Abstract
With the dramatic increase in the volume of experimental results in every domain of life sciences, assembling pertinent data and combining information from different fields has become a challenge. Information is dispersed over numerous specialized databases and is presented in many different formats. Rapid access to experiment-based information about well-characterized proteins helps predict the function of uncharacterized proteins identified by large-scale sequencing. In this context, universal knowledgebases play essential roles in providing access to data from complementary types of experiments and serving as hubs with cross-references to many specialized databases. This review outlines how the value of experimental data is optimized by combining high-quality protein sequences with complementary experimental results, including information derived from protein 3D-structures, using as an example the UniProt knowledgebase (UniProtKB) and the tools and links provided on its website (http://www.uniprot.org/). It also evokes precautions that are necessary for successful predictions and extrapolations.
Keywords: Data flood, Annotation, Swiss-Prot, Knowledgebase, UniProtKB, Proteomics, Structural genomics, Protein 3D-structure
Introduction
Combining vast amounts of data with the aim of understanding the complexity of living beings, e.g., in the context of systems biology, is a central issue of modern biology. The technological progress of the last few years has led to a literal explosion in the quantity of available data in life sciences, starting with the number of nucleotide and protein sequences, but also data from proteomic and transcriptomic studies. Likewise, the number of protein 3D-structures that are deposited at wwPDB (http://www.wwpdb.org/) and integrated via its sites at RCSB PDB, PDBe (formerly MSD), PDBj, and BMRB [1–4] has increased dramatically, and ever more protein structures are being solved. Complementary central databases and knowledge repositories, such as the protein structure initiative structural genomics knowledgebase (PSI-SGKB) [5] and the UniProt Knowledgebase (UniProtKB) [6, 7], play essential roles in simplifying access to information about proteins and protein structures, and in combining results from experiments with functional annotation.
Much of the recent data are from large-scale studies, and most new nucleotide sequences code for otherwise uncharacterized proteins from a wide range of species, from mammals to microbes, virus isolates, and environmental samples. For the correct prediction of the function of individual proteins and for the automated annotation of entire genome sequences, one needs central knowledge resources that provide information about characterized proteins. For successful predictions, it is essential to use a maximum of validated experimental findings from complementary experiments, and to take account of the sources of the information. The Universal Protein Resource KnowledgeBase (UniProtKB) (http://www.uniprot.org/) provides the scientific community with one such resource. It gives rapid access to high-quality, reliable information, has excellent search tools for the retrieval of specific sets of proteins, and puts emphasis on information that is directly derived from experimental evidence. At the same time, it serves as a hub providing links to other databases, allowing access to information and data which are stored in many different formats (Fig. 1). This facilitates the interpretation of novel experimental results and provides a solid basis for predictions and for planning new experiments. Small datasets can be directly downloaded from the UniProtKB web site by following the download link on any search result page. For downloading complete datasets, it is recommended to use the UniProt FTP site (ftp://ftp.uniprot.org/). UniProtKB values feedback from the scientific community, with each entry displaying the appropriate external links.
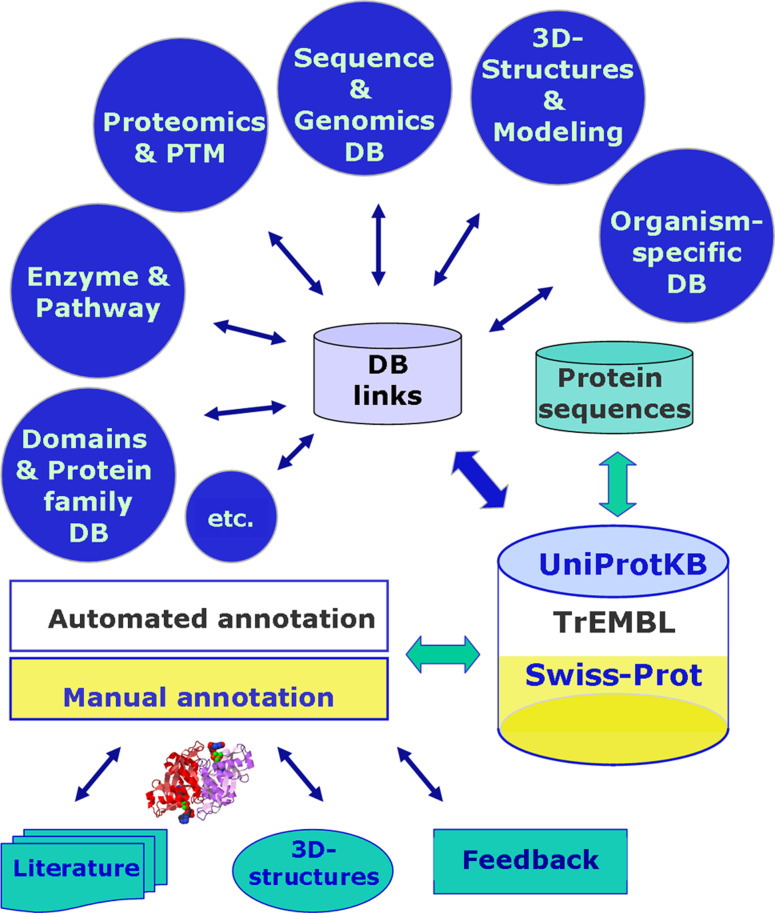
Fig. 1.
UniProtKB serves as a knowledge repository and as a central hub that provides links to numerous other databases. New protein sequences are integrated in UniProtKB/TrEMBL and annotated by an automated procedure. UniProtKB/Swiss-Prot entries are manually annotated, combining carefully checked protein sequences with information from the scientific literature, protein 3D-structures, and specialised databases, together with feedback from the scientific community
UniProtKB contains two mutually exclusive, non-redundant sections that together give access to all the protein sequences which are available to the public. However, UniProtKB excludes protein sequences for most non-germline immunoglobulins and T-cell receptors, patent application sequences, synthetic sequences, short fragments, pseudogenes, and fusion proteins. More than 99% of the protein sequences provided by UniProtKB come from the translations of coding sequences (CDS) submitted to the EMBL-Bank/GenBank/DDBJ nucleotide sequence resources. New protein sequences are integrated in UniProtKB/TrEMBL, together with information provided by the submitting authors concerning the species and the protein and/or gene name. Highly automated tools are used for further annotation. Proteins are classified using protein signatures, and assigned to families and domains. The major protein signature databases are available through the InterPro database [8, 9], the main tool for characterizing and classifying UniProtKB sequences. Depending on the entry, further information may be added by automated annotation, using automated and manually curated annotation rules from the UniProt RuleBase. Thus, while users have access to high-quality automated annotation and cross-references to numerous databases, including PDB, annotation is mostly restricted to the description of sequence-based similarity. In the same vein, the protein name is often derived from a clone identifier, and further efforts are required to establish the identity of the protein. When sequences differ from existing sequences, UniProtKB/TrEMBL creates separate entries for the gene products from a given organism. For popular or highly expressed genes, a huge number of slightly different sequences exists for the products of each gene, e.g., due to polymorphisms or alternative splicing events. This gives rise to a large number of individual UniProtKB/TrEMBL entries, making it difficult to keep track of the differences and identify the most relevant sequence.
In contrast, UniProtKB/Swiss-Prot contains manually annotated protein sequences, where annotators add information gathered from scientific publications and from protein 3D-structures, and check the output from bioinformatics tools. When several different protein sequences are available for the products of one gene from a given species, the sequences are carefully analyzed, and a master sequence is selected by annotators. Other sequences are then merged, and differences are carefully documented, with the result that one UniProtKB/Swiss-Prot entry represents the products of one gene for a given species. UniProtKB/Swiss-Prot puts emphasis on showing experimental evidence, and displays in-depth information. Within this context, protein 3D-structures are highly valuable sources of information; they give detailed information on interactions with other macromolecules or with small ligands, and contribute to elucidating enzyme mechanisms. Likewise, they can provide a basis for understanding the molecular causes of disease, elucidate the interactions between pathogens and their hosts, and help with the targeted design of new drugs and inhibitors. 3D-structures can reveal the details of post-translational modifications, such as disulfide bonds, or the covalent attachment of cofactors, sugars, or lipids. Protein 3D-structures help to classify proteins, assign proteins with low sequence similarity to known families, or identify new folds. The challenge is then to combine knowledge derived from protein structures with high-quality information about the protein sequence and its variants, complement it with results from other types of experiments, such as site-directed mutagenesis and biochemical analyses, and make the cumulated information accessible to the scientific community. This is the goal of UniProtKB/Swiss-Prot, a manually annotated knowledge resource that facilitates access to data from multiple sources, and brings together results from protein 3D-structures and biochemical and genetic analyses, and provides cross-references to numerous other databases. Information gathered for well-characterized proteins is used for propagation to uncharacterized family members, applying stringent rules. This is accomplished by highly trained annotators; indeed, this type of work takes expert knowledge and constant vigilance.
Focus on model organisms and pathogens
UniProtKB/Swiss-Prot prioritizes annotation of proteins from model organisms and from important pathogens, with particular emphasis on proteins with known 3D-structure. Since autumn 2008, UniProtKB/Swiss-Prot entries are available for all 20,330 human protein-coding genes; keeping pace with the information flood and continuing to add all the relevant information to the entries is now the next major challenge [10]. Another major priority is the annotation of important human pathogens from all branches of life, with particular emphasis on bacteria, such as Mycobacterium tuberculosis and Staphylococcus aureus, and on viruses. A dedicated web portal, ViralZone (http://www.expasy.org/viralzone/), simplifies access to information about viruses and viral proteins, and the associated 3D-structures. For species where the entire proteome has been annotated in UniProtKB, the complete set of entries can be retrieved using the keyword “Complete proteome”, e.g., for Saccharomyces cerevisiae and Schizosaccharomyces pombe, and for numerous bacteria and archaea, such as Escherichia coli, Mycoplasma pneumoniae and Methanococcus jannaschii.
UniProtKB/Swiss-Prot release 57.11 (November 2009) contains 512,994 entries, and out of these, 15,223 contain a database link to PDB. This corresponds to 48,904 PDB entries—the numerical discrepancy arises from the fact that often several structures are determined for a single protein. In addition, a single PDB entry may contain several distinct protein chains corresponding to several UniProt entries. Reciprocal cross-references between UniProtKB, PDB, and PDBSum [11] facilitate access to experimental data and further tools. In UniProtKB, adding information from protein 3D-structures has high priority, meaning that the majority of proteins with experimental 3D-structures are in the manually annotated UniProtKB/Swiss-Prot section. The keyword “3D-structure” can be used to retrieve all the corresponding UniProtKB/Swiss-Prot entries, and in combination with other search terms permits to recover the entries of interest.
Even when the structure of a protein has not been determined, 3D-structures may be available for related proteins, serving as templates for homology-based modeling. Based on the assumption that high-quality models can be obtained for proteins with at least 30% sequence identity, homology-based modeling is possible for about 50% of all proteins in UniProtKB/Swiss-Prot [12, 13]. Access to automatically generated 3D-models is furnished by database cross-references to the Swiss-Model Repository (SMR) [13] and ModBase [14], while links to HSSP [15] help to find suitable templates. Obviously, the quality of a model depends not only on protein similarity, but also on human input, and thus in UniProtKB/Swiss-Prot, a threshold of 40% sequence identity is set for adding links to automatically generated models from SMR.
Protein nomenclature: problems and solutions
Human beings are highly sociable and thrive on the exchange of news and ideas. They are also adverse to imposed rules and nomenclature systems, and yet, the use of standardized vocabularies and nomenclatures greatly facilitates sharing ideas and finding pertinent information. Thus, organism-specific nomenclature systems for genes and proteins have been created. For humans and vertebrates, recommended gene names are based on the work of the HUGO gene nomenclature committee that assigns unique, standardized, and user-friendly gene symbols to human genes [16]. For microbes, the use of standard gene names and ordered locus tags is common practice, but this is unfortunately not the case for mammalian genes and proteins, in spite of long-standing appeals for using standardized nomenclature [17, 18]. Many authors prefer to coin their own names for the proteins that they are working on, even when an official gene name already exists, with the consequence that it is difficult to find all the information related to a given protein. Worse, authors sometimes name their favorite protein using a term that is already used to design another protein or gene, or they choose names that make it extremely hard to find relevant information, e.g., the gene name “Light” used as synonym for mouse Tnfsf14 (Q9QYH9). This goes without mentioning all the proteins that are known as “p35”, e.g., annexin A1 (P07150), sororin (Q96FF9), cyclin-dependent kinase 5 activator 1 (P61809), etc. Even people working in the field may miss information about their favorite protein when it is published using an alternative name. In an era where vast amounts of data are generated, people need rapid and accurate information on proteins they are not familiar with, and the use of non-standard nomenclature makes it very difficult to find all the relevant information.
To alleviate these problems and provide access to essential information, UniProtKB/Swiss-Prot lists not only the recommended names for genes and proteins, but also the synonyms that are found in the literature. It is also possible to retrieve protein entries using PDB identifiers or sequence identifiers, such as AY037155 (for the nucleotide sequence), or AAK67645 (for the protein sequence). Indeed, more and more journals insist that authors cite a sequence identifier from a public database, such as UniProtKB, EMBL-Bank [19], or GenBank [20], to indicate unambiguously the protein and organism they used for their experiments.
Finding relevant protein sequence information in a sea of data
Currently, most protein sequences are deduced from the nucleotide sequence of the corresponding gene or cDNA, and proteins are often engineered in order to investigate a particular phenomenon or to determine the 3D-structure of an enzyme with bound substrate. Since the advent of recombinant DNA technology, it is rare to study a protein, or determine a 3D-structure, when the corresponding gene has not been cloned. Exceptions exist, but these are mostly directly-sequenced small proteins, e.g., snake venom neurotoxin P59276. Reliable sequence information is an essential basis for a large part of modern life sciences, and access to high-quality protein sequence data is taken for granted.
For eukaryotes, a single gene often gives rise to many protein sequences, due to alternative splicing, alternative initiation, or alternative promoter usage. Additional complexity is created by polymorphisms and disease mutations. Likewise, many similar sequences are submitted for popular microbial proteins. As a consequence, for a single gene, many different sequences and many sequence database entries may co-exist, making it difficult to keep an overview and determine what is the most relevant sequence. Different strategies exist to deal with this problem; for example, RefSeq [21] reduces this complexity by displaying carefully chosen reference sequences. For alternatively spliced genes, RefSeq provides separate entries for individual isoforms. UniProtKB/Swiss-Prot employs an alternative strategy and groups the protein sequences derived from a single gene from a given organism into a single entry. Differences are clearly documented, indicating whether these are due to alternative splicing events, polymorphisms, or possible sequencing errors. From one UniProtKB/Swiss-Prot entry, it is possible to create all the splice variant sequences, so that these can be analyzed independently. This helps in interpreting BLAST searches, and in keeping the overview in a sea of sequence data. Grouping all the submitted sequence data, all the commonly used names, and all the corresponding cross-references to the PDB archive in a single entry facilitates the rapid retrieval of complementary information associated with a protein.
For human, several groups and consortia endeavor to find the most representative sequence for each protein-coding gene, based on the genome sequence and information about prevalent polymorphisms and isoforms, e.g., RefSeq and CCDS [22]. This also corresponds to the goals of UniProtKB/Swiss-Prot regarding the protein world, and constant efforts are being made to show both the most relevant sequence and its variants. Database cross-references are provided to resources dealing with sequences and gene models, such as Ensembl [23] and RefSeq, and sequences are constantly being reviewed in collaboration with CCDS and Refseq. Information about polymorphisms and human disease mutations is added from scientific publications, and by integrating validated polymorphisms from dbSNP [24]. For human and mouse, the average entry contains, respectively, 6.8 and 4.3 cross-references to EMBL, thereby ensuring the reliability of the shown sequence, and providing information about sequence variation due to alternative splicing events or polymorphisms. Further information about human proteins and defects that are linked to genetic diseases is found in the Online Mendelian Inheritance in Man (OMIM) database [25].
Structured annotation helps find relevant information
When confronted with an unfamiliar protein, it is important to have rapid access to a maximum of reliable information about its function, the pathways it is involved in, known interaction partners, its subcellular location, etc. This includes information about the role of individual residues, e.g., in binding a specific ligand or as active site residues in catalysis. Another important angle is to identify proteins that are still uncharacterized as targets for future research. Finding information takes time, due to the fact that information is dispersed in the scientific literature and in many specialized databases. As a universal protein knowledgebase, UniProtKB/Swiss-Prot combines carefully checked protein sequences with detailed information from multiple sources, including protein 3D-structures, and presents this in a highly structured and user-friendly manner (Fig. 2). The “general annotation” section indicates the function of a protein, its subunit structure, subcellular location, possible involvement in human disease, and other such general information. Information about the roles of individual residues is found in the “sequence annotation” section, where the use of dedicated “feature keys” simplifies finding specific pieces of information, e.g., about residues that bind metal ions or that are involved in catalysis. Dedicated “feature keys” are also used to indicate the extents of cleavable signal or targeting sequences of propeptides, of the mature protein chains, or of particular domains or repeats, as illustrated by human prostate-specific antigen KLK3 (P07288). Dedicated keywords, such as “Signal”, “Secreted”, “Serine protease”, and “Zymogen” facilitate rapid classification and the retrieval of a set of similar proteins. Experimental qualifiers are added when specific information is propagated from a related entry or is derived from a prediction. Thus, when the N-terminus of a mature protein has been determined, as for KLK3 (P07288), no special comment is added. “By similarity” means that there is experimental evidence for a closely related protein, while “Potential” indicates data derived from the use of bioinformatics tools, e.g., for the prediction of cleavable signal sequences or transmembrane segments, where experimental evidence is not always available. This distinction is important; analysis of experimentally determined signal sequences makes it possible to constantly refine and improve prediction tools.
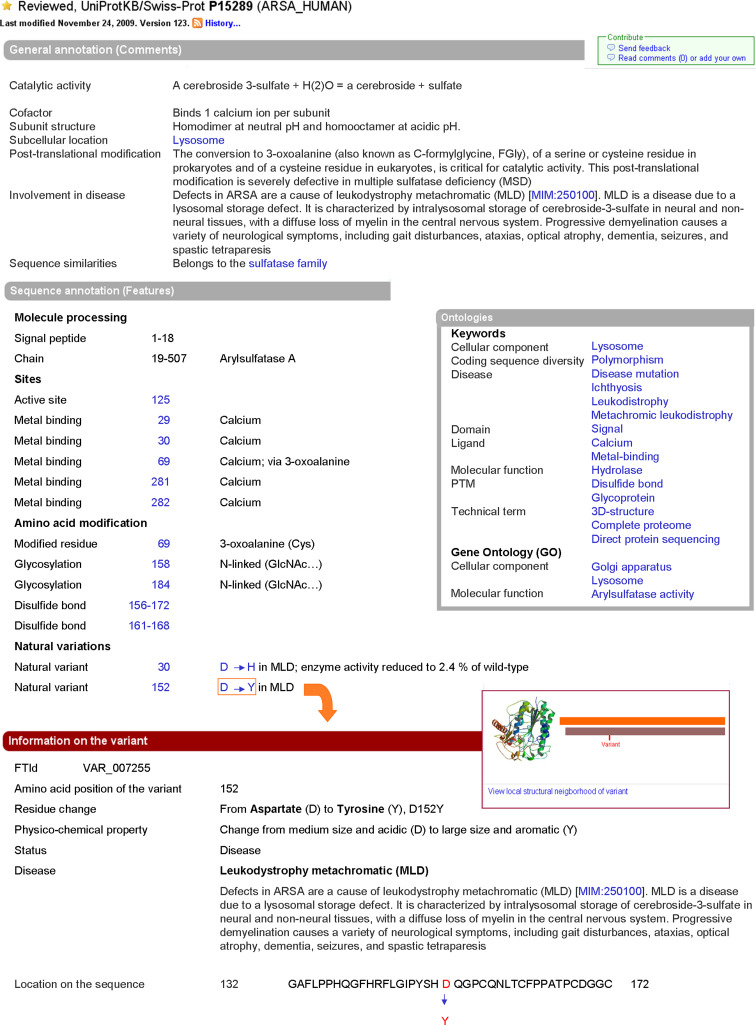
Fig. 2.
Extracts from the UniProtKB/Swiss-Prot entry for arylsulfatase A (P15289), showing selected parts of the General annotation, Sequence annotation and Ontologies section, and of one of the summary pages that are linked to individual “variant” lines. The General annotation section indicates the catalytic activity of a protein, its subunit structure, subcellular location, sequence similarities, etc., and explains post-translational modifications and the involvement in human disease. The Sequence annotation section indicates the roles of individual residues with specific “feature keys” displaying the extents of signal peptide and mature chain, active site and metal-binding residues, amino acid modifications and natural variants. For each variant, clicking on the amino acid substitution leads to a specific summary page including, when available, data from 3D-structure models. Keywords and GO terms complement the annotation
More and more of this information is presented under the form of controlled vocabularies and ontologies, to simplify data retrieval and computer-parsing. When interpreting data and building models, it is important to use direct experimental evidence as far as possible, to know the origins of the information, and to have access to the original data. Thus, UniProtKB/Swiss-Prot entries indicate the publications from which the information was taken, and provide database cross-references to specialized source databases.
Rapid identification is essential, and key information is already present in the lines devoted to the protein names. This includes the “recommended name”, the EC number for enzymes, and commonly used “alternative names” found in the literature, plus abbreviations derived from the recommended and alternative names. Likewise, keywords and links to GO terms [26] permit a rapid classification of proteins, regarding their molecular function, the process they are involved in, or in which cellular component they reside, and can be used to retrieve particular protein sets. While GO terms are extremely popular and easy to use, one should not forget to distinguish terms that are inferred from direct assay (IDA) or from a traceable author statement (TAS) from terms that are inferred from electronic annotation (IEA), e.g., based upon InterPro matches. The fact that a protein belongs to the pectinesterase family or contains a pectinesterase domain does not necessarily mean that it has pectinase activity, as exemplified by the E. coli protein ybhC (P46130).
From primary to quaternary structure
The primary structure of a protein, i.e., its amino acid sequence, contains all the information that is required to determine its final 3D-structure, and hence its biological activity [27]. While many small proteins fold as a single unit, the tertiary structure of larger proteins is formed by the assembly of several structural domains, motifs, or repeats, a striking example being the 34,350-residue-long scaffold protein titin (Q8WZ42). Two widely used databases, CATH [28] and SCOP [29], partition proteins into domains and classify these in a hierarchical manner. CATH classifies proteins according to class, architecture, topology, and homologous superfamily, while SCOP sorts domains into classes, folds, superfamilies, and families. Domains and repeats are the basic building blocks of proteins, and the combination of several such modules contributes to the evolution of functional diversity in proteins [30]. These are catalogued in the InterPro database [9], which integrates predictive models or ‘signatures’ representing protein domains, families, and functional sites from multiple, diverse member databases (HAMAP [31], Pfam [32], PROSITE [33], ProDom [34], SMART [35], TIGRFAMs [36], PIRSF [37], SUPERFAMILY [38], Gene3D [39], and PANTHER [40]). In all relevant UniProtKB entries, the InterPro member databases are cross-referenced in the “Family and domain databases” subsection. In addition, UniProtKB/Swiss-Prot indicates the presence of particular domains or repeats in the “general annotation” section under the heading “sequence similarities”. The exact extents of such domains, repeats, and sequence motifs are displayed in the “sequence annotation” section.
In addition to the many proteins and domains that have a characteristic native fold, many others remain essentially unstructured in the absence of their cognate ligand [41], as shown for the major prion protein (P04156), alpha-synuclein (P37840), and islet amyloid polypeptide (P10997). Database cross-references to DisProt help to access information about such unstructured proteins and domains [42].
Protein interactions are key elements in signaling pathways, and they can directly modulate protein function and activity. Many protein–protein interactions are identified by small-scale studies and are published in the scientific literature. Considering the huge number of proteins in a living cell, many of which are still uncharacterized, the contribution of large-scale proteomics studies is vital to identify protein complexes and chart protein interaction networks. Thus, several groups have studied the interactome of model organisms from yeast [43, 44] to Caenorhabditis elegans [45] and human [46]. Interpreting these findings and comparing results from different groups is not easy, due to differences in design and evaluation of the experiments, and because some studies aim to identify binary interactions, while others investigate protein complexes [47–49]. Novel and unexpected interactions may represent artefacts, or may indeed shed new light on the function of a protein. Data validation using several different technologies, and combination of interaction data with studies on the subcellular location and coexpression of putative interaction partners is essential. More and more often, large-scale protein interaction data are deposited in a public protein interaction database, using standardized format and protein identifiers [50]. This is essential for recovery of such information, and for comparing results from different studies. Collaboration between interaction databases, such as BIND [51], DIP [52], IntAct [53], and MINT [54], aims to speed up data integration, including data mined from the scientific literature, and to simplify public access to these findings [50]. In UniProtKB, information about protein interactions is gathered from the scientific literature and shown in the “general annotation” section under the heading “subunit”. There, one finds detailed information about binary interactions between proteins, but also about the composition of protein complexes, and about factors, such as protein phosphorylation, that modulate protein interactions. Manual evaluation is time-consuming, and thus, in UniProtKB/Swiss-Prot, additional information about binary protein interactions is imported from the IntAct database, which contains both manually annotated interactions from small-scale studies and information derived from large-scale protein interaction studies. The information is presented in the form of a table under the heading “binary interactions”, and links to the IntAct annotation provide access to the experimental details.
Ligand-binding sites and catalytic residues
A key issue for understanding the mode of action of a protein is the identification of physiologically relevant ligand binding sites and catalytic residues, and here protein 3D-structures are essential. Prior knowledge and human evaluation are then required to identify ligands that are physiologically relevant, whether these be metal ions, nucleotides, or various organic compounds. Common molecules, such as phosphate, citrate, or acetate, may, or may not, occupy the binding sites of physiological substrates or inhibitors. Likewise, inorganic ions may occupy their cognate binding sites, but in other cases the observed ionic interactions may simply reflect the buffer composition. Synthetic compounds may represent transition state analogs or substances that are of pharmaceutical interest and occupy physiologically relevant binding sites, or they may be irrelevant buffer components. Likewise, heavy metals may have been included for technical reasons, but may also occupy physiologically significant binding sites, as in the case of the cadmium-sensitive HTH-type transcriptional regulator cmtR (P67731). In the PDB archive, inorganic ions contribute the largest number of ligand binding sites, with almost as many binding sites being occupied by the large and heterogeneous class of synthetic inhibitors and non-canonical biological molecules [55]. Dedicated tools facilitate retrieving this information from PDB. There, scientists have access to all the data and can extract information about the ligands they are interested in. Public protein–ligand databases and tools, such as ReLiBase [56], Binding MOAD [57], and SRS 3D [58], help find structures with particular ligands and analysis of protein–ligand interactions. Likewise, PDBSum provides access to excellent tools and links. Even so, recovering relevant information takes time. In UniProtKB/Swiss-Prot, annotators identify physiologically relevant ligands and display information about their binding sites in text format. Combining these data with complementary results from other types of experiments, e.g., mutagenesis studies, enhances the value of these findings. Dedicated “feature keys” indicate residues that interact with specific classes of ligands, such as metal ions, nucleotides, and other small molecules, and such fine-grained annotation helps retrieval of specific datasets. Likewise, a specific “feature key” indicates active site residues that are directly involved in catalysis. There is no universally accepted definition of the term “active site”, and sometimes it is used in a very broad sense, grouping residues that are directly involved in catalysis with others that position the substrate, bind metal cofactors, or simply line the active site pocket. In UniProtKB/Swiss-Prot, the term “active site” is reserved for residues that are directly involved in catalysis, and dedicated “feature keys” are used to indicate the roles of other important residues, as illustrated by human arylsulfatase A (P15289) (Fig. 2). Precise annotation rules ensure that the same criteria are used throughout.
UniProtKB/Swiss-Prot annotation aims to show the physiological situation, meaning that a ligand is named “ATP”, or “substrate” even when a synthetic analog was used. Frequently, several alternative names are used in the literature for a single chemical entity, such as S-adenosyl-l-methionine, which does not help find all the relevant information. Using standardized vocabulary is one way to guarantee the retrieval of information. The use of tools that permit searches for specific chemical structures is another solution, and PDB implements both. UniProtKB/Swiss-Prot aims to use a single name for every ligand, with a preference for terms found in CHEBI [59]. Likewise, the use of generic ligand names, e.g., “substrate” for enzymes, or “carbohydrate” in the case of lectins, helps find the relevant information, and at the same time facilitates propagation to other family members.
The importance of post-translational modifications
While complex multicellular organisms, such as humans or mice, can live with about 20,000 protein-coding genes, the number of proteins is much higher, due to alternative splicing events, but also due to post-translational modifications. Thus, the total number of human proteins may be somewhere between 100,000 and 1,000,000 [60]. The chemical nature of post-translational modifications is extremely diverse, ranging from proteolytic cleavage to methylation, phosphorylation of specific residues to the formation of disulfide bonds, and other cross-links. This multitude of modifications results in an equally wide spectrum of biological effects: targeting proteins to specific cellular compartments, modulating protein–protein interactions, or regulating protein function and turnover. For some enzymes, e.g., human arylsulfatase A (P15289), post-translational modification is essential for catalytic activity (Fig. 2). Currently, large-scale proteomics studies make a major contribution to the identification of protein glycosylation [61] and phosphorylation sites, e.g., during mitosis [62]. Because of their biological importance, UniProtKB/Swiss-Prot prioritizes annotation of post-translational protein modifications [63], using data from the scientific literature and from protein 3D-structures.
As always, it is essential to present the information in a user-friendly, simple, and accurate manner. General information about post-translational modifications is shown under the appropriate heading in the “general annotation” section, while the exact position and the chemical nature of the modifications are shown in the “sequence annotation” section. Many of these modifications are also linked to specific ontologies and keywords, e.g., “Glycoprotein” or “Phosphoprotein”. Dedicated “feature keys”, controlled vocabularies, and strictly standardized annotation are indispensable to show unambiguously the exact chemical nature of each protein modification and the resulting mass change, and this is achieved in collaboration with the RESID database [64]. This database contains a comprehensive collection of pre-, co- and post-translational protein modifications and cross-links. It provides systematic and alternative names, formulas, and structure diagrams, and indicates the mass changes associated with each modification. This in turn is required for the correct identification of modified peptides by mass spectrometry.
Both RESID and UniProtKB/Swiss-Prot prioritize the annotation of proteins involved in post-translational modifications. In UniProtKB/Swiss-Prot, the 480-odd classical and up to 24 atypical protein kinases now believed to exist in the human and mouse genome have been recently updated and extensively annotated, providing access to up-to-date and in-depth annotation of these proteins, plus access to additional external resources by links from within each entry [65].
Membrane-spanning domains: facts and predictions
Membrane proteins are essential for the manifold functions exerted by biological membranes. They enable the ion gradients required to drive energy metabolism, and permit the uptake of nutrients and the export of signaling molecules, waste products, and toxic compounds. Integral membrane proteins serve as receptors that participate in signaling cascades, and play important roles in host–pathogen interactions, both in invasion by pathogens and in mediating defense responses. Membrane proteins constitute about one-third of the human proteome, but they are major targets of medical drugs, and the subject of numerous pharmaceutical studies, as illustrated by the G-protein coupled receptors (GPCRs) [66]. Thus, there is a huge interest in elucidating the molecular mode of action of membrane proteins. In spite of significant progress made in the last few years, integral membrane proteins are still severely underrepresented in structural databases. More often, the structures of isolated soluble domains have been determined, as exemplified by the mammalian toll-like receptors. Still, more and more such structures are determined, e.g., the crystal structure of the E. coli rhomboid protease glpG (P09391) [67, 68] or the solution structure of human VDAC1 (P21796) [69]. Specialized databases, such as the protein data bank of transmembrane proteins (PDBTM) [70], help find membrane proteins of known 3D-structure.
In the absence of a 3D-structure, it is technically quite difficult to determine which part of a protein is buried in a membrane. Thus, for most proteins, putative transmembrane domains are predicted by bioinformatics tools. In contrast to the situation for soluble domains, transmembrane domains are either all helical or all beta-strand, plus eventual connecting loops. Numerous proteins cross the membrane as beta-barrel structures, and protein 3D-structures have been essential for developing dedicated prediction tools [71]. In UniProtKB/Swiss-Prot, these proteins can be retrieved using the Keyword “Porin”, e.g., E. coli maltoporin (P02943) and the mitochondrial voltage-gated anion channel VDAC1 (P21796). Most prediction tools have been developed to predict alpha-helical transmembrane domains, and they basically assume that a hydrophobic stretch of the polypeptide chain crosses the membrane by the shortest path, burying about 18–20 amino acids in the membrane. This may be true for proteins with a single transmembrane domain. For multipass membrane proteins, helices frequently cross the membrane at a pronounced angle, or are kinked, and so the path becomes longer, and a larger number of amino acid residues are buried in the membrane. This is illustrated by GPCRs, the largest group of integral membrane proteins with over 1,000 family members in human. By now, structures for several family members have been determined, from bovine rhodopsin (P02699) to human ADORA2A (P29274), ADRB2 (P07550), and turkey ADRB1 (P07700). These proteins all have an extracellular N-terminus, seven transmembrane helices, and a cytoplasmic C-terminus. Several of the transmembrane helices have distinct kinks and are tilted up to 20° with respect to the plane of the membrane.
For other integral membrane proteins, such as human aquaporin-1 (P29972) or Clc channel family members (P37019), the topology is highly complex. In addition to the expected transmembrane helices, there are short in-membrane helices that are followed by an in-membrane loop structure, where the protein chain enters and leaves on the same side of the membrane without crossing the lipid bilayer. Again, a protein 3D-structure is the prerequisite for determining the membrane topology, and manual evaluation is required to arrive at a correct result. In the same vein, for leukotriene C4 synthase (Q16873) and the arachidonate 5-lipoxygenase-activating protein FLAP (P20292), two proteins involved in leukotriene biosynthesis, prediction programs consistently detect three transmembrane helices—the experimental structures clearly show that there are four. Moreover, prediction tools generally fail to detect transmembrane segments that contain polar or charged amino acid residues, and yet this is a common phenomenon for integral membrane proteins that are involved in active or passive transport of hydrophilic solutes. One extreme case is represented by the voltage-gated potassium channels, such as Q9YDF8 and P62483, where a helical segment with a basic amino acid residue in every third position is buried in the membrane [72, 73].
Erroneous prediction of transmembrane domains is not restricted to transporters and pore-forming proteins, as exemplified by the caveolins (Q03135). The topology of these proteins is known with both N-terminus and C-terminus being in the cytoplasm [74]. In between is a membrane-embedded “hairpin” structure that is interpreted as a single transmembrane region by prediction programs, leading to an erroneous prediction of the topology.
Establishing a correct prediction is further complicated by the existence of certain proteins that on the one hand exist as soluble, globular proteins, but on the other hand can insert into lipid membranes, form pores and kill target cells. Such proteins are essential constituents of the complement membrane attack complex of cytolytic T-cells (P07357, P02748), but are also produced by microbes as cell-lysing toxins, e.g., Vibrio cholerae hemolysin (P09545). Needless to say, predicting the extent of transmembrane helices for such proteins is extremely difficult, and 3D-structures are essential to elucidate the conformation changes involved in membrane insertion and pore formation [75, 76]. In short, predictions of transmembrane segments should be accepted as highly useful tools for generating a working hypothesis, knowing that the prediction may differ from reality in several essential points. Some transmembrane helices will be predicted correctly, for others the extents will differ significantly, and in other cases a transmembrane segment will not be detected, or a hydrophobic stretch will be erroneously predicted as transmembrane segment, meaning that the predicted topology may be wrong. Taking account of all the available experimental data is essential to arrive at a correct result, and in this context protein 3D-structures are uniquely powerful sources of information for establishing the correct transmembrane topology, and the extents of the transmembrane segments. In UniProtKB/Swiss-Prot, general information about the subcellular location of a protein is shown in the “general annotation” section, and the “sequence annotation” section shows the precise details about the predicted or experimentally validated topological domains. Manual annotation of transmembrane domains based upon protein 3D-structures is time-consuming, but UniProtKB/Swiss-Prot endeavors to use these data to show the correct membrane topology.
The role of protein structures in health and disease
In spite of the low error rates associated with DNA replication, transcription, and translation, mutations occur with a given low frequency. Large-scale alterations include the deletion of entire genes and chromosomal cross-overs that generate hybrid proteins, as shown for the proto-oncogene tyrosine-protein kinase ABL1 (P00519), where defects due to a chromosomal translocation with BCR (P11274) are a cause of chronic and acute myeloid leukemia (CML and AML) and of acute lymphoblastic leukemia (ALL). Protein 3D-structures show how this fusion leads to a constitutively active kinase, and illustrate the mode of action of inhibitors [77]. In UniProtKB/Swiss-Prot, the keyword “Chromosomal rearrangement” can be used to retrieve such proteins. Likewise, the keyword “Proto-oncogene” indicates proteins where mutations that alter its normal, regulated activity or expression pattern convert the gene product into a cancer-promoting oncogene. Such information about disease mutations and their consequences is displayed in the “general annotation” section. Cross-references to the OMIM database provide access to further information about such a disease.
Missense mutations, i.e., the substitution of one base for another, are the most common type of mutation [78]. The consequence then obviously depends on the nature and the position of the amino acid substitution. Neutral polymorphisms are most often conservative substitutions in a non-essential part of the protein. On the contrary, any mutation that affects a catalytic residue in an essential polypeptide, or residues essential for proper protein folding, may cause a disease phenotype. This information is dispersed throughout the scientific literature and specialized databases. UniProtKB/Swiss-Prot, as a universal protein knowledgebase, aims to facilitate access to information about polymorphisms and disease mutations. The data are gathered from the literature, and in the case of polymorphisms, also from dbSNP. Each variant has its own identifier and is linked to a summary page that displays the relevant information [79]. It describes the nature of the mutation and shows it in the sequence context, provides information about the disease that is linked to this mutation, and lists relevant publications, and for proteins where 3D-structures are available or modeling is possible, it includes 3D-structure models and interactive views of the mutated residue in its 3D environment (Fig. 2).
Human disease can also be caused by protein misfolding and misassembly that results in the formation of insoluble fibrils and toxic aggregates, as shown for beta-2-microglobulin (P61769). Natural mutations that favor formation of amyloid fibrils have been found in a number of proteins, including the amyloid beta A4 protein (P05067), the prion protein (P04156), transthyretin (P02766), and islet amyloid polypeptide (P10997), and the keyword “Amyloid” can be used to retrieve such proteins. 3D-structure analysis of amyloid fibrils from several proteins reveals variations of a common, characteristic cross-beta sheet structure [80]. Such fibrils can be formed by various short peptides, provided they have self-complementary sequences compatible with the formation of a dry steric zipper. Typically, amyloidogenic peptides have low-complexity sequences with residues of similar size, e.g., the sequence NNQQNY found in yeast sup35 (P05453) [81]. Thus, the propensity towards amyloidogenesis can be predicted [82]. Different mechanisms for amyloidogenesis have been proposed, where initially well-folded proteins would undergo structural fluctuations and partial unfolding, leading to the exposure of hydrophobic residues, while formation of helical elements in intrinsically disordered proteins would serve as starting point for protein interactions and the formation of aggregates [83–85]. In the case of transthyretin (P02766), 3D-structures have shown how small ligands can stabilize the native tetrameric conformation and prevent amyloid formation [86], indicating the way for a possible preventive treatment.
The relationship between protein sequence, structure and function
The protein sequence determines the native fold of a protein, and protein structure determines the function of a protein, be this an enzyme, a cell-surface receptor, or a cytoplasmic scaffolding protein. Thus, it is generally assumed that similar protein sequences give rise to similar 3D-structures, and hence to similar functions [87–89]. This paradigm generally holds true for orthologous proteins, and provides the basis for the annotation of uncharacterized proteins in newly sequenced genomes. In UniProtKB/Swiss-Prot, annotators group orthologous proteins based on sequence similarity and alignments. Information about function, cofactors, subcellular location, protein–protein interactions, etc., is then gathered from the scientific literature and from protein 3D-structures. When annotating a group of related proteins, publications and 3D-structures for orthologous proteins from several organisms are used to establish annotation rules and determine the limits of propagation. This approach is used for the annotation of proteins from all branches of life, and is the basis of automated and manual annotation using HAMAP family rules [31]. The use of such automated annotation tools is essential for keeping up with the constant arrival of freshly sequenced microbial genomes, and new microbial protein sequences. In November 2009, the HAMAP family database comprised over 1,600 manually curated protein families, providing the basis for the annotation of over 306,000 microbial proteins in UniProtKB/Swiss-Prot. Obviously, the higher the degree of sequence identity, the higher is the level of confidence, the determining factor being the conservation of known key residues, such as residues involved in catalysis or in ligand binding. Thus, it is not a problem to assign the correct function to orthologous housekeeping enzymes, such as cytosolic phosphoenolpyruvate carboxylase, where the residues involved in catalysis and substrate binding are absolutely conserved, from bacteria (Q9AEM1) and archaea (Q9UY53) to vertebrates (P05153) and human (P35558), in spite of overall sequence divergence.
While one can infer function with high confidence when there is only one copy of the gene for an essential protein, much more caution is required when dealing with large gene families, and when gene duplications have given rise to paralogs. One gene copy may keep the ancestral function, while other copies may evolve towards a new function, or may accumulate deleterious mutations and become inactive over time [87, 88, 90]. As long as the active site residues are not mutated, the general function may be conserved, but the substrate specificity may be somewhat different. In other cases, family members may have lost the original activity, in spite of high sequence similarity, and this even when the active site residues are conserved. Thus, within the sulfatase modifying factor family, SUMF2 (Q8NBJ7) lacks enzyme activity and serves as regulatory subunit that modulates the activity of SUMF1 (Q8NBK3), even though essential residues are conserved [91]. Likewise, similar structures do not guarantee similar function. Again, one has to distinguish orthologs from paralogs, and that is not always easy. Thus, 5-hydroxyisourate hydrolases were first identified as transthyretin-related proteins, before their enzyme activity was established. Human transthyretin (P02766, 3bt0) and zebrafish 5-hydroxyisourate hydrolase (Q06S87, 2h6u) have 35% sequence identity, and their structures are at first sight almost identical. Both proteins are homotetramers, where four subunits delimit a tunnel-shaped central cavity. Nevertheless, this high degree of similarity is not a proof that the proteins are orthologs and have similar functions. During early vertebrate evolution, a duplication of the gene encoding 5-hydroxyisourate hydrolase, an enzyme involved in the breakdown of uric acid, gave rise to the gene for transthyretin, a protein that transports thyroid hormone in the blood stream. 5-hydroxyisourate hydrolase is found from bacteria to mammals, and key residues are remarkably conserved between prokaryotes and eukaryotes, but the gene has been lost in the human lineage. Strikingly, the residues that are necessary for 5-hydroxyisourate hydrolase activity are not conserved in transthyretin [92].
So, precisely how much do you have to change a protein before ending up with a different function? The answer is: not at all, as exemplified by the duck eye lens crystallins, where enzymes, such as argininosuccinate lyase (P24058), have been recruited to contribute to the optical properties of the crystallin [93]. Gephyrin (Q9NQX3) presents another striking example of such a “moonlighting” protein: it functions as microtubule-associated protein involved in membrane protein–cytoskeleton interactions and is thought to anchor the inhibitory glycine receptor (GLYR) to subsynaptic microtubules, but is also involved in molybdenum cofactor biosynthesis and is required for the transfer of molybdenum to molybdopterin. These examples serve as a reminder that protein function depends critically on the biological context, and that predictions based on sequence or structural similarity have their limits. In the end, even the best prediction cannot replace experimental characterization. Then, databases, such as UniProtKB/Swiss-Prot, have an essential role in integrating information and promoting access to experimental data.
The role of protein 3D-structures for functional characterization of novel proteins
Large-scale nucleotide sequencing has led to the prediction of numerous novel protein-coding genes, and many of these are entirely uncharacterized, and their function is not known. Biochemical characterization is not easy, when nothing is known about the physiological role of a protein. Here, protein 3D-structures can help predict a possible function for otherwise uncharacterized proteins. One initial goal of the structural genomics initiatives was to provide experimental protein structures for every type of fold, so that high-confidence 3D-models could be generated for most other proteins [12]. As a consequence, structural genomics projects and individual scientists have targeted proteins of biomedical interest, but also uncharacterized microbial proteins with less than 30% sequence similarity to already characterized proteins. This resulted in numerous new 3D-structures for proteins both from mammals and from microbial model organisms, notably E. coli, M. tuberculosis and Bacillus subtilis, but also from archaea and pathogenic viruses [94–98]. Since its beginnings, structural genomics has made a large contribution to the identification of novel folds by determining unique structures [12, 99, 100], but during the same period, the protein universe has considerable expanded due to large-scale sequencing of ever more microbial genomes. This yielded a majority of proteins that can be assigned to known families, but also a host of unique predicted proteins with very low sequence similarity to already known protein families.
Structural genomics provides a starting point for further structural and functional characterization, particularly when combined with other data. This is exemplified by the M. tuberculosis protein Rv1846c (P95163), a transcription regulator of previously unknown function. Its 3D-structure showed strong similarity to S. aureus BlaI (P0A042) and MecI (P68261), two repressors involved in beta-lactam antibiotic resistance. In-depth functional characterization of Rv1846c confirmed its function as transcriptional regulator for genes involved in antibiotic resistance analogous to BlaI, and it has been renamed accordingly [101].
Other proteins are still waiting for biochemical characterization. In some cases, it is possible to predict a general function based on the structure and the genomic context; for example, E. coli protein ydhR (P0ACX3), a putative monooxygenase that may play a role in the metabolism of aromatic compounds [102]. For other proteins, the presence of a known domain, or of a fortuitously bound ligand, such as NAD, can suggest a general function, in the latter case that the protein may have enzyme activity and may function as an oxido-reductase. Still, numerous small microbial proteins present novel folds without significant similarity to characterized proteins, and their structures are devoid of informative ligands. The prediction of at least an approximate function for such uncharacterized proteins requires careful manual evaluation of all the available data, including sequence and structural similarities, genomic context, and regulation of gene expression, as well as data and predictions about subcellular location and post-translational modifications. Strategies, prediction tools, and their limitations have been the subject of several excellent publications [87, 89, 103–105]. Tools that combine several types of queries, such as ProFunc [106], facilitate this task, but do not eliminate the need for characterization and human effort. Often, predictions give clues towards several possible functions, and in depth biochemical characterization is required to establish the precise physiological role of such proteins. Databases, such as UniProtKB and the PSI-SGKB, have essential roles in promoting access to existing experimental data and helping to identify new targets.
Conclusions
The present avalanche of sequence and structural data requires central knowledgebases with rapidly accessible, reliable, and up-to-date information that provide a solid base for the interpretation of new results, and a starting point for planning further experiments. Efficient harnessing of knowledge derived from protein 3D-structures, and from genetic and biochemical analyses, requires that these data are easily accessible, and the value of experimental results is optimized by the combination of complementary biological information. Exchange of data and active collaboration between different types of databases, but also between data producers and databases, is necessary. The decreased cost of producing data and the technical progress has led to an explosion in the amount of available data. Now we need efficient means for handling these data and making them publicly accessible.
Acknowledgments
Many thanks to all the UniProtKB/Swiss-Prot team, especially to Janet James and Jules Jacobsen for critically reading this manuscript and helpful suggestions, and to Laurent Bollondi, Salvo Paesano and Gregoire Rossier for help with the illustrations. UniProt is mainly supported by Award Number U01HG02712 from the National Human Genome Research Institute. Additional support for the EBI’s involvement in UniProt comes from the European Commission contract SLING grant (226073) and from the NIH grant (2P41HG02273-07). UniProtKB/Swiss-Prot activities at the SIB are supported in addition from the Swiss Federal Government through the Federal Office of Education and Science and from the European Commission contract SLING (226073). PIR activities are also supported by the NIH grants and contracts on HHSN266200400061C, NCI-caBIG and 5R01GM080646-04, and the Department of Defense grant W81XWH0720112.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Appendix
UniProt has been prepared by: Rolf Apweiler, Maria Jesus Martin, Claire O’Donovan, Michele Magrane, Yasmin Alam-Faruque, Ricardo Antunes, Daniel Barrell, Benoit Bely, Mark Bingley, David Binns, Lawrence Bower, Paul Browne, Wei Mun Chan, Emily Dimmer, Ruth Eberhardt, Alexander Fedotov, Rebecca Foulger, John Garavelli, Rachael Huntley, Julius Jacobsen, Michael Kleen, Kati Laiho, Rasko Leinonen, Duncan Legge, Quan Lin, Wudong Liu, Jie Luo, Sandra Orchard, Samuel Patient, Diego Poggioli, Manuela Pruess, Matt Corbett, Giuseppe di Martino, Mike Donnelly and Pieter van Rensburg at the European Bioinformatics Institute (EBI); Amos Bairoch, Lydie Bougueleret, Ioannis Xenarios, Severine Altairac, Andrea Auchincloss, Ghislaine Argoud-Puy, Kristian Axelsen, Delphine Baratin, Marie-Claude Blatter, Brigitte Boeckmann, Jerven Bolleman, Laurent Bollondi, Emmanuel Boutet, Silvia Braconi Quintaje, Lionel Breuza, Alan Bridge, Edouard de Castro, Luciane Ciapina, Danielle Coral, Elisabeth Coudert, Isabelle Cusin, Fabrice David, Gwennaelle Delbard, Mikael Doche, Dolnide Dornevil, Paula Duek Roggli, Severine Duvaud, Anne Estreicher, Livia Famiglietti, Marc Feuermann, Sebastien Gehant, Nathalie Farriol-Mathis, Serenella Ferro, Elisabeth Gasteiger, Alain Gateau, Vivienne Gerritsen, Arnaud Gos, Nadine Gruaz-Gumowski, Ursula Hinz, Chantal Hulo, Nicolas Hulo, Janet James, Silvia Jimenez, Florence Jungo, Thomas Kappler, Guillaume Keller, Corinne Lachaize, Lydie Lane-Guermonprez, Petra Langendijk-Genevaux, Vicente Lara, Philippe Lemercier, Damien Lieberherr, Tania de Oliveira Lima, Veronique Mangold, Xavier Martin, Patrick Masson, Madelaine Moinat, Anne Morgat, Anais Mottaz, Salvo Paesano, Ivo Pedruzzi, Sandrine Pilbout, Violaine Pillet, Sylvain Poux, Monica Pozzato, Nicole Redaschi, Catherine Rivoire, Bernd Roechert, Michel Schneider, Christian Sigrist, Karin Sonesson, Sylvie Staehli, Eleanor Stanley, Andre Stutz, Shyamala Sundaram, Michael Tognolli, Laure Verbregue, Anne-Lise Veuthey, Lina Yip and Luiz Zuletta at the Swiss Institute of Bioinformatics (SIB) and the Biochemistry and Structural Biology Department of the University of Geneva; Cathy Wu, Cecilia Arighi, Leslie Arminski, Winona Barker, Chuming Chen, Yongxing Chen, Zhang-Zhi Hu, Hongzhan Huang, Raja Mazumder, Peter McGarvey, Darren A. Natale, Jules Nchoutmboube, Natalia Petrova, Nisha Subramanian, Baris E. Suzek, Uzoamaka Ugochukwu, Sona Vasudevan, C. R. Vinayaka, Lai Su Yeh and Jian Zhang at the Protein Information Resource (PIR).
Footnotes
The full author list is shown under Appendix.
References
- 1.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 2.Dutta S, Burkhardt K, Young J, Swaminathan GJ, Matsuura T, Henrick K, Nakamura H, Berman HM. Data deposition and annotation at the worldwide protein data bank. Mol Biotechnol. 2009;42:1–13. doi: 10.1007/s12033-008-9127-7. [DOI] [PubMed] [Google Scholar]
- 3.Boutselakis H, Dimitropoulos D, Fillon J, Golovin A, Henrick K, Hussain A, Ionides J, John M, Keller PA, Krissinel E, McNeil P, Naim A, Newman R, Oldfield T, Pineda J, Rachedi A, Copeland J, Sitnov A, Sobhany S, Suarez-Uruena A, Swaminathan J, Tagari M, Tate J, Tromm S, Velankar S, Vranken W. E-MSD: the European Bioinformatics Institute Macromolecular Structure Database. Nucleic Acids Res. 2003;31:458–462. doi: 10.1093/nar/gkg065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H, Markley JL. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berman HM, Westbrook JD, Gabanyi MJ, Tao W, Shah R, Kouranov A, Schwede T, Arnold K, Kiefer F, Bordoli L, Kopp J, Podvinec M, Adams PD, Carter L, Minor W, Nair R, Baer J. The protein structure initiative structural genomics knowledgebase. Nucleic Acids Res. 2009;37:D365–D368. doi: 10.1093/nar/gkn790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Uni Prot Consortium The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jain E, Bairoch A, Duvaud S, Phan I, Redaschi N, Suzek BE, Martin MJ, McGarvey P, Gasteiger E. Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics. 2009;10:136. doi: 10.1186/1471-2105-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mulder NJ, Kersey P, Pruess M, Apweiler R. In silico characterization of proteins: UniProt, InterPro and Integr8. Mol Biotechnol. 2008;38:165–177. doi: 10.1007/s12033-007-9003-x. [DOI] [PubMed] [Google Scholar]
- 9.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, Finn RD, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Laugraud A, Letunic I, Lonsdale D, Lopezm R, Madera M, Maslen J, McAnulla C, McDowall J, Mistry J, Mitchell A, Mulder N, Natale D, Orengo C, Quinn AF, Selengut JD, Sigrist CJ, Thimma M, Thomas PD, Valentin F, Wilson D, Wu CH, Yeats C. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mueller M, Martens L, Apweiler R. Annotating the human proteome: beyond establishing a parts list. Biochim Biophys Acta. 2007;1774:175–191. doi: 10.1016/j.bbapap.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Laskowski RA. PDBsum new things. Nucleic Acids Res. 2009;37:D355–D359. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grabowski M, Joachimiak A, Otwinowski Z, Minor W. Structural genomics: keeping up with expanding knowledge of the protein universe. Curr Opin Struct Biol. 2007;17:347–353. doi: 10.1016/j.sbi.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pieper U, Eswar N, Webb BM, Eramian D, Kelly L, Barkan DT, Carter H, Mankoo P, Karchin R, Marti-Renom MA, Davis FP, Sali A. MODBASE, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2009;37:D347–D354. doi: 10.1093/nar/gkn791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dodge C, Schneider R, Sander C. The HSSP database of protein structure-sequence alignments and family profiles. Nucleic Acids Res. 1998;26:313–315. doi: 10.1093/nar/26.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruford EA, Lush MJ, Wright MW, Sneddon TP, Povey S, Birney E. The HGNC Database in 2008: a resource for the human genome. Nucleic Acids Res. 2008;36:D445–D448. doi: 10.1093/nar/gkm881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White J, Wain H, Bruford E, Povey S. Promoting a standard nomenclature for genes and proteins. Nature. 1999;402:347. doi: 10.1038/46405. [DOI] [PubMed] [Google Scholar]
- 18.Tamames J, Valencia A. The success (or not) of HUGO nomenclature. Genome Biol. 2006;7:402. doi: 10.1186/gb-2006-7-5-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cochrane G, Akhtar R, Bonfield J, Bower L, Demiralp F, Faruque N, Gibson R, Hoad G, Hubbard T, Hunter C, Jang M, Juhos S, Leinonen R, Leonard S, Lin Q, Lopez R, Lorenc D, McWilliam H, Mukherjee G, Plaister S, Radhakrishnan R, Robinson S, Sobhany S, Hoopen PT, Vaughan R, Zalunin V, Birney E. Petabyte-scale innovations at the European Nucleotide Archive. Nucleic Acids Res. 2009;37:D19–D25. doi: 10.1093/nar/gkn765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, Hart E, Suner MM, Landrum MJ, Aken B, Ayling S, Baertsch R, Fernandez-Banet J, Cherry JL, Curwen V, Dicuccio M, Kellis M, Lee J, Lin MF, Schuster M, Shkeda A, Amid C, Brown G, Dukhanina O, Frankish A, Hart J, Maidak BL, Mudge J, Murphy MR, Murphy T, Rajan J, Rajput B, Riddick LD, Snow C, Steward C, Webb D, Weber JA, Wilming L, Wu W, Birney E, Haussler D, Hubbard T, Ostell J, Durbin R, Lipman D. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Eyre T, Fitzgerald S, Fernandez-Banet J, Gräf S, Haider S, Hammond M, Holland R, Howe KL, Howe K, Johnson N, Jenkinson A, Kähäri A, Keefe D, Kokocinski F, Kulesha E, Lawson D, Longden I, Megy K, Meidl P, Overduin B, Parker A, Pritchard B, Prlic A, Rice S, Rios D, Schuster M, Sealy I, Slater G, Smedley D, Spudich G, Trevanion S, Vilella AJ, Vogel J, White S, Wood M, Birney E, Cox T, Curwen V, Durbin R, Fernandez-Suarez XM, Herrero J, Hubbard TJ, Kasprzyk A, Proctor G, Smith J, Ureta-Vidal A, Searle S. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrell D, Dimmer E, Huntley RP, Binns D, O’Donovan C, Apweiler R. The GOA database in 2009—an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anfinsen CB, Haber E, Sela M, White FH. The kinetics of formation of native ribonuclease during oxidation of the reduced polypeptide chain. Proc Natl Acad Sci USA. 1961;47:1309–1314. doi: 10.1073/pnas.47.9.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuff A, Redfern OC, Greene L, Sillitoe I, Lewis T, Dibley M, Reid A, Pearl F, Dallman T, Todd A, Garratt R, Thornton J, Orengo C. The CATH hierarchy revisited-structural divergence in domain superfamilies and the continuity of fold space. Structure. 2009;17:1051–1062. doi: 10.1016/j.str.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andreeva A, Howorth D, Chandonia J, Brenner SE, Hubbard TJP, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore AD, Björklund AK, Ekman D, Bornberg-Bauer E, Elofsson A. Arrangements in the modular evolution of proteins. Trends Biochem Sci. 2008;33:444–451. doi: 10.1016/j.tibs.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Lima T, Auchincloss AH, Coudert E, Keller G, Michoud K, Rivoire C, Bulliard V, de Castro E, Lachaize C, Baratin D, Phan I, Bougueleret L, Bairoch A. HAMAP: a database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot. Nucleic Acids Res. 2009;37:D471–D478. doi: 10.1093/nar/gkn661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz H, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJA. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bru C, Courcelle E, Carrère S, Beausse Y, Dalmar S, Kahn D. The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res. 2005;33:D212–D215. doi: 10.1093/nar/gki034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu CH, Nikolskaya A, Huang H, Yeh LL, Natale DA, Vinayaka CR, Hu Z, Mazumder R, Kumar S, Kourtesis P, Ledley RS, Suzek BE, Arminski L, Chen Y, Zhang J, Cardenas JL, Chung S, Castro-Alvear J, Dinkov G, Barker WC. PIRSF: family classification system at the Protein Information Resource. Nucleic Acids Res. 2004;32:D112–D114. doi: 10.1093/nar/gkh097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson D, Pethica R, Zhou Y, Talbot C, Vogel C, Madera M, Chothia C, Gough J. SUPERFAMILY–sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 2009;37:D380–D386. doi: 10.1093/nar/gkn762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeats C, Lees J, Reid A, Kellam P, Martin N, Liu X, Orengo C. Gene3D: comprehensive structural and functional annotation of genomes. Nucleic Acids Res. 2008;36:D414–D418. doi: 10.1093/nar/gkm1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–D252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 42.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, Obradovic Z, Dunker AK. DisProt: the Database of Disordered Proteins. Nucleic Acids Res. 2007;35:D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gavin A, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 44.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual JF, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C, de Smet AS, Motyl A, Hudson ME, Park J, Xin X, Cusick ME, Moore T, Boone C, Snyder M, Roth FP, Barabási AL, Tavernier J, Hill DE, Vidal M. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simonis N, Rual J, Carvunis A, Tasan M, Lemmens I, Hirozane-Kishikawa T, Hao T, Sahalie JM, Venkatesan K, Gebreab F, Cevik S, Klitgord N, Fan C, Braun P, Li N, Ayivi-Guedehoussou N, Dann E, Bertin N, Szeto D, Dricot A, Yildirim MA, Lin C, de Smet AS, Kao HL, Simon C, Smolyar A, Ahn JS, Tewari M, Boxem M, Milstein S, Yu H, Dreze M, Vandenhaute J, Gunsalus KC, Cusick ME, Hill DE, Tavernier J, Roth FP, Vidal M. Empirically controlled mapping of the Caenorhabditis elegans protein–protein interactome network. Nat Methods. 2009;6:47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rual J, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 47.Cusick ME, Klitgord N, Vidal M, Hill DE. Interactome: gateway into systems biology. Hum Mol Genet. 2005;14:R171–R181. doi: 10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- 48.Köcher T, Superti-Furga G. Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat Methods. 2007;4:807–815. doi: 10.1038/nmeth1093. [DOI] [PubMed] [Google Scholar]
- 49.Wodak SJ, Pu S, Vlasblom J, Séraphin B. Challenges and rewards of interaction proteomics. Mol Cell Proteomics. 2009;8:3–18. doi: 10.1074/mcp.R800014-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Orchard S, Salwinski L, Kerrien S, Montecchi-Palazzi L, Oesterheld M, Stümpflen V, Ceol A, Chatr-aryamontri A, Armstrong J, Woollard P, Salama JJ, Moore S, Wojcik J, Bader GD, Vidal M, Cusick ME, Gerstein M, Gavin AC, Superti-Furga G, Greenblatt J, Bader J, Uetz P, Tyers M, Legrain P, Fields S, Mulder N, Gilson M, Niepmann M, Burgoon L, De Las Rivas J, Prieto C, Perreau VM, Hogue C, Mewes HW, Apweiler R, Xenarios I, Eisenberg D, Cesareni G, Hermjakob H. The minimum information required for reporting a molecular interaction experiment (MIMIx) Nat Biotechnol. 2007;25:894–898. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- 51.Alfarano C, Andrade CE, Anthony K, Bahroos N, Bajec M, Bantoft K, Betel D, Bobechko B, Boutilier K, Burgess E, Buzadzija K, Cavero R, D’Abreo C, Donaldson I, Dorairajoo D, Dumontier MJ, Dumontier MR, Earles V, Farrall R, Feldman H, Garderman E, Gong Y, Gonzaga R, Grytsan V, Gryz E, Gu V, Haldorsen E, Halupa A, Haw R, Hrvojic A, Hurrell L, Isserlin R, Jack F, Juma F, Khan A, Kon T, Konopinsky S, Le V, Lee E, Ling S, Magidin M, Moniakis J, Montojo J, Moore S, Muskat B, Ng I, Paraiso JP, Parker B, Pintilie G, Pirone R, Salama JJ, Sgro S, Shan T, Shu Y, Siew J, Skinner D, Snyder K, Stasiuk R, Strumpf D, Tuekam B, Tao S, Wang Z, White M, Willis R, Wolting C, Wong S, Wrong A, Xin C, Yao R, Yates B, Zhang S, Zheng K, Pawson T, Ouellette BF, Hogue CW. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xenarios I, Salwínski L, Duan XJ, Higney P, Kim S, Eisenberg D. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, Kohler C, Khadake J, Leroy C, Liban A, Lieftink C, Montecchi-Palazzi L, Orchard S, Risse J, Robbe K, Roechert B, Thorneycroft D, Zhang Y, Apweiler R, Hermjakob H. IntAct—open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lopez G, Valencia A, Tress M. FireDB—a database of functionally important residues from proteins of known structure. Nucleic Acids Res. 2007;35:D219–D223. doi: 10.1093/nar/gkl897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hendlich M, Bergner A, Günther J, Klebe G. Relibase: design and development of a database for comprehensive analysis of protein–ligand interactions. J Mol Biol. 2003;326:607–620. doi: 10.1016/s0022-2836(02)01408-0. [DOI] [PubMed] [Google Scholar]
- 57.Benson ML, Smith RD, Khazanov NA, Dimcheff B, Beaver J, Dresslar P, Nerothin J, Carlson HA. Binding MOAD, a high-quality protein–ligand database. Nucleic Acids Res. 2008;36:D674–D678. doi: 10.1093/nar/gkm911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O’Donoghue SI, Meyer JEW, Schafferhans A, Fries K. The SRS 3D module: integrating structures, sequences and features. Bioinformatics. 2004;20:2476–2478. doi: 10.1093/bioinformatics/bth260. [DOI] [PubMed] [Google Scholar]
- 59.Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcántara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boeckmann B, Blatter M, Famiglietti L, Hinz U, Lane L, Roechert B, Bairoch A. Protein variety and functional diversity: Swiss-Prot annotation in its biological context. CR Biol. 2005;328:882–899. doi: 10.1016/j.crvi.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 61.Wollscheid B, Bausch-Fluck D, Henderson C, O’Brien R, Bibel M, Schiess R, Aebersold R, Watts JD. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat Biotechnol. 2009;27:378–386. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Farriol-Mathis N, Garavelli JS, Boeckmann B, Duvaud S, Gasteiger E, Gateau A, Veuthey A, Bairoch A. Annotation of post-translational modifications in the Swiss-Prot knowledge base. Proteomics. 2004;4:1537–1550. doi: 10.1002/pmic.200300764. [DOI] [PubMed] [Google Scholar]
- 64.Garavelli JS. The RESID Database of Protein Modifications as a resource and annotation tool. Proteomics. 2004;4:1527–1533. doi: 10.1002/pmic.200300777. [DOI] [PubMed] [Google Scholar]
- 65.Braconi Quintaje S, Orchard S. The annotation of both human and mouse kinomes in UniProtKB/Swiss-Prot: one small step in manual annotation, one giant leap for full comprehension of genomes. Mol Cell Proteomics. 2008;7:1409–1419. doi: 10.1074/mcp.R700001-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Topiol S, Sabio M. X-ray structure breakthroughs in the GPCR transmembrane region. Biochem Pharmacol. 2009;78:11–20. doi: 10.1016/j.bcp.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 67.Wang Y, Zhang Y, Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–180. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- 68.Ben-Shem A, Fass D, Bibi E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc Natl Acad Sci USA. 2007;104:462–466. doi: 10.1073/pnas.0609773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321:1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tusnády GE, Dosztányi Z, Simon I. Transmembrane proteins in the Protein Data Bank: identification and classification. Bioinformatics. 2004;20:2964–2972. doi: 10.1093/bioinformatics/bth340. [DOI] [PubMed] [Google Scholar]
- 71.Bagos PG, Liakopoulos TD, Hamodrakas SJ. Evaluation of methods for predicting the topology of beta-barrel outer membrane proteins and a consensus prediction method. BMC Bioinformatics. 2005;6:7. doi: 10.1186/1471-2105-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. X-ray structure of a voltage-dependent K + channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 73.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K + channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 74.Bauer M, Pelkmans L. A new paradigm for membrane-organizing and -shaping scaffolds. FEBS Lett. 2006;580:5559–5564. doi: 10.1016/j.febslet.2006.08.077. [DOI] [PubMed] [Google Scholar]
- 75.Hadders MA, Beringer DX, Gros P. Structure of C8alpha-MACPF reveals mechanism of membrane attack in complement immune defense. Science. 2007;317:1552–1554. doi: 10.1126/science.1147103. [DOI] [PubMed] [Google Scholar]
- 76.Olson R, Gouaux E. Crystal structure of the Vibrio cholerae cytolysin (VCC) pro-toxin and its assembly into a heptameric transmembrane pore. J Mol Biol. 2005;350:997–1016. doi: 10.1016/j.jmb.2005.05.045. [DOI] [PubMed] [Google Scholar]
- 77.Cowan-Jacob SW, Fendrich G, Floersheimer A, Furet P, Liebetanz J, Rummel G, Rheinberger P, Centeleghe M, Fabbro D, Manley PW. Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia. Acta Crystallogr D Biol Crystallogr. 2007;63:80–93. doi: 10.1107/S0907444906047287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Antonarakis SE, Cooper DN. Mutations in human genetic disease. In: Cooper DN, editor. Encyclopedia of the human genome. London: Nature Publishing Group; 2003. pp. 227–253. [Google Scholar]
- 79.Yip YL, Famiglietti M, Gos A, Duek PD, David FPA, Gateau A, Bairoch A. Annotating single amino acid polymorphisms in the UniProt/Swiss-Prot knowledgebase. Hum Mutat. 2008;29:361–366. doi: 10.1002/humu.20671. [DOI] [PubMed] [Google Scholar]
- 80.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, Madsen AØ, Riekel C, Eisenberg D. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 81.Nelson R, Sawaya MR, Balbirnie M, Madsen AØ, Riekel C, Grothe R, Eisenberg D. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ivanova MI, Thompson MJ, Eisenberg D. A systematic screen of beta(2)-microglobulin and insulin for amyloid-like segments. Proc Natl Acad Sci USA. 2006;103:4079–4082. doi: 10.1073/pnas.0511298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gerber R, Tahiri-Alaoui A, Hore PJ, James W. Oligomerization of the human prion protein proceeds via a molten globule intermediate. J Biol Chem. 2007;282:6300–6307. doi: 10.1074/jbc.M608926200. [DOI] [PubMed] [Google Scholar]
- 84.Chiti F, Dobson CM. Amyloid formation by globular proteins under native conditions. Nat Chem Biol. 2009;5:15–22. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 85.Wiltzius JJW, Sievers SA, Sawaya MR, Eisenberg D. Atomic structures of IAPP (amylin) fusions suggest a mechanism for fibrillation and the role of insulin in the process. Protein Sci. 2009;18:1521–1530. doi: 10.1002/pro.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johnson SM, Connelly S, Wilson IA, Kelly JW. Toward optimization of the linker substructure common to transthyretin amyloidogenesis inhibitors using biochemical and structural studies. J Med Chem. 2008;51:6348–6358. doi: 10.1021/jm800435s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Redfern OC, Dessailly B, Orengo CA. Exploring the structure and function paradigm. Curr Opin Struct Biol. 2008;18:394–402. doi: 10.1016/j.sbi.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Todd AE, Orengo CA, Thornton JM. Evolution of function in protein superfamilies, from a structural perspective. J Mol Biol. 2001;307:1113–1143. doi: 10.1006/jmbi.2001.4513. [DOI] [PubMed] [Google Scholar]
- 89.Juncker A, Jensen L, Pierleoni A, Bernsel A, Tress M, Bork P, von Heijne G, Valencia A, Ouzounis C, Casadio R, Brunak S. Sequence-based feature prediction and annotation of proteins. Genome Biol. 2009;10:206. doi: 10.1186/gb-2009-10-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chothia C, Gough J. Genomic and structural aspects of protein evolution. Biochem J. 2009;419:15–28. doi: 10.1042/BJ20090122. [DOI] [PubMed] [Google Scholar]
- 91.Zito E, Fraldi A, Pepe S, Annunziata I, Kobinger G, Di Natale P, Ballabio A, Cosma MP. Sulphatase activities are regulated by the interaction of sulphatase-modifying factor 1 with SUMF2. EMBO Rep. 2005;6:655–660. doi: 10.1038/sj.embor.7400454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zanotti G, Cendron L, Ramazzina I, Folli C, Percudani R, Berni R. Structure of zebra fish HIUase: insights into evolution of an enzyme to a hormone transporter. J Mol Biol. 2006;363:1–9. doi: 10.1016/j.jmb.2006.07.079. [DOI] [PubMed] [Google Scholar]
- 93.Piatigorsky J, O’Brien WE, Norman BL, Kalumuck K, Wistow GJ, Borras T, Nickerson JM, Wawrousek EF. Gene sharing by delta-crystallin and argininosuccinate lyase. Proc Natl Acad Sci USA. 1988;85:3479–3483. doi: 10.1073/pnas.85.10.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Markley JL, Aceti DJ, Bingman CA, Fox BG, Frederick RO, Makino S, Nichols KW, Phillips GN, Primm JG, Sahu SC, Vojtik FC, Volkman BF, Wrobel RL, Zolnai Z. The Center for Eukaryotic Structural Genomics. J Struct Funct Genomics. 2009;10:165–179. doi: 10.1007/s10969-008-9057-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fogg MJ, Alzari P, Bahar M, Bertini I, Betton JM, Burmeister WP, Cambillau C, Canard B, Corrondo MA, Coll M, Daenke S, Dym O, Egloff MP, Enguita FJ, Geerlof A, Haouz A, Jones TA, Ma Q, Manicka SN, Migliardi M, Nordlund P, Owens RJ, Peleg Y, Schneider G, Schnell R, Stuart DI, Tarbouriech N, Unge T, Wilkinson AJ, Wilmanns M, Wilson KS, Zimhony O, Grimes JM. Application of the use of high-throughput technologies to the determination of protein structures of bacterial and viral pathogens. Acta Crystallogr D Biol Crystallogr. 2006;62:1196–1207. doi: 10.1107/S0907444906030915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gileadi O, Knapp S, Lee WH, Marsden BD, Müller S, Niesen FH, Kavanagh KL, Ball LJ, von Delft F, Doyle DA, Oppermann UC, Sundström M. The scientific impact of the Structural Genomics Consortium: a protein family and ligand-centered approach to medically-relevant human proteins. J Struct Funct Genomics. 2007;8:107–119. doi: 10.1007/s10969-007-9027-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shin DH, Hou J, Chandonia J, Das D, Choi I, Kim R, Kim S. Structure-based inference of molecular functions of proteins of unknown function from Berkeley Structural Genomics Center. J Struct Funct Genomics. 2007;8:99–105. doi: 10.1007/s10969-007-9025-4. [DOI] [PubMed] [Google Scholar]
- 98.Matte A, Sivaraman J, Ekiel I, Gehring K, Jia Z, Cygler M. Contribution of structural genomics to understanding the biology of Escherichia coli . J Bacteriol. 2003;185:3994–4002. doi: 10.1128/JB.185.14.3994-4002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dessailly BH, Nair R, Jaroszewski L, Fajardo JE, Kouranov A, Lee D, Fiser A, Godzik A, Rost B, Orengo C. PSI-2: structural genomics to cover protein domain family space. Structure. 2009;17:869–881. doi: 10.1016/j.str.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nair R, Liu J, Soong T, Acton TB, Everett JK, Kouranov A, Fiser A, Godzik A, Jaroszewski L, Orengo C, Montelione GT, Rost B. Structural genomics is the largest contributor of novel structural leverage. J Struct Funct Genomics. 2009;10:181–191. doi: 10.1007/s10969-008-9055-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sala C, Haouz A, Saul FA, Miras I, Rosenkrands I, Alzari PM, Cole ST. Genome-wide regulon and crystal structure of BlaI (Rv1846c) from Mycobacterium tuberculosis . Mol Microbiol. 2009;71:1102–1116. doi: 10.1111/j.1365-2958.2008.06583.x. [DOI] [PubMed] [Google Scholar]
- 102.Revington M, Semesi A, Yee A, Shaw GS. Solution structure of the Escherichia coli protein ydhR: a putative mono-oxygenase. Protein Sci. 2005;14:3115–3120. doi: 10.1110/ps.051809305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Punta M, Ofran Y. The rough guide to in silico function prediction, or how to use sequence and structure information to predict protein function. PLoS Comput Biol. 2008;4:e1000160. doi: 10.1371/journal.pcbi.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rentzsch R, Orengo CA. Protein function prediction—the power of multiplicity. Trends Biotechnol. 2009;27:210–219. doi: 10.1016/j.tibtech.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 105.Addou S, Rentzsch R, Lee D, Orengo CA. Domain-based and family-specific sequence identity thresholds increase the levels of reliable protein function transfer. J Mol Biol. 2009;387:416–430. doi: 10.1016/j.jmb.2008.12.045. [DOI] [PubMed] [Google Scholar]
- 106.Watson JD, Sanderson S, Ezersky A, Savchenko A, Edwards A, Orengo C, Joachimiak A, Laskowski RA, Thornton JM. Towards fully automated structure-based function prediction in structural genomics: a case study. J Mol Biol. 2007;367:1511–1522. doi: 10.1016/j.jmb.2007.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]