Abstract
An intercross between atherosclerosis susceptible (C57BL/6J ApoE0) and resistant (FVB/N ApoE0) mice revealed a susceptibility locus on chromosome 10 (11 cM, logarithm of odds 7.8). Surprisingly, the genotypic means for this locus revealed that heterozygosity or homozygosity for the C57BL/6J allele was associated with decreased atherosclerosis. A candidate gene in this region is A20, which is involved in the feedback suppression of NFκB activation induced by tumor necrosis factor α (TNFα). We sequenced the A20 gene coding region from the parental strains and found a single-nucleotide polymorphism resulting in a single amino acid exchange, Glu627Ala (C57BL/6J vs. FVB/N). This mutation introduces a putative casein kinase 2 phosphorylation site in C57BL/6J-A20 not present in FVB/N-A20. NFκB reporter gene assays showed that this amino acid change results in less effective termination of TNFα-stimulated NFκB activation by C57BL/6J-A20. In accordance, the TNFα-induced expression of NFκB target genes (A20, IκBα) in vascular smooth muscle cells was prolonged in cells isolated from C57BL/6J compared with FVB/N mice. In light of the genotypic means for atherosclerosis at the chromosome 10 locus in F2 mice from this intercross, the observations now reported suggest that prolonged expression of genes induced by NFκB might be antirather than proatherogenic.
Atherosclerotic coronary heart disease is the most common cause of death in westernized countries. It is a complex genetic disease, and disease occurrence is determined by genetic factors, environmental exposure, and important gene–environment interactions. A number of risk factors have been identified, including cigarette smoking, dyslipidemia, hypertension, diabetes, obesity, and physical inactivity (1, 2). However, for a given level of each of these risk factors, individuals vary in their susceptibility to coronary heart disease; presumably on a genetic basis (3). This variation in artherosclerosis susceptibility can be modeled in the mouse. In this species, several induced mutations that sufficiently raise non-high-density lipoprotein cholesterol levels have been shown to result in the development of human-like atherosclerotic lesions (4). However, large differences in lesion size occur depending on the genetic background of the mouse strain, implying the presence of modifier genes that influence susceptibility either by changing known risk factors or by influencing other pathways important in lesion formation, such as how the blood vessel wall responds to insults like hyperlipidemia, hypertension, or diabetes.
In previous work, we showed that targeted disruption of the apolipoprotein E (ApoE) gene greatly elevated non-high-density lipoprotein cholesterol levels and resulted in the formation of atherosclerotic lesions at the aortic sinus, proximal coronary arteries, brachiocephalic artery, lesser curvature of the aorta, and aortic branch points (5, 6). These lesions progressed from fatty streaks to fibrous plaques, and lesion progression could be accelerated by feeding a western-type diet. Breeding the ApoE knockout trait onto the C57BL/6J and FVB/N backgrounds, weaning these mice to a chow diet, and killing them at 16 weeks of age revealed 7- to 9-fold larger lesions in the aortic sinus in the former compared with the latter (7). An intercross was performed between these two strains, and quantitative trait locus mapping for atherosclerotic lesion area in the F2 mice revealed three atherosclerosis susceptibility loci. The most significant locus on proximal chromosome 10 had a logarithm of odds (lod) score of 7.8 and accounted for 19% of the variance of lesion area in the F2s. The effect of this locus was independent of plasma cholesterol levels. The genotypic means for the marker at the peak of the lod, D10Mit213 (11 cM), indicated that homozygotes for the FVB/N allele had 2-fold more atherosclerosis than either heterozygotes or homozygotes for the C57BL/6J allele (8). Although this finding was counter to our expectations based on the phenotypes of the parentals, it does commonly occur in mouse intercrosses and suggests interactions between C57BL/6J alleles in other parts of the genome and the FVB/N atherosclerosis susceptibility allele at proximal chromosome 10.
A candidate gene for atherosclerosis susceptibility that maps to mouse chromosome 10 at 13 cM is A20, also called tumor necrosis factor α induced protein 3 (TNFaip3). One function of A20 is to terminate the activation of NFκB that follows stimulation by various agents, including TNFα (9, 10). NFκB is a transcription factor that regulates the expression of inflammatory and survival genes and might play an important role in atherosclerosis (11). Thus far, it is not clear what role this transcription factor plays during lesion development. Although frequent associations have been found between the activation of NFκB and/or its target genes and atherosclerosis, one published study using TNF receptor p55 knockout mice indicated the opposite (12). NFκB is maintained in an inactive state in the cytoplasm bound to the inhibitory protein inhibitor of NFκBα (IκBα), which masks its nuclear localization domain (13). Phosphorylation marks IκBα for degradation by the proteasome. This process is mediated by the IκB kinase (IKK) complex (14, 15). NFκB terminates its own activation by inducing the expression of its inhibitor IκBα, which then binds, inactivates, and translocates the transcription factor back to the cytosol (16). NFκB also induces the expression of A20, which inhibits IKK-complex activity, thus supporting the reaccumulation of IκBα in the cytosol (10, 17). In fact, A20 knockout mice develop severe inflammation and tissue damage in multiple organs and die prematurely at 3–6 weeks of age. Cells isolated from these mice fail to terminate the TNFα-induced NFκB activation due to a lack of IκBα protein reaccumulation (10).
In the current study, we assessed the chromosome 10 candidate gene A20 in C57BL/6J and FVB/N mice. Sequencing A20 cDNA revealed a difference between the strains, resulting in an amino acid exchange Glu627Ala (C57BL/6J vs. FVB/N), which introduces a putative casein kinase 2 (CKII) phosphorylation site in C57BL/6J. In transfection assays using A20 expression plasmids and an NFκB reporter gene, C57BL/6J-A20 was less effective in shutting down TNFα-induced NFκB activity than FVB/N-A20. Compatible with this decreased potency of C57BL/6J-A20 were studies in vascular smooth muscle cells (VSMCs) showing that TNFα stimulation resulted in prolonged expression of A20 as well as IκBα in cells isolated from C57BL/6J compared with FVB/N mice. Taken together with our previous finding of the genotypic means for atherosclerosis in the F2s, the results now reported suggest that the C57BL/6J, which allows prolonged NFκB activation, is actually antiatherogenic.
Methods
Animals. Wild-type C57BL/6J (stock no. 000664) and FVB/N (stock no. 001800) were purchased from The Jackson Laboratory. ApoE heterozygous knockouts generated as C57BL/6J×129Sv/ter F1s were backcrossed to either wild-type C57BL/6J or FVB/N mice for 10 generations and then bred to homozygosity. The animals were housed in The Rockefeller University Laboratory Animal Research Center in a specific pathogen-free environment in rooms with a 7:00 a.m. to 7:00 p.m. light/dark cycle. All procedures involving mice were approved by The Rockefeller University's Institutional Animal Care and Use Committee. Two-month-old mice on a chow diet were used for all studies.
Isolation of Total RNA and cDNA Generation for Expression Analysis by Real-Time PCR and Sequencing. RNA was isolated from tissue and cell samples by using the Trizol reagent (Invitrogen) according to the manufacturer's protocol. To remove contaminating DNA, the DNA-free Kit from Ambion (Austin, TX) was used. cDNA was generated from total RNA with the SuperScript II Rnase H– Reverse Transcriptase Kit (Invitrogen) by using 1–2 μg of total RNA as template and a 1:1 mixture of Oligo(dT) and Hexamer primers. The resulting cDNA pool was used for real-time PCR. The cDNA pool generated from the liver of mice treated with TNFα was used to amplify the A20 coding region, which was then cloned into the pCR2.1 Topo expression plasmid (Invitrogen). Three independent colonies were sequenced over the entire coding region (2,328 bp) per strain. The region corresponding to the coding sequence difference between the strains was sequenced in genomic DNA from five C57BL/6J and five FVB/N mice and from purchased genomic DNA of the following strains: 129, AKR, BALB/C, DBA, CBA, SJL, and SWR (The Jackson Laboratory).
Real-Time PCR. A20 and IκBα expression was quantified by using cDNAs from tissue and cell samples by real-time PCR. The Sequence Detection System 7700 (Applied Biosystems) was used for amplification and specific sequence detection. Forward and reverse PCR primers were used at a final concentration of 300 nM, and probes, containing a 5′ fluorophor (6-FAM) and a 3′ quencher (TAMRA), were used at a final concentration of 100 nM. Expression of A20 and IκBα was evaluated relative to the mRNA expression of the housekeeping gene β-actin in the same sample. Cycling parameters were: 2 min, 50°C; 10 min, 95°C; and 40 times: 30 s, 95°C; 1 min, 60°C. Primers and probes were purchased from Applied Biosystems.
A20 Expression Plasmid Construction and Transfection into Human Embryonic Kidney (HEK)293 Cells in an NFκB Reporter Gene Assay. The A20 coding region of both strains was cut out of the pCR2.1 Topo cloning vector and the resulting DNA fragments cloned into a pcDNA3 mammalian expression vector by using T4-DNA Ligase (NEB, Beverly, MA). The pcDNA3 vector introduced a hemagglutinin and a Flag tag at the N terminus of A20 just after the translation start ATG. Two mutant forms of A20 were also generated. In both the C57BL/6J-A20 and FVB/N-A20 expression vectors, a protein kinase C (PKC) phosphorylation site was deleted by changing serine 626 to alanine. The original expression vectors and the two mutated ones were used to perform NFκB reporter gene assays with a vector containing luciferase driven by five NFκB responsive sites (Stratagene). HEK293 cells purchased from American Type Culture Collection were grown to 70% confluence on collagen-coated 12-well plates. The cells were cotransfected with 0.05 μg of a β-galactosidase (β-gal) normalization vector; 0.15 μgofaNFκB reporter gene; and 0.01, 0.02, 0.05, or 0.1 μg of each of the four expression vectors by using Lipofectamine Plus (Invitrogen) according to the manufacturer's instructions. Twenty hours posttransfection, human TNFα was added to a final concentration of 30 ng/ml. Four hours later, the cells were harvested in Reporter Lysis Buffer (Promega). Luciferase activity was measured by using the Luciferase Detection System (Promega) and β-gal activity determined.
VSMC Culture. Mice anesthetized with Nembutol (Abbott) were perfused through the left ventricle with 10 ml of PBS. The thoracic aorta was visualized to remove fat and connective tissue, and the vessel opened up longitudinally in situ, removed, and placed in sterile Hanks' balanced salt solution (HBSS). Two thoracic aortas from each strain were then placed in sterile HBSS containing 175 units/ml Type II collagenase (Worthington) and incubated in a temperature-controlled air oven for 15 min at 37°C. The intimal surface was then gently scraped with a sterilized Q-Tip to remove endothelial cells and the adventitia peeled off with forceps under a dissecting microscope. The remaining thoracic aorta was placed in 10% FBS–DMEM supplemented with penicillin/streptomycin and incubated in a tissue culture incubator overnight. The vessel was then digested in sterile HBSS containing 175 units/ml Type II collagenase and 0.5 mg/ml Type I Elastase (Sigma) in a temperature-controlled air oven for 90 min at 37°C. During the incubation, the solution containing cells was drawn up and down several times into a Pasteur pipette and at the end transferred into 10 ml of 10% FBS–DMEM to stop the enzymatic digestion. After spinning (5 min, 150 × g, room temperature), VSMCs were resuspended in 20% FBS–DMEM containing penicillin/streptomycin and plated in a 60-mm Petri dish. The medium was changed every 2 d. The cells started to divide within 3 d and reached 70% confluence in 6 d, when they were trypsinized and split 1:3. After the second passage, the medium was switched to 10% FBS–DMEM supplemented with penicillin/streptomycin. Cells between passages four and nine were used for experiments. The purity of each preparation was verified by immunostaining for α-actin (anti-human smooth muscle actin/horseradish peroxidase, DAKO). In all experiments, the medium FBS concentration was reduced to 1%, and the cells were incubated in 30 ng/ml human TNFα.
Western Blotting. For Western blotting, HEK293 cells were lysed in 1× Reporter Lysis Buffer (Promega), cytosolic protein chromatographed by SDS/PAGE, and the separated proteins transferred onto a nitrocellulose membrane (Invitrogen) by electroblotting. The amount of cytosolic protein loaded per lane was normalized to β-gal expression. The primary monoclonal (mouse) antihemagluttinin tag antibody (Sigma) was diluted (1:1,000) in TBS containing 0.1% Tween and applied overnight at 4°C. In the case of the VSMCs, the same procedure was followed, except that 20 μg of cytosolic protein was used per lane, and the primary polyclonal (goat) anti IκBα antibody (Santa Cruz Biotechnology) was diluted (1:150) in casein blocker (Pierce) and applied overnight at 4°C. Secondary antibodies conjugated to horseradish peroxidase (Calbiochem) were used at a 1:10,000 dilution. For visualization, the Western Lightning substrate (Perkin–Elmer) was used.
The sequences of the real-time PCR primers and probes and sequencing primers are in Supporting Data Set, which is published as supporting information on the PNAS web site.
Results
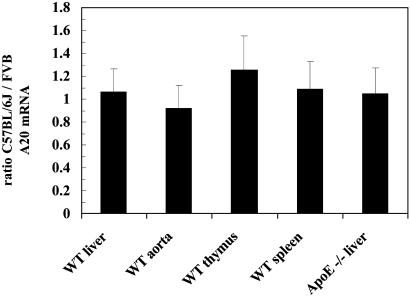
Basal A20 Expression in Tissues. The level of A20 expression was compared between C57BL/6J and FVB/N mice. A20 mRNA levels were quantified by real-time PCR in liver, aorta, thymus, and spleen of wild-type and in liver of ApoE knockout mice. The ratios of the mRNA levels in C57BL/6J to FVB/N are shown in Fig. 1. No difference in A20 expression was found in any of the tissues examined.
Fig. 1.
Expression of A20 mRNA in different tissues from parental mice. Ratio (C57BL/6J to FVB/N) of A20 mRNA expression levels in various tissues determined by real-time PCR and normalized to β-actin. Averages of three independent experiments ± SD are shown.
TNFα Induction of A20 Expression in Liver. Basal expression of A20 is low but can be induced quickly through NFκB activation by a proinflammatory signal such as TNFα. To compare A20 mRNA induction in C57BL/6J and FVB/N mice, five mice per strain were injected with either TNFα (0.1 μg/g of body weight) or PBS as a control and killed 2 h postinjection. TNFα injection caused similar inductions (40-fold) of A20 expression in livers of both strains (data not shown).
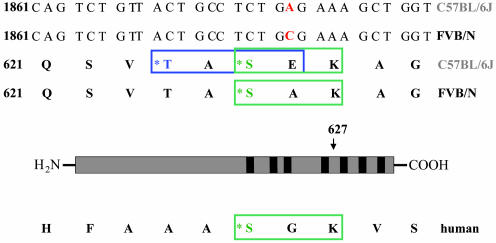
A20 Coding Region Sequence. To sequence the coding region of A20 from both strains, liver cDNA from C57BL/6J and FVB/N mice was generated and cloned into the pCR2.1 expression plasmid (Invitrogen). Sequencing of the complete coding region of A20 (2,328 bp) from three independent clones per strain revealed a difference at base pair 1880 resulting in an amino acid exchange Glu627Ala (C57BL/6J vs. FVB/N) (Fig. 2). The coding difference was located between zinc fingers 4 and 5 and generated a putative CKII phosphorylation site in C57BL/6J adjacent to a PKC phosphorylation site. There is high-sequence variability in this region between mouse and human (Fig. 2). In humans, the lack of a threonine at the residue corresponding to 624 in the mouse precludes the introduction of a second phosphorylation site in this region. However, the PKC site is conserved between the two species. Genomic DNA from 129, AKR, BALB/C, DBA, CBA, SJL, and SWR mice was sequenced in the region of the A20 gene containing the amino acid exchange Glu627Ala (C57BL/6J vs. FVB/N). All strains examined showed the FVB/N form of the gene.
Fig. 2.
Sequence comparison of C57BL/6J-, FVB/N-, and human A20 cDNA. The amino acid residue 627 that differs between C57BL/6J-A20 and FVB/N-A20 is labeled in red. The putative CKII phosphorylation site introduced in the C57BL/6J sequence is marked by a blue square, whereas the conserved PKC site is framed in green. The asterisks indicate the amino acids that could be phosphorylated by the respective kinases.
Functionality of the A20 Coding Difference. To compare the ability of C57BL/6J-A20 and FVB/N-A20 variants to diminish TNFα-induced NFκB activation, subconfluent HEK293 cells were transfected with increasing amounts of either C57BL/6J-A20 or FVB/N-A20 expression vector and an NFκB reporter plasmid for 20 h. The cultures were then incubated for 4 h with30ng/ml human TNFα and NFκB activity assessed. Because A20 does not interfere with TNFα-induced NFκB nuclear translocation and DNA binding, this transfection assay was felt to be the best way of comparing the abilities of different forms of A20 to shut down TNFα-stimulated NFκB activity (18, 19). A dose–response curve was observed with increasing amounts of A20 expression plasmid resulting in more inhibition (Fig. 3). C57BL/6J-A20 was less effective than FVB/N-A20, which was most apparent at 0.01 μg of A20 expression plasmid, where C57BL/6J-A20 reduced NFκB activity 50%, whereas FVB/N-A20 achieved an 80% inhibition. Increasing the amounts of the A20 expression plasmids further decreased NFκB activity and gradually narrowed the difference between C57BL/6J and FVB/N until it was no longer significant at 0.1 μg. From the shape of the dose–response curves, we estimate that C57BL/6J-A20 has one-fifth the potency of FVB/N-A20 in this assay.
Fig. 3.
Activity of A20 with mutated phosphorylation sites. HEK293 cells were transfected with the indicated amounts of C57BL/6J-A20, FVB/N-A20, CKII-A20, or noPhos-A20 in addition to an NFκB reporter and a β-gal normalization plasmid. After 4 h of stimulation with TNFα (30 ng/ml), β-gal activity was determined, and NFκB activity was quantified by using a commercial luciferase assay. Shown are averages of nine independent experiments ± SD. (Inset) A20 expression normalized to total protein content (0.01 μg of A20 DNA), visualized by Western blotting, to demonstrate equal expression of different A20 mutants. Lane 1, C57BL/6J-A20; lane 2, CKII-A20; lane 3, FVB/N-A20; lane 4, noPhos-A20. *, P < 0.05; **, P < 0.001.
To further elucidate the importance of each phosphorylation site for A20-mediated termination of NFκB expression, two mutant A20 expression vectors were generated. One of these contained neither the CKII nor the PKC phosphorylation site (noPhos-A20), and the other had a sole CKII phosphorylation site (CKII-A20). In the NFκB reporter gene assay described above, the noPhos-A20 mutant was as effective in terminating the TNFα-induced NFκB activity as FVB/N-A20. On the other hand, the CKII-A20 mutant functionally resembled C57BL/6J-A20 (Fig. 3). These results suggest that the presence of the CKII phosphorylation site diminishes the ability of A20 to decrease NFκB activity, whereas the presence or absence of the PKC phosphorylation site has no effect.
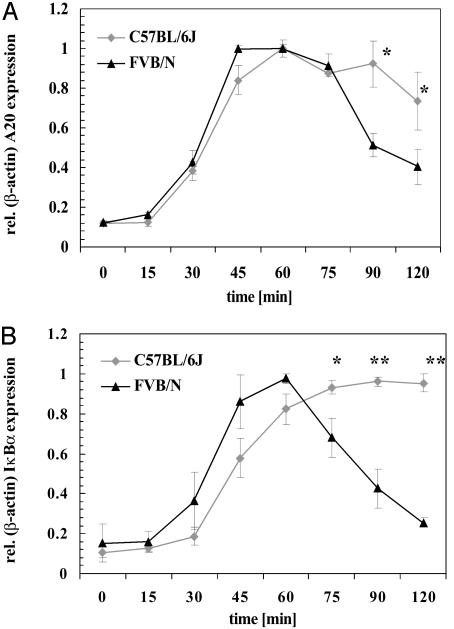
TNFα Induction of A20 and IκBα Expression in VSMCs. TNFα stimulated activation of NFκB results in increased expression of its target genes, A20 and IκBα, both of which then feed back to inhibit NFκB activation. Thus prolonged expression of A20 and IκBα indicates prolonged activity of the NFκB transcription factor. The functional differences revealed in Fig. 3 between C57BL/6J-A20 and FVB/N-A20 could affect this feedback mechanism and alter expression of these target genes. Therefore, the time course of TNFα stimulation of A20 and IκBα expression was studied in cultures of VSMCs isolated from each strain. Cultures were incubated with 30 ng/ml TNFα for up to 2 h. The initial induction of A20 mRNA was similar for both strains through 75 min, including the maximal expression level at ≈40–60 min. Subsequently, at 90 and 120 min, A20 mRNA levels were higher in C57BL/6J compared with FVB/N VSMCs (Fig. 4A). IκBα mRNA measurements showed an almost identical expression pattern (Fig. 4B). Both results indicate that NFκB activity is prolonged after TNFα stimulation in C57BL/6J compared with FVB/N VSMCs. The maximum amount of A20 mRNA found in VSMCs after TNFα induction is similar to that found in 293 cells transfected with 0.01 μg of either C57BL/6J-A20 or FVB/N-A20 cDNA (data not shown).
Fig. 4.
Expression of A20 and IκBα after TNFα stimulation in VSMCs. VSMCs isolated from C57BL/6J and FVB/N mice were stimulated with TNFα (30 ng/ml) for the indicated times before preparation of RNA and determination of A20 (A) or IκBα (B) expression levels via real-time PCR. Data shown represent mean ± SD of three independent experiments, i.e., three different cell preparations followed by TNFα incubation, RNA isolation, and expression analysis. *, P < 0.05; **, P < 0.001.
TNFα Regulation of IκBα Protein Levels in VSMCs. NFκB is transcriptionally active when it is not bound to its inhibitor IκBα. Degradation of IκBα is initiated via phosphorylation mediated by the IKK complex. The IKK complex is activated by a variety of stimuli, including TNFα. As part of the feed-back regulation after NFκB activation IκBα mRNA expression is induced thus leading to reaccumulation of IκBα protein and to inactivation of NFκB. A20 deficient mice fail to reaccumulate IκBα after being stimulated with TNFα due to a persistently active IKK complex. This suggests that A20 exerts its NFκB inhibitory function by supporting the reaccumulation of IκBα protein in the cytosol (10).
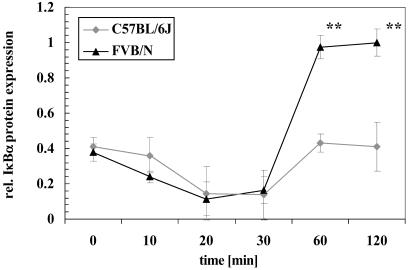
We next tested whether the reduced ability of C57BL/6J-A20 compared with FVB/N-A20 to terminate TNFα-induced NFκB activation results in differences in the reaccumulation of IκBα protein. VSMCs of both strains were incubated with TNFα for different time intervals and cytosolic protein extracts subjected to Western blotting with an IκBα specific antibody (Fig. 5). The degradation of the IκBα protein follows the same time course in VSMCs from both strains reaching a nadir 20 min after addition of TNFα. This is followed by recovery of cytosolic IκBα protein by 60 min to much higher levels in FVB/N compared with C57BL/6J VSMCs. This may reflect increased turnover of IκBα in the latter due to weaker A20.
Fig. 5.
IκBα protein level after TNFα stimulation in VSMCs. VSMCs isolated from C57BL/6J or FVB/N mice were incubated with TNFα (30 ng/ml) for the indicated times. Afterward, cells were harvested and lysed, and the resulting cell extracts were subjected to Western blot analysis by using an antibody specific for IκBα. Protein expression was quantified by laser densitometry. The data represent means ± SD of three independent experiments. *, P < 0.05; **, P < 0.001.
Discussion
The NFκB-regulated gene A20, which is involved in the feedback termination of NFκB activity, is located at an atherosclerosis susceptibility locus on mouse chromosome 10. This locus was revealed by quantitative trait locus mapping by using second-generation mice of an intercross between atherosclerosis-sensitive C57BL/6J ApoE0 and atherosclerosis-resistant FVB/N ApoE0 mice (8). In the current study, a coding difference leading to an amino acid exchange Glu627Ala (C57BL/6J vs. FVB/N) was identified between the two strains. In an NFκB reporter gene assay, the C57BL/6J form of A20 was shown to be less potent in shutting down TNFα-stimulated NFκB activity than the FVB/N form. Compatible with this, expression of the NFκB target genes A20 and IκBα was prolonged in VSMCs isolated from C57BL/6J mice in comparison to cells from FVB/N mice. Taken together, these findings suggest that NFκB activation is extended in C57BL/6J mice in comparison to FVB/N mice due to a reduction in A20 functionality.
The A20 gene encodes an 80-kDa protein that contains 7 Cys-2/Cys-2 zinc finger structures in its C-terminal domain (20). This region of A20 was shown to mediate NFκB inhibition, A20 homodimerization, and A20 interaction with other proteins such as IKKγ and A20-binding inhibitors of NFκB activation (9). The amino acid exchange Glu627Ala (C57BL/6J vs. FVB/N) identified in this study is located in the C terminus of A20 between zinc fingers 4 and 5 and generates a potential CKII phosphorylation site in C57BL/6J-A20 adjacent to a PKC phosphorylation site. The use of two mutant forms of A20 in addition to C57BL/6J-A20 and FVB/N-A20 in an NFκB reporter gene assay demonstrated that the sole presence of the putative CKII phosphorylation site is sufficient to attenuate the ability of A20 to terminate TNFα-induced NFκB activation. The actual phosphorylation of this CKII site or the alterations in size and charge of residue 627 by changing alanine (FVB/N) to glutamic acid (C57BL/6J) could result in an A20 conformational change, reducing its ability to interact with proteins necessary for terminating NFκB activity.
TNFα stimulation causes a rapid transient induction of A20 gene expression that is mediated by several conserved NFκB consensus motifs in the promoter region of this gene (17). Basal A20 mRNA levels did not differ between C57BL/6J and FVB/N mice, nor did TNFα-induced A20 mRNA levels differ in livers from these mice. However, a careful time course performed in VSMCs derived from the two strains showed prolonged IκBα and A20 expression after TNFα stimulation in C57BL/6J, likely due to decreased potency of A20 leading to prolonged NFκB activity. By using tissue culture systems, NFκB activity has been shown to be a strictly regulated oscillatory process, which would make it difficult to demonstrate prolonged expression in an in vivo system (21). Thus more careful examination of the expression patterns of the genes involved seems important for tightly regulated systems. The failure to observe differences in A20 mRNA levels in tissues from C57BL/6J and FVB/N mice indicates that gene expression arrays would not have detected the alteration of A20 expression resulting from the difference in A20 functionality between the strains.
A20-deficient mice suffer from severe inflammation and are hypersensitive to TNFα. These mice fail to terminate TNFα-induced NFκB activity as a consequence of a persistently active IKK complex, which inhibits reaccumulation of IκBα protein. Hence NFκB remains active in the nucleus, leading to severe inflammation. These data suggest that A20 might terminate NFκB activity by assisting reaccumulation of IκBα via interacting with proteins involved in TNFα signaling upstream of IκBα degradation (9, 10). In the current study, we demonstrated that IκBα protein does not reaccumulate as efficiently in VSMCs isolated from C57BL/6J mice as it does in cells from FVB/N mice. Thus it is tempting to speculate that what was seen in the A20 knockout mice is partially true for C57BL/6J mice. The amino acid exchange in C57BL/6J-A20 might attenuate the protein's ability to terminate the activity of the IKK complex, thereby delaying the reaccumulation of IκBα and prolonging the transcriptional activity of NFκB.
The genomes of common laboratory strains of mice, including C57BL/6J and FVB/N, are derived from domesticus and Asian mice (primarily musculus and molossinus). Compatible is the observation of two predominant haplotypes in 22 of 27 regions (500–1,000 bp in length) sequenced from nine strains. In comparing any inbred laboratory strain with C57BL/6J, two-thirds of the genome was found to have a low polymorphism rate, indicating its origin from the same ancestors. The other one-third showed divergence with one single-nucleotide polymorphism (SNP) every 200 bp, indicating it arose from a different ancestor than C57BL/6J (22). In the current study, 400 bp surrounding the SNP responsible for the A20 amino acid change Glu627Ala (C57BL/6J vs. FVB/N) were sequenced in 129, DBA and SWR, atherosclerosis-susceptible strains, AKR and BALB/C, atherosclerosis-resistant strains, and CBA and SJL, which have not been characterized with regard to atherosclerosis. The A20 coding change SNP was found only in C57BL/6J mice. There are two interpretations of these findings. It is possible that the A20 gene in C57BL/6J mice is derived from a different ancestor than all of the other strains. However, in this case, one might have expected to find more SNPs in this region. Alternatively, the more likely scenario is that the SNP in A20 identified in this study is a specific mutation that arose in C57BL/6J at an early stage of inbreeding in a region of the genome otherwise shared between the common laboratory strains. To distinguish between these possibilities, more comparative sequencing will have to be done in the A20 region. Nevertheless, this functional mutation would not have been discovered by simply comparing atherosclerosis-sensitive and resistant strains in this region.
Activation of the NFκB pathway has been correlated with atherosclerosis development and progression. In various studies, this correlation has been attributed to NFκB increasing the expression of target genes in the vessel wall, such as cytokines, chemokines, and leukocyte adhesion molecules. In addition, NFκB also increases the expression of genes whose products antagonize apoptosis and promote cell proliferation, thus leading to cell survival. Many people believe that NFκB is a major proatherosclerotic pathway by one or both of these mechanisms (11, 23). However, surprisingly, studies using knockout animals have indicated the opposite. For example, C56BL6/J mice lacking the TNF receptor p55 developed 2.3-fold larger aortic sinus atherosclerotic lesion areas than wild-type mice when fed a high-cholesterol high-fat atherogenic diet, despite similar plasma lipid levels (12). In addition, low-density lipoprotein receptor knockout mice with induced macrophage specific deficiency of IKK2, one of the kinases that phosphorylate the inhibitor IκBα leading to its degradation, showed 60% larger atherosclerotic lesions that were more severe and displayed increased necrosis compared with controls (24). In addition, crossing a knockout of the p50 subunit of NFκB, which was shown to diminish NFκB activity (25), to the ApoE knockout background increased atherosclerotic lesion area 2-fold (H.M.D. and J.L.B, unpublished data). If the candidate gene A20 proves responsible for the atherosclerosis susceptibility locus on chromosome 10 revealed in the C57BL/6J FVB/N ApoE knockout cross, then according to the genotypic means for lesion area at this locus, the C57BL/6J form is antiatherogenic. Because this form of A20 is shown in the current study to terminate NFκB activity less efficiently, taken together, these findings would be compatible with increased NFκB activity being antiatherogenic and would agree with other results obtained with knockout mice of the TNF-NFκB pathway mentioned above. A possible model to explain these results would be that, at least at a certain stage of lesion development, NFκB activation of survival mechanisms overrides its proinflammatory effects and protects against atherosclerosis.
In conclusion, A20 appears to be a promising candidate gene at the chromosome 10 locus for playing a role in the differing susceptibilities to atherosclerosis of C57BL/6J and FVB/N ApoE knockout mice. An amino acid exchange Glu627Ala (C57BL/6J vs. FVB/N) was found to introduce a new potential CKII phosphorylation site in C57BL/6J-A20 and to reduce its ability to terminate NFκB activity. This mutation is likely to be responsible for the prolonged expression of NFκB target genes leading to extended NFκB transcriptional activity in VSMCs from C57BL/6J compared with FVB/N mice after TNFα stimulation.
Supplementary Material
Acknowledgments
This work was supported by the German Academic Exchange Service (Deutscher Akademischer Austauschdienst) (to S.I.) and by National Institutes of Health Grant HL54591–08.
Abbreviations: TNF, tumor necrosis factor; CKII, casein kinase 2; lod, logarithm of odds; IκBα, inhibitor of NFκBα; IKK, IκB kinase; ApoE, apolipoprotein E; VSMC, vascular smooth muscle cell; β-gal, β-galactosidase; HEK, human embryonic kidney; SNP, single-nucleotide polymorphism.
References
- 1.Glass, C. K. & Witztum, J. L. (2001) Cell 104, 503–516. [DOI] [PubMed] [Google Scholar]
- 2.Lusis, A. J. (2000) Nature 407, 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fox, C. S., Polak, J. F., Chazaro, I., Cupples, A., Wolf, P. A., D'Agostino, R. A. & O'Donnell, C. J. (2003) Stroke 34, 397–401. [DOI] [PubMed] [Google Scholar]
- 4.Smith, J. D. (1998) Lab. Anim. Sci. 48, 573–579. [PubMed] [Google Scholar]
- 5.Plump, A. S., Smith, J. D., Hayek, T., Aalto-Setala, K., Walsh, A., Verstuyft, J. G., Rubin, E. M. & Breslow, J. L. (1992) Cell 71, 343–353. [DOI] [PubMed] [Google Scholar]
- 6.Nakashima, Y., Plump, A. S., Raines, E. W., Breslow, J. L. & Ross, R. (1994) Arterioscler. Thromb. 14, 133–140. [DOI] [PubMed] [Google Scholar]
- 7.Dansky, H. M., Charlton, S. A., Sikes, J. L., Heath, S. C., Simantov, R., Levin, L. F., Shu, P., Moore, K. J., Breslow, J. L. & Smith, J. D. (1999) Arterioscler. Thromb. Vasc. Biol. 19, 1960–1968. [DOI] [PubMed] [Google Scholar]
- 8.Dansky, H. M., Shu, P., Donavan, M., Montagno, J., Nagle, D. L., Smutko, J. S., Roy, N., Whiteing, S., Barrios, J., McBride, T. J., et al. (2002) Genetics 160, 1599–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beyaert, R., Heyninck, K. & Van Huffel, S. (2000) Biochem. Pharmacol. 60, 1143–1151. [DOI] [PubMed] [Google Scholar]
- 10.Lee, E. G., Boone, D. L., Chai, S., Libby, S. L., Chien, M., Lodolce, J. P. & Ma, A. (2000) Science 289, 2350–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins, T. & Cybulsky, M. I. (2001) J. Clin. Invest. 107, 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schreyer, S. A., Peschon, J. J. & LeBoeuf, R. C. (1996) J. Biol. Chem. 271, 26174–26178. [DOI] [PubMed] [Google Scholar]
- 13.Baldwin, A. S., Jr. (1996) Annu. Rev. Immunol. 14, 649–683. [DOI] [PubMed] [Google Scholar]
- 14.Israel, A. (2000) Trends Cell Biol. 10, 129–133. [DOI] [PubMed] [Google Scholar]
- 15.Karin, M. & Ben-Neriah, Y. (2000) Annu. Rev. Immunol. 18, 621–663. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh, S. & Karin, M. (2002) Cell 109, Suppl., S81–S96. [DOI] [PubMed] [Google Scholar]
- 17.Krikos, A., Laherty, C. D. & Dixit, V. M. (1992) J. Biol. Chem. 267, 17971–17976. [PubMed] [Google Scholar]
- 18.Heyninck, K., De Valck, D., Vanden Berghe, W., Van Criekinge, W., Contreras, R., Fiers, W., Haegeman, G. & Beyaert, R. (1999) J. Cell Biol. 145, 1471–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zetoune, F. S., Murthy, A. R., Shao, Z., Hlaing, T., Zeidler, M. G., Li, Y. & Vincenz, C. (2001) Cytokine 15, 282–298. [DOI] [PubMed] [Google Scholar]
- 20.Opipari, A. W., Jr., Boguski, M. S. & Dixit, V. M. (1990) J. Biol. Chem. 265, 14705–14708. [PubMed] [Google Scholar]
- 21.Hoffmann, A., Levchenko, A., Scott, M. L. & Baltimore, D. (2002) Science 298, 1241–1245. [DOI] [PubMed] [Google Scholar]
- 22.Wade, C. M., Kulbokas, E. J., III, Kirby, A. W., Zody, M. C., Mullikin, J. C., Lander, E. S., Lindblad-Toh, K. & Daly, M. J. (2002) Nature 420, 574–578. [DOI] [PubMed] [Google Scholar]
- 23.Libby, P. (2002) Nature 420, 868–874. [DOI] [PubMed] [Google Scholar]
- 24.Kanters, E., Gijbels, M., Vergouwe, M., Fijneman, R., Pasparakis, M., Kraal, G., Hofker, M. H. & de Winter, M. P. (2002) Circulation Suppl. II, 120.
- 25.Sha, W. C., Liou, H. C., Tuomanen, E. I. & Baltimore, D. (1995) Cell 80, 321–330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.