIM genome. The algorithm used here was PSORTb. Limiting this to the proteins clearly assigned to distinct 2D gel spots, the coverage was roughly 25% for the periplasm, 20% for the cytoplasm, 1% for the IM and 25% for the OM. The prediction of subcellular proteomes is incomplete because assignments are not made for all ORFs (e.g., 45% of the 4086 Y. pestis KIM ORFs using PSORTb). Many proteins were not profiled quantitatively. However, subcellular fractionation allowed us to increase the number of surveyed proteins and the dynamic range of abundance measurements.
Proteome profiles derived from iron-starved and iron-replete growth conditions, often abbreviated as '-Fe and +Fe conditions' from here on, were compared. When cells were harvested, they were in the stationary phase for at least 3 h (+Fe conditions) or near complete growth arrest due to the lack of iron (-Fe conditions). This is visualized in growth curves at the temperatures of 37°C and 26°C provided in the graphics of Additional File 1. Cells grown in the absence of iron at 37°C consistently reached a 10-20% higher OD600 than those grown at 26°C. Earlier growth time points (exponential phase) would have been of interest, but were not included due to already extensive proteomic profiling efforts. Our rationale was that the greater difference in cell doubling times during the exponential phase (-Fe vs. +Fe) would have confounded identification of iron starvation-specific protein changes more than that for the late growth stage.
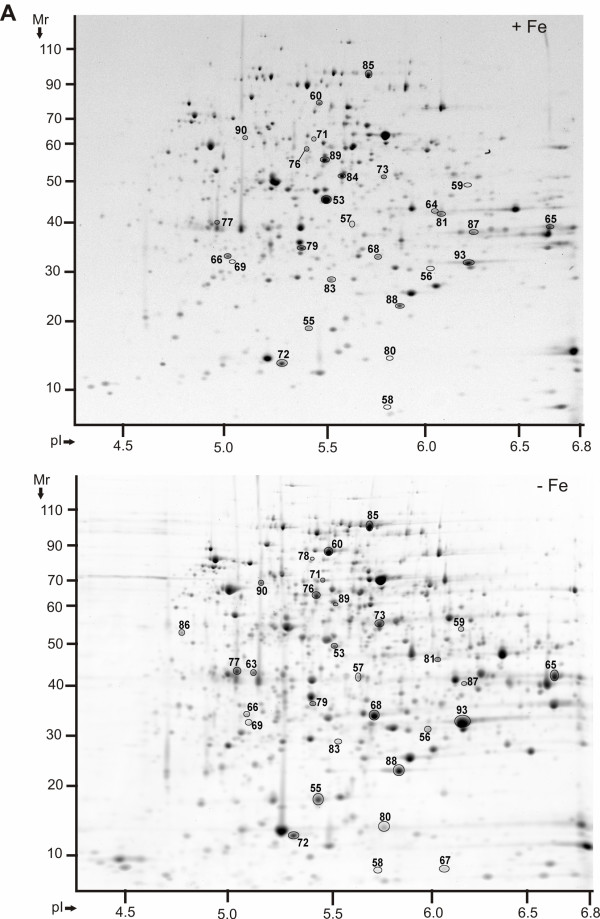
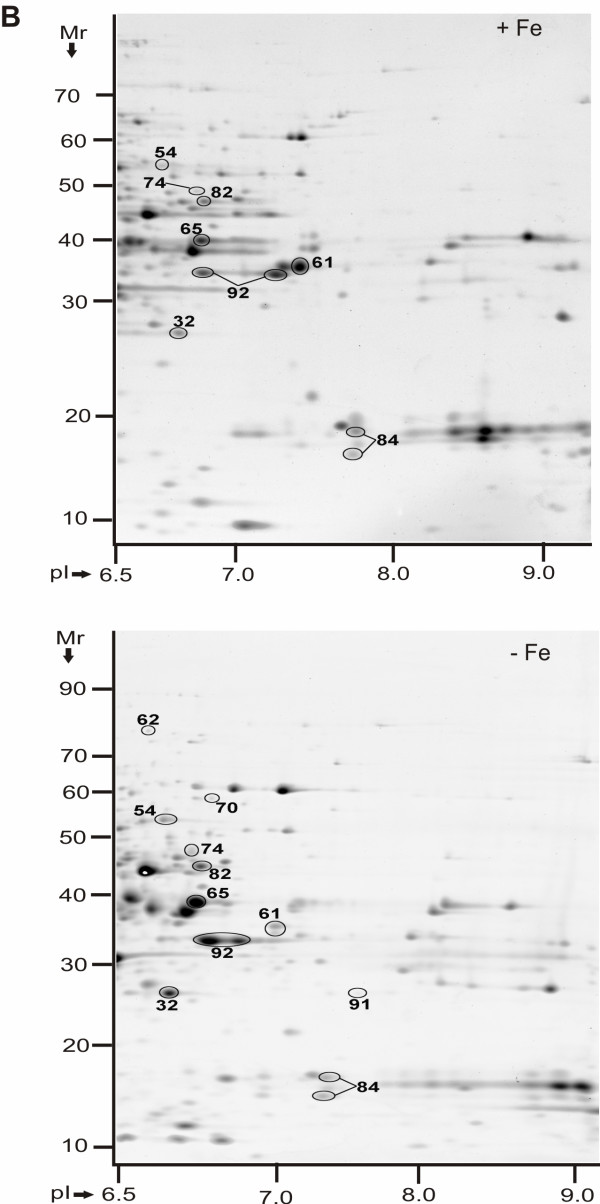
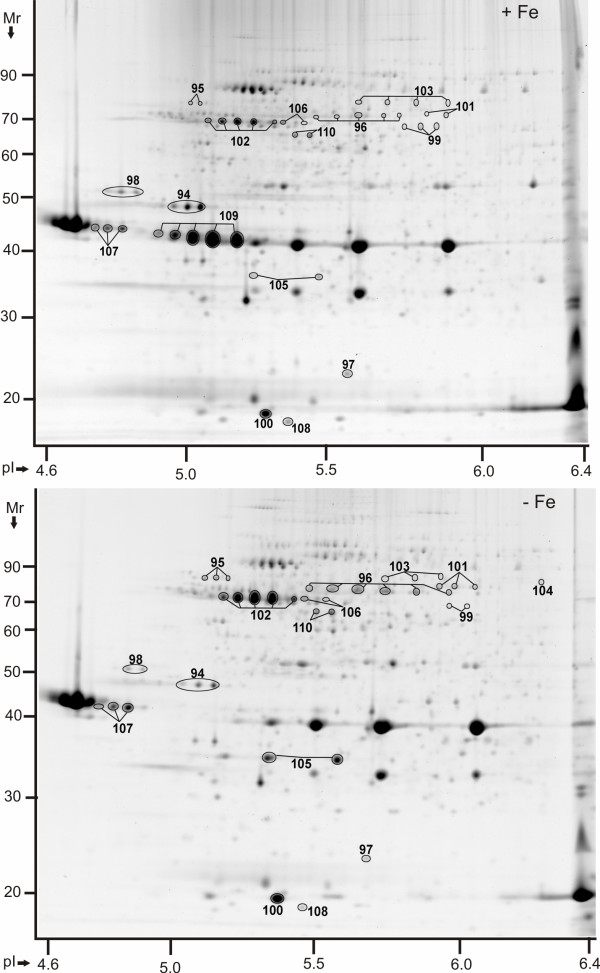
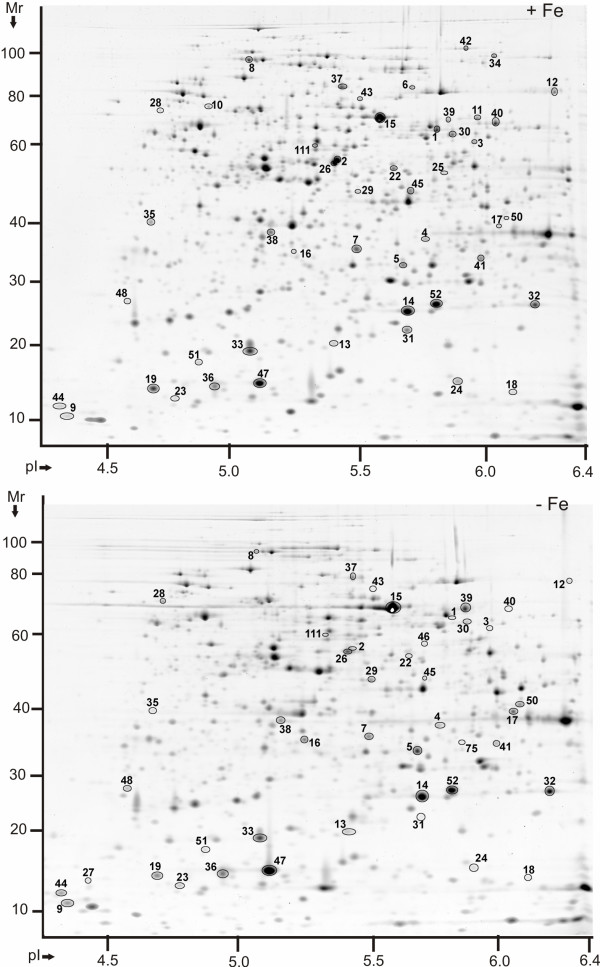
Differential display experiments were focused on the pH range 4-7 in 2D gels because the majority of mature proteins have pI values ranging from 4 to 7. The removal of basic N-terminal signal sequences from exported proteins, which are displayed in the periplasmic and mixed membrane fractions, often result in a shift towards more acidic pIs. Few integral IM proteins, typically those with low Mr values, were quantitatively profiled because TMD proteins are too hydrophobic to be sufficiently solubilised or resolved as spots in 2D gels. Periplasmic fractions consistently showed contamination with cytoplasmic proteins which was attributed to partial lysis of Y. pestis spheroplasts during the fractionation. The outcome of this cross-contamination was a moderately decreased depth of analysis for periplasmic proteins. Of nearly 250 statistically significant spot abundance changes with confident protein identifications, observed at 26°C and/or 37°C, some were associated with spot trains. Particularly the 2D profile of the usb-MBR fraction featured extensive spot trains. Based on MS2 data from MALDI-TOFTOF and ESI-ion trap experiments, we have evidence that Asn and Gln amino acid side chain deamidations, which lead to acidic 2D spot shifts of proteins, account for many of the spot trains [48]. The Table of Additional File 2 lists all significant spot abundance changes (-Fe vs. +Fe conditions). Comprehensive MS and MS2 datasets are provided in the Table of Additional File 3. The concise protein lists in the Tables 1, 2 and 3 are of particular interest in the context of iron homeostasis. Only if protein abundance ratios differed substantially comparing the -Fe vs. +Fe datasets at 26°C and 37°C, the temperature dependency was pointed out in the following paragraphs.
Table 1.
Abundance differences of Y. pestis proteins profiled in periplasmic fractions of iron-rich vs. iron-starved cells
| Spot No a) |
Gene locus b) |
gene name c) |
Protein description c) | Subc. Loc. d) |
Fur/RyhB e) | Mascot Score f) |
exp Mr (Da) | exp pI | 26°C, Vs (-Fe) g) | 26°C, Vs (+Fe) h) | 26-ratio -Fe/+Fe i) | 26°C P-value j) | 37-ratio -Fe/+Fe k) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 53 | y0028 | malE | periplasmic maltose-binding protein | PP | 2150 | 43937 | 5.53 | 0.72 | 5.98 | 0.121 | 0.000 | 0.760 | ||
| 54 | y0137 | degQ | serine endoprotease | PP | 1077 | 55588 | 6.43 | 0.39 | 0.11 | 2.41 | 0.0177 | 0.900 | ||
| 55 | y0291 | - | putative tospovirus resistance protein D | U | 486 | 18721 | 5.44 | 2.05 | 0.47 | 4.320 | 0.000 | N.D. | ||
| 56 | y0541 | hmuT | hemin-binding periplasmic protein | PP | Fur | 228 | 27164 | 5.85 | 0.46 | 0.11 | 4.328 | 0.000 | > 20 | |
| 57 | y0542 | hmuS | hemin uptake system component | U | Fur | 989 | 38188 | 5.56 | 0.53 | 0.19 | 2.780 | 0.000 | 2.091 | |
| 58 | y0869 | cybC | cytochrome b(562) | PP | Fur | 626 | 5035 | 5.64 | 0.13 | 0.03 | 4.746 | N.D. | 3.160 | |
| 59 | y0964 | frsA | fermentation/respiration switch protein | U | 586 | 51326 | 5.98 | 0.15 | 0.07 | 2.208 | 0.000 | 1.875 | ||
| 60 | y1128 | bglX | putative beta-glucosidase | PP | 2324 | 81506 | 5.43 | 3.01 | 0.52 | 5.822 | 0.000 | 1.740 | ||
| 61 | y1189 | gltI | solute-binding periplasmic protein of glutamate/aspartate ABC transporter | PP | 2512 | 35927 | 7.20 | 0.41 | 2.91 | 0.141 | 0.005 | N.D. | ||
| 62 | y1223 | nrdE | ribonucleoside-diphosphate reductase 2, alpha subunit | U | Fur | 198 | 79914 | 6.32 | 0.03 | - | > 20 | N.D. | N.D. | |
| 63 | y1222 | nrdF | ribonucleoside-diphosphate reductase 2, beta chain | U | Fur | 561 | 39335 | 5.11 | 0.77 | - | > 20 | N.D. | > 20 | |
| 64 | y1430 | - | putative putative periplasmic iron-binding signal peptide protein | U | 3359 | 41211 | 6.09 | - | 0.57 | < 0.05 | N.D. | < 0.05 | ||
| 65 | y1526 | yfuA | putative solute-binding protein for iron ABC transporter | PP | Fur | 1979 | 39620 | 6.65 | 2.36 | 1.46 | 1.618 | 0.061 | N.D. | |
| 66 | y1607 | hisJ | histidine-binding periplasmic protein of high-affinity histidine transport system | PP | 1494 | 31529 | 5.01 | 0.29 | 0.93 | 0.309 | 0.000 | 0.350 | ||
| 67 | y1744 | - | hypothetical protein y1744 | CY | 324 | 5183 | 5.92 | 0.38 | - | > 20 | N.D. | 4.510 | ||
| 68 | y1897 | yfeA | periplasmic-binding protein for iron and manganese ABC transporter | CM | Fur | 1201 | 31395 | 5.80 | 2.87 | 0.63 | 4.576 | 0.000 | 4.780 | |
| 69 | y1936 | sufC | iron-sulfur cluster assembly protein SufC, ATPase component | ML | Fur | 726 | 28460 | 5.10 | 0.16 | 0.02 | 7.514 | 0.000 | > 20 | |
| 70 | y1937 | sufD | cysteine desulfurase activator complex subunit SufD | U | Fur | 369 | 60476 | 6.76 | 0.06 | - | > 20 | N.D. | N.D. | |
| 71 | y2358 | hmsF | hemin storage system, HmsF protein | U | 542 | 61790 | 5.47 | 0.40 | 0.12 | 3.467 | 0.000 | 1.480 | ||
| 72 | y2368 | - | putative ferrous iron transport protein | U | 532 | 13556 | 5.29 | 1.71 | 1.64 | 1.030 | 0.390 | 2.330 | ||
| 73 | y2394 | ybtS | anthranilate synthase | CY | Fur | 1323 | 50265 | 5.82 | 1.65 | 0.36 | 4.538 | 0.000 | > 20 | |
| 74 | y2401 | ybtU | thiazolinyl-S-HMWP1 reductase of Ybt system | U | Fur | 351 | 48765 | 6.63 | 0.33 | 0.11 | 3.057 | 0.006 | N.D. | |
| 76 | y2403 | ybtE | salicyl-AMP ligase | CY | Fur | 1205 | 58276 | 5.43 | 2.04 | 0.31 | 6.660 | 0.000 | 7.060 | |
| 77 | y2451 | efeO | putative ferrous iron transport protein | U | 998 | 38614 | 4.96 | 1.71 | 0.90 | 1.896 | 0.000 | 1.274 | ||
| 78 | y2638 | ysuG | siderophore biosynthetic protein of the Ysu system | U | Fur | 182 | 77918 | 5.36 | 0.06 | - | > 20 | N.D. | N.D. | |
| 79 | y2662 | mglB | periplasmic D-galactose-binding ABC transport protein | PP | 1440 | 33113 | 5.40 | 0.51 | 1.53 | 0.330 | 0.000 | 0.251 | ||
| 80 | y2828 | pheA | putative chorismate mutase | PP | 630 | 14433 | 5.88 | 0.86 | 0.05 | 19.293 | 0.000 | 2.817 | ||
| 81 | y2842 | - | putative periplasmic binding protein of iron/siderophore ABC transporter | U | 1096 | 51189 | 5.97 | 0.62 | 1.87 | 0.332 | 0.000 | 0.501 | ||
| 82 | y2875 | yiuA | solute-binding periplasmic protein of iron/siderophore ABC transporter | U | Fur | 1690 | 46030 | 6.69 | 0.73 | 0.37 | 1.957 | 0.002 | N.D. | |
| 83 | y3037 | modA | molybdate-binding periplasmic protein of molybdate ABC transporter | PP | 2136 | 27031 | 5.55 | 0.17 | 0.72 | 0.234 | 0.000 | 2.089 | ||
| 84 | y0815 | sodC | periplasmic superoxide dismutase (Cu-Zn) | PP | 695 | 16562 | 7.54 | 0.55 | 0.63 | 0.89 | 0.4490 | N.D. | ||
| 85 | y3165 | ptr | protease III | PP | 1794 | 96878 | 5.60 | 2.71 | 1.86 | 1.454 | 0.001 | 1.032 | ||
| 86 | y3676 | - | putative type VI secretion system protein | CY | 375 | 50035 | 4.81 | 0.29 | - | > 20 | N.D. | N.D. | ||
| 87 | y3772 | lsrB | putative periplasmic autoinducer II-binding protein | U | 917 | 36377 | 6.30 | 0.31 | 1.96 | 0.159 | 0.000 | N.D. | ||
| 88 | y3812 | dsbA | protein disulfide isomerase I | PP | 1587 | 22454 | 5.91 | 2.57 | 1.18 | 2.176 | 0.000 | 0.910 | ||
| 89 | y3825 | dppA | periplasmic dipeptide transport protein of ABC transporter | PP | 1253 | 54903 | 5.52 | 0.68 | 2.46 | 0.277 | 0.000 | 0.696 | ||
| 90 | y3837 | yhjJ | predicted zinc-dependent peptidase | U | 1215 | 62177 | 5.10 | 0.44 | 0.17 | 2.613 | 0.000 | 0.720 | ||
| 91 | y3956 | crp | cAMP-regulatory protein | CY | 220 | 26494 | 7.82 | 0.06 | - | > 20 | N.D. | N.D. | ||
| 92 | y3977 | fkpA | FKBP-type peptidyl-prolyl cis-trans isomerase | PP | 2031 | 33670 | 6.94 | 5.50 | 3.45 | 1.594 | 0.007 | N.D. | ||
| 93 | y4125 | - | putative solute-binding periplasmic protein precursor for ABC transporter | PP | 2766 | 30250 | 6.27 | 6.09 | 3.67 | 1.661 | 0.001 | 2.264 | ||
a) spot number as denoted in Figures 1 and 2; b) protein accession number and locus tag as listed in Y. pestis KIM genome database (NCBI); c) gene name and protein description from the KIM database or a conserved E. coli K12 ortholog http://www.ecocyc.org, if >65 pct. sequence identity; d) subcellular localization based on PSORTb data: CY, cytoplasm; ML: multiple localizations; CM: inner membrane; PP, periplasm; U: unknown; e) proven or putative regulation by Fur or a Fur-dependent small RNA (e.g. RyhB); f) highest Mascot score for a protein from LC-MS/MS or MALDI data; g) Vs (-Fe): average spot volume (n ≥ 3) in 2D gels for iron-depleted growth conditions at 26°C as shown in Figures 1-2; h) Vs (+Fe): average spot volume (n ≥ 3) in 2D gels for iron-supplemented growth conditions at 26°C; i) spot volume ratio (-Fe/+Fe) at 26°C, N.D.: not determined; -: no spot detected; j) two-tailed t-test p-value for spot abundance change at 26°C; 0.000 stands for < 0.001; k) average spot volume ratio (-Fe/+Fe) at 37°C; additional data for the statistical spot analysis at 37°C are part of Additional Table 1. exp.pI/Mr = experimental pI and Mr values.
Table 2.
Abundance differences of Y. pestis proteins profiled in membrane fractions of iron-rich vs. iron-starved cells
| Spot No a) |
Gene locus b) |
gene name c) |
Protein description c) | Subc. Loc. d) |
Fur/RyhB e) | Mascot Score f) | exp Mr (Da) | exp pI | 26°C, Vs (-Fe) g) | 26°C, Vs(+Fe) h) | 26-ratio -Fe/+Fe i) | 26°C P-value j) | 37-ratio -Fe/+Fe k) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 94 | y0032 | lamB | Maltoporin | OM | 331 | 48645 | [4.95 - 5.09] | 0.76 | 1.49 | 0.516 | 0.000 | 1.27 | ||
| 95 | y0543 | hmuR | hemin outer membrane receptor | OM | Fur | 1064 | 76570 | 5.05 | 0.25 | 0.10 | 2.600 | 0.000 | 4.665 | |
| 96 | y0850 | - | putative iron/chelate outer membrane receptor | OM | Fur | 57 | 70302 | [5.5 - 6.0] | 1.54 | 0.22 | 6.978 | 0.000 | 2.430 | |
| 97 | y1355 | - | hypothetical inner membrane protein y1355 | U | 53 | 22715 | 5.59 | 0.32 | 0.57 | 0.560 | 0.000 | 0.820 | ||
| 98 | y1577 | fadL | long-chain fatty acid transport protein (OM receptor) | OM | 1008 | 51392 | [4.77 - 4.87] | 0.37 | 0.81 | 0.460 | 0.000 | 0.370 | ||
| 99 | y1632 | nuoC | NADH dehydrogenase I chain C, D | CY | 654 | 68079 | [5.79 - 5.9] | 0.07 | 0.18 | 0.367 | 0.000 | 0.578 | ||
| 100 | y1682 | ompX | outer membrane protein X | OM | 389 | 18271 | 5.31 | 5.65 | 3.08 | 1.859 | 0.000 | 0.557 | ||
| 101 | y1919 | arnA | bifunctional UDP-glucuronic acid decarboxylase/UDP-4-amino-4-deoxy-L-arabinose formyltransferase | U | 346 | 72392 | [5.86 - 5.92] | 0.76 | 0.20 | 3.748 | 0.000 | > 20 | ||
| 102 | y2404 | psn | pesticin/yersiniabactin outer membrane receptor | OM | Fur | 148 | 67582 | [5.20 - 5.45] | 6.80 | 1.46 | 4.862 | 0.000 | 2.656 | |
| 103 | y2556 | fcuA | ferrichrome receptor, TonB dependent | OM | Fur | 801 | 76097 | [5.64 - 5.94] | 0.20 | 0.18 | 1.070 | 0.710 | 0.860 | |
| 104 | y2633 | ysuR | outer membrane iron/siderophore receptor | OM | Fur | 202 | 73135 | 6.30 | 0.11 | 0.04 | 2.790 | 0.001 | N.D. | |
| 105 | y2735 | ompA | outer membrane porin A, N-t. fragment | OM | 686 | 34018 | [5.52 - 5.75] | 5.05 | 0.70 | 7.245 | 0.000 | 3.390 | ||
| 106 | y2872 | yiuR | putative iron/siderophore outer membrane receptor | OM | Fur | 133 | 67256 | 5.55 | 0.65 | 0.29 | 2.260 | 0.000 | N.D. | |
| 107 | y2966 | ompC | outer membrane porin protein C | OM | 1110 | 43707 | [4.78 - 4.88] | 2.18 | 1.45 | 1.500 | 0.010 | 0.487 | ||
| 108 | y2980 | yfaZ | hypothetical protein y2980 | CM | 96 | 20054 | 5.48 | 0.30 | 0.66 | 0.459 | 0.000 | 0.202 | ||
| 109 | y2983 | phoE | putative outer membrane porin | OM | 65 | 41703 | [4.94 - 5.22] | - | 14.60 | < 0.05 | N.D. | < 0.05 | ||
| 110 | y3674 | - | putative type VI secretion system protein | U | 350 | 63614 | [5.52 - 5.58] | 0.72 | 0.44 | 1.620 | 0.002 | N.D. | ||
a) spot number as denoted in Figure 3; b) protein accession number and locus tag as listed in Y. pestis KIM genome database (NCBI); c) gene name and protein description from the KIM database or a conserved E. coli K12 ortholog http://www.ecocyc.org, if >65 pct. sequence identity; d) subcellular localization based on PSORTb data: CY, cytoplasm; CM: inner membrane; OM: outer membrane; U: unknown; e) proven or putative regulation by Fur or a Fur-dependent small RNA (e.g. RyhB); f) highest Mascot score for a protein from LC-MS/MS or MALDI data; g) Vs (-Fe): average spot volume (n ≥ 3) in 2D gels for iron-depleted growth conditions at 26°C as shown in Figure 3; h) Vs (+Fe): average spot volume (n ≥ 3) in 2D gels for iron-supplemented growth conditions at 26°C; i) spot volume ratio (-Fe/+Fe) at 26°C, N.D.: not determined; -: no spot detected; j) two-tailed t-test p-value for spot abundance change at 26°C, 0.000 stands for < 0.001; k) average spot volume ratio (-Fe/+Fe) at 37°C; additional data for the statistical spot analysis at 37°C are part of Additional Table 1. Experimental pI values span a pH range because proteins were visualized as spot trains.
Table 3.
Abundance differences of Y. pestis proteins profiled in cytoplasmic fractions of iron-rich vs. iron-starved cells
| Spot No a) | Gene locus b) |
gene name c) |
Protein description c) | Subc. Loc. d) |
Fur/RyhB e) | Mascot Score f) |
exp Mr (Da) | exp pI | 26°C, Vs (-Fe) g) | 26°C, Vs (+Fe) h) | 26-ratio -Fe/+Fe i) | 26°C P-value j) | 37-ratio -Fe/+Fe k) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | y0015 | aceB | malate synthase A | CY | Fur | 688 | 63974 | 5.86 | 0.06 | 1.73 | 0.036 | 0.000 | 0.421 | |
| 2 | y0016 | aceA | isocitrate lyase | CY | 741 | 54571 | 5.47 | 0.38 | 4.19 | 0.090 | 0.000 | 0.408 | ||
| 3 | y0047 | glpK | glycerol kinase | CY | 828 | 60235 | 6.01 | 0.07 | 0.33 | 0.198 | 0.000 | 0.570 | ||
| 4 | y0320 | oxyR | DNA-binding transcriptional regulator OxyR | CY | 510 | 36649 | 5.82 | 0.49 | 0.40 | 1.237 | 0.004 | 0.791 | ||
| 5 | y0548 | metF2 | putative methylenetetrahydrofolate reductase | U | 321 | 31848 | 5.73 | 1.77 | 1.06 | 1.677 | 0.000 | 0.543 | ||
| 6 | y0617 | frdA | fumarate reductase, anaerobic, flavoprotein subunit | PP | 437 | 80764 | 5.77 | - | 0.23 | < 0.05 | N.D. | 0.339 | ||
| 7 | y0668 | mdh | malate dehydrogenase | ML | 2170 | 34545 | 5.55 | 1.03 | 1.80 | 0.576 | 0.001 | 1.253 | ||
| 8 | y0771 | acnB | aconitate hydrase B | CY | RyhB | 1408 | 95757 | 5.13 | 0.22 | 0.98 | 0.220 | 0.000 | 0.229 | |
| 9 | y0801 | erpA | iron-sulfur cluster insertion protein ErpA | U | 76 | 10730 | 4.41 | 1.41 | 0.57 | 2.492 | 0.008 | 1.260 | ||
| 10 | y0818 | cysJ | sulfite reductase subunit alpha | U | 340 | 72332 | 4.97 | - | 0.20 | < 0.05 | N.D. | < 0.05 | ||
| 11 | y0854 | fumA | fumarase A | CY | RyhB | 255 | 68184 | 6.02 | - | 0.50 | < 0.05 | N.D. | < 0.05 | |
| 12 | y0870 | katY | catalase; hydroperoxidase HPI(I) | U | 768 | 78569 | 6.32 | 0.11 | 0.48 | 0.231 | 0.000 | 0.081 | ||
| 13 | y0888 | luxS | predicted S-ribosylhomocysteinase | CY | 670 | 19733 | 5.46 | 0.79 | 0.30 | 2.617 | 0.000 | 2.164 | ||
| 14 | y0988 | ahpC | putative peroxidase | CY | 898 | 24298 | 5.75 | 5.02 | 6.10 | 0.823 | 0.202 | 1.376 | ||
| 15 | y1069 | ymt | murine toxin | U | 7052 | 67771 | 5.64 | 13.61 | 9.61 | 1.415 | 0.143 | 1.359 | ||
| 16 | y1069 | ymt | murine toxin, C-t. fragment | U | 245 | 33893 | 5.30 | 0.89 | 0.19 | 4.634 | 0.000 | N.D. | ||
| 17 | y1069 | ymt | murine toxin, N-t. fragment | U | 164 | 39074 | 6.11 | 0.84 | 0.17 | 4.860 | 0.000 | N.D. | ||
| 18 | y1208 | fur | ferric uptake regulator | CY | 95 | 13425 | 6.16 | 0.11 | 0.17 | 0.651 | 0.055 | N.D. | ||
| 19 | y1282 | yfiD | formate acetyltransferase, glycyl radical cofactor GrcA | CY | 521 | 13866 | 4.75 | 1.27 | 2.27 | 0.560 | 0.000 | 0.456 | ||
| 20 | y1334 | iscS | selenocysteine lyase/cysteine desulfurase | U | 408 | 51519 | 5.96 | - | - | N.D. | N.D. | 0.59 | ||
| 21 | y1339 | hscA | chaperone protein HscA | CY | 384 | 49149 | 5.53 | - | - | N.D. | N.D. | 0.48 | ||
| 22 | y1342 | pepB | aminopeptidase B | U | 855 | 52355 | 5.69 | 0.12 | 0.75 | 0.153 | 0.000 | 0.681 | ||
| 23 | y1452 | ypeA | predicted acyltransferase | CY | 188 | 12771 | 4.83 | 0.39 | 0.14 | 2.844 | 0.000 | 3.300 | ||
| 24 | y1677 | dps | DNA starvation/stationary phase protection protein | U | 724 | 14844 | 5.94 | 0.27 | 0.80 | 0.337 | 0.000 | 0.808 | ||
| 25 | y1791 | pepT | putative peptidase T | CY | 310 | 51106 | 5.89 | - | 0.18 | < 0.05 | N.D. | N.D. | ||
| 26 | y1802 | icdA | isocitrate dehydrogenase, specific for NADP+ | CY | 459 | 53760 | 5.46 | 0.92 | 1.80 | 0.511 | 0.002 | 1.238 | ||
| 27 | y1934 | sufA | iron-sulfur cluster assembly scaffold protein SufA | U | Fur | 156 | 13330 | 4.48 | 0.13 | - | > 20 | N.D. | 2.170 | |
| 28 | y1935 | sufB | cysteine desulfurase activator complex subunit SufB | U | Fur | 330 | 70431 | 4.69 | 0.25 | 0.06 | 4.022 | 0.000 | 3.836 | |
| 29 | y1938 | sufS | selenocysteine lyase | U | Fur | 369 | 46479 | 5.55 | 0.65 | 0.15 | 4.294 | 0.000 | 2.420 | |
| 30 | y1944 | pykF | pyruvate kinase I | CY | 525 | 62400 | 5.93 | 0.38 | 1.23 | 0.309 | 0.525 | 1.265 | ||
| 31 | y1951 | sodB | superoxide dismutase, iron | U | RyhB | 285 | 21541 | 5.75 | 0.16 | 0.94 | 0.172 | 0.000 | >20 | |
| 32 | y1968 | gst | glutathionine S-transferase | CY | 1326 | 25438 | 6.25 | 3.15 | 2.14 | 1.471 | 0.054 | 1.247 | ||
| 33 | y1990 | tpx | thiol peroxidase | U | 479 | 18655 | 5.13 | 3.02 | 3.06 | 0.986 | 0.816 | 1.198 | ||
| 34 | y2063 | acnA | aconitate hydratase A | CY | RyhB | 565 | 97825 | 6.08 | - | 0.22 | < 0.05 | N.D. | < 0.05 | |
| 35 | y2255 | yebC | hypothetical protein y2255 | U | 219 | 39957 | 4.74 | 0.11 | 0.40 | 0.285 | 0.000 | 0.777 | ||
| 36 | y2524 | ftnA | ferritin iron storage complex protein | CY | RyhB | 223 | 14143 | 4.99 | 2.67 | 1.61 | 1.656 | 0.000 | 1.275 | |
| 37 | y2790 | pflB | formate acetyltransferase 1 | CY | 804 | 80979 | 5.49 | 0.63 | 1.38 | 0.454 | 0.000 | 0.980 | ||
| 38 | y2802 | trxB | thioredoxin reductase | ML | 702 | 37892 | 5.21 | 0.96 | 0.99 | 0.967 | 0.446 | 1.037 | ||
| 39 | y2821 | poxB | pyruvate oxidase | CY | 448 | 67362 | 5.91 | 1.89 | 0.33 | 5.722 | 0.000 | 3.710 | ||
| 40 | y2981 | katE | catalase; hydroperoxidase HPII(III) | CY | RyhB | 481 | 66313 | 6.09 | 0.04 | 1.20 | 0.032 | 0.000 | 0.113 | |
| 41 | y3064 | sucD | succinyl-CoA synthetase, alpha subunit | CY | 597 | 33015 | 6.04 | 0.33 | 0.91 | 0.363 | 0.000 | 0.472 | ||
| 42 | y3067 | sucA | 2-oxoglutarate dehydrogenase (decarboxylase component) | CY | 1153 | 102739 | 5.98 | - | 0.43 | < 0.05 | N.D. | 0.277 | ||
| 43 | y3069 | sdhA | succinate dehydrogenase, flavoprotein subunit | ML | RyhB | 965 | 75497 | 5.56 | 0.05 | 0.21 | 0.248 | 0.000 | 0.207 | |
| 44 | y3142 | fldA3 | predicted flavodoxin | CY | 267 | 11842 | 4.37 | 0.93 | 0.39 | 2.395 | 0.003 | 1.502 | ||
| 45 | y3499 | yqhD | NADP-dependent dehydrogenase | CY | 369 | 46727 | 5.76 | 0.35 | 1.922 | 0.179 | 0.001 | 1.404 | ||
| 46 | y3600 | uxaC | D-glucuronate/D-galacturonate isomerase | U | 842 | 56072 | 5.75 | 0.09 | - | > 20 | N.D. | 2.383 | ||
| 47 | y3673 | hcp1 | hemolysin-coregulated protein | U | 508 | 14459 | 5.16 | 8.35 | 4.38 | 1.908 | 0.001 | N.D. | ||
| 48 | y3675 | - | putative type VI secretion protein | CY | 392 | 25923 | 4.62 | 0.43 | 0.16 | 2.735 | 0.001 | N.D. | ||
| 49 | y3802 | bipA | putative GTP-binding factor | CY | 435 | 82945 | 5.27 | - | - | N.D. | N.D. | 4.096 | ||
| 50 | y3966 | tauD | taurine dioxygenase | U | 228 | 40946 | 6.12 | 0.50 | 0.16 | 3.129 | 0.001 | N.D. | ||
| 51 | y3988 | bfr | bacterioferritin, iron storage and detoxification protein | CY | RyhB | 143 | 17087 | 4.92 | 0.22 | 0.29 | 0.779 | 0.006 | 0.927 | |
| 52 | y4080 | sodA | superoxide dismutase, manganese | U | 597 | 25405 | 5.86 | 4.11 | 5.10 | 0.805 | 0.074 | 0.877 | ||
| 75 | y2402 | ybtT | yersiniabactin thioesterase | U | Fur | 123 | 34389 | 5.88 | 0.10 | - | > 20 | N.D. | 12.08 | |
| 111 | y3066 | sucB | 2-oxoglutarate dehydrogenase (dihydrolipoyltranssuccinase E2 component) | CY | 789 | 58844 | 5.38 | 0.29 | 0.76 | 0.38 | 0.0468 | 0.38 | ||
a) spot number as denoted in Figure 4; b) protein accession number and locus tag as listed in Y. pestis KIM genome database (NCBI); c) gene name and protein description from the KIM database or a conserved E. coli K12 ortholog http://www.ecocyc.org, if >65 pct. sequence identity; d) subcellular localization based on PSORTb data: CY, cytoplasm; ML: multiple localizations; PP, periplasm; U: unknown; e) proven or putative regulation by Fur or a Fur-dependent small RNA (e.g. RyhB); f) highest Mascot score for a protein from LC-MS/MS or MALDI data; g) Vs (-Fe): average spot volume (n ≥ 3) in 2D gels for iron-depleted growth conditions at 26°C as shown in Figure 4; h) Vs (+Fe): average spot volume (n ≥ 3) in 2D gels for iron-supplemented growth conditions at 26°C; i) spot volume ratio (-Fe/+Fe) at 26°C, N.D.: not determined; -: no spot detected; j) two-tailed t-test p-value for spot abundance change at 26°C, 0.000 stands for < 0.001; k) average spot volume ratio (-Fe/+Fe) at 37°C; additional data for the statistical spot analysis at 37°C are part of Additional Table 1.
Y. pestis iron acquisition systems
Proteomic profiling of characterized Y. pestis iron/siderophore and heme transporters (Ybt, Yfe, Yfu, Yiu and Hmu) was in good agreement with negative regulation of the respective operons by Fur and iron [15,16,20,49,50]. The subscript number following a protein name represents the spot number displayed in Figures 1, 2, 3 and 4, and is also denoted in the left-most column of Tables 1, 2 and 3. Periplasmic binding proteins of four of the ABC transporters (YfeA#68, YfuA#65, YiuA#82 and HmuT#56; Figures 1 and 2) were increased in abundance in iron-starved cells. The integral IM proteins YbtP and YbtQ were identified from streaky 2D spots of the usb-MBR fraction of iron-depleted cells, but could not be differentially quantitated. Two of these five transporters have an OM receptor responsible for iron/yersiniabactin or heme uptake (Psn#102 and HmuR#95, respectively; Figure 3), both of which were increased in iron-starved cells. Y0850#96 (Figure 3) is hypothesized to be a TonB-dependent OM receptor with Fe3+/siderophore uptake activity. This protein was also more abundant in iron-depleted cells. Detection in the usb-MBR fraction, its Mr of ca. 75-85 kDa and the presence of a highly conserved Fur-box upstream of the gene's transcriptional start site (AATGATAATTGATATCATT, -100 to -82) with a position weight matrix score of 13.2 using the patser-matrix tool [51] further supported the assignment as a Fur-regulated TonB-dependent OM receptor. Fur#18 was also detected in the cytoplasm, but not altered in abundance (Figure 4).
Figure 1.
Protein display in 2D gels of Y. pestis KIM6+ periplasmic fractions in the pI range 4-7 (-Fe vs. +Fe conditions). Proteins were derived from cell growth in the presence of 10 μM FeCl3 at 26°C (top) or the absence of FeCl3 at 26°C (bottom). Gels (20 × 25 cm) were stained with Coomassie Brilliant Blue G250 (CBB), with five gel replicates representing each group, and subjected to differential display analysis using the software Proteomweaver v.4.0. Protein assignment to a spot required validation by MS data from at least two representative gels. The denoted spot numbers are equivalent to those listed in Table 1 with their '-Fe vs. +Fe' protein abundance ratios and other data.
Figure 2.
Protein display in 2D gels of Y. pestis KIM6+ periplasmic fractions in the pI range 6.5-9 (-Fe vs. +Fe conditions). Proteins were derived from cell growth in the presence of 10 μM FeCl3 at 26°C (top) or absence of FeCl3 at 26°C (bottom). Gels (20 × 25 cm) were stained with CBB, with three gel replicates representing each group, and subjected to differential display analysis using the software Proteomweaver v.4.0. Protein assignment to a spot required validation by MS data from at least two representative gels. The denoted spot numbers are equivalent to those listed in Table 1 with their '-Fe vs. +Fe' protein abundance ratios and other data.
Figure 3.
Protein display in 2D gels of Y. pestis KIM6+ membrane fractions in the pI range 4-7 (-Fe vs. +Fe conditions). Proteins were derived from cell growth in the presence of 10 μM FeCl3 at 26°C (top) or absence of FeCl3 at 26°C (bottom). Gels (20 × 25 cm) were stained with CBB, with five gel replicates representing each of the groups, and subjected to differential display analysis using the software Proteomweaver v.4.0. Protein assignments to a spot (or a spot train) required validation by MS data from at least two representative gels. The denoted spots and spot trains are equivalent to those listed in Table 2 with their '-Fe vs. +Fe' protein abundance ratios and other data.
Figure 4.
Protein display in 2D gels of Y. pestis KIM6+ cytoplasmic fractions in the pI range 4-7 (-Fe vs. +Fe conditions). Proteins were derived from cell growth in the presence of 10 μM FeCl3 at 26°C (top) or the absence of FeCl3 at 26°C (bottom). Gels (20 × 25 cm) were stained with CBB, with four gel replicates representing each group, and subjected to differential display analysis using the software Proteomweaver v.4.0. Protein assignment to a spot required validation by MS data from at least two representative gels. The denoted spot numbers are equivalent to those listed in Table 3 with their '-Fe vs. +Fe' protein abundance ratios and other data.
Abundance increases in iron-starved cells were observed for the multifunctional yersiniabactin synthase subunits HMWP1 and HMWP2 (products of the irp1 and irp2 genes, respectively) and other enzymes contributing to yersiniabactin biosynthesis (YbtS#73, YbtT#75, YbtE#76 and YbtU#74). The high Mr proteins HMWP1 and HMWP2 were reliably quantitated only from SDS-PAGE gels (data not shown). The ysu locus encodes an OM receptor (YsuR/Y2633), an ABC transporter (Y2634-Y2637) and a suite of siderophore biosynthetic enzymes (Y2638-Y2641). Two subunits, Y2638#78 (Figure 1) and YsuR#104 (Figure 3) were of low abundance and detected only in iron-starved cells at 26°C. Biosynthesis of a Ysu siderophore has not been proven, and a siderophore biosynthetic pseudogene precedes the y2633-y2637 locus [18]. The OM β-barrel ferrichrome receptor FcuA#103 (Y2556) was identified as a protein of moderate abundance in usb-MBR fractions at 26°C (Figure 3) and 37°C, but not significantly altered in abundance comparing -Fe vs. +Fe conditions. Many membrane proteins ascribed to have putative functions in iron transport were not detected, e.g. the OM receptors Y3948 and IutA/Y3385 and the transport systems FitA-D (Y4043-Y4046), Y2837-Y2842 and FepB/Y3477. Our data support the notion of a hierarchy of iron (Fe3+)/siderophore transporters [15], with the Ybt and Yfe systems being dominant compared to the Yfu, Yiu and Hmu systems. Periplasmic subunits of two ferrous iron (Fe2+) transporters, EfeO/Y2451 and Y2368, were also profiled in 2D gels (Figure 1). The low Mr protein Y2368#72 was increased in iron-starved cells at 37°C. The tripartite Fe2+ transport protein EfeO#77 was increased in abundance in iron-starved cells at 26°C.
The energy metabolism of Y. pestis is affected by iron starvation
Lower growth rates of Y. pestis in deferrated medium followed by growth arrest at OD600s between 0.5 and 0.9 suggest perturbations of energy generation pathways. Many oxidoreductive processes are catalyzed by enzymes containing Fe-S clusters or heme, and we sought to understand the consequences of limited iron availability as it pertains to the Y. pestis energy metabolism. The EcoCyc database http://www.ecocyc.org with its extensive data on E. coli energy metabolic pathways and iron cofactors of proteins was a useful resource in this context. Y. pestis aconitases A and B (AcnA#34 and AcnB#8; Figure 4) have functions in the TCA cycle and were decreased in abundance or detected only in iron-starved cells. So were subunits of two other TCA cycle enzymes harboring Fe-S clusters (SdhA#43 and FumA#11; Figure 4). Some TCA cycle enzymes devoid of Fe-S clusters were decreased at moderate levels under -Fe conditions (IcdA#26, SucA#42, SucD#41 and SucB#111; Figure 4). Strongly decreased abundances were denoted for AceA#2 and AceB#1 (Figure 4), enzymes which catalyze the glyoxalate bypass reaction of the TCA cycle and are regulated by the catabolite repressor protein (CRP). Glycerol kinase, also regulated by CRP, was more moderately decreased in iron-starved cells (GlpK#3, Figure 4). GlpK catalyzes the rate-limiting step of glycerol utilization and feeds its metabolites into the glycolytic pathway. CRP#91 itself was identified with low abundance in the periplasmic fraction (Figure 2). In summary, the data suggested reduced pyruvate metabolism via the citrate cycle when iron resources are exhausted in Y. pestis cells. Aconitase activity assays supported this assumption; the reaction rates were 2.8-fold and 2-fold higher in lysates derived from iron-replete cells vs. those from iron-starved cells at 26°C (stationary and exponential phase, respectively; Table 4).
Table 4.
Reaction rates for four Y. pestis enzyme classes comparing -Fe vs. +Fe conditions
| Reaction ratea) (nmol min-1 mL-1); (U mL-1)b) |
Reaction ratea) (nmol min-1 mL-1); (U mL-1)b) |
|||||
|---|---|---|---|---|---|---|
| Enzyme | +Fe, exp, n = 4e) | -Fe, early, n = 5e) | p-valuef) | +Fe, stat, n = 4e) | -Fe, late, n = 5e) | p-valuef) |
| Aconitase c) | 2.31 | 1.14 | 0.019 | 4.98 | 1.82 | 0.008 |
| Pyruvate oxidase c) | 167.5 | 1307 | 0.0001 | 463.0 | 2405 | 0.0001 |
| Catalase d) | 82.5 | 31.8 | 0.0001 | 93.4 | 29.0 | 0.0001 |
| Superoxide dismutase d) | 887.8 | 426.9 | 0.002 | 448.5 | 312.5 | 0.234 |
a) Spectrophotometric assays in 96-well microtiter plates were used for the determination of enzyme reaction rates. Total protein concentrations in crude cell lysates were the same for all samples used in a given enzyme assay: aconitase, 0.5 mg/mL; pyruvate oxidase, 0.4 mg/mL; catalase, 0.15 mg/mL; superoxide dismutase, 1.1 μg/mL. b) Units ml-1 was the definition for the superoxide dismutase reaction rate. All assays were performed in duplicate. c) Reaction rates from the linear part of the slope of the absorbance change over time. d) Reaction rates from endpoint assays. e) Number of biological replicates of cell lysates (n); exp: abbreviation for exponential, early: early growth phase equivalent to exp. phase (-Fe); average OD600 = 0.66 (+Fe) and OD600 = 0.47 (-Fe); stat: abbreviation for stationary growth phase, late: late growth phase equivalent to stat. phase (-Fe); average OD600 = 2.0 (+Fe) and OD600 = 0.81 (-Fe). True exponential and stationary growth phases were not observed for cell cultures in iron-free media. f) p-values were calculated from to comparison of reaction rates (+ Fe vs. -Fe) using a two-tailed t-test method.
The question arose whether iron-starved Y. pestis cells activated a different metabolic route of pyruvate degradation able to produce reducing equivalents (NADH and UQH2) for the electron transport chain. Pyruvate oxidase (PoxB) degrades pyruvate to acetate and is a flavin-dependent, iron-independent enzyme that generates UQH2 [52]. The pyruvate oxidase pathway indeed appeared to be important, as judged by the strong abundance increase of PoxB#39 (Figure 4) under -Fe conditions. The flavin cofactor may be recruited from redox activities of two flavodoxins. FldA3#44 was quite abundant and moderately increased in iron-depleted cells (Figure 4). FldA was identified in faint 2D spots and not reproducibly quantitated. PoxB activity measurements revealed excellent correlation between enhanced abundances and increased reaction rates in iron-starved cells. PoxB activities were 5.3-fold and 7.8-fold higher in lysates of iron-starved cells than in lysates of iron-replete cells at 26°C (stationary and exponential phase, respectively; Table 4).
Electron transport chains are localized in the IM, a fact that compromised the analysis of subunits of these IM protein complexes in 2D gels. NuoCD#99, a peripheral membrane protein of the NADH:ubiquinone oxidoreductase, was moderately decreased in abundance in iron-depleted cells (Figure 3). The E. coli NuoCD sub-complex is important for binding of some of the six Nuo-integrated Fe-S clusters [53]. Subunits of Fe-S cluster proteins with roles in two anaerobic energy metabolism branches were also less abundant in iron-depleted cells. This pertained to PflB#37 and YfiD#19, proteins of the formate-pyruvate lyase complex, and FrdA#6, which is part of the terminal electron acceptor fumarate reductase (Figure 4). Decreased abundances of metabolically active Fe-S cluster enzymes were a notable feature of iron-starved Y. pestis proteome profiles, while the abundance and activity of PoxB suggested that this enzyme was important to maintain the aerobic energy metabolism and iron cofactor-independent generation of UQH2 in iron-deficient Y. pestis cells.
Oxidative stress response in Y. pestis under iron starvation conditions
Oxidative stress is caused by various oxygen radicals and H2O2, and catalyzed by redox enzymes in non-specific reactions. While the presence of free intracellular iron aggravates oxidative stress via the Fenton reaction, it is mitigated by cytoplasmic proteins that scavenge free iron, e.g. Dps and the ferritins FtnA and Bfr [54]. The question arose how aerobically growing, iron-deficient Y. pestis cells coped with oxidative stress. One of the main E. coli global regulators of the oxidative stress response, the Fe-S cluster protein SoxR, is not encoded in the Y. pestis genome [2]. The other global oxidative stress response regulator is OxyR. OxyR#4 (Figure 4) was not altered in abundance in Y. pestis comparing -Fe and+Fe conditions. Among the enzymes deactivating H2O2 and oxygen radicals are catalases/peroxidases and superoxide dismutases (SODs). Y. pestis produces two catalases with heme cofactors in high abundance. KatE#40 (Y2981) was predominantly expressed at 26°C (Figure 4) and KatY#12 (Y0870) at 37°C. Cytoplasmic SODs include SodB#31, which has an iron cofactor, and SodA#52, which has a manganese cofactor (Figure 4). Periplasmic SodC#84 has a copper/zinc cofactor (Figure 2). Iron availability-dependent patterns of abundance changes reminiscent of enzymes with functions in energy metabolism were observed. Only the iron-dependent proteins KatE, KatY and SodB were strongly diminished in abundance in iron-depleted cells (Table 3). We also determined overall catalase and SOD activities. Catalase reaction rates were 3.2-fold and 2.6-fold higher in lysates derived from iron-replete vs. iron-starved cells at 26°C (stationary and exponential phase, respectively; Table 4). SOD reaction rates were 2-fold higher in the exponential phase, but not significantly altered in the stationary phase (Table 4). This data was in good agreement with differential abundance data, although individual activities of SodA, SodB and SodC could not be discerned with the assay.
AhpC#14, Tpx#33, TrxB#38 and Gst#32, all of which are involved in redox homeostasis and deactivation of oxidative compounds, were similarly abundant in iron-rich vs. iron-starved Y. pestis cells (Figure 4). These enzymes contain either disulfide- or flavin-based redox centers. Dps#24, an iron-scavenging protein important for the protection and repair of DNA under general stress conditions, was moderately decreased in abundance under -Fe conditions, but only at 26°C. The OxyR H2O2-response system of E. coli was reported to restore Fur in its ability to repress gene expression in the presence of iron by increasing the protein's synthesis during oxidative stress [32], a mechanism that may be applicable to Y. pestis. We conclude that the bacterium adjusts its repertoire of oxidative stress response proteins when iron is in short supply, by reducing the abundance of those proteins that require iron cofactors for functional activity.
Iron storage and iron-sulfur cluster biosynthesis in Y. pestis
High concentrations of free Fe3+ are toxic to bacterial cells and require sequestration by proteins. FtnA and Bfr are the main cytoplasmic iron storage proteins. FtnA#36 was slightly increased in iron-depleted cells at 26°C (Figure 4), but not at 37°C. Bfr#51 (Figure 4) was of considerably lower abundance than FtnA and not significantly changed in abundance comparing -Fe vs. +Fe conditions. The Y. pestis KIM genome harbors two gene clusters orthologous to those of the E. coli isc and suf operons (y1333-y1341 and y1934-1939, respectively). The gene products are responsible for Fe-S cluster assembly under normal growth and stress conditions, respectively. E. coli sufABCDSE expression was reported to be controlled by the regulators OxyR (oxidative stress) and Fur (iron starvation) [55]. Protein profiling revealed that the Y. pestis Suf proteins were considerably increased or detected only in iron-depleted cells (SufC#69 and SufD#70, Figure 1; SufA#27, SufB#28 and the cysteine desulfurase SufS#29; Figure 4). Four Y. pestis Isc subunits (IscS, NifU, HscA and HscB) were detected at very low abundance in cytoplasmic fractions. The cysteine desulfurase IscS#20 and the chaperone HscA#21 were diminished in abundance in iron-starved cells at 37°C (Table 3). In contrast, an ortholog of the E. coli essential respiratory protein A (ErpA#9) was increased in abundance in iron-starved cells, particularly at 26°C (Figure 4). This low Mr Fe-S cluster protein was proposed to serve in the transfer of Fe-S moieties to an enzyme involved in isoprenoid biosynthesis [56]. Its expression was described to be under the control of E. coli IscR, the regulator of the isc gene locus. However, the abundance changes of Y. pestis ErpA (-Fe vs. +Fe) resemble those of the Suf rather than the Isc subunits.
The question arose whether sulfur-mobilizing proteins were also altered in abundance comparing -Fe and +Fe conditions, in order to support a Fe-S cluster rebalancing effort among proteins localized in the Y. pestis cytoplasm. Periplasmic binding proteins of sulfate/thiosulfate ABC transporters (Sbp, CysP) were not altered in abundance (data not shown). The flavoprotein subunit of sulfate reductase (CysJ#10) was strongly decreased in abundance under iron-limiting conditions (Figure 4). CysI, the Fe-S cluster subunit, was not detected. Taurine dioxygenase however (TauD#50, Figure 4), which utilizes aliphatic sulfonates as a sulfur source, was increased in iron-starved Y. pestis cells. The E. coli dioxygenase TauD seems to require iron for activity according to a note in the EcoCyc database. Whether the activity of TauB is linked to Fe-S cluster biosynthesis or repair remains to be shown. In summary, our data supported a functional role of the Y. pestis Suf system in Fe-S cluster assembly when cells are deprived of iron. Data related to CysIJ suggested that Fe-S cluster proteins active in pathways unrelated to energy metabolism were also down-regulated upon intracellular iron starvation.
Protein abundance changes less obviously linked to iron homeostasis
Iron is an essential cofactor for many cellular processes, and a network of global regulators (CRP, OxyR and Fur/RyhB; Figure 5) are affected by or implicated in responses to iron deficiency. We expected to detect protein abundance changes less obviously linked to iron homeostasis. S-ribosylhomocysteinase (LuxS#13) is an enzyme of central importance in the activated methyl cycle and plays a role in autoinducer 2-mediated quorum sensing in E. coli [57]. The enzyme harbors tetrahedrally coordinated Fe2+ in its catalytic center. LuxS was moderately increased in iron-depleted cells at 26°C (Figure 4). In contrast, LsrB#87 whose E. coli ortholog facilitates periplasmic transport of the autoinducer 2 following cellular re-uptake was decreased in abundance in iron-starved cells (Figure 1), similar to YebC#35, a protein hypothesized to be involved in quorum sensing regulation [58]. Y. pestis has been shown to produce the autoinducer 2, although genes controlled by this system have not been identified [59]. Slightly increased abundances of four subunits of a putative type VI secretion system (T6SS) were also observed in iron-deficient vs. iron-rich cells. The proteins HCP1#47 and Y3675#48 (Figure 4), Y3676#86 (Figure 1) and Y3674#110 (Figure 3) were not at all detected in Y. pestis protein profiles at 37°C. The T6SS is temperature-regulated. The flea survival factor Ymt#15 (Figure 4) was moderately increased in iron-starved cells at 26°C. It was one of the most abundant proteins in cells grown at 26°C. N- and C-terminal fragments of Ymt, each ca. 30 kDa in size and with a single cleavage site between V300 and I304 (Ymt#16 and Ymt#17, respectively; Figure 4), were also increased under -Fe vs. +Fe conditions. There is no evidence for a connection between the functional roles of Ymt or the T6SS and the iron starvation response.
Figure 5.
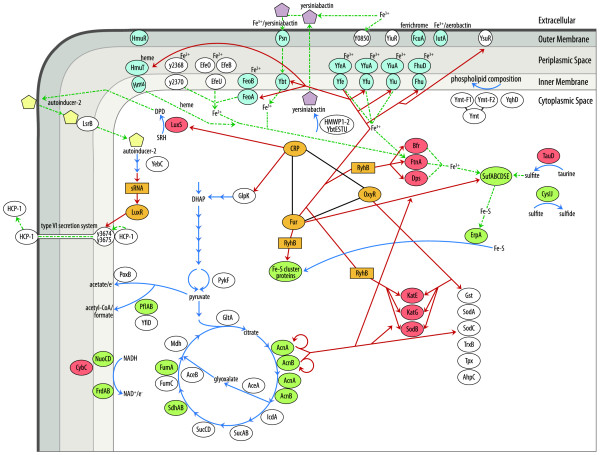
Iron homeostasis in Y. pestis. The center of the schematic depicts a network of regulators (orange color), known or potentially involved in iron homeostasis. Details are provided in the text. CRP (carbon metabolism); OxyR (oxidative stress); Fur and small RNAs like RyhB (iron homeostasis). Red lines/arrows show which genes (or mRNAs) are controlled by these regulators. Additional arrows symbolize enzymatic reactions (blue line) or small molecule transport processes (dotted green line). The lower/left side of the schematic depicts components of the energy metabolism. It includes glycolytic steps from dihydroxyacetone phosphate (DHAP) to pyruvate, the TCA/glyoxalate bypass cycle and on the left side alternative pyruvate metabolism branches generating acetate or acetyl-CoA. Subunits of electron transport systems (NuoCD, FrdAB and CybC) are also displayed. The top/left side of the schematic pertains to quorum sensing. LuxS converts S-ribosylhomocysteine (SRH) to 4,5-dihydroxy-2,3-pentanedione (DPD) which is a precursor of autoinducer-2 (yellow pentagon). In E. coli, the autoinducer-2 is exported and imported via periplasmic LsrB into different cells followed by activation of LuxR via small RNA regulators. The precise functional role of YebC in quorum sensing is not known. LuxR influences the expression of virulence factors in pathogenic E. coli strains. The role of LuxR in the regulation of the type VI secretion system is speculative, but both iron starvation [73] and the T6SS [74] have been linked to quorum sensing in other organisms. In the upper part of the schematic, iron transporter subunits are placed according their predicted or known subcellular localizations. Transporters with a blue color background are known to be functional in Y. pestis. On the center/right side, iron storage proteins, the Suf Fe-S cluster assembly system and putative sulfur-mobilizing enzymes (TauD and CysIJ) are displayed. The bottom/right part of the schematic features oxidative stress response proteins. Finally, the top/right part of the schematic displays the flea survival factor Ymt and its fragments, as well as the protein YqhD. These proteins may be implicated in the enzymatic modifications of IM phospholipids. Proteins with a red and green background harbor iron/heme and Fe-S cluster cofactors, respectively.
Periplasmic subunits of ABC transporters for amino acids, sugars and phosphate, various diffusion porins of the OM allowing passage of nutrients into the periplasm, and various amino acid tRNA-synthetases and enzymes implicated in amino acid biosynthesis were significantly increased in abundance in iron-replete cells. These observations were consistent with the notion that Y. pestis cells grown to stationary phase under +Fe conditions were depleted of various nutrients and induced the expression of high affinity transport systems for their import into the cell. Examples were the phosphate-specific OM porin PhoE#109 (Figure 3) and the periplasmic maltose-binding protein MalE#53 (Figure 1), each of which was much more abundant under +Fe vs. -Fe conditions.
Discussion
Regulation of iron homeostasis
Fur in Y. pestis, as in many other Gram-negative bacteria, is a central transcriptional regulator responding to the cellular iron status [20,50], as indicated in the schematic of Figure 5. Many iron uptake systems are transcriptionally repressed during iron-replete growth conditions to reduce accumulation of intracellular iron. Evidence has emerged that small RNA regulators are implicated in bacterial stress responses [22]. These small RNAs act by base-pairing with specific mRNAs whose translation they stimulate or inhibit in the presence of a unique protein, the RNA chaperone Hfq. A small RNA of 90 nucleotides determined to regulate genes involved in iron homeostasis in E. coli [23] and Pseudomonas aeruginosa [24] was termed RyhB. It is negatively regulated by Fur and was shown to down-regulate the translation of many of the same iron-dependent enzymes we detected as decreased in iron-starved Y. pestis cells (SdhA, AcnA, FumA, FrdA, SodB, KatE and KatY) [23]. We hypothesize that one or both of the conserved Y. pestis homologs of RyhB [22] co-regulate Y. pestis iron homeostasis and selectively decrease translation of mRNAs whose protein products depend on or store iron, as illustrated in Figure 5. Such a mechanism may restrict the use of scarce intracellular iron to processes pivotal to bacterial survival. Some of the encoding genes (e.g. ftnA, katE and sodB) may also be positively controlled by Fur as suggested by Yang et al. [35]. Gel shift assays revealed binding of recombinant Fur to promoter regions upstream of the genes ftnA and katE [20].
Several of the enzymes decreased in abundance in iron-deficient Y. pestis harbor Fe-S clusters. Expression of the respective genes did not appear to be altered under conditions sequestering or depleting iron in Y. pestis according to two DNA microarray studies [33,35] and suggests post-transcriptional mechanisms. The involvement of RyhB in controlling the abundances of proteins with iron cofactors when cells are iron-deficient needs to be verified. Since our data were derived from proteomic comparisons of Y. pestis cells harvested at different cell densities (OD600s of ~2.0 for stationary phase cells vs. OD600s of ~0.8 for growth arrested, iron-starved cells), the argument can be made that population density differences account for some of the protein abundance changes. Unpublished data (Pieper, R.) and a previous study analyzing the Y. pestis periplasmic proteome in the context of two growth phases [39] allow us to largely refute this notion. Among the proteins with iron or Fe-S cofactors, only PflB and KatE were increased in stationary vs. exponential phase proteomic profiles with ratios comparable to those observed in iron-rich vs. iron-starved cells. FtnA and Bfr are iron storage proteins and, via regulation by RyhB, were reported to be quantitatively decreased when iron supplies are limited in E. coli [23]. Our data on the FtnA and Bfr orthologs of Y. pestis were not consistent with the results of the aforementioned studies, nor with two Y. pestis transcriptional profiling studies where increased bfr expression and, in one case, decreased ftnA expression were reported for iron-limiting growth environments [33,35].
Post-transcriptional regulatory functions in iron-deficient cells have also been attributed to aconitases. In fact, eukaryotic AcnA has been termed iron-responsive protein 1 (IRP-1) [60]. Apo-enzyme versions of E. coli aconitases stabilize their cognate mRNAs and influence the expression of sodA. AcnA enhanced sodA transcript stability and was induced by iron starvation and oxidative stress in E. coli [61,62]. These findings could not be easily reconciled with our data onAcnA and AcnB abundance changes in Y. pestis. AcnA and AcnB were decreased in abundance, as were the combined aconitase activities, in iron-depleted cells. SodA abundance was not significantly affected by either growth phase [39] or iron depletion. The response of Y. pestis to iron starvation and cellular stress resulting from the loss of this metal ion seems to implicate a network of regulators, as presented in Figure 5. Indeed, functional relationships between Fur and OxyR [32], Fur and CRP [31] and Fur and apo-aconitases [62] were previously reported for E. coli.
Iron starvation stress responses
Numerous E. coli genes encoding oxidative stress response proteins are co-regulated by SoxR, Fur and OxyR according to information in the EcoCyc database. The OxyR H2O2-response system restored Fur repression in iron-replete media during oxidative stress in E. coli [32], a mechanism that we think is also relevant in Y. pestis. Strong abundance decreases in iron-starved Y. pestis cells were observed for three iron-dependent proteins, SodB, KatE and KatY. The three enzymes detoxify peroxides and radicals formed during oxidative stress. Proteins with similar functions but cofactors other than iron (e.g. SodA and AhpC) were not markedly changed in abundance. Functional assays supported such proteomic data; SOD activities in iron-depleted cells dropped markedly less than catalase activities. In conclusion, our data strongly support the notion that Y. pestis adapts its repertoire of oxidative stress response enzymes by limiting the expression of iron cofactor-dependent enzymes, when iron is in short supply. The coordination of bacterial responses to iron limitation and the defence against oxidative stress was proposed earlier [63].
Iron acquisition systems
All Y. pestis biovars have several proven iron acquisition systems, and transcriptional control by Fur has been demonstrated [18,64]. The genes and operons for putative iron transporters (e.g. Ysu, Fit, Fhu, Iuc, Has) also feature conserved 19-nt Fur-binding sites to which recombinant Fur binds [20]. Our data indicated that none of the abovementioned iron transporters were expressed at levels similar to those observed for subunits of proven iron transport systems (Ybt, Yfe, Yfu, Yiu and Hmu) under iron-deficient conditions. Two microarray studies, however, reported increased transcript abundances for many of the putative iron transporters when iron was complexed with dipyridyl [35] or sequestered by iron-binding proteins in blood plasma [33]. 2D gel analysis has known limitations pertaining to protein detection sensitivity and the resolution of hydrophobic IM-localized proteins, e.g. many nutrient transporters. Except Ysu subunits, unproven iron transporters were also not profiled employing a peptide-based LC-MS/MS analysis approach with Y. pestis lysates [47,65]. These lysates were derived from iron-replete growth conditions. Only functional iron transporters are presented in the schematic of Figure 5 and appear to follow a hierarchy of importance in the order of Ybt, Yfe (each important for virulence in a bubonic plague model), Yfu and Yiu [15]. The delivery of Fe3+ or Fe2+ from the extracellular milieu to periplasmic binding proteins of the ABC transporters Yfe, Yfu and Yiu is unclear, although a YiuR surface receptor was expressed according to our data. The Hmu transporter acquires heme from blood plasma proteins such as myoglobin, hemoglobin and hemopexin [16]. Three Fe2+ transport systems (EfeUOB, Y2368-Y2370 and FeoAB, Figure 5) were shown to be functional in either Y. pestis [17] or other bacteria [66-68]. We identified the subunits EfeO and Y2368 as periplasmic proteins, and their abundance increases in iron-deficient cells appeared to be moderately temperature-dependent. There is no evidence to date for their regulation by Fur. FeoB was recently identified in Y. pestis membrane proteome surveys [47,65]. A protein highly abundant in membrane fractions of iron-depleted Y. pestis cells but not characterized in the context of iron transport was the orphan TonB-dependent OM receptor Y0850. The protein is a candidate for Fur regulation and the contribution to iron uptake, but its exact function remains to be elucidated. A conserved Fur box upstream of the gene and sequence similarity of Y0850 to Bordetella bronchiseptica BfrA and Campylobacter coli CfrA [69,70] were established. Our proteomic surveys did not support the activation of specific iron uptake pathways at only one of the physiologically relevant temperatures. Based on multivariate transcriptional profiling data for Y. pestis (28°C vs. 37°C, iron-supplemented cell growth vs. iron sequestration in plasma), Carniel et al. [33] suggested that the Ybt system and the TonB protein are of particular importance for iron acquisition at 37°C.
Fe-S cluster biosynthesis and energy metabolism in iron-starved Y. pestis
Growth of iron-depleted Y. pestis cells was arrested at an OD600 of ~0.8, indicative of the inability of iron-dependent enzymes to perform essential metabolic functions. In addition to the already discussed impact of iron depletion on oxidative stress response enzymes and aconitases, we explored how Fe-S cluster assembly systems and other energy metabolism enzymes were affected. Iron-sulfur clusters are critical to the function of many redox enzymes in bacteria [27]. Incorporation of Fe-S into proteins requires Fe-S cluster assembly systems, which were named Suf and Isc in E. coli. Our data showed that SufA, SufB, SufC and SufS, four of the six subunits of the Suf complex, were more abundant under iron starvation conditions. Regulation of the Y. pestis suf operon by Fur and a functional Fur-binding site were reported previously [20]. The cysteine desulfurase subunits of the Suf and Isc systems (SufS and CsdA, respectively) were quantitatively changed in opposite directions (-Fe vs. +Fe), suggesting that Suf functionally replaces Isc at the onset of iron starvation in Y. pestis. Mobilization of sulfur from cysteine appears to be catalyzed by SufS in E. coli [71]. The increased abundance of TauD, an enzyme that mobilizes sulfite from taurine, in iron-depleted Y. pestis cells was intriguing. TauD is a dioxygenase, harbors a Fe2+ cofactor and was reported to be induced under sulfate starvation conditions in E. coli [72]. We speculate that TauD plays an accessory role in sulphur mobilization for Fe-S cluster assembly via the Suf pathway. Furthermore, the Y. pestis ortholog of a recently discovered Fe-S cluster protein ErpA was also increased under iron-limiting conditions. Since ErpA was proposed to transfer Fe-S clusters to apo-enzymes [56], we hypothesize that Y. pestis ErpA may perform such activities cooperatively with the Suf system. Transcriptional data on erpA and tauD expression changes for -Fe vs. +Fe growth conditions are not available. Mammalian hosts starve Y. pestis of iron and, therefore, the Suf complex constitutes a good target for inhibitory drug design.
Enzymes with Fe-S clusters in their catalytic cores, many of them in the TCA cycle, are also displayed in Figure 5. Although in different ratios, subunits of such enzyme complexes (e.g. FumA, SdhA, FrdA and CysJ) were invariably decreased in abundance in iron-starved Y. pestis cells. Most of these quantitative decreases appear to be unrelated to population density differences, because they were not observed in cells cultured to stationary vs. exponential phase in iron-replete PMH2 medium(Pieper, R., unpublished data). A decreased pyruvate metabolism rate should be the consequence of the loss of Fe-S cluster enzyme activities in the TCA cycle and may be followed by reduced production of ATP and NADPH reducing equivalents in the electron transport chain. Furthermore, a decreased turnover of citrate may lead to its accumulation in the cytoplasm, which could chelate iron and exacerbate iron starvation [30]. A highly interesting observation was the dramatic abundance and activity increase of PoxB in iron-starved Y. pestis cells, both at 26°C and 37°C. PoxB activity increases were independent of Y. pestis cell densities during growth in chemically defined media. poxB expression was reported to be moderately enhanced in Y. pestis cells grown in human plasma vs. LB media [33]. We suggest that the metabolism of pyruvate via the PoxB route compensates for reduced activities of Fe-S cluster enzymes in the TCA cycle. The pathway catalyzed by PoxB is iron-independent. The E. coli ortholog, a thiamin/flavin-dependent enzyme activated by binding to IM phospholipids, was shown to feed electrons directly from the cytosol to the respiratory chain [52]. To our knowledge, this is the first report linking enhanced PoxB activities in bacteria specifically to iron starvation. PoxB is a potential drug target in the context of intracellular pathogens surviving in environments where iron is sequestered.
Conclusions
Proteomic surveys of Y. pestis subcellular fractions grown under iron-replete vs. iron-starved conditions supported the physiological importance of the iron acquisition systems Ybt, Yfe, Yfu, Yiu and Hmu. An uncharacterized TonB-dependent OM receptor, Y0850, was also highly abundant in iron-depleted cells, appeared to be Fur-regulated and may participate in iron uptake. Numerous enzymes harboring iron and Fe-S cluster cofactors were significantly decreased in abundance in iron-starved cells, suggesting a regulatory process shifting the metabolism of Y. pestis to iron-independent pathways when the supply of this metal ion is limited. Small Fur-regulated RNAs termed RyhB in E. coli may be involved in this process. Finally, this study revealed biochemical pathways likely essential for the iron starvation response in Y. pestis. Examples are the energy metabolism via the pyruvate oxidase route and Fe-S cluster assembly mediated by the Suf system.
Abbreviations
2D: 2-dimensional gel electrophoresis; CBB: Coomassie Brilliant Blue G250; Fe-S: iron-sulfur; IM: inner membrane; usb-MBR: urea/thiourea/amidosulfobetaine-14-extracted membrane; IPG: immobilized pH gradient; OM: outer membrane; SOD: superoxide dismutase; T3SS: type III secretion system; T6SS: type VI secretion system; TMD: transmembrane domain; VS: spot volume.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
RP: primary role in designing the study, analyzing and interpreting the data, performing the enzyme assays, writing the article; STH: quantitative and bioinformatic data analysis, database queries, generation of Figures and Tables for the article; PPP: sample preparation, 2D gel experiments and proof-reading; DJC: acquisition of the LC-MS/MS data; HA: acquisition of the MALDI-MS data; RDF: generated the framework for the performance of this study; RDP: major role in the design and initial experiments of the study, biological interpretation of the data, writing parts of the article and its review; SNP: major role in the biological data interpretation and the review of the article.
Supplementary Material
Yersinia pestis growth curves in PMH2 medium. Growth curves (OD600) are displayed in graphical form for Y. pestis KIM6+ cell cultures in iron rich and iron-depleted media, at 26°C and at 37°C.
Comprehensive list of differentially displayed Yersinia pestis proteins comparing iron-replete and iron starvation conditions. A variety of qualitative and quantitative data are provided for differentially displayed proteins derived from + Fe vs. -Fe growth conditions, from cell cultures at 26°C and at 37°C.
Comprehensive list of MS and MS2 data for Y. pestis KIM6+ proteins. For all proteins listed in the Tables 1, 2 and 3 and in the Additional File 2, MS and MS2 data were parsed from MALDI-TOFTOF and LC-nESI-LC-MS/MS datasets.
Contributor Information
Rembert Pieper, Email: rpieper@jcvi.org.
Shih-Ting Huang, Email: shuang@jcvi.org.
Prashanth P Parmar, Email: pparmar@jcvi.org.
David J Clark, Email: dclark@jcvi.org.
Hamid Alami, Email: talami@jcvi.org.
Robert D Fleischmann, Email: rdfleisc@jcvi.org.
Robert D Perry, Email: rperry@pop.uky.edu.
Scott N Peterson, Email: scottp@jcvi.org.
Acknowledgements
This work was performed under the Pathogen Functional Genomics Resource Center contract (contract No. N01-AI15447), funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health. We thank Jasmine Pollard for the graphic presented in Figure 4, Christine Bunai for the development of the mass spectrometry analysis platform and John Braisted for advice on statistical data analysis methods.
References
- Brubaker RR, Sussman M. Molecular Medical Microbiology. Vol. 3. London, UK: Academic Press; 2002. Yersinia pestis; pp. 2033–2058. full_text. [Google Scholar]
- Deng W, Burland V, Plunkett G, Boutin A, Mayhew GF, Liss P, Perna NT, Rose DJ, Mau B, Zhou S. Genome sequence of Yersinia pestis KIM. J Bacteriol. 2002;184(16):4601–4611. doi: 10.1128/JB.184.16.4601-4611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Elliott J, McCready P, Skowronski E, Garnes J, Kobayashi A, Brubaker RR, Garcia E. Structural organization of virulence-associated plasmids of Yersinia pestis. J Bacteriol. 1998;180(19):5192–5202. doi: 10.1128/jb.180.19.5192-5202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindler LE, Plano GV, Burland V, Mayhew GF, Blattner FR. Complete DNA sequence and detailed analysis of the Yersinia pestis KIM5 plasmid encoding murine toxin and capsular antigen. Infect Immun. 1998;66(12):5731–5742. doi: 10.1128/iai.66.12.5731-5742.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch BJ. In: Yersinia Molecular and Cellular Biology. Carniel EaH BJ, editor. Norfolk, U.K.: Horizon Bioscience; 2004. The evolution of flea-borne transmission of Yersinia pestis; pp. 49–73. [Google Scholar]
- Perry RD, Fetherston JD. Yersinia pestis--etiologic agent of plague. Clin Microbiol Rev. 1997;10(1):35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett CO, Deak E, Isherwood KE, Oyston PC, Fischer ER, Whitney AR, Kobayashi SD, DeLeo FR, Hinnebusch BJ. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J Infect Dis. 2004;190(4):783–792. doi: 10.1086/422695. [DOI] [PubMed] [Google Scholar]
- Perry RD, Bobrov AG, Kirillina O, Jones HA, Pedersen L, Abney J, Fetherston JD. Temperature regulation of the hemin storage (Hms+) phenotype of Yersinia pestis is posttranscriptional. J Bacteriol. 2004;186(6):1638–1647. doi: 10.1128/JB.186.6.1638-1647.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaible UE, Kaufmann SH. Iron and microbial infection. Nat Rev Microbiol. 2004;2(12):946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- Bearden SW, Fetherston JD, Perry RD. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect Immun. 1997;65(5):1659–1668. doi: 10.1128/iai.65.5.1659-1668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetherston JD, Bertolino VJ, Perry RD. YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol Microbiol. 1999;32(2):289–299. doi: 10.1046/j.1365-2958.1999.01348.x. [DOI] [PubMed] [Google Scholar]
- Fetherston JD, Lillard JW Jr, Perry RD. Analysis of the pesticin receptor from Yersinia pestis: role in iron-deficient growth and possible regulation by its siderophore. J Bacteriol. 1995;177(7):1824–1833. doi: 10.1128/jb.177.7.1824-1833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearden SW, Perry RD. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol Microbiol. 1999;32(2):403–414. doi: 10.1046/j.1365-2958.1999.01360.x. [DOI] [PubMed] [Google Scholar]
- Gong S, Bearden SW, Geoffroy VA, Fetherston JD, Perry RD. Characterization of the Yersinia pestis Yfu ABC inorganic iron transport system. Infect Immun. 2001;69(5):2829–2837. doi: 10.1128/IAI.67.5.2829-2837.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirillina O, Bobrov AG, Fetherston JD, Perry RD. Hierarchy of iron uptake systems: Yfu and Yiu are functional in Yersinia pestis. Infect Immun. 2006;74(11):6171–6178. doi: 10.1128/IAI.00874-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JM, Jones HA, Perry RD. Molecular characterization of the hemin uptake locus (hmu) from Yersinia pestis and analysis of hmu mutants for hemin and hemoprotein utilization. Infect Immun. 1999;67(8):3879–3892. doi: 10.1128/iai.67.8.3879-3892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RD, Mier I Jr, Fetherston JD. Roles of the Yfe and Feo transporters of Yersinia pestis in iron uptake and intracellular growth. Biometals. 2007;20(3-4):699–703. doi: 10.1007/s10534-006-9051-x. [DOI] [PubMed] [Google Scholar]
- Perry RD, Fetherston JD. In: Yersinia Molecular and Cellular Biology. Carniel EaH BJ, editor. Norfolk, U.K.: Horizon Bioscience; 2004. Iron and Heme Uptake Systems; pp. 257–283. [Google Scholar]
- Hantke K. Iron and metal regulation in bacteria. Curr Opin Microbiol. 2001;4(2):172–177. doi: 10.1016/S1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- Gao H, Zhou D, Li Y, Guo Z, Han Y, Song Y, Zhai J, Du Z, Wang X, Lu J. The iron-responsive Fur regulon in Yersinia pestis. J Bacteriol. 2008;190(8):3063–3075. doi: 10.1128/JB.01910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V, Perez-Martín J, Escolar L, Pesole G, Bertoni G. Mode of binding of the Fur protein to target DNA: negative regulation of iron-controlled gene expression. Washington D.C.: ASM Press; 2004. [Google Scholar]
- Gottesman S, McCullen CA, Guillier M, Vanderpool CK, Majdalani N, Benhammou J, Thompson KM, FitzGerald PC, Sowa NA, FitzGerald DJ. Small RNA regulators and the bacterial response to stress. Cold Spring Harb Symp Quant Biol. 2006;71:1–11. doi: 10.1101/sqb.2006.71.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2002;99(7):4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci USA. 2004;101(26):9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadner RJ. Regulation by iron: RNA rules the rust. J Bacteriol. 2005;187(20):6870–6873. doi: 10.1128/JB.187.20.6870-6873.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Salvail H, Desnoyers G, Arguin M. Small RNAs controlling iron metabolism. Curr Opin Microbiol. 2007;10(2):140–145. doi: 10.1016/j.mib.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Kiley PJ, Beinert H. The role of Fe-S proteins in sensing and regulation in bacteria. Curr Opin Microbiol. 2003;6(2):181–185. doi: 10.1016/S1369-5274(03)00039-0. [DOI] [PubMed] [Google Scholar]
- Cheng VW, Ma E, Zhao Z, Rothery RA, Weiner JH. The iron-sulfur clusters in Escherichia coli succinate dehydrogenase direct electron flow. J Biol Chem. 2006;281(37):27662–27668. doi: 10.1074/jbc.M604900200. [DOI] [PubMed] [Google Scholar]
- Flint DH, Emptage MH, Guest JR. Fumarase a from Escherichia coli: purification and characterization as an iron-sulfur cluster containing enzyme. Biochemistry. 1992;31(42):10331–10337. doi: 10.1021/bi00157a022. [DOI] [PubMed] [Google Scholar]
- Varghese S, Tang Y, Imlay JA. Contrasting sensitivities of Escherichia coli aconitases A and B to oxidation and iron depletion. J Bacteriol. 2003;185(1):221–230. doi: 10.1128/JB.185.1.221-230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Gosset G, Barabote R, Gonzalez CS, Cuevas WA, Saier MH Jr. Functional interactions between the carbon and iron utilization regulators, Crp and Fur, in Escherichia coli. J Bacteriol. 2005;187(3):980–990. doi: 10.1128/JB.187.3.980-990.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese S, Wu A, Park S, Imlay KR, Imlay JA. Submicromolar hydrogen peroxide disrupts the ability of Fur protein to control free-iron levels in Escherichia coli. Mol Microbiol. 2007;64(3):822–830. doi: 10.1111/j.1365-2958.2007.05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvaux S, Rosso ML, Frangeul L, Lacroix C, Labarre L, Schiavo A, Marceau M, Dillies MA, Foulon J, Coppee JY. Transcriptome analysis of Yersinia pestis in human plasma: an approach for discovering bacterial genes involved in septicaemic plague. Microbiology. 2007;153(Pt 9):3112–3124. doi: 10.1099/mic.0.2007/006213-0. [DOI] [PubMed] [Google Scholar]
- Han Y, Qiu J, Guo Z, Gao H, Song Y, Zhou D, Yang R. Comparative transcriptomics in Yersinia pestis: a global view of environmental modulation of gene expression. BMC Microbiol. 2007;7:96. doi: 10.1186/1471-2180-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Qin L, Han Y, Qiu J, Chen Z, Li B, Song Y, Wang J, Guo Z, Zhai J. Global analysis of iron assimilation and fur regulation in Yersinia pestis. FEMS Microbiol Lett. 2006;258(1):9–17. doi: 10.1111/j.1574-6968.2006.00208.x. [DOI] [PubMed] [Google Scholar]
- Fetherston JD, Perry RD. The pigmentation locus of Yersinia pestis KIM6+ is flanked by an insertion sequence and includes the structural genes for pesticin sensitivity and HMWP2. Mol Microbiol. 1994;13(4):697–708. doi: 10.1111/j.1365-2958.1994.tb00463.x. [DOI] [PubMed] [Google Scholar]
- Lillard JW Jr, Bearden SW, Fetherston JD, Perry RD. The haemin storage (Hms+) phenotype of Yersinia pestis is not essential for the pathogenesis of bubonic plague in mammals. Microbiology. 1999;145(Pt 1):197–209. doi: 10.1099/13500872-145-1-197. [DOI] [PubMed] [Google Scholar]
- Lucier TS, Fetherston JD, Brubaker RR, Perry RD. Iron uptake and iron-repressible polypeptides in Yersinia pestis. Infect Immun. 1996;64(8):3023–3031. doi: 10.1128/iai.64.8.3023-3031.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper R, Huang ST, Clark DJ, Robinson JM, Parmar PP, Alami H, Bunai CL, Perry RD, Fleischmann RD, Peterson SN. Characterizing the dynamic nature of the Yersinia pestis periplasmic proteome in response to nutrient exhaustion and temperature change. Proteomics. 2008;8(7):1442–1458. doi: 10.1002/pmic.200700923. [DOI] [PubMed] [Google Scholar]
- Chang YY, Cronan JE Jr. Mapping nonselectable genes of Escherichia coli by using transposon Tn10: location of a gene affecting pyruvate oxidase. J Bacteriol. 1982;151(3):1279–1289. doi: 10.1128/jb.151.3.1279-1289.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose IA, O'Connell EL. Mechanism of aconitase action. I. The hydrogen transfer reaction. J Biol Chem. 1967;242(8):1870–1879. [PubMed] [Google Scholar]
- Johansson LH, Borg LA. A spectrophotometric method for determination of catalase activity in small tissue samples. Anal Biochem. 1988;174(1):331–336. doi: 10.1016/0003-2697(88)90554-4. [DOI] [PubMed] [Google Scholar]
- Peskin AV, Winterbourn CC. A microtiter plate assay for superoxide dismutase using a water-soluble tetrazolium salt (WST-1) Clin Chim Acta. 2000;293(1-2):157–166. doi: 10.1016/S0009-8981(99)00246-6. [DOI] [PubMed] [Google Scholar]
- Pieper R, Huang ST, Robinson JM, Clark DJ, Alami H, Parmar PP, Perry RD, Fleischmann RD, Peterson SN. Temperature and growth phase influence the outer-membrane proteome and the expression of a type VI secretion system in Yersinia pestis. Microbiology. 2009;155(Pt 2):498–512. doi: 10.1099/mic.0.022160-0. [DOI] [PubMed] [Google Scholar]
- Gatlin CL, Pieper R, Huang ST, Mongodin E, Gebregeorgis E, Parmar PP, Clark DJ, Alami H, Papazisi L, Fleischmann RD. Proteomic profiling of cell envelope-associated proteins from Staphylococcus aureus. Proteomics. 2006;6(5):1530–1549. doi: 10.1002/pmic.200500253. [DOI] [PubMed] [Google Scholar]
- Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. PRED-TMBB: a web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Res. 2004. pp. W400–404. [DOI] [PMC free article] [PubMed]
- Pieper R, Huang ST, Clark DJ, Robinson JM, Alami H, Parmar PP, Suh MJ, Kuntumalla S, Bunai CL, Perry RD. Integral and peripheral association of proteins and protein complexes with Yersinia pestis inner and outer membranes. Proteome Sci. 2009;7:5. doi: 10.1186/1477-5956-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh M-J, Alami H, Clark DJ, Parmar PP, Robinson JM, Huang S-T, Fleischmann RD, Peterson SN, Pieper R. Widespread Occurrence of Non-Enzymatic Deamidations of Asparagine Residues in Yersinia pestis Proteins Resulting from Alkaline pH Membrane Extraction Conditions. Open Proteomics J. 2008;1:106–115. doi: 10.2174/1875039700801010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RD, Abney J, Mier I Jr, Lee Y, Bearden SW, Fetherston JD. Regulation of the Yersinia pestis Yfe and Ybt iron transport systems. Adv Exp Med Biol. 2003;529:275–283. doi: 10.1007/0-306-48416-1_53. full_text. [DOI] [PubMed] [Google Scholar]
- Staggs TM, Perry RD. Fur regulation in Yersinia species. Mol Microbiol. 1992;6(17):2507–2516. doi: 10.1111/j.1365-2958.1992.tb01427.x. [DOI] [PubMed] [Google Scholar]
- van Helden J. Regulatory sequence analysis tools. Nucleic Acids Res. 2003;31(13):3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P, Weidner A, Pech A, Stubbs MT, Tittmann K. Structural basis for membrane binding and catalytic activation of the peripheral membrane enzyme pyruvate oxidase from Escherichia coli. Proc Natl Acad Sci USA. 2008;105(45):17390–17395. doi: 10.1073/pnas.0805027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belevich G, Euro L, Wikstrom M, Verkhovskaya M. Role of the conserved arginine 274 and histidine 224 and 228 residues in the NuoCD subunit of complex I from Escherichia coli. Biochemistry. 2007;46(2):526–533. doi: 10.1021/bi062062t. [DOI] [PubMed] [Google Scholar]
- Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- Outten FW, Djaman O, Storz G. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol Microbiol. 2004;52(3):861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- Loiseau L, Gerez C, Bekker M, Ollagnier-de Choudens S, Py B, Sanakis Y, Teixeira de Mattos J, Fontecave M, Barras F. ErpA, an iron sulfur (Fe S) protein of the A-type essential for respiratory metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2007;104(34):13626–13631. doi: 10.1073/pnas.0705829104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. Making 'sense' of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol. 2005;3(5):383–396. doi: 10.1038/nrmicro1146. [DOI] [PubMed] [Google Scholar]
- Liang H, Li L, Dong Z, Surette MG, Duan K. The YebC family protein PA0964 negatively regulates the Pseudomonas aeruginosa quinolone signal system and pyocyanin production. J Bacteriol. 2008;190(18):6217–6227. doi: 10.1128/JB.00428-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrov AG, Bearden SW, Fetherston JD, Khweek AA, Parrish KD, Perry RD. Functional quorum sensing systems affect biofilm formation and protein expression in Yersinia pestis. Adv Exp Med Biol. 2007;603:178–191. doi: 10.1007/978-0-387-72124-8_15. full_text. [DOI] [PubMed] [Google Scholar]
- Cairo G, Pietrangelo A. Iron regulatory proteins in pathobiology. Biochem J. 2000;352(Pt 2):241–250. doi: 10.1042/0264-6021:3520241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Guest JR. Direct evidence for mRNA binding and post-transcriptional regulation by Escherichia coli aconitases. Microbiology. 1999;145(Pt 11):3069–3079. doi: 10.1099/00221287-145-11-3069. [DOI] [PubMed] [Google Scholar]
- Tang Y, Quail MA, Artymiuk PJ, Guest JR, Green J. Escherichia coli aconitases and oxidative stress: post-transcriptional regulation of sodA expression. Microbiology. 2002;148(Pt 4):1027–1037. doi: 10.1099/00221287-148-4-1027. [DOI] [PubMed] [Google Scholar]
- Touati D. Iron and oxidative stress in bacteria. Arch Biochem Biophys. 2000;373(1):1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- Zhou D, Han Y, Yang R. Molecular and physiological insights into plague transmission, virulence and etiology. Microbes Infect. 2006;8(1):273–284. doi: 10.1016/j.micinf.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Hixson KK, Adkins JN, Baker SE, Moore RJ, Chromy BA, Smith RD, McCutchen-Maloney SL, Lipton MS. Biomarker candidate identification in Yersinia pestis using organism-wide semiquantitative proteomics. J Proteome Res. 2006;5(11):3008–3017. doi: 10.1021/pr060179y. [DOI] [PubMed] [Google Scholar]
- Cao J, Woodhall MR, Alvarez J, Cartron ML, Andrews SC. EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E. coli O157:H7. Mol Microbiol. 2007;65(4):857–875. doi: 10.1111/j.1365-2958.2007.05802.x. [DOI] [PubMed] [Google Scholar]
- Dubbels BL, DiSpirito AA, Morton JD, Semrau JD, Neto JN, Bazylinski DA. Evidence for a copper-dependent iron transport system in the marine, magnetotactic bacterium strain MV-1. Microbiology. 2004;150(Pt 9):2931–2945. doi: 10.1099/mic.0.27233-0. [DOI] [PubMed] [Google Scholar]
- Grosse C, Scherer J, Koch D, Otto M, Taudte N, Grass G. A new ferrous iron-uptake transporter, EfeU (YcdN), from Escherichia coli. Mol Microbiol. 2006;62(1):120–131. doi: 10.1111/j.1365-2958.2006.05326.x. [DOI] [PubMed] [Google Scholar]
- Beall B, Hoenes T. An iron-regulated outer-membrane protein specific to Bordetella bronchiseptica and homologous to ferric siderophore receptors. Microbiology. 1997;143(Pt 1):135–145. doi: 10.1099/00221287-143-1-135. [DOI] [PubMed] [Google Scholar]
- Guerry P, Perez-Casal J, Yao R, McVeigh A, Trust TJ. A genetic locus involved in iron utilization unique to some Campylobacter strains. J Bacteriol. 1997;179(12):3997–4002. doi: 10.1128/jb.179.12.3997-4002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer G, Gaddam SA, Ayala-Castro CN, Ollagnier-de Choudens S, Lascoux D, Fontecave M, Outten FW. SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly. J Biol Chem. 2007;282(18):13342–13350. doi: 10.1074/jbc.M608555200. [DOI] [PubMed] [Google Scholar]
- Ploeg JR van der, Weiss MA, Saller E, Nashimoto H, Saito N, Kertesz MA, Leisinger T. Identification of sulfate starvation-regulated genes in Escherichia coli: a gene cluster involved in the utilization of taurine as a sulfur source. J Bacteriol. 1996;178(18):5438–5446. doi: 10.1128/jb.178.18.5438-5446.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oglesby AG, Farrow JM, Lee JH, Tomaras AP, Greenberg EP, Pesci EC, Vasil ML. The influence of iron on Pseudomonas aeruginosa physiology: a regulatory link between iron and quorum sensing. J Biol Chem. 2008;283(23):15558–15567. doi: 10.1074/jbc.M707840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Coulthurst SJ, Pritchard L, Hedley PE, Ravensdale M, Humphris S, Burr T, Takle G, Brurberg MB, Birch PR. Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog. 2008;4(6):e1000093. doi: 10.1371/journal.ppat.1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Yersinia pestis growth curves in PMH2 medium. Growth curves (OD600) are displayed in graphical form for Y. pestis KIM6+ cell cultures in iron rich and iron-depleted media, at 26°C and at 37°C.
Comprehensive list of differentially displayed Yersinia pestis proteins comparing iron-replete and iron starvation conditions. A variety of qualitative and quantitative data are provided for differentially displayed proteins derived from + Fe vs. -Fe growth conditions, from cell cultures at 26°C and at 37°C.
Comprehensive list of MS and MS2 data for Y. pestis KIM6+ proteins. For all proteins listed in the Tables 1, 2 and 3 and in the Additional File 2, MS and MS2 data were parsed from MALDI-TOFTOF and LC-nESI-LC-MS/MS datasets.