Abstract
Epstein-Barr virus (EBV) has been causally associated with at least five human malignancies. The exact contributions made by EBV to these cancers remain unknown. We demonstrate that one viral protein found in all EBV-associated malignancies, Epstein-Barr nuclear antigen 1 (EBNA-1), is required for survival of one of these cancers, EBV-positive Burkitt's lymphoma. Inhibition of EBNA-1 decreases survival of these tumor cells by inducing apoptosis. Expression of EBNA-1 in uninfected cells also can inhibit apoptosis induced by expression of p53 in the absence of the EBV genome. Our findings demonstrate that EBNA-1 is critical for the continued survival of EBV-associated Burkitt's lymphoma, and, by extension, for the other B cell tumors with which EBV is associated. Efficient inhibitors of EBNA-1's functions would likely prove useful in the therapy of EBV-associated malignancies.
Fifteen percent of human cancers are now causally associated with tumor viruses. This fraction will likely increase as more cancers are associated with known viruses and new human tumor viruses are identified. A driving force to identify these viral cancers is the potential for the tumor virus to maintain some tumor phenotypes and thereby provide a nonhost target for therapeutic interventions. Epstein-Barr virus (EBV) causes Burkitt's lymphoma (BL), which is endemic in Africa, and, as we demonstrate, promotes survival of the tumor cells even long after their explantation.
BL is an aggressive B cell malignancy with a high proliferative rate that may be fatal within months if not treated promptly (1). Activation of the c-myc oncogene through reciprocal chromosomal translocations that juxtapose c-Myc to one of the Ig loci characterizes most BLs (2). Additionally, many BLs carry point mutations in the p53 tumor suppressor gene or other defects in the p14ARF-MDM2-p53 pathway, or have p16INK4a genes inactivated by promoter methylation or homozygous deletion (3, 4). Thus, BL involves multiple genetic events likely to promote cellular proliferation and inhibit apoptosis. In areas where BL is endemic, virtually all cases are associated with EBV (5). Multiple viral genes are used by EBV to induce and maintain proliferation of infected B cells, but most of these genes are not expressed in BL tumors (6), making it difficult to know what, if anything, EBV contributes to the survival of BL tumors. We have found that functions of the only viral protein consistently expressed in BL cells, Epstein-Barr nuclear antigen 1 (EBNA-1), are required for their survival.
Materials and Methods
Cell Culture and Transient Transfection. EBV-negative B cell lines used in these experiments include BJAB (7) and DG75 (8). EBV-positive B cell lines used in these experiments include 721 (9), Oku1 (10), AG-876 (11), 11/17-3 (12), Akata (13), GG68 (14), and Sav1 (15). The EBV-positive cell lines, RPMI 1788 (CCL-156), Daudi (CCL-213), and Namalwa (CRL-1432), are described in the ATCC catalog. DLD1 cells (CCL-221), an EBV-negative colon carcinoma cell line, and SAOS2 (HTB-85), a human osteosarcoma cell line, are available from the ATCC. The VM-10 cell line (16, 17) was kindly provided by A. Levine (Institute for Advanced Studies, Princeton). VM10 cells were grown at 39°C and apoptosis was induced by a temperature shift to 32°C.
Retroviral Production and Infection. The dominant-negative EBNA-1 (Dom Neg 1) was described (18, 19). The derivative of EBNA-1 lacking amino acids 65-89 (Dom Neg 2) inhibits transcription mediated by wild-type EBNA-1, but supports replication of an oriP plasmid with efficiency similar to that of wild-type EBNA-1 (20). Retroviruses were produced and purified as described (20).
Analyses of Cells. Infected cells were sorted by a fluorescence-activated cell sorter (FACS) Vantage or DIVA (Becton Dickinson), and the population of cells expressing the highest levels of GFP (top 50%) was collected. Different numbers (1-240) of cells were directly deposited into multiple wells of 96-well plates containing 200 μl of culture medium with or without irradiated human fibroblast feeder layers. Wells containing surviving and proliferating cells were identified visually at 10-14 days after sorting. The cloning efficiency was determined by using the Poisson distribution where the cloning efficiency = [-1n(number of negative wells)]/number of cells plated per well. γ-Irradiated, human fibroblast feeder layers increased the cloning efficiency of AG876 and Oku1 cells 10- to 15-fold but were shown to have no effect on the fraction of cells with decreased survival mediated by Dom Neg 1 or 2.
Apoptosis Assays. To measure apoptosis after cell sorting, cells were collected and stained with Hoechst 33342 and propidium iodide (Sigma); the total number of cells and apoptotic cells were counted on a hemocytometer by fluorescent microscopy. In all cases, 100-200 GFP-positive cells were examined microscopically.
Labeling of DNA strand breaks by terminal deoxynucleotidyltransferase was performed on both DLD1 and Oku1 cells according to the in situ cell death detection kit, TMR red (Roche Molecular Biochemicals, Indianapolis). In all cases, 100-200 green cells were counted and scored for the number of cells TUNEL-positive by using a Zeiss Axiovert 200M.
PCR. The different methods based on PCR have been described (21, N. Lam and B.S., unpublished work). The sequences of primers are available on request.
Western Blot, Southern Blot, and Statistical Analysis. The methods for detecting EBNA-1, LMP1, and EBV-viral DNA and statistical analyses have been described (20, 22). The Wilcoxon rank sum test was used unless otherwise stated.
Results
Inhibition of EBNA-1 Results in Decreased Survival of EBV-Positive Cells. Retroviral vectors expressing a Dom Neg derivative of EBNA-1, Dom Neg 1, and enhanced GFP (EGFP) expressed from an internal ribosomal entry site (IRES) were used to infect EBV-positive and -negative, normal and tumor B cells (Fig. 1). Control retroviruses used the same bicistronic message with EGFP expressed from the IRES but, in place of Dom Neg 1, had either β-galactosidase (LacZ) or a polylinker sequence. Dom Neg 1 has been shown to inhibit two functions of EBNA-1, its support of extrachromosomal replication and of transcription, to 3% of wild-type levels (18). Dom Neg 1's ability to inhibit EBNA-1's functions is primarily dependent on its ability to bind DNA site-specifically and competitively displace wild-type EBNA-1 from the DNA (18). Infected cells were identified by their expression of EGFP, sorted by FACS, and tested for survival by colony formation after limiting dilutions. Two EBV-negative and ten EBV-positive cell lines, including seven BL tumor lines, were tested, and survival data are shown in Table 1. When the cell lines 11/17-3, Akata, Daudi, GG68, Namalwa, and SavI were analyzed as a group, we found that the inhibition of EBNA-1 by infection with a retrovirus expressing Dom Neg 1 results in a statistically significant decrease in survival compared with cells infected with a control retrovirus (P = 0.007, Wilcoxon rank sum test). We infected 721, RPMI 1788, Oku1, and AG876 multiple times with a retrovirus expressing Dom Neg 1 and found that inhibition of EBNA-1 in these cell lines significantly decreased survival (P = 0.0004, 0.04, 0.04, and 0.02, respectively).
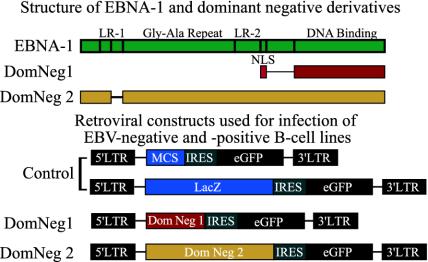
Fig. 1.
The structure of EBNA-1 and the Dom Neg derivatives used are represented schematically at the top. Dom Neg 1 consists only of the nuclear localization sequence (NLS) and the DNA-binding domain of EBNA-1. Dom Neg 2 has a deletion of 25 residues from linking region 1 (LR1) of EBNA-1 and is otherwise intact. (Bottom) The retroviruses used for expression of the EBNA-1 derivatives (Dom Neg 1 and 2).
Table 1. Survival of EBV-positive B cells is significantly reduced by infection with Dom Neg 1 or 2.
| Survival, %
|
||||
|---|---|---|---|---|
| Cell line | Dom Neg 1 | Dom Neg 2 | N | EBV status |
| BJAB | 95 ± 5 | 123 ± 11 | 6 | Negative |
| DG75 | 100 | 1 | Negative | |
| 721 | 3 ± 2 | 10 ± 6 | 7* | Positive |
| RPMI 1788 | 33 ± 3 | 6* | Positive | |
| Oku1 | 14 ± 10 | 15 ± 9 | 3* | Positive |
| AG-876 | 51 ± 23 | 42 ± 10 | 6* | Positive |
| 11/17-3 | 10 | 1 | Positive | |
| Akata | 30 | 1 | Positive | |
| Daudi | 45 ± 11 | 3 | Positive | |
| GG68 | 40 ± 7 | 2 | Positive | |
| Namalwa | 34 ± 13 | 2 | Positive | |
| Savl | 30 ± 3 | 2 | Positive | |
Survival of each cell line infected with a control retrovirus was normalized to 100% and is not shown. Survival of cells infected with Dom Neg 1 or 2 was calculated by multiplying, by 100, the cloning efficiency of cells infected with the Dom Neg-expressing virus divided by the cloning efficiency of cells infected with the control virus. The mean survival and SE of the means are shown. N, the number of observations made for each cell line.
Statistical analysis using the Wilcoxon rank sum test reveals P values <0.05, indicating a significant decrease in survival when these cell lines were infected with retroviruses expressing Dom Neg 1 or 2. The remaining cell lines were grouped according to their EBV status, and statistical analysis revealed a significant decrease (P = 0.007) in survival mediated by infection of these EBV-positive cells with a retrovirus expressing Dom Neg 1
Whereas EBV DNA is generally maintained extrachromosomally, a few tumor cell lines have their viral DNA integrated into host chromosomes. One such cell line is Namalwa, whose survival was reduced by 50-85% by inhibiting EBNA-1's functions (Table 1). This finding likely indicates that EBNA-1's support of transcription, not just its support of extrachromosomal replication, is important for survival of EBV-positive BL cells. We used a derivative of EBNA-1 that has been shown to inhibit EBNA-1's ability to activate transcription (20), but supports replication of an EBV-derived plasmid at wild-type levels (ref. 23 and our unpublished observations) to test whether EBNA-1's activation of transcription likely contributes to survival of EBV-positive BL cells. This derivative, termed Dom Neg 2, was inserted into a retroviral vector and, along with Dom Neg 1, was used to infect one EBV-negative cell line, BJAB, and three EBV-positive cell lines 721, Oku1, and AG876. Cells from the 721 line have been derived by infection of normal B cells in vitro, whereas Oku1 and AG876 cells have been derived from BL tumors. We found that expression of Dom Neg 2 inhibited survival in all EBV-positive cell lines tested as efficiently as does Dom Neg 1 (Table 1). Remarkably, a significant reduction in survival was seen in Oku1 and AG876 cells, which, we have confirmed, fail to express LMP1, a viral gene required for proliferation of normal EBV-positive cells (P < 0.05; refs. 24 and 25 and our unpublished observations). Inhibiting EBNA-1 with its Dom Neg derivative that inhibits transcriptional activation mediated by EBNA-1 inhibits survival of EBV-positive normal and BL cells.
Inhibition of EBNA-1 Decreased Survival of Cells in a Dose-Dependent Manner. In our experiments, some cells transduced with the Dom Neg derivatives of EBNA-1 survived (Table 1). We were interested to learn whether their survival reflects differences in the level of expression of the inhibitory genes, or an ability of a subset of EBV-positive cells to survive independent of EBV, as has been demonstrated for atypical BL-derived cell lines such as Akata (26). Infection of EBV-positive Akata cells with the retrovirus expressing Dom Neg 1 does select survivors that have lost EBV DNA (Fig. 2B). However, 721 cells that survive infection with the retrovirus expressing Dom Neg 1 retain viral DNA and express low levels of the inhibitor (Fig. 2). We therefore tested whether other cells survive because the level of expression of the Dom Neg 1 or 2 derivatives is insufficient to inhibit EBNA-1's functions. Because the level of expression of Dom Neg 1 and 2 proteins detected in sorted populations corresponded to the intensities of EGFP used to sort the populations (Fig. 3A), the level of expression of GFP could be used to select cells with different levels of these inhibitors of EBNA-1. We infected EBV-negative BJAB cells and three EBV-positive cell lines, 721, Oku1, and AG876, with the control, Dom Neg 1, or 2 retroviruses. These infected populations of cells were then FACS sorted for those with the top 15%, middle 15%, or bottom 15% intensities of GFP, and the sorted cells were plated in limiting dilutions. The inhibition of survival by both Dom Neg 1 and 2 is dose-dependent in the three EBV-positive cell lines tested (P < 0.05 in all cases, Jonckheere-Terpstra test; Fig. 3B), whereas varying levels of expression of the control virus had no effect on their survival (data not shown). Survival of the EBV-negative BJAB cells was independent of the levels of expression of any of the retroviruses (data not shown). Inhibiting EBNA-1 inhibits survival of EBV-positive normal and tumor cells in a dose-dependent manner, holding promise that the effective inhibition of EBNA-1's functions in patients would kill EBV-positive tumors efficiently.
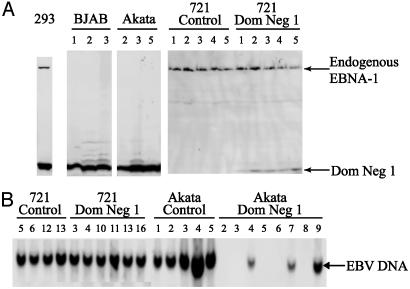
Fig. 2.
Inhibition of EBNA-1 by Dom Neg 1 can select for loss of EBV DNA in the EBV-positive Akata cell line. (A) The EBV-negative 293/EBNA-1 and BJAB and the EBV-positive Akata and 721 cell lines were infected with a retrovirus expressing Dom Neg 1. Infected cells were sorted and plated in limiting dilutions. Clones that grew after 14 days were expanded, and Western analyses were performed. Wild-type EBNA-1 is detected in 293/EBNA-1 (293) and 721 cell lines, but not in BJAB cells or in Akata clones 2, 3, and 5. Dom Neg 1 can be detected in all cell lines infected with the Dom Neg 1-expressing retrovirus. (B) DNA was harvested from each of the surviving clones, and Southern analyses were performed. EBV DNA readily can be detected in the EBV-positive cell line 721 infected with either the control or Dom Neg 1-expressing retrovirus. However, EBV-DNA has been lost from the Akata clones 2, 3, 5, 6, and 8 that were infected with Dom Neg 1, which correlates with the loss of EBNA-1 expression in clones 2, 3, and 5 (see A above). EBV DNA from the Akata clones infected with control retrovirus was readily detected.
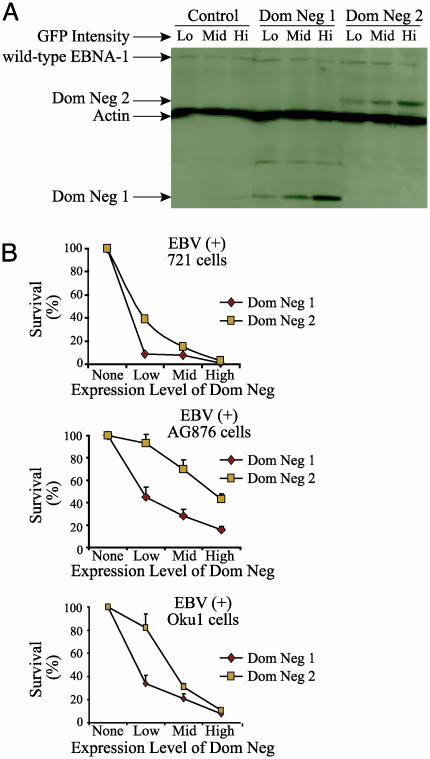
Fig. 3.
Inhibition of EBNA-1 leads to a dose-dependent decrease in survival. (A) The EBV-positive, normal B cell line, 721, was infected with the control retrovirus or ones expressing Dom Neg 1 or 2. Infected cells were sorted 72 h postinfection for the top 15%, middle 15%, and bottom 15% of green intensity. The sorted cells were collected, and Western blots were performed. (B) Cells were infected and sorted for the top 15% green, middle 15% green, and bottom 15% of green expression and were assayed for survival. The decrease in survival between low, middle, and high GFP expression of cells infected with Dom Neg 1 or 2 is statistically significant in all three of the EBV-positive cell lines tested (P < 0.05, Jonckheere-Terpstra test).
Dom Neg 1 or 2 Do Not Affect Survival of EBV-Negative Cells as Can EBNA-1. EBNA-1 is the only viral protein consistently expressed in all EBV-associated tumors (27). Its protein level of expression in EBV-positive cells appears to be regulated, because the number of molecules of EBNA-1 per cell is not proportional to the viral DNA present in those cells (28). Furthermore, we have found that transient, high-level expression of EBNA-1 in cells can limit their ability to proliferate (Fig. 4). These findings make it essential to test whether the Dom Neg derivatives of EBNA-1 share this phenotype. They do not; for example, when BJAB cells were infected with control, Dom Neg 1-, or 2-expressing retroviruses, sorted 48 h later for the 15% most, 15% middle, and 15% least green, and plated in limiting dilutions, all survived to proliferate similarly (Fig. 4 and data not shown). However, BJAB cells in parallel experiments infected with a retrovirus expressing wild-type EBNA-1 survived less well than when infected with the other three retroviruses (Fig. 4). Efficient expression of EBNA-1 resulted in the accumulation of large, multinucleated cells, whereas cells infected with either Dom Neg 1 or 2 appeared to be phenotypically normal (data not shown). Therefore, cells infected with a retrovirus expressing wild-type EBNA-1 do not appear to be dying through necrosis or apoptosis; rather, the cells appear to be arrested in mitosis with a disruption in cytokinesis, resulting in the accumulation of large, multinucleated cells. The Dom Neg derivatives of EBNA-1 are not inhibiting survival of EBV-negative cells, as can the efficient expression of wild-type EBNA-1.
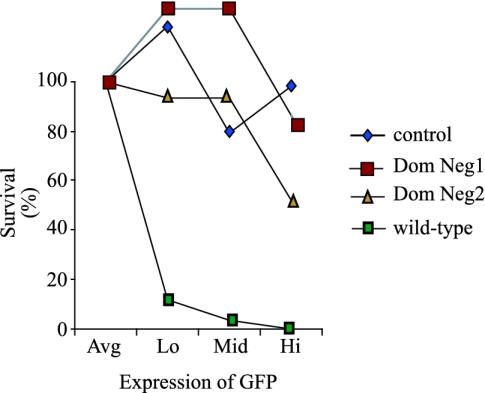
Fig. 4.
The Dom Neg derivatives of EBNA-1 do not inhibit growth of EBV-negative B cells, whereas efficient expression of wild-type EBNA-1 does. The EBV-negative B cell line, BJAB, was infected with retroviruses expressing control, Dom Neg 1 or 2, or wild-type EBNA-1. Infected cells were sorted 48 h postinfection for the highest 15%, middle 15%, and lowest 15% intensity of GFP expression, plated in limiting dilutions, and scored for survival. The average survival of cells infected with the control virus was calculated and was set at 100% survival (average). The survival of cells infected with wild-type EBNA-1 sorted for low, middle, and high was 11%, 3%, and 0.1%, respectively. Survival of cells infected with Dom Neg 1 sorted for low, middle, and high was 129%, 129%, and 86%, respectively. Survival of cells infected with Dom Neg 2 sorted for low, middle, and high was 93%, 93%, and 51%, respectively.
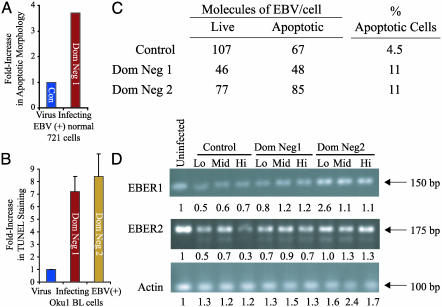
Inhibition of EBNA-1 Results in Apoptosis of EBV-Positive Cells. We examined the fate of the EBV-positive cells in which EBNA-1 was inhibited to determine the mechanism by which the cells fail to survive. We assayed the survival of EBV-negative BJAB cells and EBV-positive 721, RPMI 1788, AG876, Daudi, and Oku1 cells by infecting them with control or Dom Neg 1 virus, sorting for infected cells, and monitoring their proliferation in bulk cultures. BJAB cells infected with either virus proliferated similarly, whereas the EBV-positive cells infected with Dom Neg 1 virus grew more slowly than when infected with the control virus (data not shown). EBV-positive cells would be killed were EBV to undergo productive infection in them. We therefore assayed cells for the expression of EBV lytic antigens and detected a background staining in 1.5% and 1.9% in control and Dom Neg 1-virus-infected 721 cells at 3 days postinfection, respectively. This similar background level of staining indicates that the inhibition of EBNA-1 does not induce EBV's lytic cycle. Analysis of the nuclear morphology of these normal, EBV-positive cells was revealing, however; staining them with DNA-specific dye, Hoechst 33342, and propidium iodide for apoptotic bodies showed that cells infected with Dom Neg 1 had a 4-fold increase in apoptosis relative to cells infected with a control virus (Fig. 5A). These initial observations were confirmed by detecting apoptotic cells stained with an antibody to active caspase 9 and by detecting annexin V binding by FACS (data not shown). To extend our observations to an EBV-positive BL cell line, we infected Oku1 cells with a control virus or either Dom Neg 1 or 2. Seventy-two hours postinfection, the cells were scored for the presence of apoptosis by terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) assay. We found that the fraction of cells found positive by the TUNEL assay was increased 7- or 8-fold in cells infected with either Dom Neg 1 or 2 compared with cells infected with control virus (Fig. 5B, P < 0.05). This apoptosis could result by inhibiting EBNA-1 directly or by EBNA-1's inhibition, leading to loss of viral DNA, and, thereby, other EBV functions. To determine whether loss of the EBV genome contributed to the death of the EBV-positive cells, Oku1 cells were infected with the control retrovirus or a retrovirus encoding Dom Neg 1 or 2. Green, apoptotic cells were identified by staining with annexin V 72 h postinfection and separated from green, nonapoptotic cells by FACS. DNA from the cell populations was harvested, and the average number of EBV DNA molecules measured per cell by using real-time PCR. The number of viral genomes in those cells undergoing apoptosis resulting from the inhibition of EBNA-1 was similar to that in nonapoptotic cells (Fig. 5C) and unlikely to underlie the observed apoptosis. These cells, therefore, are not dying as a result of loss of the viral genome 72 h after the inhibition of EBNA-1. This conclusion is consistent with our findings that the level of endogenous EBNA-1 is unchanged in 721 cells infected with high levels of Dom Neg 1 or 2 for 72 h (Fig. 3A). Inhibiting EBNA-1 in EBV-positive normal and tumor cells inhibits their survival, at least in part, by resulting in apoptosis, whereas loss of the EBV genome is not required for their death.
Fig. 5.
Inhibition of EBNA-1's functions by Dom Neg 1 or 2 leads to apoptosis of EBV-positive cells without their prior loss of viral DNA or EBERs. (A) The EBV-positive normal B cell line, 721, was infected with retroviruses expressing either control or Dom Neg 1. Morphologic evidence of apoptosis was present in 5-25% of 721 cells infected with Dom Neg 1 in three independent assays, which was 4-fold greater than the evidence of apoptosis identified in cells infected with control virus (P < 0.05). (B) Seventy-two hours postinfection, the EBV-positive BL cell line, Oku1, was scored for the fraction of green cells staining positively for apoptosis by the TUNEL assay. In three independent infections, each performed in duplicate, the fraction of cells infected with Dom Neg 1 or 2 that stained positively by the TUNEL assay ranged from 20% to 45%, which was seven to eight times higher than that of cells infected with the control retrovirus (P < 0.05). (C) Oku1 cells were infected, and, at 72 h, were stained with annexin V conjugated to Alexa Flour 647. Green cells were sorted plus or minus annexin V, their DNA was isolated, and the number of molecules of EBV DNA was assayed by real-time PCR and normalized to values of actin. The number of EBV DNA molecules does not change significantly in cells that are undergoing apoptosis. (D) Oku1 cells were infected, and cells were sorted 72 h postinfection for the top 15%, middle 15%, and bottom 15% of green intensity. Reverse transcription followed by PCR was performed on each fraction to determine relative expression levels of the EBER1, EBER2, and actin genes. Below each lane are shown expression levels of each of the bands adjusted for the intensity of the actin amplification product relative to the intensity of the actin product from the uninfected cells. The levels of EBER1 and EBER2 are not related to the survival of the infected Oku1 cells.
The observation that inhibiting EBNA-1's functions results in apoptosis of EBV-positive cells indicates that EBNA-1 itself directly or indirectly through other EBV genes inhibits apoptosis. Most EBV-positive cells, for example, express two small nonpolyadenylated RNAs, the EBERs, which can affect apoptosis (21, 29-31). To determine whether loss of the expression of the EBERs underlies the induction of apoptosis of the EBV-positive BL cell line, Oku1, we infected cells with the control retrovirus or a retrovirus encoding Dom Neg 1 or 2 and sorted 72 h postinfection for the top 15% green, middle 15% green, and the bottom 15% of green expression. Total RNA was extracted from the cells and reverse transcription, followed by PCR (21), was performed in a linear range of the assay to determine relative levels of EBER expression (Fig. 5D). Increasing expression of Dom Neg inhibitors of EBNA-1 had no effect on levels of expression of EBERs. Therefore, loss of expression of EBERs cannot explain the 10-fold decrease in survival resulting from the inhibition of EBNA-1 in Oku1 cells (Fig. 3B).
EBNA-1 Can Inhibit Apoptosis in the Absence of the Viral Genome. Apoptosis was induced by efficient expression of p53 in two p53-negative cell lines, SAOS2 and DLD1, as well as in VM10 cells in the presence or absence of transfected EBNA-1. EBNA-1 reduced the number of TUNEL-positive cells by 80% in the DLD 1 cell line (Fig. 6A, P < 0.05). Importantly, both Dom Neg 1 and 2 failed to inhibit apoptosis (Fig. 6A). The apoptosis induced by transfection of a p53-expressing vector in SAOS2 cells, and by temperature shift in VM10 cells, which carry a ts-allele of p53 (16, 17), was inhibited by ≈50% by expression EBNA-1 (data not shown). An analogue of EBNA-1, LANA-1, encoded by ORF73 of Kaposi's sarcoma herpes virus (32), has been found to inhibit p53-mediated apoptosis and served as a positive control in these assays (33). Furthermore, we demonstrated that expression of EBNA-1 in the presence of p53 leads to a statistically significant increased survival of transfected cells up to 96 h posttransfection (Fig. 6B), indicating that the diminution of TUNEL-positive cells is not a result of EBNA-1's inhibiting their survival. EBNA-1 in the absence of other EBV genes can inhibit apoptosis. Most EBV-positive BL cells are functionally p53-negative (34); therefore, EBNA-1's ability to inhibit apoptosis is not limited to the inhibition of p53, whereas LANA-1 inhibits p53 directly, and, thus, inhibits only p53-dependent apoptosis (33).
Fig. 6.
Expression of EBNA-1 in DLD1 cells inhibits p53's ability to kill cells. (A) The p53-negative human colon carcinoma cell line, DLD1, was transfected with DNA encoding p53 to induce apoptosis in the presence of a DNA-encoding EGFP, and an empty vector (control) or DNAs encoding wild-type EBNA-1, Dom Neg 1 or 2, or LANA-1. Forty-eight hours posttransfection, the cells were stained by the TUNEL assay, and the fraction of TUNEL-positive, green cells was measured in three independent transfections performed in duplicate. EBNA-1 inhibited p53's ability to induce apoptosis 5-fold (P < 0.05). Importantly, Dom Neg 1 and 2 had no effect on p53's ability to induce apoptosis. LANA-1 was found to inhibit p53's ability to induce apoptosis by 50%, which was consistent with what has been reported (33). (B) DLD1 cells were transfected with plasmid DNAs expressing EGFP, with or without cells expressing p53 and EBNA-1. Approximately 24 h posttransfection, cells were trypsinized and plated on gridded tissue culture plates. The number of cells expressing EGFP per grid (low power field) was counted 48- and 96-h post-transfection, and 100-200 cells were counted per plate. There is a statistically significant reduction in the number of cells expressing EGFP and transfected with p53 by 48 h (15 EGFP-expressing cells per field) compared with control (25 EGFP-expressing cells per field) or EBNA-1 (24 EGFP-expressing cells per field) (P = 0.04). This reduction was also seen at 96 h posttransfection (P = 0.02). We found fewer green cells transfected with p53 and EBNA-1 (15 EGFP-expressing cells per field) when compared with control (P = 0.08) at 48 h. By 96 h, EBNA-1, when coexpressed with p53, increased the number of green cells per field (15 EGFP-expressing cells per field) compared with cells transfected with p53 alone (seven EGFP-expressing cells per field) (P = 0.02). At 96 h, there is no statistically significant difference in the number of green cells transfected with GFP from those transfected with GFP plus EBNA-1 (P = 0.5), indicating that under these experimental conditions in which EBNA-1 inhibits apoptosis, EBNA-1 alone is not inhibiting survival or proliferation of DLD1 cells.
Discussion
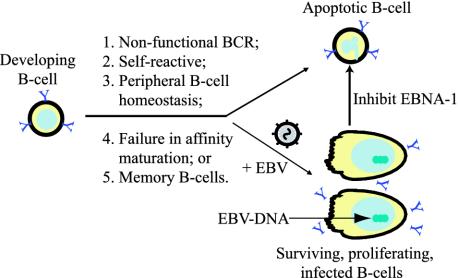
That EBV, and EBNA-1 in particular, can inhibit apoptosis in B cells likely contributes to the persistence of EBV-infected cells in vivo. EBV can infect and induce B cells to proliferate during most stages of their development, including pro-B, preB, immature, and mature B cells (35). It appears that the majority of the B cells progressing through these stages of development die through apoptosis (ref. 36 and Fig. 7). Infection of these cells with EBV would require EBV to inhibit apoptosis for infected cells to persist in vivo. A striking example of EBV's potentially rescuing such cells from apoptosis is the identification of Hodgkin and Reed-Sternberg cells, which have nonfunctional rearrangements of their Ig loci but survive in the presence of EBV infection (37). EBV infection also can drive the expansion of a large fraction of peripheral B cells (39) and be maintained in memory B cells (37, 38), cases again in which EBNA-1's inhibition of apoptosis would allow EBV to persist in vivo. In addition to EBNA-1's inhibiting apoptosis in infected cells, LMP1, another viral gene usually expressed in EBV-infected B cells, has been thought to inhibit apoptosis. Recent findings with conditional mutants, however, indicate that LMP1's functioning drives proliferation of B cells, whereas its absence leads to quiescence but not to apoptotic death (U. Dirmeier, B.S., and W. Hammerschmidt, unpublished findings). Additionally, when the EBV-positive, normal B cell line 721 was infected with Dom Neg 1 and 2, the expression of LMP1 was found to be independent of their survival (data not shown). Thus EBNA-1's ability to block apoptosis likely allows EBV to persist in vivo and is a survival factor for BL.
Fig. 7.
Outlined are multiple events in the normal development of B cells that can lead to apoptosis (36). Infection with EBV leads to survival of B cells with nonfunctional B cell receptors (BCR), avoiding apoptosis associated with peripheral B cell homeostasis, and results in latent infection within long-term memory B cells (37, 38). Inhibition of EBNA-1 results in apoptosis in all of the EBV-infected B cells we have tested.
Others have used the conditional expression of a Dom Neg derivative of EBNA-1 in one cell line in which EBV DNA is integrated (IB4) and failed to detect an effect of expression of the Dom Neg EBNA-1 on cell survival (40). The use of a retrovirus to deliver Dom Neg derivatives to multiple cell lines allowed us to detect a decrease in survival mediated by the inhibition of EBNA-1's functions. Our collected findings indicate that EBV is required to sustain BLs long after they have evolved to accumulate cellular mutations that render them independent of normal cellular controls. EBV shares this characteristic with human papillomaviruses that cause cervical carcinomas (41, 42). Tumors caused by these two human tumor viruses should be treatable by specific antiviral therapies. For EBV-associated BL and, perhaps, for other EBV-associated malignancies, targeting EBNA-1 successfully should provide a potent therapy for these human cancers.
Acknowledgments
We thank Jackie Perrigoue and Julia D'Alo for their expert technical assistance; Kathy Schell and the staff of the University of Wisconsin Comprehensive Cancer Center flow cytometry facility for their assistance; and Norman Drinkwater, Paul Ahlquist, Paul Lambert, Paul Friesen, and members of the Sugden laboratory for their critical reviews of this manuscript. This work was supported by Public Health Service Grants CA-22443, CA-07175, and T32-CA-009614-13. Bill Sugden is an American Cancer Society Research Professor.
Abbreviations: EBV, Epstein-Barr virus; EBNA-1, Epstein-Barr nuclear antigen 1; BL, Burkitt's lymphoma; Dom Neg 1, dominant-negative EBNA-1; Dom Neg 2, the derivative of EBNA-1 lacking amino acids 65-89; EGFP, enhanced GFP; FACS, fluorescence-activated cell sorter.
References
- 1.Evens, A. M. & Gordon, L. I. (2002) Curr. Treat. Options Oncol. 3, 291-305. [DOI] [PubMed] [Google Scholar]
- 2.Boxer, L. M. & Dang, C. V. (2001) Oncogene 20, 5595-5610. [DOI] [PubMed] [Google Scholar]
- 3.Lindstrom, M. S., Klangby, U. & Wiman, K. G. (2001) Oncogene 20, 2171-2177. [DOI] [PubMed] [Google Scholar]
- 4.Lindstrom, M. S. & Wiman, K. G. (2002) Semin. Cancer Biol. 12, 381-387. [DOI] [PubMed] [Google Scholar]
- 5.Niedobitek, G., Meru, N. & Delecluse, H. J. (2001) Int. J. Exp. Pathol. 82, 149-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowe, M., Rowe, D. T., Gregory, C. D., Young, L. S., Farrell, P. J., Rupani, H. & Rickinson, A. B. (1987) EMBO J. 6, 2743-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein, G., Lindahl, T., Jondal, M., Leibold, W., Menezes, J., Nilsson, K. & Sundstrom, C. (1974) Proc. Natl. Acad. Sci. USA 71, 3283-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-Bassat, H., Goldblum, N., Mitrani, S., Goldblum, T., Yoffey, J. M., Cohen, M. M., Bentwich, Z., Ramot, B., Klein, E. & Klein, G. (1977) Int. J. Cancer 19, 27-33. [DOI] [PubMed] [Google Scholar]
- 9.Kavathas, P., Bach, F. H. & DeMars, R. (1980) Proc. Natl. Acad. Sci. USA 77, 4251-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nonkwelo, C., Ruf, I. K. & Sample, J. (1997) J. Virol. 71, 6887-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pizzo, P. A., Magrath, I. T., Chattopadhyay, S. K., Biggar, R. J. & Gerber, P. (1978) Nature 272, 629-631. [DOI] [PubMed] [Google Scholar]
- 12.Sugden, B., Phelps, M. & Domoradzki, J. (1979) J. Virol. 31, 590-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takada, K., Horinouchi, K., Ono, Y., Aya, T., Osato, T., Takahashi, M. & Hayasaka, S. (1991) Virus Genes 5, 147-156. [DOI] [PubMed] [Google Scholar]
- 14.Weigel, R. & Miller, G. (1983) Virology 125, 287-298. [DOI] [PubMed] [Google Scholar]
- 15.Ruf, I. K., Rhyne, P. W., Yang, H., Borza, C. M., Hutt-Fletcher, L. M., Cleveland, J. L. & Sample, J. T. (1999) Mol. Cell. Biol. 19, 1651-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu, X., Bayle, J. H., Olson, D. & Levine, A. J. (1993) Genes Dev. 7, 1126-1132. [DOI] [PubMed] [Google Scholar]
- 17.Wu, X. & Levine, A. J. (1994) Proc. Natl. Acad. Sci. USA 91, 3602-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirchmaier, A. L. & Sugden, B. (1997) J. Virol. 71, 1766-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mackey, D. & Sugden, B. (1999) Mol. Cell. Biol. 19, 3349-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennedy, G. & Sugden, B. (2003) Mol. Cell. Biol. 23, 6901-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nanbo, A., Inoue, K., Adachi-Takasawa, K. & Takada, K. (2002) EMBO J. 21, 954-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimizu, N., Yoshiyama, H. & Takada, K. (1996) J. Virol. 70, 7260-7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu, H., Kapoor, P. & Frappier, L. (2002) J. Virol. 76, 2480-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rickinson, A. B., Young, L. S. & Rowe, M. (1987) J. Virol. 61, 1310-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly, G., Bell, A. & Rickinson, A. (2002) Nat. Med. 8, 1098-1104. [DOI] [PubMed] [Google Scholar]
- 26.Shimizu, N., Tanabe-Tochikura, A., Kuroiwa, Y. & Takada, K. (1994) J. Virol. 68, 6069-6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leight, E. R. & Sugden, B. (2000) Rev. Med. Virol. 10, 83-100. [DOI] [PubMed] [Google Scholar]
- 28.Sternas, L., Middleton, T. & Sugden, B. (1990) J. Virol. 64, 2407-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komano, J., Maruo, S., Kurozumi, K., Oda, T. & Takada, K. (1999) J. Virol. 73, 9827-9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komano, J., Sugiura, M. & Takada, K. (1998) J. Virol. 72, 9150-9156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitagawa, N., Goto, M., Kurozumi, K., Maruo, S., Fukayama, M., Naoe, T., Yasukawa, M., Hino, K., Suzuki, T., Todo, S. & Takada, K. (2000) EMBO J. 19, 6742-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rainbow, L., Platt, G. M., Simpson, G. R., Sarid, R., Gao, S. J., Stoiber, H., Herrington, C. S., Moore, P. S. & Schulz, T. F. (1997) J. Virol. 71, 5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friborg, J., Jr., Kong, W., Hottiger, M. O. & Nabel, G. J. (1999) Nature 402, 889-894. [DOI] [PubMed] [Google Scholar]
- 34.Farrell, P. J., Allan, G. J., Shanahan, F., Vousden, K. H. & Crook, T. (1991) EMBO J. 10, 2879-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nilsson, K. (1992) Hum. Cell 5, 25-41. [PubMed] [Google Scholar]
- 36.Defrance T., Casamayor-Palleja, M. & Krammer, P. H. (2002) Adv. Cancer Res. 86, 195-225. [DOI] [PubMed] [Google Scholar]
- 37.Kuppers, R., Rajewsky, K. & Kanzler, H. (1997) Blood 89, 1288-1298. [PubMed] [Google Scholar]
- 38.Babcock, G. J., Decker, L. L., Volk, M. & Thorley-Lawson, D. A. (1998) Immunity 9, 395-404. [DOI] [PubMed] [Google Scholar]
- 39.Robinson, J., Smith, D. & Niederman, J. (1980) Nature 287, 334-335. [DOI] [PubMed] [Google Scholar]
- 40.Kang, M. S., Hung, S. C. & Kieff, E. (2001) Proc. Natl. Acad. Sci. USA 98, 15233-15238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodwin, E. C., Yang, E., Lee, C. J., Lee, H. W., DiMaio, D. & Hwang, E. S. (2000) Proc. Natl. Acad. Sci. USA 97, 10978-10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodwin, E. C. & DiMaio, D. (2001) Cell Growth Differ. 12, 525-534. [PubMed] [Google Scholar]