Abstract
Clinical outcome prognostication in oncology is a guiding principle in therapeutic choice. A wealth of qualitative empirical evidence links disease progression with tumor morphology, histopathology, invasion, and associated molecular phenomena. However, the quantitative contribution of each of the known parameters in this progression remains elusive. Mathematical modeling can provide the capability for quantifying the connection between variables governing growth, prognosis and treatment outcome. We apply a biologically founded, multi-scale, mathematical model to identify and quantify tumor biologic and molecular properties relating to clinical and morphological phenotype and to demonstrate that tumor growth and invasion are predictable processes governed by biophysical laws, and regulated by heterogeneity in phenotypic, genotypic, and microenvironmental parameters. This heterogeneity drives migration and proliferation of more aggressive clones up cell substrate gradients within and beyond the central tumor mass, while often also inducing loss of cell adhesion. The model predicts that this process triggers a gross morphologic instability that leads to tumor invasion via individual cells, cell chains, strands or detached clusters infiltrating into adjacent tissue producing the typical morphologic patterns seen, e.g., in the histopathology of glioblastoma multiforme. The model further predicts that these different morphologies of infiltration correspond to different stages of tumor progression regulated by heterogeneity. By quantifying the link between the tumor boundary morphology and the invasive phenotype, this work provides a quantitative tool for the study of tumor progression and diagnostic/prognostic applications. This establishes a framework for monitoring system perturbation towards development of therapeutic strategies and correlation to clinical outcome for prognosis.
Keywords: tumor invasion, clinical outcome prognostication, computer simulation
INTRODUCTION
Prognosis of clinical outcome in oncology determines treatment decisions in patients with early and advanced cancer. Variables currently used include epidemiological information, tumor type, molecular characterization, and clinical parameters such as tumor size and the presence of nodal and extranodal metastasis (tumor-node-metastasis staging). Quantitative histopathologic analysis is often limited to mitotic rates (number of mitosis per high-power field) and size and depth of invasive fingers (usually in microns). Degree of pleiomorphism and nuclear atypia are also used as prognosticators, although no quantitative definition has been generally accepted and hence this is often subjective to the pathologist. Animal models have been used to gain a molecular handle on which parameters indicate tumors with poor prognosis. New methodologies are needed to integrate and quantify these variables and enable prediction of outcomes, selection of existing therapies, and development of new treatments, possibly on a personalized individual basis.
Correlations between morphology and cellular dynamics such as mitosis and motility are of fundamental importance here, since these dynamics produce the various morphologies that generate the patterns pathologists use for diagnosis with proven diagnostic power. Morphological analysis based on histopathology, though, is more art than science. While pathologists describe different features in tumors, the relative impact of each feature on patient outcome and tumor progression remains unknown. To involve epidemiology of human tumors, either in retrospective studies, which often exclude necessary data, or multi-year prospective studies, is cumbersome and time consuming. Hence, determining the clinical predictive value from histological epidemiology remains limited. Nevertheless, the value of histopathologic analysis is that it does not rely on any single feature alone and thus obtains a comprehensive view of the entire morphological behavior of any particular tumor at the time of biopsy or excision.
Mathematical modeling can provide a rigorous, more precise approach for quantifying correlations between tumor parameters, prognosis, and treatment outcomes. Integration of these elements in a computational model of tumor progression would be an important tool to advance clinical decision-making. Tumors are complex systems dominated by large numbers of processes with highly nonlinear dynamics spanning a wide range of dimensions. Typically such complex systems can be understood only through complementary experimental investigation and mathematical modeling. Thus, there is a critical need for biologically realistic and predictive multiscale and multivariate models of tumor growth and invasion, and much recent effort has been directed towards this goal [1–7].
We previously described quantitative multiscale models [2,8–13] to determine precise functional relationships among quantifiable parameters from analyses of specific phenotypic or genetic alterations in a tumor, and from in vitro experiments [10] and clinical observations [2,11] of tumor morphology such as cell arrangement patterns at the tumor boundary. These models predict that morphologic instability of a tumor mass, i.e., morphology resulting in “roughness” or harmonic content [14,15] of the tumor margin, may provide a powerful tissue-invasion mechanism since it allows tumor cells to escape growth limitations imposed by diffusion (even in vitro [10,16]) and invade the host independently of the extent of angiogenesis [9,10]. Experiments with various glioma models in vivo [17,18,19,20,21] also support these findings. For example, recently published images of rat glioblastoma in vivo [20] showed that while the bulk tumor is perfused by blood, infiltrative cell clusters are much less perfused or not at all. These may be universal considerations that apply to tumor invasion across many different tissue types [22,23].
Changes in tumor cells at the biochemical and genetic levels are also implicated in tumor progression. Mutations in genes that regulate cell cycle and adhesion result in unrestrained proliferation, invasion, and accumulation of further genetic damage characteristic of high-grade disease [24–26]. In particular, in glioblastoma the oncogene EGFR is frequently overexpressed, amplified or mutated [27], and promotes mitosis [25], tumor progression in vivo [28], and inhibits apoptosis [29]. See for example [30–32] for recent mathematical models on the effect of EGFR gene expression on tumor growth patterns and [33] for phenomenological modeling of multiple mutations. The tumor suppressor genes TP53 and Rb down-regulate cell division [25] and, secondarily affect oxygen/nutrient consumption, while PTEN controls angiogenesis, migration, and invasiveness [34]. These genes are inactivated in most malignant brain tumors [35].
In the present study, we use a biologically founded, multi-scale, mathematical model of tumor progression [8,11–13] in 3-D (Supplemental Fig. 1) to demonstrate that molecular phenomena regulating cell proliferation, migration, and adhesion forces (including those associated with genetic evolution from lower- to higher-grade tumors) generate, in a predictable and quantifiable way, heterogeneous proliferation and oxygen/nutrient demand (and suppression of apoptosis) across the three-dimensional tumor mass, determining its morphology. The model describes physical conservation laws (e.g., of mass, momentum), with conserved variables representing known characteristics of tumor behavior, and hypothesizes phenomenological functional relationships linking genetic and phenotypic effects, the microenvironment, and tissue-scale growth and morphology.
Variables that characterize the biophysics of tumor growth can be considered in the model, and applied to determine the probabilistic behavior of tumors given their molecular biology and pathologic appearance. By solving the model equations numerically we predict the combination of variables most likely leading towards tumor invasiveness. At any given time during tumor growth, the model outputs the computed values of all relevant variables at every location within the three-dimensional tumor tissue, e.g., the spatial distributions of cell substrates and tumor cells. Rather than relying on one variable as the sole indicator, the multi-parameter computational model enables a more systematic search to be performed for those values of specific variables that reproduce and explain observed tumor behavior. Variables include mutations and phenotypic changes affecting proliferation, apoptosis, nutrient uptake, tumor-cell adhesion and motility, and their collective tumor mass effects. By quantitatively linking the invasive phenotype with the observable morphology of the tumor boundary, the model provides a tool for quantitative study of tumor progression as well as diagnostic and prognostic applications.
MATERIALS AND METHODS
Histopathology of human glioma
Four archived autopsied brains (Brown University-Rhode Island Hospital Brain Bank) and 16 surgical specimens (Columbia University Brain Bank and Cooperative Human Tissue Network) were received according to Institutional Review Board approval, and examined in haemotoxylin-eosin stained paraffin sections prepared according to standard autopsy procedures. Diagnosis of glioblastoma multiforme was confirmed by two neuropathologists, and morphology at the tumor-brain interface imaged on a Zeiss AxioImager by standard bright field and by fluorescence using FITC and rhodamine filters [11]. Selective fluorescence in the rhodamine channel of hemoglobin in red blood cells combined with autofluorescence of connective tissue in the FITC channel greatly enhances detection of vasculature patterns in H&E sections of archived material [11]. Entire brain-tumor interface was imaged for each specimen, although many, especially the surgical specimens, were received in multiple fragments. Even in the autopsied brains, the tumor rendered the tissue friable making it difficult to embed as a block. Representative images were selected for presentation here as comparison to model predictions. All patterns present in the group of tumors were included.
Multiscale tumor model
The model considers genotype, phenotype, and morphologic parameters (Quick-Guide, Supplement), and accounts for feedback from the microenvironment, i.e., mutations or phenotypic changes induced by hypoxia [36], as local levels of oxygen/nutrients induce changes in the mutation function. The model also allows for the development of a (hypoxia-induced) migratory phenotype. At the tumor scale, the model is based on first principles describing conservation laws of mass and momentum. Local mass fractions of tumor clones, necrotic and host tissues are described. Phenomenological parameters describe cell adhesion. Tumor cell migration velocity depends on proliferation-driven mechanical pressure in the tissue, chemotaxis and haptotaxis due to gradients of chemokines. Cell substrate delivery from neo-vasculature (via convection and diffusion [37,8,11]) and cellular uptake, and nutrient/oxygen diffusion through tumor tissue ([8,12], references) are modeled. The effects of each parameter on outcome can be tested individually or in combination.
QUICK-GUIDE TO EQUATIONS AND ASSUMPTIONS
| Equation (1) |
This partial differential equation is derived from the conservation of mass [12,11]. 3D in vivo environment is modeled as a mixture of two viable tumor species (each with volume fraction ϕi, i=1,2), dead tumor (ϕ3) and host tissue (ϕ4), and interstitial fluid (given indirectly by and assumed to move freely) flowing through the ECM, which is treated as a porous medium. From left to right: change of volume fraction with respect to time; bulk transport by tumor mass with local velocity ui; fluxes Ji that account for mechanical interactions among cell species (based on a generalized Fick’s Law [12]); and net tissue source Si from cell proliferation, death and mutation.
In Words
Temporal rate of change in a species at any tumor location equals amount transported by the bulk tumor motion and cell adhesion, plus net result of mass creation/loss due to cell proliferation/death.
Major Assumptions
Tumor is a mixture of cells, interstitial fluid, and ECM. Cell adhesion is modeled through flux J using an approach from continuum thermodynamics [12].
| Equation (2) |
Cell velocity ui of species i is a function of tissue oncotic (solid) pressure and cell mobility due to chemotaxis and haptotaxis. Right side: changes in pressure p create motion counteracted by cell adhesion γi mediated through an energy variation (for specific forms of this energy E and its variation, see [12]); chemotaxis χn due to soluble gradients of cell substrates and oxygen n ≤ nV; haptotaxis χh due to insoluble gradients of ECM molecules f ≤ fH . Motility k reflects cellular response to pressure gradients.
In Words
Species movement depends on oncotic pressure from cell proliferation, adhesion forces, and relative strengths of chemotaxis/haptotaxis.
Major Assumptions
Tumor is a viscous, inertia-less fluid; interstitial fluid and cell motion through ECM is as fluid flow in a porous medium. Cells move with a mass-averaged velocity arising from a generalized Darcy-type constitutive law for velocity from excess forces due to chemotaxis and haptotaxis. Cells prefer to adhere to one another rather than the host, modeled by the energy as a function of total solid tumor fraction. Tumor/host interface is well delineated. Although the model is general, here total solid and liquid volume fractions are assumed constant. Therefore, separate fluid hydrostatic pressure and mechanical oncotic pressure due to cell-cell interactions are calculated. Energy E is a function of total tumor volume.
| Equation (3) |
Equations specify net sources of mass for two tumor viable species, directly linking Eq. (1) to the cell phenotype through hypothesized phenomenological functional relationships involving cell substrates (local oxygen or nutrient concentration n) through tumor interstitium [12]. For species S1, right side terms represent, from left to right, volume fraction gained from mitosis (rate λM,1), and lost to apoptosis (λA,1), necrosis (λN), and mutation to become Species S2 (rate λTR). For species S2, terms respectively represent gain in volume fraction from Species S1 through mutation (rate λTR) and mitosis (λM,2), and loss to apoptosis (λA,2) and necrosis (λN). Mutation rate λTR is a biased random function 0 ≤ frand ≤1 of position and time within clone 1. Heaviside function ℋ, smoothed over a region of biologically realistic thickness (10–100 µm), models necrosis as a result of substrate depletion below a level nN [11,12,10]. Rates are inverse time.
In Words
Mass of original species increases through cell proliferation and decreases through apoptosis, necrosis, and mutation to the mutated species. Mass of mutated species increases through mitosis and mutation (from original species), and decreases through apoptosis and necrosis.
Major Assumptions
Cells are composed entirely of water [12]. Mitosis and necrosis are proportional to substrate concentration n [44]. As mitosis occurs, an appropriate amount of water from interstitial fluid is converted into cell mass [12]. Lysis represents a loss of solid mass converted into water that is absorbed into interstitial fluid. Necrosis occurs only at sufficiently low nutrient.
| Equation (4) |
This partial differential equation describes cell substrate concentration n across tumor tissue. Vessels originate randomly from existing vasculature (not shown) around the growing tumor in response to VEGF produced by hypoxic tissue. First term on the right side models diffusion of substrates n (with coefficient D) into tumor tissue, second term represents the source of substrates at the vasculature denoted by δC as function of distance from the vessels (1 − n/nV) and vascular pressure (1 − p/pV), and third term represents uptake ηi by tumor species ϕi. Nutrient n is normalized with respect to the vasculature level nV.
In Words
Steady-state cell substrate concentration across a tumor region equals amount that diffuses into the region plus the production from the vasculature minus the amount uptaken by tumor cells.
Major assumptions
Nutrient diffusion occurs on a shorter time scale (minutes) than cell proliferation (day); hence, there is no time derivative on the left side, indicating a quasi-steady state.
RESULTS
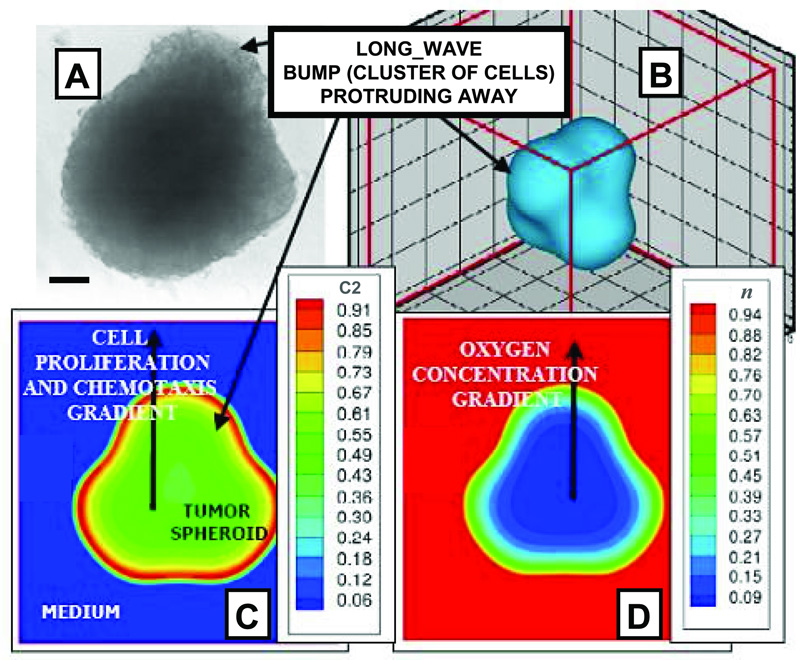
Fig. 1 shows the onset of diffusion-driven morphologic instability [14,15,9,10] from our simulations. Perturbations arise in the spatial arrangement of cells at the periphery of human glioma spheroids in culture (Fig 1A) and are consistently replicated by our model (Fig. 1B). Once this shape asymmetry is created, local cell substrate gradients (Fig. 1D) cause spatially heterogeneous cell proliferation and migration (Fig. 1C), as cells that are exposed to more substrates proliferate more. Mechanical forces, e.g., cell-cell and cell-matrix adhesion, which are in general stabilizing [22,14,10], were in this case not strong enough to prevent morphologic instability.
Figure 1.
Onset of morphological instability in glioblastoma is consistently replicated by the mathematical model. Collective motion of cells of clone 2 “C2” (ℳ = {0,1}) is shown due to heterogeneous proliferation (and possibly chemotaxis) up concentration gradients and under weak cell adhesion (γ2 < 6.7λM,2, Supplement). (A) In vitro glioblastoma spheroid (bar, 130 µm), and (B–D) computer simulation. (B) Representation of observed tumor shape. (C) Local mass fraction “C2” of viable tumor cells and (D) oxygen concentration “n” (n=1 in the medium). Note hypoxic core (D) and corresponding low cell viability levels (C) within, and high viability at outer rim. [A: Adapted from [10]; reprinted from Frieboes et al., Cancer Res., Vol. 66, p. 1602, (2006), with permission from the American Association for Cancer Research].
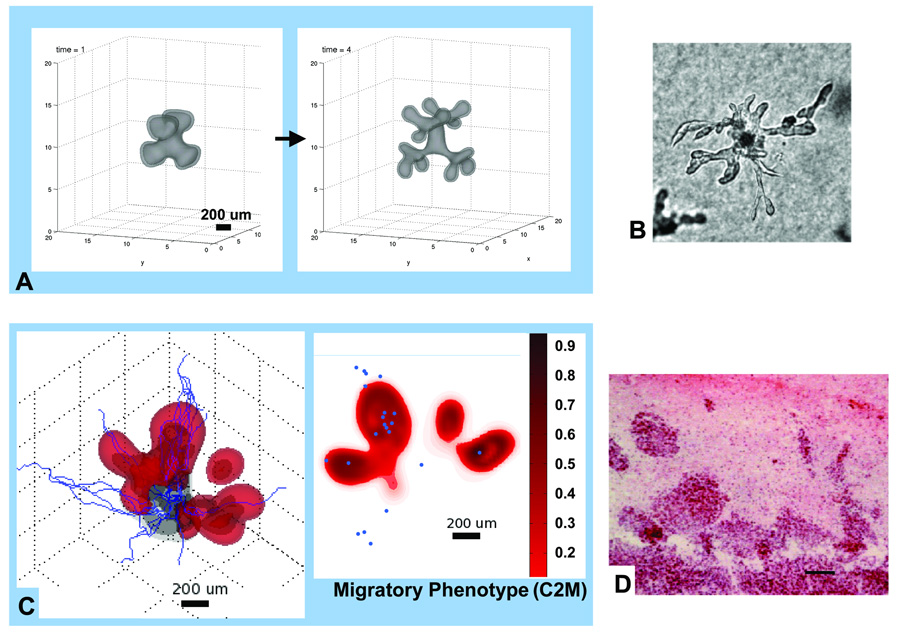
When the instability persists, it leads to proliferative growth of bud-like clusters or “bumps” of cells (Fig. 1,Fig.2). In vitro, these eventually may detach as sub-spheroids from the parent spheroid [10], analogous to micro-satellites in vivo, and also represent the initial stage of the growth of cell chains, strands or detached clusters [2,22] observed in vitro and in vivo (Figs. 2B,D). Simulations reveal that when chemotaxis or haptotaxis are dominant, e.g., if mitosis is downregulated, protrusions begin as high-frequency perturbations (linear stability1 predicts the growth of short-wave-lengths [14,13]) on the tumor surface and develop into cell chains and strands [22], Fig. 2A. When proliferation is the prevailing pro-invasion mechanism, the buds grow into round fingers, Fig. 1B, which may detach as clusters [10] (linear stability predicts the growth of long-wave-length perturbations [9,14,15]). This is clearly seen in Fig. 2C, where cells acquire a hypoxia-induced migratory phenotype. These simulations are supported by experimental observations under hypoxic conditions (Fig. 2B,D) [16,17].
Figure 2.
Variability and persistence of morphologic patterns predicted by the mathematical model simulating heterogeneity in vitro (A) [10,40] and in vivo (C) [12,11]. In both cases, only the higher-grade clone 2 is simulated (no mutations) although the clone is allowed to acquire a hypoxic-induced migratory phenotype (Supplement). (A): Proliferation is downregulated and the clone migrates up oxygen gradients towards the far-field boundary (computational box not shown) (arrow: time direction). (C): red denotes migratory phenotype of tumor clone 2 (C2M) while gray denotes less motile clone 2 (C2). Proliferation of both clones is regulated by oxygen levels. Darker interior region of 3D graph denotes necrotic region. 2D horizontal slice shows distribution of C2M; small blue circles indicate position of mature, blood-conducting vessels. Morphological instability occurs in both simulations because this clone’s cell adhesion is low, resulting in cell strands in (A) and fingers and detached clusters in (C). Simulations are supported by experimental observations revealing morphologic instability after inducing hypoxia in spheroids in vitro (B) [Reprinted from Cancer Cell, Vol. 3, Pennacchietti et al. [16], p. 354, Copyright (2003), with permission from Elsevier] and in xenografts in vivo (D) [Adapted from [17]; bar = 80 µm; reprinted with permission from Rubenstein et al., Neoplasia, Vol. 2, p. 311, (2000). Copyright © Neoplasia].
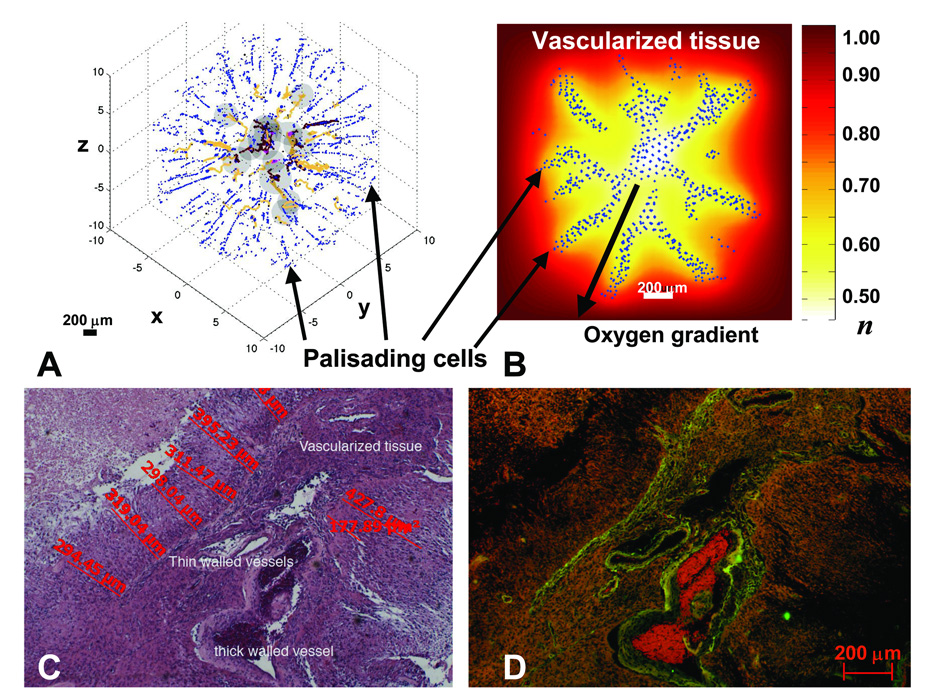
As necrosis and substrate gradients develop within the tumor, proliferation may be downregulated and motility upregulated. While various substrate components (e.g., oxygen, glucose, growth factors, metabolites) may be determinants of these phenotypic changes, we focus on oxygen because of its well-established role in regulating cell proliferation and motility (e.g., [16]). Fig. 3 shows a migratory or hypoxic growth stage, where palisading cells forming “Indian files” are seen from simulations (A,B) and from histology (C,D). In the simulations (hybrid continuum-discrete approach, Supplement), individual cells originate in the peri-necrotic region (center) and have undergone the phenotypic changes described above. In particular, the cells down-regulate proliferation and migrate via chemotaxis up oxygen gradients and haptotaxis up bound chemokines in the extracellular matrix (ECM), which the cells also degrade and remodel. The resulting morphology (Fig. 3A,B) compares with pathology data surprisingly well (Fig. 3C,D) (see also [11]). As strands of cells move away from the peri-necrotic regions and into the brain stroma, they may trigger angiogenesis, a wound -healing process, thereby increasing nutrient availability in the microenvironment. Increased oxygen and nutrients would result in up-regulation of proliferation and down-regulation of motility, creating the micro-satellite cell clusters in Fig. 2D (mouse brain) and Fig. 5B (human specimen, below).
Figure 3.
Palisading glial cells invade vascularized tissue as predicted by the model (A,B) and observed in histology (C,D). A: Computer simulation showing palisading cells escaping from the peri-necrotic region (dark gray) by undergoing a hypoxia-induced phenotypic change to upregulate motility and downregulate adhesion and proliferation. Cell-migration occurs via chemotaxis and haptotaxis in response to gradients of oxygen and ECM concentration, respectively (Methods). Brown: conducting vessels; yellow: non-conducting. B: Background shows distribution of oxygen concentration (“n” in legend). n=1 in vascularized tissue and is lower in the tumor (white/yellow: peri-necrotic region). C: 4x magnification of a high-grade glioblastoma interior. D: Corresponding fluorescent image showing vessels (green, red). As predicted by the simulations (A,B), palisading malignant glial cells (C) invade the vascularized tissue (D), amidst a tangle of thick and thin-walled large vessels and a few smaller vessels, from two areas of necrosis. Also note the distances between necrosis and vessels (C), corroborating our choice for diffusion penetration length L2 (Supplement). Further phenotypic changes (e.g. down-regulation of motility and up-regulation of proliferation) may occur when migrating cells reach tissue regions richer in substrates, leading to the morphologies described in Fig. 4D,5A–D.
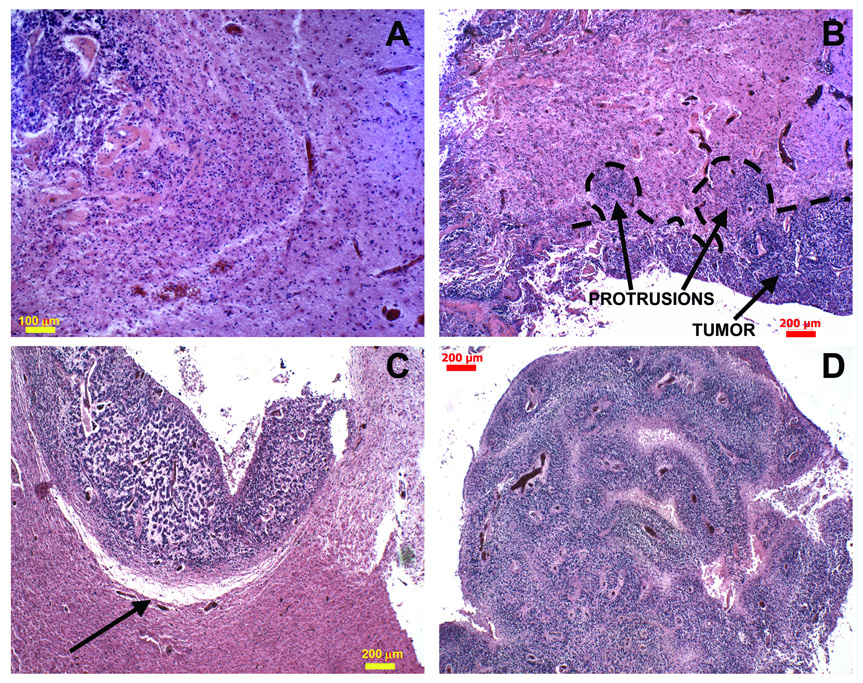
Figure 5.
Histology sections reveal infiltrative patterns predicted by simulations in the proliferative growth stage. A: Higher magnification of invading finger shown in Fig. 4D. B: Two invasive protrusions are seen emanating from the tumor mass. C: Boundary of another protrusion from a different specimen is indicated (arrow); there also appears to be ECM degradation. D: Morphology details of one large bulb-shaped tumor protrusion from (B). Interior reveals clusters of viable cells surrounding blood vessels, and areas depleted of cells farther away, indicating increased substrate availability in the microenvironment. Additional histology sections reveal similar features (Supplement). Altogether, these data suggest the proliferative growth stage predicted by the model for this invading morphology. [Panels A and D were adapted from NeuroImage, Vol. 37, Frieboes et al. [11], pp. S65–6, Copyright (2007), with permission from Elsevier].
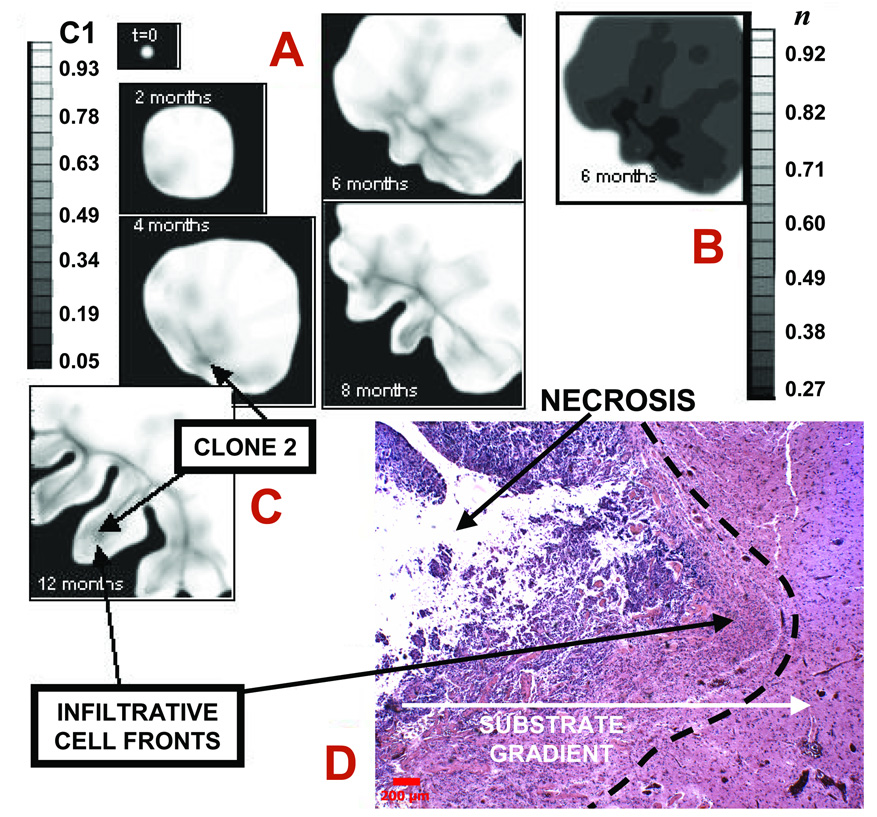
Computer simulation of a human glioma over time in the proliferative growth stage produces finger-like extensions (Fig. 4). Expression of oncogenes and absence of tumor suppressor pathways initially results in the net growth of a relatively low-grade type of morphology. After two months, the tumor expands to approximately 3 mm by co-opting the vasculature (which is not shown) while retaining a compact shape with negligible necrosis (Fig. 4A). However, increased nutrient demand generates hypoxic and other substrate gradients pointing radially outwards from the lesion (not shown). A second, more proliferative clone is generated by ongoing hypoxia-driven [36] mutations and starts to grow (lower left corner, shaded area). Its higher cellular uptake introduces perturbations in the spatial gradients of oxygen further enhancing local hypoxia. These gradients generate spatially heterogeneous cell proliferation and migration. After four months, this perturbation triggers a morphologic instability, which noticeably deforms the tumor mass (lower left). Hypoxia and necrosis are present within the regions where the more malignant clone grows. Shape instability leads to clusters of clone 2 protruding “finger-like” (darker regions) into the mass of clone 1 first, and the host brain later, growing at the expense of the less proliferative clone and the host tissue. We have also observed that detachment of these clusters may occur in our model (cf. Fig. 2C, [10]). These fingers grow away from the bulk tumor and tend to follow substrate gradients.
Figure 4.
Infiltration of a high-grade glioma. A: Computer simulation in proliferative growth stage (field of view=6–10 mm). For each time-snapshot, two-dimensional slices depict spatial distribution of two different clones: genotype ℳ = {1,0} (lower-grade clone 1, “C1”) evolving to ℳ = {0,1} (higher-grade clone 2, “C2” = 1-C1) (Materials and Methods). Legend: local mass fraction of C1. Arrows pointing to darker areas in the tumor indicate C2. B: Simulated oxygen 23 concentration (“n” in legend) at 6 months, indicating hypoxic gradients (n=1 in normal brain and is lower in tumor). The larger oxygen uptake of C2 enhances local hypoxia (e.g., lower-left tumor corner), and leads to shape instability whereby clusters of C2 cells protrude “finger-like” into the tumor mass of C1 first, and host brain later. The model predicts that C2 expansion is the main determinant of tumor morphology. C: Tumor detail at 12 months showing invading fingers. D: Histology section of a tumor front (left) showing an invading finger into more normal brain (bar, 200 µm). Normal brain (white matter) has fewer cell bodies and more abundant amorphous matrix (right). Invading malignant astrocytes (left) have pleiomorphic nuclei and an irregular distribution. Note the clearly demarcated margin (left of dashed line) between tumor and more normal brain. Neo-vascularization (and inflammation) at tumor-brain interface is visible as darker spots to the right in brain parenchyma, implying that substrate gradients drive collective tumor cell infiltration into the brain. Morphology and size of the invading finger are consistent with simulation predictions (A,B) suggesting the proliferative growth stage.
In about six months’ time, the aggressive, invasive proliferation of clone 2 (darker regions) enables it to infiltrate almost all regions of the tumor, in particular around the boundary, and leads to a higher-grade lesion. A bud-like protrusion emerges on the tumor lower left. Hypoxic, necrotic areas continue to expand (Fig. 4B). In eight months, the glioma aggressively infiltrates the surrounding brain tissue. Clone 1 is being confined by competition with clone 2. Extensive necrosis is present. Additional buds have appeared, and the initial (middle) bud has grown into an invasive finger. Strands and clusters of clone 2 drive growth of the finger and buds (extent of the darker area). Clone 1 has been mostly eliminated from this region of the tumor, and remains stagnant. In twelve months, the surrounding brain has been severely compromised. Expansion of clone 2, accompanied by continued necrosis, is now the main determinant of tumor morphology. The lesion reaches a size of approximately 4 cm in a little over one year of simulated time, consistent with human glioma progression (Fig. 4C shows only a tumor portion at one year).
Different tumors are likely to have different genomic instability factors—different types and mutation rates. The idealized tumor in Fig. 4A–C is “programmed” to exhibit progressive appearance of one highly malignant clone (clone 2). In reality, multiple clones may arise with varying degrees of malignancy.
A histology section of glioblastoma from one patient reveals the tip of a round invading finger (Fig. 4D), consistent with the morphology and size of these infiltrative cell clusters predicted by simulations during the proliferative growth stage (cf. Fig. 4C at 12 months, where the tip of one protruding cell front is about 2 mm in size). The fact that tumor cells rely on vessels beyond the protrusions—and may grow towards blood vessels that they stimulate [38,39,11]—also suggests the proliferative growth stage since these vessels increase substrate availability in the microenvironment. Older tumor vessels may have thicker walls that are not as permeable for nutrient/oxygen exchange, and may become occluded due to increased pressure from the tumor mass (Supplement), further promoting substrate gradients.
Additional histology sections from four glioblastomas (Fig. 5) reveal protruding fronts of cells pointing away from a necrotic area into an area of the host brain where neo-vascularization is evident. These invading fronts are also consistent with the tumor boundary morphology predicted by simulation in the proliferative growth regime (Fig. 4A–C). While infiltrative shapes were consistently observed in histological sections (Supplement), the model predicts that their size may vary based on the stage of growth. For example, these shapes can be extremely slender in the hypoxic growth regime, down to single rows of palisading cells migrating up substrate concentration gradients (Fig. 3A,B), and thus away from hypoxic regions, as seen in histology (Fig. 3C,D).
Supplemental Fig. 2 reports additional histologies showing invasive fingering. Such morphologies are predicted by the model to occur in the proliferative growth stage, where there is increased substrate availability. This is confirmed by the presence of viable vessels in the histopathology acting as sources of substrates around the invasive fingers.
DISCUSSION
We have used a biologically founded, mathematical model to demonstrate that tumor progression can be described as a predictable process dependent on biophysical laws. Conservation laws were posed with variables that account for genetic and phenotypic changes that influence cell proliferation, apoptosis, adhesion, motility, and uptake of substrates. Quantitative functional relationships were hypothesized linking genetic and phenotypic effects, the microenvironment, and tissue-scale growth and morphology. Although the model was trained with in vitro, in vivo, and clinical data for glioma, these considerations may also apply to tumor invasion beyond glioma [22,23]. All known forms of collective cell migration (chains, strands, detached clusters) [22] observed in experiments and histopathology were consistently reproduced by the mathematical model (e.g., [2,10–14]) for different values of the molecular and microenvironmental parameters, with heterogeneity in cellular genotype, phenotype, and the microenvironment being the regulating feature. Multi-clonality, abnormal angiogenesis, or hypoxia-inducing local injuries, and even therapeutic intervention such as anti-angiogenic therapy [9,10,17–19] can be sources of this heterogeneity.
The central finding of this work is that tumor growth and invasion are not erratic or unpredictable, or solely explained through genomic and molecular events, but rather are predictable processes obeying biophysical laws, driven by microenvironmental substrate gradients, and regulated by genotypic, phenotypic, and microenvironmental parameters (e.g., cell adhesion, proliferation, motility). The model enables quantitative study of invading cell clusters as functional units that move as complex systems. Substrate gradients, e.g., of nutrient, oxygen, growth factors, and metabolites, result from diffusion, cellular activity, and heterogeneous delivery and removal. This leads to local hypoxia, nutrient starvation, acidosis, necrosis, and the pleiomorphic appearance of tumors. The underlying physical mechanism of collective cell migration, namely a gross tumor morphologic instability [2,8,9,10,12,13,14,40], maximizes cell exposure to substrates by evading a compact, nearly spherical morphology in favor of infiltrating shapes moving up gradients of substrates such as oxygen. Phenotypic changes that increase nutrient uptake and augment cell proliferation (and also increase cell motility and reduce adhesion) have a quantifiable effect on morphology at the tumor scale. In particular, they trigger invasive fingering into host tissue while inducing necrosis and angiogenesis for certain parameter conditions.
The model predicts that different morphologies of infiltration are associated with phenotypic and genotypic changes, which depend on cell substrate gradients and microenvironmental parameters, and reflect the growth stage of the tumor. In regions of hypoxia, these changes include down-regulation of proliferation and up-regulation of motility, which under low cell adhesion results in palisading cells forming “Indian files” (Fig. 3A,B). As cells localize in tissue regions that are richer in substrates, i.e., better-vascularized, further phenotypic changes occur in which motility is downregulated and proliferation is upregulated. This leads to the formation of wave-like patterns of cell rearrangements at the tumor boundary and round infiltrative fingers that can detach from the tumor bulk as clusters (Fig. 4A–C). Actually, different tumor regions may exhibit different growth regimes at the same time depending on heterogeneity in the local microenvironment and in cell geno-/phenotypes. In our simulations proliferation and collective migration of more aggressive clones or phenotypes drives tumor infiltration, as observed in patient biopsies (e.g., [41,42] and this study).
Model predictions of infiltrative morphologies in the hypoxic and proliferative growth stages compare well with histopathology of human glioblastoma. Tumor microenvironment conditions were similar to those predicted in the model with respect to vascular distribution, implied cell substrate availability, and palisading of malignant cells between vascular and avascular regions. These findings are further supported by published correlations of invasion with hypoxia and necrosis induced in glioma by characteristically abnormal, inadequate and hemorrhagic vasculature or by anti-angiogenic therapy [17,18,36,43,44]. Formal comparison between observations and model predictions, e.g., measured via a “merit function,” could be performed as the next stage to confirm the model hypotheses.
By quantifying the close connection between the observable morphology of the tumor boundary and cellular/molecular dynamics, the work presented here provides a quantitative tool for the study of tumor progression and diagnostic/prognostic applications. This connection is important because the dynamics that give rise to various tumor morphologies also control invasiveness. In particular, by describing morphology as a function of parameters dependent on cellular and environmental phenomena [45,46], the model quantifies, under the unifying umbrella of morphologic stability analyses, the often seemingly diverse and unrelated morphologies and invasive phenotypes, e.g., palisading cells, round fingers, and clusters. It is generally observed [22] that pre-metastatic phenotypic transitions (e.g., epithelial-mesenchymal transitions) follow a collective-migration stage, and are regulated by the environment (e.g., local hypoxia) [45,46]. Invasive characteristics may strongly influence whether a tumor can be effectively treated by local resection and may suggest specific treatment options [33,1,47]. Observation of tumor morphology, for example, could indicate the presence of hypoxia and, therefore, the potential to respond to oxygen-dependent treatments such as radiation therapy and certain chemotherapy treatments.
The model may be used to study system perturbations by therapeutic intervention and may aid in the design of novel clinical endpoints in therapeutic trials. By integrating the model with patient data for key tumor phenotypic and microenvironmental parameters [2], model results could be used to enhance clinical outcome prognostication. Initial conditions regarding tumor physical location, structure, and vasculature, e.g., obtained from contrast-enhanced MRI2, and possibly coupled with CT2 would be translated using a computer program to the model coordinate system [2], e.g., a finite-element computational mesh discretizing the space occupied by tumor and host tissues [12]. Viable region spatial information and microvasculature structure would be obtained from histopathology [11]. Vasculature-specific information could be defined from DCE-CT2, yielding blood volume, flow, and microvascular permeability parameters [2]. Other input data include cell-scale parameters (e.g., proliferation rates). The model then calculates local tumor growth, angiogenesis, and response to treatment under various conditions by solving in time and space the conservation and other equations at the tissue scale.
The model allows predictions of cellular and molecular perturbations that may alter invasiveness and that can be measured through changes in tumor morphology. Therefore, morphologies obtained from the model could be used to both understand the underlying cellular physiology and predict subsequent invasive behavior. For example, novel individualized therapeutic strategies could be designed in which microenvironment and cellular factors are manipulated to decrease invasiveness and promote well-defined tumor margins—an outcome that would also benefit treatment by improving local tumor control through surgery or radiation. In addition to existing strategies that act on relevant cellular behaviors (e.g., promotion of tumor cell adhesiveness [22,14,10]), or that target oncogenes such as EGFR, tumor morphological stability could be enhanced by improving nutrient supply [9,40], thus enforcing a more homogeneous microenvironment and normoxic conditions. This could be achieved through “vascular normalization” [9,48] or uniform nanoparticle delivery [49], e.g., releasing oxygen and anti-angiogenic drugs. Further, by maintaining microenvironmental homogeneity, effects of genetic mutations that lead to morphological instability may be minimized (e.g., [50]), without direct intervention at the genotype level.
Applying biologically founded, mathematical modeling to quantify the connections between the microenvironment, tumor morphology, genotype, and phenotype may direct prognosis beyond the limitations of current methodologies, and suggest new directions in the way we think about cancer growth and invasion.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge Robert Gatenby (Moffitt Cancer Center) for useful discussions, Xiangrong Li (UC-Irvine) for Fig. 2A, Ed Stopa and Pathology Dept. (Rhode Island Hospital) for autopsied specimens, Aleksey Novikov and Bryan Kinney (ELB’s lab) for technical assistance, and Henry Hirschberg (UC-Irvine) for information about recent results [20].
Grant support: The Cullen Trust of Health Care, NSF-DMS 0818104, National Cancer Institute, Department of Defense (V Cristini); NSF Division of Mathematical Sciences and NIH-P50GM76516 for a Center of Excellence in Systems Biology at University of California, Irvine (J Lowengrub); NIGMS-GM47368 and NINDS-NS046810 (E Bearer); NCI U54 Center for Cancer Nanotechnology Excellence-TR CA119367 (DB Agus).
Footnotes
REFERENCES
- 1.Sanga S, et al. Mathematical modeling of cancer progression and response to chemotherapy. Expert Rev Anticancer Ther. 2006;6:1361–1376. doi: 10.1586/14737140.6.10.1361. [DOI] [PubMed] [Google Scholar]
- 2.Sanga S, et al. Predictive oncology: a review of multidisciplinary, multiscale in silico modeling linking phenotype, morphology and growth. NeuroImage. 2007;37:S120–S134. doi: 10.1016/j.neuroimage.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araujo R, McElwain D. A History of the Study of Solid Tumour Growth: The Contribution of Mathematical Modelling. Bull Math Biol. 2004;66:1039–1091. doi: 10.1016/j.bulm.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Hatzikirou H, Deutsch A, Schaller C, Simon M, Swanson K. Mathematical Modeling of Glioblastoma Tumour Development: A Review. Math Models Meth Appl Sci. 2005;15:1779–1784. [Google Scholar]
- 5.Byrne HM, Alarćon T, Owen MR, Webb SW, Maini PK. Modeling Aspects of Cancer Dynamics: A Review. Phi. Trans. R. Soc. A. 2006;364:1563–1578. doi: 10.1098/rsta.2006.1786. [DOI] [PubMed] [Google Scholar]
- 6.Roose T, Chapman SJ, Maini PK. Mathematical Models of Avascular Tumor Growth. SIAM Review. 2007;49:179–208. [Google Scholar]
- 7.Anderson ARA, Quaranta V. Integrative mathematical oncology. Nat Rev Cancer. 2008;8:227–244. doi: 10.1038/nrc2329. [DOI] [PubMed] [Google Scholar]
- 8.Zheng X, Wise SM, Cristini V. Nonlinear Simulation of Tumor Necrosis, Neo-Vascularization and Tissue Invasion via an Adaptive Finite-Element/Level-Set Method. Bull Math Biol. 2005;67:211–259. doi: 10.1016/j.bulm.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Cristini V, et al. Morphologic Instability and Cancer Invasion. Clin Cancer Res. 2005;11:6772–6779. doi: 10.1158/1078-0432.CCR-05-0852. [DOI] [PubMed] [Google Scholar]
- 10.Frieboes HB, et al. An Integrated Computational/Experimental Model of Tumor Invasion. Cancer Res. 2006;66:1597–1604. doi: 10.1158/0008-5472.CAN-05-3166. [DOI] [PubMed] [Google Scholar]
- 11.Frieboes HB, et al. Computer Simulation of Glioma Growth and Morphology. NeuroImage. 2007;37:S59–S70. doi: 10.1016/j.neuroimage.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wise SM, Lowengrub JS, Frieboes HB, Cristini C. Three-dimensional multispecies nonlinear tumor growth-- I. Model and numerical method. J Theor Biol. 2008;253:524–543. doi: 10.1016/j.jtbi.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cristini V, Li X, Lowengrub JS, Wise SM. Nonlinear simulations of solid tumor growth using a mixture model: invasion and branching. J Math Biol. 2009;58:723–763. doi: 10.1007/s00285-008-0215-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cristini V, Lowengrub J, Nie Q. Nonlinear Simulation of Tumor Growth. J Math Biol. 2003;46:191–224. doi: 10.1007/s00285-002-0174-6. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Cristini V, Nie Q, Lowengrub JS. Nonlinear Three-Dimensional Simulation of Solid Tumor Growth. Disc Dyn Contin Dyn Syst B. 2007;7:581–604. [Google Scholar]
- 16.Pennacchietti S, Michieli P, Galluzzo M, Giordano S, Comoglio P. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell. 2003;3:347–361. doi: 10.1016/s1535-6108(03)00085-0. [DOI] [PubMed] [Google Scholar]
- 17.Rubenstein JL, et al. Anti-VEGF antibody treatment of glioblastoma prolongs survival but results in increased vascular cooption. Neoplasia. 2000;2:306–314. doi: 10.1038/sj.neo.7900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunkel P, et al. Inhibition of Glioma Angiogenesis and Growth in Vivo by Systemic Treatment with a Monoclonal Antibody against Vascular Endothelial Growth Factor Receptor-2. Cancer Res. 2001;61:6624–6628. [PubMed] [Google Scholar]
- 19.Bello L, et al. Combinatorial Administration of Molecules That Simultaneously Inhibit Angiogenesis and Invasion Leads to Increased Therapeutic Efficacy in Mouse Models of Malignant Glioma. Clin Cancer Res. 2004;10:4527–4537. doi: 10.1158/1078-0432.CCR-04-0194. [DOI] [PubMed] [Google Scholar]
- 20.Madsen SJ, et al. Photodynamic therapy of newly implanted glioma cells in the rat brain. Lasers Surg Med. 2006;38:540–548. doi: 10.1002/lsm.20274. [DOI] [PubMed] [Google Scholar]
- 21.Lamszus K, Kunkel P, Westphal M. Invasion as Limitation to Anti-angiogenic Glioma Therapy. Acta Neurochir Suppl. 2003;88:169–177. doi: 10.1007/978-3-7091-6090-9_23. [DOI] [PubMed] [Google Scholar]
- 22.Friedl P, Wolf K. Tumor cell invasion and migration: diversity and escape mechanisms. Nature Rev Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 23.Debnath J, Brugge J. Modelling glandular epithelial cancers in three-dimensional cultures. Nature Rev Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 24.Maher EA, et al. Malignant glioma: genetics and biology of a grave matter. Genes & Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- 25.Benjamin R, Capparella J, Brown A. Classification of glioblastoma multiforme in adults by molecular genetics. The Cancer Journal. 2003;9:82–90. doi: 10.1097/00130404-200303000-00003. [DOI] [PubMed] [Google Scholar]
- 26.Merlo A. Genes and pathways driving glioblastomas in humans and murine disease models. Neurosurg Rev. 2003;26:145–158. doi: 10.1007/s10143-003-0267-8. [DOI] [PubMed] [Google Scholar]
- 27.Lal A, et al. Mutant Epidermal Growth Factor Receptor Up-Regulates Molecular Effectors of Tumor Invasion. Cancer Res. 2002;62:3335–3339. [PubMed] [Google Scholar]
- 28.Nishikawa R, et al. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci USA. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagane M, Levitzki A, Gazit A, Cavenee WK, Huang HJS. Drug resistance of human glioblastoma cells conferred by a tumor-specific mutant epidermal growth factor receptor through modulation of Bcl-X L and caspase-3-like proteases. Proc Natl Acad Sci USA. 1998;95:5724–5729. doi: 10.1073/pnas.95.10.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Athale CA, Deisboeck TS. Development of a three-dimensional multiscale agent-based tumor model: simulating gene-protein interaction profiles, cell phenotypes and multicellular patterns in brain cancer. J Theor Biol. 2007;244:96–107. doi: 10.1016/j.jtbi.2006.06.034. [DOI] [PubMed] [Google Scholar]
- 31.Stein AM, Demuth T, Mobley D, Berens M, Sander LM. A mathematical model of glioblastoma tumor spheroid invasion in a three-dimensional in vitro experiment. Biophys J. 2007;92:356–365. doi: 10.1529/biophysj.106.093468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Athale CA, Deisboeck TS. The effects of EGF-receptor density on multiscale tumor growth patterns. J Theor Biol. 2006;238:771–779. doi: 10.1016/j.jtbi.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 33.Anderson ARA, Weaver AM, Cummings PT, Quaranta V. Tumor Morphology and Phenotypic Evolution Driven by Selective Presure from the Microenvironment. Cell. 2006;127:905–915. doi: 10.1016/j.cell.2006.09.042. [DOI] [PubMed] [Google Scholar]
- 34.Tysnes BB, Mahesparan R. Biological mechanisms of glioma invasion and potential therapeutic targets. J Neurooncol. 2001;53:129–147. doi: 10.1023/a:1012249216117. [DOI] [PubMed] [Google Scholar]
- 35.Ishii N, et al. Frequent co-alterations of TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human glioma cell lines. Brain Pathol. 1999;9:469–479. doi: 10.1111/j.1750-3639.1999.tb00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jensen RL. Hypoxia in the tumorigenesis of gliomas and as a potential target for therapeutic measures. Neurosurg Focus. 2006;20:E24. doi: 10.3171/foc.2006.20.4.16. [DOI] [PubMed] [Google Scholar]
- 37.Chaplain MAJ. Avascular Growth, Angiogenesis and Vascular Growth in Solid Tumours: The Mathematical Modelling of the Stages of Tumour Development. Math Comp Modeling. 1996;23:47–87. [Google Scholar]
- 38.Bartels U, et al. Vascularity and angiogenesis as predictors of growth in optic pathway/hypothalamic gliomas. J Neurosurg. 2006;104 5 Suppl:314–320. doi: 10.3171/ped.2006.104.5.314. [DOI] [PubMed] [Google Scholar]
- 39.Preusser M, et al. Histopathologic assessment of hot-spot microvessel density and vascular patterns in glioblastoma: Poor observer agreement limits clinical utility as prognostic factors: a translational research project of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Cancer. 2006;107:162–170. doi: 10.1002/cncr.21973. [DOI] [PubMed] [Google Scholar]
- 40.Macklin P, Lowengrub JS. Nonlinear Simulation of the Effect of Microenvironment on Tumor Growth. J Theor Biol. 2007;245:677–704. doi: 10.1016/j.jtbi.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 41.Okada Y, et al. Selection Pressures of TP53 Mutation and Microenvironmental Location Influence Epidermal Growth Factor Receptor Gene Amplification in Human Glioblastomas. Cancer Res. 2003;63:413–416. [PubMed] [Google Scholar]
- 42.Hu B, et al. Angiopoietin-2 induces human glioma invasion through the activation of matrix metalloprotease-2. Proc Natl Acad Sci USA. 2003;100:8904–8909. doi: 10.1073/pnas.1533394100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plate KH, Risau W. Angiogenesis in malignant gliomas. Glia. 1995;15:339–347. doi: 10.1002/glia.440150313. [DOI] [PubMed] [Google Scholar]
- 44.Rong Y, Durden DL, Van Meir EG, Brat DJ. 'Pseudopalisading' Necrosis in Glioblastoma: A Familiar Morphologic Feature That Links Vascular Pathology, Hypoxia, and Angiogenesis. J Neuropath Exp Neurol. 2006;65:529–539. doi: 10.1097/00005072-200606000-00001. [DOI] [PubMed] [Google Scholar]
- 45.Sierra A. Metastases and their microenvironments: linking pathogenesis and therapy. Drug Resist Updates. 2005;8:247–257. doi: 10.1016/j.drup.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 46.van Kempen LCLT, Ruiter DJ, van Muijen GNP, Coussens LM. The tumor microenvironment: a critical determinant of neoplastic evolution. Eur J Cell Biol. 2003;82:539–548. doi: 10.1078/0171-9335-00346. [DOI] [PubMed] [Google Scholar]
- 47.Sinek J, et al. Predicting drug pharmacokinetics and effect in vascularized tumors using computer simulation. J Math Biol. 2009;58:485–510. doi: 10.1007/s00285-008-0214-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jain RK. Normalizing tumor vasculature with anti-angiogenic therapy: a new paradigm for combination therapy. Nature Med. 2001;7:987–989. doi: 10.1038/nm0901-987. [DOI] [PubMed] [Google Scholar]
- 49.Ferrari M. Cancer nanotechnology: opportunities and challenges. Nature Rev Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 50.Kenny PA, Lee GY, Bissell MJ. Targeting the tumor microenvironment. Front Biosci. 2007;12:3468–3474. doi: 10.2741/2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.