Abstract
In our previous studies, we have stated to build a new strategy for developing defective, pseudoinfectious flaviviruses (PIVs) and applying them as a new type of vaccine candidates. PIVs combined the efficiency of live vaccines with the safety of inactivated or subunit vaccines. The results of the present work demonstrate further development of chimeric PIVs encoding dengue virus 2 (DEN2V) glycoproteins and yellow fever virus (YFV)-derived replicative machinery as potential vaccine candidates. The newly designed PIVs have synergistically functioning mutations in the prM and NS2A proteins, which abolish processing of the latter proteins and make the defective viruses capable of producing either only noninfectious, immature and/or subviral DEN2V particles. The PIV genomes can be packaged to high titers into infectious virions in vitro using the NS1-deficient YFV helper RNAs, and both PIVs and helpers can then be passaged as two-component genome viruses at an escalating scale.
Keywords: pseudoinfectious flaviviruses, chimeric flavivirus, vaccines, packaging
INTRODUCTION
The Flavivirus genus in the Flaviviridae family contains a spectrum of major human and animal pathogens. Some of the flaviviruses produce diseases ranging from mild febrile illness to meningoencephalitis and hemorrhagic fever (Burke and Monath, 2001). More than a half of the world population lives in the areas that have circulating yellow fever (YFV), Japanese encephalitis, West Nile (WNV), dengue (DENV) and tick-borne encephalitis viruses. In nature, flaviviruses are maintained through continuous circulation between arthropod vectors, such as mosquitoes and ticks, and amplifying hosts, which are mainly represented by birds and mammals. In arthropods, they cause a persistent, life-long infection that leads to accumulation of the virus in the salivary gland and its transmission to vertebrate hosts during the blood meal (Burke and Monath, 2001). Infected hosts develop an acute infection characterized by high titer viremia, sufficient for infecting new mosquitoes or ticks and subsequent furthering of virus circulation.
DENV infections are the great public health concern. More than 2 billion people live in the risk areas, and an estimated annual number of human cases approaches 50–100 million (Halstead, 2007). Moreover, dengue viruses continue to expand their circulation range, and cause outbreaks that correlate with Ae. aegypti and Ae. albopictus mosquito habitats (Effler et al., 2005; Halstead, 2007). DENV infection in humans results in dengue fever and life-threatening dengue haemorrhagic fever (DHF) and shock syndromes (DSS) (Halstead, 2003). The detailed mechanism of haemorrhagic fever development needs yet to be characterized fully; however, it is known that the DHF and DSS occurrences are mediated by antibodies induced by previous DENV infection(s). These antibodies are incapable of neutralizing the secondary infection with another DENV serotype, but frequently promote it via a so-called immune enhancement (IE) mechanism and induce more severe symptoms. Thus, co-circulation of different DENV serotypes and the existence of the IE phenomenon make development of DENV vaccine very challenging and suggest that a universal vaccine must induce neutralizing antibodies to all four serotypes at the same time (Widman et al., 2008).
One of the promising strategies for development of multivalent DEN vaccines is based on the application of infectious cDNA clones of flavivirus genomes. They can be used for the construction of infectious, chimeric flaviviruses encoding the replicative machinery and capsid-coding gene of highly attenuated viruses, such as YFV 17D (Chambers et al., 1999; Guirakhoo et al., 2002; Pugachev et al., 2003), DEN2V PDK-53 (Huang et al., 2000), or DEN4V (Bray and Lai, 1991; Pletnev and Men, 1998). The envelope glycoprotein-coding genes, prM/E, can be derived from the heterologous flaviviruses, such as DENV1-4 (Guirakhoo et al., 2002; Guirakhoo et al., 2004). These chimeric flaviviruses demonstrate high safety and efficacy; however, possibility of their further evolution to more pathogenic phenotype cannot be completely ruled out. Inactivated (INV) or subunit vaccines to DEN1-4 infections can be advantageous from the safety standpoint. However, the induction of neutralizing antibodies by INV is less efficient than that of replicating viruses and repeated vaccinations are required to achieve long-term protection (Widman et al., 2008). This, in turn, necessitates a large-scale production and purification of infectious viruses. Thus, vaccinations can be lengthy and expensive.
In our previous studies, we and others have made an attempt to develop defective flaviviruses as a new type of vaccine candidates that combine the efficiency of live vaccines and the safety of inactivated or subunit vaccines (Aberle et al., 2005; Kofler, Heinz, and Mandl, 2004; Mason, Shustov, and Frolov, 2006; Shustov, Mason, and Frolov, 2007). The genomes of the designed pseudoinfectious viruses (PIV) encode no capsid protein, and thus, upon delivery into the cells, they only develop one round of replication. Such viruses do not produce infectious, genome-containing virions, but the infected cells release subviral particles (SVPs), which serve as efficient immunogens. For delivery into the cells in vivo, YFV and WNV PIV genomes were packaged into infectious viral particles in vitro and propagated at an escalating scale by using the capsid-producing cell lines (Ishikawa et al., 2008; Mason, Shustov, and Frolov, 2006).
Another means of production of packaged PIV genomes is based on the application of defective helper genomes, which can autonomously replicate and produce the capsid protein, but not prM/E (Shustov, Mason, and Frolov, 2007). The entire set of structural proteins required for infectious virion formation is expressed only in the cells containing both capsid-deficient PIV and prM/E-deficient helper RNAs. Both of these defective genomes are packaged and released from the cells as two-component genome virus, characterized by packaging of PIV and helper RNAs into separate viral particles (Shustov, Mason, and Frolov, 2007). At a multiplicity of the infection (MOI) above ~1 inf.u/cell, the two-component genome viruses could be propagated at an escalating scale. High titers sufficient for achieving such MOIs could be readily achieved during their passaging in vitro.
The capsid-producing cell lines and the helper genome-based systems efficiently packaged YFV and WNV PIV genomes. No homologous or nonhomologous recombinations leading to formation of the infectious virus have been detected throughout numerous experiments in vivo and in vitro, highlighting the high safety levels of the designed packaging systems. However, in this study, packaging of chimeric PIV genomes, having DEN2V prM/E and YFV-derived replicative machinery, was found to be below 107 inf.u/ml, and such low titers made the possibility of large-scale production questionable. Therefore, to achieve packaging of chimeric, defective DEN2/YFV PIV, we have designed a new type of PIV genomes having mutations in prM- and/or NS2A-coding genes. Their replication leads to release of DEN2V noninfectious viral particles, and that can be efficiently propagated in vitro as a two-component genome virus. Their packaging to high titers into infectious virions was achieved by using defective helper genomes encoding YFV structural proteins, but having NS1 deleted. These new two-component genome viruses efficiently propagated in vitro and could be passaged at an escalating scale. This strategy of defective virus design and trans-complementation of their defects for packaging into infectious virions in vitro can be applied for other members of the Flaviviridae family.
RESULTS
Replication of chimeric DEN2/YF viruses
In our previous studies, we began to build a new strategy for designing defective-in-replication pseudoinfectious WN and YF viruses, PIVs, which only develop a single round of infection in vivo (Mason, Shustov, and Frolov, 2006; Shustov, Mason, and Frolov, 2007). Upon delivery into the cells, YFV and WNV PIV genomes expressed all of the viral proteins, except capsid protein, and were able to produce only highly immunogenic SVPs (Mason, Shustov, and Frolov, 2006; Shustov, Mason, and Frolov, 2007), but not infectious virions. For immunization experiments, packaging of PIV genomes into infectious viral particles was performed in vitro using either the capsid protein-producing cell lines or by using another defective helper genome, which encoded capsid protein, but had prM/E genes deleted. Such PIV-based strategy was expected to be applicable for the development of a new type of vaccine candidates against a variety of flavivirus infections.
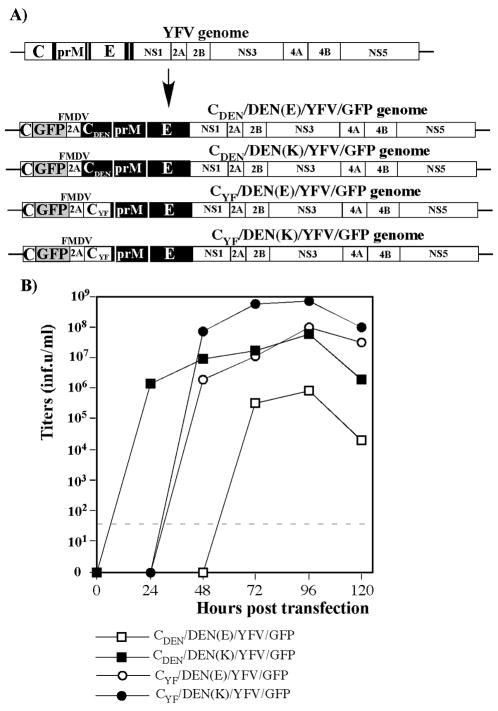
In this study, we were interested in expanding the PIV-based technology from WNV and YFV to other viruses, such as DENV, and therefore DEN2V was selected as a model. In vitro, dengue viruses replicate to titers that are significantly lower than those of WNV and YFV, and it was unlikely that after deletion of the capsid-coding sequence, such defective genomes could be packaged to high titers into infectious particles in the capsid-producing, trans-complementing cell lines or by using prM/E-deficient helper RNAs. Therefore, we focused our efforts on development of chimeric DEN2/YF PIV, which encoded efficient, YFV 17D-derived replicative machinery and DEN2V structural proteins. However, from the available published data (Guirakhoo et al., 2002; Guirakhoo et al., 2004), it was unclear whether the YFV-specific capsid or that derived from DEN2V would be the one more efficient in infectious virion formation when supplied in trans during the packaging procedure. Therefore, the initial experiments were aimed at defining the optimal capsid and assessing the ability of different DEN2V prM/E envelope proteins to form infectious virions. To achieve this, we designed chimeric viral genomes in which the 5′ terminal cyclization sequence and the sequence encoding functional capsid capable of RNA packaging were separated (see Fig. 1A for details). The amino terminal 25 codons of the YFV capsid gene, containing the cyclization signal, were left in the natural position in the beginning of the open reading frame (ORF). The ORF continued into the GFP gene, followed by the FMDV 2A protease-coding sequence and the entire DEN2V or YFV capsid genes, DEN2V prM/E and YFV NS1-NS5 polypeptide. The tested capsid proteins were expected to contain an additional amino terminal proline, which was required for the FMDV 2A protease-mediated proteolytic cleavage. To avoid the possibility of intramolecular recombination between two different capsid-coding sequences, both the YFV and DEN2V full-length capsid-coding sequences were synthesized, using the codon frequency of the most efficiently translated cellular templates (Haas, Park, and Seed, 1996). As a result, the genes strongly differed from their natural counterparts. The GFP gene insertions were made to simplify titer assessments of the released viruses and evaluation the rates of infection propagation in tissue culture. In all of the recombinants, the prM signal peptide was derived from YFV, because it was previously identified as the most efficient in infectious virus production (data not shown). The presented constructs differed not only in capsid, but also in the aa 126 of E-protein, which was either Lys or Glu (K126 or E126). The previously published data indicated that DEN2V infectivity is strongly dependent on the presence of E or K in this position (Bray et al., 1998).
FIG. 1.
Replication of chimeric DEN2V/YFV viruses. (A) Schematic representation of the viral genomes. YFV-specific sequences are indicated by open boxes. DEN2V-specific sequences are indicated by filled boxes. FMDV 2A indicates position of FMDV 2A protease. (B) Replication of chimeric viruses in BHK-21 cells electroporated with 5 μg of in vitro-synthesized RNA. Electroporated cells were seeded into 100-mm dishes and incubated at 37°C. At the indicated time points, media were replaced, and titers of the recombinant viruses were determined by plaque assay on BHK-21 cells as described in the Materials and Methods. Dashed line indicates the limit of detection.
Equal amounts of the in vitro-synthesized RNAs were transfected into BHK-21 cells by electroporation. All of the RNAs demonstrated similar infectivity, evaluated by the number of GFP-positive cells. At 24 h post transfection, before the second round of infection has started, ~50% of the cells were GFP-positive. However, the rescued recombinants viruses differed in their replication efficiencies (Fig. 1B). The recombinant expressing DEN2V capsid protein and having E126 in E protein {CDEN/DEN(E)/YFV/GFP} demonstrated the lowest titers. The replacements of dengue virus capsid gene by that of YFV or E126 in E-coding gene by K126 were beneficial, and CDEN/DEN(K)/YFV/GFP and CYF/DEN(E)/YFV/GFP variants demonstrated higher infectious titers. However, combination of CYF and K126 in E protein led to the highest titers. Chimeric CYF/DEN(K)/YFV/GFP virus was capable of forming plaques in BHK-21 cells, and its titers approached almost 109 inf.u/ml. Thus, the combination of YFV capsid and DEN2V-specific, K126 E protein genes was used for the development of all of the constructs described in the following sections.
Packaging of chimeric PIV genomes into virions with different flavivirus envelops
In the next experiments, we designed a chimeric PIV genome (DEN/YFV/GFP) (Fig. 2A) derived from the above-described CYF/DEN(K)/YFV/GFP, but, in order to make virus defective in replication, the capsid-coding sequence was deleted. DEN/YFV/GFP PIV RNA was packaged into infectious viral particles by using the CYF-producing stable cell line that contains the persistently replicating, CYF-expressing VEEV replicon (VEErep/CYF/Pac) (Mason, Shustov, and Frolov, 2006). However, packaging was very inefficient, and titers of packaged PIV genomes did not exceed 106 inf.u/ml (data not shown). The attempts to use CDEN-producing BHK-21 or Vero cells lines for packaging of the DEN/YFV/GFP PIV RNA were also unsuccessful (data not shown). Titers remained below 106 inf.u/ml and, at the next passage in the same capsid-expressing cells, harvested samples could not induce spreading infection. After having such discouraging results, in subsequent experiments, in order to trans-complement the capsid protein deficiency of DEN/YFV/GFP PIV, we applied helper CYF/YFV/Cherry RNA (Fig. 2A). This helper RNA encoded the entire YFV replicative machinery and was capable of self-replication. It also encoded CYF, but had prM and E genes deleted. In contrast to previous results with YFV and WNV PIVs (Shustov, Mason, and Frolov, 2007), the DEN2V prM/E-containing chimeric PIV RNA genome and helper itself were packaged inefficiently and did not develop spreading infection (Fig. 2A). This inefficient packaging was not specific to DEN2V prM/E. The chimeric WN/YFV/GFP PIV genome, expressing WNV-derived prM/E, was inefficiently packaged by the CYF/YFV/Cherry helper as well (Fig. 2B). Titers of infectious virions did not exceed 106 inf.u/ml. In contrast, by 24 h post electroporation, titers of infectious particles containing YF/YFV/GFP PIV and the same CYF/YFV/Cherry helper genomes were above 108 inf.u/ml (Fig. 2C), and their release continued up to 120 h post electroporation. The efficiency of DEN/YFV/GFP RNA packaging was not improved by the replacement of YFV-derived capsid in the helper by that of DEN2V, and titers remained below the level required for PIV passaging at an escalating scale (data not shown). Taken together, the results indicated that packaging of chimeric PIV genomes using trans-complementation with capsid protein is an inefficient process. Thus, development of DEN/YFV PIV packaging systems required new approaches.
FIG. 2.
Packaging of different PIV genomes using CYF/YFV/Cherry helper. The in vitro-synthesized PIV and helper RNAs were transfected into BHK-21 cells by electroporation (see Materials and Methods for details). Cells were seeded into 100-mm dishes in 10 ml of media and incubated at 37°C. At the indicated time points, media were replaced and titers of packaged PIV and helper genomes were measured by infecting naïve BHK-21 cells with different dilutions of the samples and evaluating the numbers of GFP- and Cherry-positive cells. YFVsp indicates YFV-specific signal peptide. CYFco indicates position of YFV-specific, synthetic capsid-coding sequence, having optimized codon usage. FMDV 2A indicates positions of FMDV-specific 2A protease. YFV-specific sequences are indicated by open boxes. WNV- and DEN2V-specific sequences are indicated by filled, grey boxes. Dashed line indicates the limit of detection. (A) The results of DEN/YFV/GFP PIV and helper genome packaging. (B) The results of WN/YFV/GFP PIV and helper genome packaging. (C) The results of YF/YFV/GFP PIV and helper genome packaging.
Packaging of chimeric PIV genomes into infectious virions using the NS1-deficient helpers
Low titers of packaged chimeric PIV genomes by trans-complementation with capsid protein contradicted to some extent results of the experiments with the infectious chimeric viruses (Fig. 1). Expression of the same YFV capsid protein in cis, from the genomic RNA, was sufficient to support virus replication to titers approaching almost 109 inf.u/ml. These data suggested that co-expression of capsid protein and DEN2V prM/E as a single polyprotein precursor from the same replicating RNA might be an important positive contributor to formation of infectious, RNA-containing viral particles. Thus, the efficiency of capsid-encoding helpers could likely be improved by making them capable of expressing the entire cassette of structural genes, C/prM/E. Therefore, we modified the YFV/GFP and chimeric infectious virus CYF/DEN(K)/YFV/GFP genomes (Fig. 1), making them capable of red fluorescent protein, Cherry, expression and having an extended, in frame deletion in the NS1 gene (Fig. 3). This helper could produce the entire set of the structural proteins, required for infectious virion formation, but its replication depended on the supplied in trans NS1 protein. The latter protein was produced by the replicating PIV. Combining both defective genomes, which encode trans-complementing proteins, in the same cell was expected to lead to a productive infection, characterized by both PIV and helper replication, and their subsequent packaging and release in the form of infectious viral particles.
FIG. 3.
Packaging of PIV genomes using NS1-deficient helper RNAs encoding C/prM/E. The in vitro-synthesized PIV and helper RNAs were transfected into BHK-21 cells by electroporation (see Materials and Methods for details). Cells were seeded into 100-mm dishes in 10 ml of media and incubated at 37°C. At the indicated time points, media were replaced and titers of packaged PIV and helper genomes were measured by infecting BHK-21 cells, containing VEErep/Pac-2A-NS1 replicon, by different dilutions of the samples and evaluating the numbers of GFP- and Cherry-positive cells. YFVsp indicates YFV-specific signal peptide. CYFco indicates position of YFV-specific, synthetic capsid-coding sequence, having optimized codon usage. FMDV 2A indicates positions of FMDV-specific 2A protease. YFV-specific sequences are indicated by open boxes. DEN2V-specific sequences are indicated by filled, grey boxes. (A) The results of YF/YFV/GFP PIV and YF/YFV/ΔNS1/Cherry helper genome packaging. (B) The results of DEN/YFV/GFP PIV and DEN/YFV/ΔNS1/Cherry helper genome packaging. (C) The results of DEN/YFV/GFP PIV and YF/YFV/ΔNS1/Cherry helper genome packaging. The figures represent the result of one of three highly reproducible experiments.
Initially, the possibility of applying this system was tested on YFV PIV genome lacking the capsid gene, YF/YFV/GFP (Fig. 3A). Packaging was achieved using the YF/YFV/ΔNS1/Cherry helper RNA, encoding all of the YFV structural proteins, but having a 106 aa-long, in frame deletion in the NS1 gene. Both in vitro-synthesized RNAs were transfected into BHK-21 cells, and titers of packaged GFP- and Cherry-coding genomes were evaluated on the stable cell line containing the VEErep/Pac-2A-NS1YF replicon expressing YFV NS1 protein from the subgenomic promoter (see Materials and Methods for details). The YFV-specific NS1, produced by this cell line, complemented the NS1 deficiency of the helper RNA, and made it capable of replication and Cherry protein expression (Lindenbach and Rice, 1997; Lindenbach and Rice, 1999). Thus, on this cell line, we could evaluate titers of both the YF/YFV/GFP PIV and YF/YFV/ΔNS1/Cherry helper genome-containing particles.
After electroporation of BHK-21 cells with in vitro-synthesized RNAs, titers of the PIV and helper genome-containing infectious virions exceeded 108 inf.u/ml (Fig. 3A), indicating efficient trans-complementation. This two-component genome virus could be passaged in tissue culture as an ordinary virus if the applied MOI remained above ~1 inf.u/ml (data not shown).
In the next round of experiments, the NS1 trans-complementation-based system was applied for packaging of the chimeric DEN/YFV/GFP PIV genomes (Figs. 3B and C). DEN/YFV/ΔNS1/Cherry and YF/YFV/ΔNS1/Cherry RNAs, encoding DEN2V and YFV prM/E, respectively, and YFV capsid protein, were used as helpers. After electroporation of the in vitro-synthesized RNAs into BHK-21 cells, replication of both pairs did not cause CPE. In multiple, reproducible experiments, by 72–96 h post electroporation, the transfected cells reached confluency, and all of them expressed both fluorescent markers, GFP and Cherry, indicating replication and spreading of both the PIV and helper genomes. These cells could be passaged and continued to produce infectious viral particles (data not shown). It should be noted that in the experiments with these constructs, we did not detect superinfection exclusion. Helper genome-containing particles appeared to be capable of infecting the cells, having already established PIV RNA replication, indicating that helper-encoded replicative complexes could utilize already synthesized NS1 protein for RNA replication. The NS1-deficient helpers packaged DEN/YFV/GFP PIV genomes more efficiently (Figs. 3B and C) than did the previously applied capsid-producing cell lines or capsid- but not prM/E-expressing helper genomes. As we noticed in the previous experiments, the DEN2V prM/E-encoding helper was less efficient in packaging (Fig. 3B) than the similar YFV prM/E-expressing construct (Fig. 3C). In the latter case, the released infectious particles were packaged mainly into YFV-derived envelope, and their infectivity was readily neutralized by YFV-specific Ab (data not shown). Thus, the data demonstrated that DEN/YFV/Cherry chimeric PIV genomes could be packaged to high titers into infectious virions.
Chimeric PIVs with inactivated furin cleavage site
The above-described experiments suggested that capsid expression in cis (as a C/prM/E polyprotein) from the replicating RNA is greatly beneficial for infectious virion formation. Therefore, it was reasonable to expect that capsid production from PIV genomes could additionally improve their packaging into infectious viral particles. On the other hand, PIVs must remain noninfectious. To achieve this, we decided to take an advantage of a flavivirus-specific mechanism of viral particles formation. The prM/M- and E-containing SVPs that lack flavivirus genetic material and the entire nucleocapsid are not the only means of expressing the noninfectious virus-like particles. During their biogenesis, flaviviruses initially form the immature, prM-containing, noninfectious virions, which attain infectivity only at the last stages of biogenesis, following furin protease-mediated prM cleavage. Therefore, we used this feature and designed a new generation of PIV genomes. The distinguishing characteristic of CYF/DEN*/YFV/GFP (Figs. 4A) was in its ability to express CYF and DEN2V-derived prM/E having furin-specific cleavage site in prM mutated (prM*). Upon delivery into the cells by electroporation, the PIV genomes were capable of replication, readily detected by GFP expression, and no spreading infection indicating formation of true or pseudorevertants has been ever detected throughout the experiments (data not shown). We termed these new defective, chimeric RNAs encoding functional capsid protein as PIV2 genomes.
FIG. 4.
Replication of chimeric PIV2 with mutated furin cleavage site in BHK-21 cells. (A) The schematic representation of CYF/DEN*/YFV/GFP PIV2 genome. The position of mutations in prM cleavage site is indicated by arrow. Identical amino acids in the prM alignment are denoted by dashes. The introduced mutations are indicated by blue color. YFV-specific sequences in PIV2 and helper genomes are indicated by open boxes. DEN2V-specific sequences are indicated by filled, grey boxes. FMDV 2A indicates position of FMDV 2A protease. (B) BHK-21 cells were electroporated by the in vitro-synthesized PIV2 and helper RNAs (see Materials and Methods for details). Cells were seeded into 100-mm dishes in 10 ml of media and incubated at 37°C. (C and D) BHK-21 cells were infected by the samples harvested at the previous passage at 96 h post transfection or infection, at an MOI of 1 inf.u/cell. Cells were seeded into 100-mm dishes in 10 ml of media and incubated at 37°C. At the indicated time points (B–D), media were replaced and titers of packaged PIV2 and helper genomes were measured by infecting BHK-21 cells, containing VEErep/Pac-2A-NS1 replicon, by different dilutions of the samples and evaluating the numbers of GFP- and Cherry-positive cells.
Packaging of capsid prM*/E-encoding PIV2 RNA into infectious virus particles was achieved by co-transfection of their in vitro-synthesized RNAs and helper RNAs. The latter helper constructs, DEN/YFV/ΔNS1/Cherry and YF/YFV/ΔNS1/Cherry, encoded functional prM/E proteins of DEN2V and YFV, respectively, YFV capsid protein, and contained an above-described deletion in NS1 gene (Fig. 4A). After electroporation of the in vitro-synthesized RNAs, packaging of PIV2 and helper genomes with YF/YFV/ΔNS1/Cherry helper was efficient (Fig. 4B), and by 48 h post electroporation, titers of the CYF/DEN*/YFV/GFP and YF/YFV/ΔNS1/Cherry genome-containing viral particles reproducibly approached 108 inf.u/ml, and were higher than those previously detected in the experiments with chimeric, capsid-deficient PIVs (Fig. 3). Importantly, the newly designed, two-component genome virus could be passaged in tissue culture at an MOI ~1 inf.u/cell without a noticeable decrease in titers of either of the packaged genomes (Figs. 4C and D). Consistently with other experiments described in the previous sections, DEN2V prM/E-expressing DEN/YFV/ΔNS1/Cherry helper was less efficient, and titers of PIV2 and helper genome-containing particles were almost two orders of magnitude lower (Fig. 4B). Further passaging of this two-component genome virus was unsuccessful (data not shown). Thus, expression of capsid from the replicating PIV2 genome had an additional positive impact on packaging efficiency.
Chimeric PIVs with mutated NS2A
One of the important issues in development of live vaccines is their safety. PIV2 genomes represent complete viral genomes having clustered mutations only in the furin cleavage site. Therefore, the defective phenotype of such viruses relies on a limited genome modification, and a possibility of generating viable infectious virus by accumulation of true or pseudoreverting mutations might be a concern. Thus, from the safety standpoint, PIV2 needed additional modifications that would reduce the possibility of reversion to the replication-competent phenotype.
The previously published data demonstrated that mutations in the YFV NS2A-specific, internal processing site make YFV incapable of forming nucleocapsid-containing virions (Kummerer and Rice, 2002). Application of such mutations for PIV development could bring in additional safety features into the two-component genome flaviviruses. Therefore, in the next round of experiments, we designed PIV2 genomes, which either contained the mutations in NS2A only or in the cleavage sites of both prM and NS2A (Figs. 5A and B). PIV RNAs were co-transfected into the cells with the YF/YFV/ΔNS1/Cherry helper. The designed PIV and helper genomes efficiently complemented each other’s defects and developed spreading infection characterized by GFP and Cherry expression. Titers of particles containing PIV2 and helper genomes were determined on the above-described YFV NS1-producing cell line. By 72 h post transfection, they approached 108 inf.u/ml. Replication of these viruses did not induce CPE, and cells continued to grow and produce PIV and helper genome-containing virions after splitting. Most importantly, in repeated experiments, both of the designed PIV/helper pairs could be passaged in cell culture at an MOI of ~1 inf.u/cell without noticeable decreases in titers (Fig. 5C). In numerous experiments, which involved either electroporation or passaging, we have not detected infectious, single-genome virus formation, suggesting that this appears to be a very rare event, if it is possible for such constructs at all (Taucher, Berger, and Mandl, 2009).
FIG. 5.
Replication of two-component genome viruses having a helper genome with a deleted NS1 and the chimeric PIV2 genomes having either a mutated NS2A or mutations in both the NS2A and furin cleavage sites. (A) The schematic representation of chimeric PIV2 genomes having either mutation in YFV NS2A (CYF/DEN/YFV*/GFP) or in both YFV NS2A and furin cleavage site of DEN2V-specific prM (CYF/DEN*/YFV*/GFP), and helper genome YF/YFV/ΔNS1/Cherry. YFV-specific sequences are indicated by open boxes. DEN2V-specific sequences are indicated by filled, grey boxes. Arrows indicate the positions of mutations introduced into prM and NS2A. (B) Alignments of amino acid sequences of the protein fragments containing furin cleavage site and the internal cleavage site of NS2A. Identical amino acids are denoted by dashes. The introduced mutations are indicated by blue. Arrows indicate positions of cleavage. (C) BHK-21 cells were electroporated by indicated PIV2 and helper genomes and seeded into 100-mm dishes. Samples harvested at 96 h post electroporation or after starting passage 1 were used to perform the next passage, to infect naïve BHK-21 cells at an MOI of 1 inf.u/cell. At the indicated time points, media were replaced and titers of packaged PIV2 and helper genomes were measured by infecting BHK-21 cells containing VEErep/Pac-2A-NS1 replicon, by different dilutions of the samples and evaluating the numbers of GFP- and Cherry-positive cells. Dashed lines indicate the limits of detection.
In additional experiments, we tested the effects of the mutations introduced into the prM cleavage site and NS2A on the density of the released particles. BHK-21 cells were electroporated with CYF/DEN(K)/YFV/GFP (representing replication-competent virus genome), CYF/DEN*/YFV/GFP PIV2 and CYF/DEN*/YFV*/GFP PIV2 RNAs. Cells were incubated in serum-free medium, and, at 2 days post transfection, the released viral particles and SVPs were concentrated by ultracentrifugation and analyzed on the discontinuous sucrose gradients. As expected, replication of CYF/DEN*/YFV/GFP RNA (having mutated prM) led to a release of high density, genome-containing particles and low-density SVPs, having a distribution similar to that found for the particles released from CDEN/DEN(K)/YFV/GFP RNA transfected cells (Fig. 6). Replicating CYF/DEN*/YFV*/GFP PIV2 RNA, having mutations in both prM and NS2A, produced mostly low-density particles, corresponding to SVPs (Fig. 6B).
Fig. 6.
Analysis of viral and subviral particles released from the cells transfected with viral and PIV2 RNAs. (A) Schematic representation of the RNAs used in the experiments. Arrows indicate positions of the mutations introduced into prM and NS2A. (B) Distribution of viral and subviral particles in the discontinuous sucrose gradients. Transfected cells were seeded into 150-mm dishes and, after 24 h incubation at 37°C, media were replaced to serum-free VP-SF medium. After an additional 24 h-long incubation, 20 ml samples of collected media were clarified by low-speed centrifugation and particles were pelleted by ultracentrifugation through the cushion of 10% sucrose in SW-28 rotor (1h, 25, 000 rpm, 4°C). Pellets were suspended in PBS, loaded onto the discontinuous sucrose gradients (see Materials and Methods for details), and centrifugation was performed at 50,000 rpm for 4 h at 4°C in SW-55 rotor of Beckman Optima L-90K ultracentrifuge. Gradients were fractionated, aliquots were diluted with PBS and viruses and VLPs were pelleted in the Beckman TLA-55 rotor. Pellets were dissolved in SDS-containing protein loading buffer having no β-mercaptoethanol. Samples were analyzed by Western blotting using D1-4G2 monoclonal antibody. Secondary goat anti-mouse antibodies were labeled with IRDye 800. Membranes were analyzed on LI-COR imager.
DISCUSSION
Recent advances in understanding the molecular mechanism of RNA-positive virus replication suggested new approaches for the development of rationally designed vaccines. Manipulations with the infectious cDNAs provide an opportunity to design vaccine candidates that greatly differ from the traditional ones: live attenuated, inactivated and subunit. In previous studies, we succeeded in designing defective WNV and YFV capsid-deficient PIVs, which expressed only SVPs, lacking any viral genetic material (Mason, Shustov, and Frolov, 2006; Shustov, Mason, and Frolov, 2007). The distinguishing feature of the PIVs was their productive replication in the capsid-producing cell lines or in the presence of capsid-expressing defective viral helper RNAs, which promoted packaging of PIV genomes into infectious virus particles. In animals, packaged PIV genomes developed only a single-round infection leading to production of SVP, which efficiently induced neutralizing antibodies. Therefore, the flavivirus PIVs demonstrated high safety levels due to their inability to develop spreading infection in vivo and also a high efficiency of inducing neutralizing antibodies due to antigen presentation in the SVP form. Large scale PIV production did not require high biocontainment conditions, because, in numerous experiments, no RNA recombination and self-replication-competent virus were ever detected.
Application of the same strategy of PIV design and propagation to chimeric viruses encoding DEN2V prM/E turned to be less successful. Packaging of DEN/YFV/GFP PIV genomes into infectious virus particles was inefficient in spite of the chimeric, infectious viruses encoding all of the structural and nonstructural proteins could replicate to high titers (Fig. 1). This ambiguity between capsid functions when supplied in cis or in trans, could be explained by the translation strategy of the flavivirus genome in which all of the viral structural and nonstructural proteins are translated from the same RNA. As a result, all of the proteins appear to be co-expressed to high concentration in the same or closely located cellular compartments. Subsequently, infectious virus genome packaging into the nucleocapsid and envelope appears to proceed reasonably efficiently even if the heterologous structural proteins were synthesized from the replicating viral genome. However, the inefficiency of infectious particle formation becomes evident when capsid and heterologous envelope proteins are expressed in trans and co-compartmentalize at lower concentration. Notably, expression of the entire DEN2V structural polyprotein from VEEV replicons in the replicating chimeric PIV genome-containing cells also failed to increase the efficiency of the PIV genome packaging (data not shown), suggesting that proper compartmentalization of heterologous DEN2V structural proteins and replicating PIV RNA might also play an important role in packaging.
Thus, the results of the initial study demonstrated that packaging of chimeric PIV genomes using capsid or capsid/prM/E-expressing cell lines was very inefficient. Moreover, the design of standard PIV genomes and capsid expressing VEEV replicons did not provide much room for modification of this packaging system. The two-component genome flaviviruses are more flexible and facilitate introduction of a wide variety of changes and testing different trans-complementation strategies. Therefore, to achieve more efficient packaging of chimeric PIV genomes into infectious viral particles, we strongly modified the design of both the PIV and the helper RNAs. Newly developed PIV2 genomes were constructed in a way to contain two sets of the mutations, and each of them was sufficient for making PIVs noninfectious despite the fact that their genomes encoded a complete set of structural proteins (C/prM/E). The mutations in the prM-specific furin cleavage site make the virus able to produce only the immature, prM-containing virions, and the mutations in NS2A make it incapable of infectious virions (but not SVP) formation (Burke and Monath, 2001). Introduction of mutations into different viral proteins added an additional level of safety to PIV application and had no negative effect on RNA replication and packaging, as evaluated by GFP expression at different times post transfection or infection. Simultaneous reversion of numerous mutations or the appearance of multiple pseudoreverting mutations in different proteins likely represents a negligible risk. Moreover, even in the event of multiple reversions occurring, the resulting virus will be a DEN2/YFV chimera, which has been previously shown to have highly attenuated phenotype (Guirakhoo et al., 2002; Guirakhoo et al., 2004).
The newly designed helpers used for PIV2 genome packaging encoded functional C/prM/E and were incapable of self-replication by the introduction of an extended deletion into the NS1-coding sequence. These NS1-deficient helper RNAs produced all of the structural proteins required for both their own and PIV2 packaging, but required the NS1 protein to be supplied in trans by the replicating PIV. (The schematic representation of trans-complementation and packaging strategies is shown in Fig. 7.) In all of the experiments, the helper genome encoding YFV structural proteins, homologous to PIV2 and helper backbones, were consistently more efficient in packaging than the same structural proteins derived from DEN2V. On one hand, this might be considered a negative characteristic of the new system, because, for large-scale production, the chimeric PIVs need to be packaged into the homologous to backbone envelope. However, we strongly believe that this is a very useful feature: for immunization, all of the chimeras can be packaged into the same glycoproteins. There is always a concern, that in the presence of subneutralizing levels of pre-existing antibodies, even replicating PIVs might induce a phenomenon similar to DHF or DSS, as do the replication-competent DEN viruses (Halstead, 2003). The chimeric PIV genome packaging into the envelope derived from the viruses belonging to a different serocomplex (i.e. YFV or WNV) provides an opportunity to avoid this problem for the most part. Application of the tetravalent live vaccine against dengue infections might also encounter some level of interference between four recombinant viruses in vivo or possible strong differences in the immunogenicity of the recombinants, which might result from variations in tissue specificities between different DENV serotypes. Again, delivery of different PIV genomes using virions with the same heterologous envelope is a means to avoid this problem. However, these assumptions need further experimental support.
FIG. 7.
Proposed replication strategies of the chimeric PIV2 and helper genome-containing viral particles at high and low MOIs. At high MOI, both genomes, the PIV2 genome (encoding C/prM*/E, which are incapable of forming infectious virions) and NS1-deficient helper genome (encoding functional capsid, prM and E), are delivered to the same cell and produce a complete set of YFV nonstructural and structural proteins required for virus replication. Cells produce a two-component genome virus that can be further passaged at an escalating scale. At low MOI, cells receive only one of the genomes, and those infected with PIV2 produce SVPs and/or immature noninfectious viral particles with the DEN2V envelope.
The designed two-component YFV backbone-based system efficiently packages chimeric PIV2 genomes. The titers reproducibly approach 108 inf.u/ml (Figs. 4 and 5). Nevertheless, the chimeric PIV2 packaging system can certainly be additionally improved. Firstly, the deletion of GFP and Cherry will lead to more natural capsid expression, and, as we detected before, to higher growth rates. Secondly, the use of more efficiently replicating backbones, such as those based on the WNV genome, might additionally increase titers of chimeric PIVs.
One of the classic questions asked regarding defective viruses is whether they can generate replication-competent variants through recombination. The strategy of the genomes of the two-component genome viruses suggests that the defective structural genes in the PIV2 RNA can be replaced by those derived from the NS1-defective helper RNA. Such recombinant events have been recently described for another flavivirus, albeit occurring with very low efficiency (Taucher, Berger, and Mandl, 2009). However, besides RNA recombination, formation of the infectious recombinant virus will also require reverting or pseudoreverting mutations to restore function of the defective NS2A. To date, in numerous experiments, we have not detected the formation of infectious, productively replicating, recombinant virus, YFV 17D, which could be capable of developing spreading infection at low MOI. In spite such recombinant is a highly attenuated vaccine strain, possibility of its formation as a result of single or double recombination event can be further strongly reduced by using modified cyclization sequences in the helper genome (Suzuki et al., 2008) and/or by using helper genomes with alternative codon frequencies in sites of possible recombination.
Taken together, the results of our study demonstrate that i) chimeric PIV genomes can be packaged to high titers into infectious virus particles using the NS1-deficient helper RNAs. ii) This efficient packaging is determined by the expression of capsid protein from both PIV2 and helper genomes and the expression of all of the viral structural genes from the helper RNA. iii) Chimeric PIV2 genomes are most efficiently packaged into the envelope proteins, which are homologous to PIV2 and helper genome backbones. iv) The newly designed PIVs and helpers can be passaged as two-component genome virus at an escalating scale in cell culture. v) The designed PIVs are capable of producing SVPs, which were previously shown to function as efficient immunogens.
MATERIALS AND METHODS
Cell cultures
The BHK-21 cells were kindly provided by Paul Olivo (Washington University, St. Louis, Mo). They were maintained at 37°C in alpha minimum essential medium (αMEM) supplemented with 10% fetal bovine serum (FBS) and vitamins.
Plasmid constructs
Standard recombinant DNA techniques were used for all plasmid constructions. The parental low-copy number plasmid pACNR/FLYF-17Dx containing infectious cDNA of the YFV 17D strain genome was described elsewhere (Bredenbeek et al., 2003) and kindly provided by Dr. Charles M. Rice (Rockefeller University, New York). DEN2V-specific genes used in the study encoded proteins corresponding to Dengue virus 2 New Guinea C isolate (NGC) (GenBank accession number M29095). pCDEN/DEN(E)/YFV/GFP and pCDEN/DEN(K)/YFV/GFP encoded 5′UTR of YFV genome, 25 amino acids (aa) of YFV capsid, fused with GFP- and foot-and-mouth disease virus (FMDV) 2A protease-coding sequences, followed by codon-optimized DEN2V capsid gene with YFV-specific signal peptide, codon-optimized DEN2V prM/E and the rest of YFV 17D genome. Plasmids differed only in the sequence coding the residue 126 of E protein, represented by either Glu or Lys (E or K). pCYF/DEN(E)/YFV/GFP and pCYF/DEN(K)/YFV/GFP had essentially the same design, but instead of DEN2V capsid, they encoded a codon-optimized YFV capsid gene with a YFV-specific signal peptide. The schematic representations of these viral genomes are shown in Fig. 1A. pDEN/YFV/GFP, pWN/YFV/GFP and pYF/YFV/GFP plasmids encoded PIV genomes, in which YFV 5′UTR was followed by 25 codons of YFV capsid, fused in frame with GFP and FMDV 2A protease genes. The FMDV 2A protease was fused with YFV-specific signal peptide, followed by either DEN2V, YFV or WNV prM/E-coding sequences. The latter structural genes were followed by YFV NS1-NS5 and the 3′ UTR. CYF/DEN*/YFV/GFP PIV2 genome-coding plasmid contained a YFV 5′UTR, 25 aa of YFV capsid, fused with GFP and FMDV 2A protease, followed by a codon-optimized YFV capsid- and a signal peptide-coding sequences, DEN2V prM/E, YFV NS1-5 and 3′UTR. It had three amino acid substitutions in the DEN2V prM sequence, which inactivated the furin cleavage site (see Fig. 4A for details). CYF/DEN/YFV*/GFP PIV2 and CYF/DEN*/YFV*/GFP PIV2 genome-coding plasmids had essentially the same design as that described above for CYF/DEN*/YFV/GFP PIV2. Both of them contained a mutation inactivating the internal processing site in NS2A (Kummerer and Rice, 2002) (see Fig. 5B for details) and encoded either wt DEN2V prM or the DEN2V prM with above described mutations in the furin cleavage site, respectively.
Plasmid pCYF/YFV/Cherry encoded a helper genome in which YFV 5′UTR was followed by 25 codons of YFV capsid, fused in frame with Cherry, FMDV 2A protease and codon-optimized YFV capsid genes, YFV prM signal peptide, 6 amino terminal codons of YFV prM, 49 carboxy terminal codons of YFV E and the rest of YFV genome, including the 3′UTR. Plasmid pYF/YFV/ΔNS1/Cherry encoded the NS1-deficient helper genome, having the YFV 5′UTR, 25 codons of YFV capsid, Cherry and FMDV 2A protease genes, followed by codon-optimized YFV capsid gene, YFV prM/E and the rest of the YFV genome, in which the sequence coding aa 167 to 272 of NS1 was deleted. pDEN/YFV/ΔNS1/Cherry had essentially the same design, but encoded DEN2V-specific instead of YFV-specific prM/E. pVEErep/Pac-2A-NS1 encoded Venezuelan equine encephalitis virus (VEEV) replicon (Petrakova et al., 2005), which contained the puromycin acetyltransferase (Pac) gene fused in frame with FMDV 2A protease gene, 23 aa-long, YFV NS1-specific signal peptide and the entire YFV NS1 under control of the subgenomic promoter. In all of the plasmids, virus-specific sequences were under the control of the SP6 RNA polymerase promoter. Maps and sequences of the plasmids are available from the authors upon request.
RNA transcriptions
All of the plasmids were purified by centrifugation in CsCl gradients. Before the transcription reaction, plasmids encoding PIVs, helpers and chimeric virus genomes were linearized by XhoI, and plasmids encoding VEEV replicons were linearized by MluI. RNAs were synthesized by SP6 RNA polymerase in the presence of cap analog as described elsewhere (Bredenbeek et al., 1993). The yield and integrity of transcripts were analyzed by gel electrophoresis under non-denaturing conditions. Aliquots of transcription reactions were used for electroporation without additional purification.
RNA transfections
Electroporation of BHK-21 cells with PIV, helper and VEEV replicons RNAs was performed under the previously described conditions (Liljeström et al., 1991). VEEV replicons-carrying cell lines were established by transfecting the in vitro-synthesized RNAs followed by puromycin selection (10 μg/ml).
Measuring the titers of infectious viral particles containing defective YFV genomes
To measure the titers of released virions containing different defective genomes, BHK-21 cells were seeded into six-well Costar dishes at a concentration of 5×105 cells/well. Four hours later, cells were infected with different dilutions of the samples, and, after 1 h incubation at 37°C in a 5% CO2 incubator, 2 ml of αMEM supplemented with 10% FBS was added. The numbers of infected cells were estimated by counting GFP- and Cherry-positive cells under an inverted UV microscope after 36 h of incubation at 37°C. The fraction of infected cells was determined by counting GFP-positive cells in multiple defined area of microscopic field. Counts for different fields were averaged and used for calculation of the titers.
Titers of replication-competent viruses were determined by standard plaque assay on BHK-21 cells (Lemm et al., 1990). After three days of incubation at 37°C, monolayers were fixed by 2.5% formaldehyde and either stained with crystal violet, or studied under a fluorescent, dissecting microscope in order to evaluate the numbers of GFP-positive foci.
Titers of viral particles containing PIV2 and NS1-deficient helper genomes were evaluated on BHK-21 cells producing YFV NS1 protein from the persistently replicating VEEV replicon. Cells were seeded into six-well Costar dishes and infected with different dilutions of the samples as described above. The numbers of infected cells were estimated by counting GFP- and Cherry-positive cells under an inverted UV microscope after 36 h of incubation at 37°C. The fraction of infected cells was determined by counting GFP- and/or Cherry-positive cells in multiple defined area of the microscopic field. Counts for different fields were averaged and used for calculation of the titers.
Passaging of two-component genome viruses
Naïve BHK-21 cells were infected with samples of packaged PIV and helper genomes at MOIs indicated in the figures. Media were replaced at the time points indicated in the figures. Titers of infectious viral particles in the harvested samples were determined as described above.
Analysis of viral and subviral particles
BHK-21 cells were transfected by 8 μg of in vitro-synthesized RNAs. Transfected cells were seeded into 150-mm dishes and incubated between 24 and 48 h in serum-free medium VP-SF (Invitrogen). 20 ml samples of collected media were clarified by low-speed centrifugation and particles were pelleted by ultracentrifugation for 1 h in the SW-28 rotor (25, 000 rpm, at 4°C).
Pellets were suspended in PBS and loaded on the discontinuous sucrose gradients (1.2 ml of 50%, 2.3 ml of 37.5% and 1 ml of 10% sucrose), and centrifugation was performed at 50,000 rpm for 4 h at 4°C in the SW-55 rotor of Beckman Optima L-90K ultracentrifuge. Gradients were fractionated, aliquots were diluted with PBS and viruses and virus-like particles were pelleted in the Beckman TLA-55 rotor at 45,000 rpm for 1 h at 4°C. Pellets were dissolved in the SDS-containing protein loading buffer having no β-mercaptoethanol.
Samples were analyzed by Western blotting using group-specific (anti-flavivirus E-protein) monoclonal Ab D1-4G2 and IRDye 800-labeled secondary antibody. Membranes were analyzed on the LI-COR imager.
Acknowledgments
We thank Dr. P. Mason for providing D1-4G2 monoclonal antibody. This work was supported by Public Health Service grant AI070207.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aberle JH, Aberle SW, Kofler RM, Mandl CW. Humoral and cellular immune response to RNA immunization with flavivirus replicons derived from tick-borne encephalitis virus. J Virol. 2005;79(24):15107–13. doi: 10.1128/JVI.79.24.15107-15113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M, Lai CJ. Construction of chimeric dengue virus by substitution of structural protein genes. Proc Natl Acad Sci USA. 1991;88:10342–10346. doi: 10.1073/pnas.88.22.10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M, Men R, Tokimatsu I, Lai CJ. Genetic determinants responsible for acquisition of dengue type 2 virus mouse neurovirulence. J Virol. 1998;72(2):1647–51. doi: 10.1128/jvi.72.2.1647-1651.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenbeek PJ, Frolov I, Rice CM, Schlesinger S. Sindbis virus expression vectors: Packaging of RNA replicons by using defective helper RNAs. J Virol. 1993;67:6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenbeek PJ, Kooi EA, Lindenbach B, Huijkman N, Rice CM, Spaan WJ. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J Gen Virol. 2003;84(Pt 5):1261–8. doi: 10.1099/vir.0.18860-0. [DOI] [PubMed] [Google Scholar]
- Burke DS, Monath TP. Flaviviruses. In: Knipe DM, Howley PM, editors. Fields’ Virology. 4. Lippincott, Williams and Wilkins; New York: 2001. pp. 1043–1125. [Google Scholar]
- Chambers TJ, Nestorowicz A, Mason PW, Rice CM. Yellow fever/Japanese encephalitis chimeric viruses: construction and biological properties. J Virol. 1999;73(4):3095–101. doi: 10.1128/jvi.73.4.3095-3101.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effler PV, Pang L, Kitsutani P, Vorndam V, Nakata M, Ayers T, Elm J, Tom T, Reiter P, Rigau-Perez JG, Hayes JM, Mills K, Napier M, Clark GG, Gubler DJ. Dengue fever, Hawaii, 2001–2002. Emerg Infect Dis. 2005;11(5):742–9. doi: 10.3201/eid1105.041063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirakhoo F, Pugachev K, Arroyo J, Miller C, Zhang ZX, Weltzin R, Georgakopoulos K, Catalan J, Ocran S, Draper K, Monath TP. Viremia and immunogenicity in nonhuman primates of a tetravalent yellow fever-dengue chimeric vaccine: genetic reconstructions, dose adjustment, and antibody responses against wild-type dengue virus isolates. Virology. 2002;298(1):146–59. doi: 10.1006/viro.2002.1462. [DOI] [PubMed] [Google Scholar]
- Guirakhoo F, Pugachev K, Zhang Z, Myers G, Levenbook I, Draper K, Lang J, Ocran S, Mitchell F, Parsons M, Brown N, Brandler S, Fournier C, Barrere B, Rizvi F, Travassos A, Nichols R, Trent D, Monath T. Safety and efficacy of chimeric yellow Fever-dengue virus tetravalent vaccine formulations in nonhuman primates. J Virol. 2004;78(9):4761–75. doi: 10.1128/JVI.78.9.4761-4775.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas J, Park EU, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Current Biology. 1996;6(3):315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- Halstead SB. Neutralization and antibody-dependent enhancement of dengue viruses. Adv Virus Res. 2003;60:421–67. doi: 10.1016/s0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- Halstead SB. Dengue. Lancet. 2007;370(9599):1644–52. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- Huang CY, Butrapet S, Pierro DJ, Chang GJ, Hunt AR, Bhamarapravati N, Gubler DJ, Kinney RM. Chimeric dengue type 2 (vaccine strain PDK-53)/dengue type 1 virus as a potential candidate dengue type 1 virus vaccine. J Virol. 2000;74(7):3020–8. doi: 10.1128/jvi.74.7.3020-3028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Widman DG, Bourne N, Konishi E, Mason PW. Construction and evaluation of a chimeric pseudoinfectious virus vaccine to prevent Japanese encephalitis. Vaccine. 2008;26(22):2772–81. doi: 10.1016/j.vaccine.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Kofler RM, Heinz FX, Mandl CW. A novel principle of attenuation for the development of new generation live flavivirus vaccines. Arch Virol Suppl. 2004;(18):191–200. doi: 10.1007/978-3-7091-0572-6_17. [DOI] [PubMed] [Google Scholar]
- Kummerer BM, Rice CM. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J Virol. 2002;76(10):4773–84. doi: 10.1128/JVI.76.10.4773-4784.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm JA, Durbin RK, Stollar V, Rice CM. Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J Virol. 1990;64(6):3001–11. doi: 10.1128/jvi.64.6.3001-3011.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljeström P, Lusa S, Huylebroeck D, Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol. 1997;71(12):9608–17. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol. 1999;73(6):4611–21. doi: 10.1128/jvi.73.6.4611-4621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PW, Shustov AV, Frolov I. Production and characterization of vaccines based on flaviviruses defective in replication. Virology. 2006;351(2):432–43. doi: 10.1016/j.virol.2006.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrakova O, Volkova E, Gorchakov R, Paessler S, Kinney RM, Frolov I. Noncytopathic replication of Venezuelan equine encephalitis virus and eastern equine encephalitis virus replicons in Mammalian cells. J Virol. 2005;79(12):7597–608. doi: 10.1128/JVI.79.12.7597-7608.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletnev AG, Men R. Attenuation of the Langat tick-borne flavivirus by chimerization with mosquito-borne flavivirus dengue type 4. Proc Natl Acad Sci USA. 1998;95:1746–1751. doi: 10.1073/pnas.95.4.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugachev KV, Guirakhoo F, Trent DW, Monath TP. Traditional and novel approaches to flavivirus vaccines. Int J Parasitol. 2003;33(5–6):567–82. doi: 10.1016/s0020-7519(03)00063-8. [DOI] [PubMed] [Google Scholar]
- Shustov AV, Mason PW, Frolov I. Production of pseudoinfectious yellow fever virus with a two-component genome. J Virol. 2007;81(21):11737–48. doi: 10.1128/JVI.01112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Fayzulin R, Frolov I, Mason PW. Identification of mutated cyclization sequences that permit efficient replication of West Nile virus genomes: use in safer propagation of a novel vaccine candidate. J Virol. 2008;82(14):6942–51. doi: 10.1128/JVI.00662-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taucher C, Berger A, Mandl CW. A trans-Complementing Recombination Trap Demonstrates a Low Propensity of Flaviviruses for Intermolecular Recombination. J Virol. 2009 doi: 10.1128/JVI.01063-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widman DG, Ishikawa T, Fayzulin R, Bourne N, Mason PW. Construction and characterization of a second-generation pseudoinfectious West Nile virus vaccine propagated using a new cultivation system. Vaccine. 2008;26(22):2762–71. doi: 10.1016/j.vaccine.2008.03.009. [DOI] [PubMed] [Google Scholar]