Abstract
Many long-lived organisms, including humans, can regenerate some adult tissues lost to physical injury or disease. Much of the previous research on mechanisms of regeneration has focused on adult stem cells, which give rise to new tissue necessary for the replacement of missing body parts. Here we report that apoptosis of differentiated cells complements stem cell division during regeneration in the planarian Schmidtea mediterranea. Specifically, we developed a whole-mount TUNEL assay that allowed us to document two dramatic increases in the rate of apoptosis following amputation – an intial localized response near the wound site and a subsequent systemic response that varies in magnitude depending on the type of fragment examined. The latter cell death response can be induced in uninjured organs, occurs in the absence of planarian stem cells, and can also be triggered by prolonged starvation. Taken together, our results implicate apoptosis in the restoration of proper anatomical scale and proportion through remodeling of existing tissues. We also report results from initial mechanistic studies of apoptosis in planarians, which revealed that a S. mediterranea homolog of the antiapoptotic gene BCL2 is required for cell survival in adult animals. We propose that apoptosis is a central mechanism working in concert with stem cell division to restore anatomical form and function during metazoan regeneration.
Keywords: Planaria, Planarian, Cell Death, Apoptosis, Regeneration, Tissue Remodeling, Cell Turnover, Tissue Homeostasis, BCL2
Introduction
Examples of regeneration have been documented throughout the animal kingdom, yet there is considerable variation in regenerative capacity from one species to the next (Gurley and Sánchez Alvarado, 2008). Human organs also differ markedly in their ability to repair damage caused by physical injury or disease. Thus, one important challenge in the field of regenerative medicine is to define the cellular mechanisms of naturally occurring regenerative phenomena, with the ultimate goal of developing treatments that stimulate these mechanisms in a clinical setting.
Previous mechanistic studies of metazoan regeneration have generally focused on the question of how new tissue is created in pathological contexts. Although proliferation of differentiated cells may be sufficient in some cases (Dor et al., 2004), the recruitment of stem or progenitor cell populations has emerged as a central and evolutionarily conserved regeneration paradigm (Gurley and Sánchez Alvarado, 2008). Adult stem cells may be formed de novo in response to injury via dedifferentiation, or they may be maintained under physiological conditions for tissue homeostasis (Gurley and Sánchez Alvarado, 2008). In either case, they serve a critical purpose in regeneration by fueling the production of various differentiated cell types that are necessary for restoring a functional anatomy.
The freshwater planarian S. mediterranea has emerged as a powerful model organism for studies of stem cell function in tissue renewal and repair. Planarians turn over most if not all of their somatic tissues throughout life (Pellettieri and Sánchez Alvarado, 2007) and have the remarkable capacity to regenerate complete individuals from small body fragments (Reddien and Sánchez Alvarado, 2004). Each of these attributes depends on a large population of adult stem cells termed neoblasts. In response to amputation, neoblasts increase their rate of division (Baguñà, 1976b; Saló and Baguñà, 1984) and migrate to the wound site (Dubois, 1949; Eisenhoffer et al., 2008; Newmark and Sánchez Alvarado, 2000), where they give rise to a mass of new tissue called the blastema. Cells in the blastema then differentiate over a period of several days to replace missing body structures (Reddien and Sánchez Alvarado, 2004).
An equally important, but much less well understood aspect of regeneration involves the remodeling of preexisting tissues. First described by T.H. Morgan in 1901 and termed ‘morphallaxis’ (Morgan, 1901), this phenomenon restores anatomical scale and proportion and allows for the integration of new and old tissues (Reddien and Sánchez Alvarado, 2004). In planarians, a similar remodeling process also occurs when animals are starved for a period of several months, resulting in ‘degrowth’, or an up to ~20-fold reduction in overall size (Baguñà and Romero, 1981; Reddien and Sánchez Alvarado, 2004; Romero and Baguñà, 1991). In contrast to the relatively well-characterized neoblast proliferation response that underlies blastema formation, the cellular mechanisms responsible for remodeling preexisting tissues have remained largely unknown.
Here we report the development of a TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling) assay allowing the visualization and quantification of apoptotic cells in whole-mounted planarians. Using this assay, we show that dynamic spatial and temporal changes in apoptosis are key features of tissue remodeling after injury and in response to prolonged starvation. We also show that planarian cell death is regulated by a homolog of BCL2, an evolutionarily conserved antiapoptotic gene that inhibits caspase activation and physical dismantling of dying cells in animals ranging from C. elegans to humans (Cotter, 2009; Danial and Korsmeyer, 2004). Our findings argue that the deletion of differentiated cells through apoptosis functions in concert with stem cell division to regulate the scale and proportion of adult tissues during regeneration.
Materials and methods
Planarian maintenance
All experiments were performed with a clonal line of asexual S. mediterranea maintained as described (Cebria and Newmark, 2005) and fed with homogenized beef liver 5 days to 2 weeks prior to the start of each experiment.
Electron microscopy
Specimens were prepared for electron microscopy using high pressure freezing – freeze substitution, as described (Hammarlund et al., 2007), with the following modifications: 1) specimens were transferred to 20% BSA prior to freezing; 2) animals were fixed in anhydrous acetone with 1% osmium tetroxide, 0.1% uranyl acetate, and 2% water during freeze-substitution; 3) specimens were embedded in epon-araldite (30% resin/acetone for 5 hrs., 70% resin/acetone for 6 hrs., 90% resin overnight, and fresh 100% resin for 8 hrs., curing at 60°C for 2 days); 4) sections were collected using an Ultracut 6 microtome (Leica). TEM specimens were stained with 2.5% uranyl acetate for 4 min. prior to imaging on a Hitachi H-7100 electron microscope equipped with a Gatan Orius CCD camera. Serial block-face scanning electron microscopy was performed as described (Denk and Horstmann, 2004), with the following modifications: 1) samples were processed as described above for TEM; 2) after substitution and embedding, samples were mounted on an aluminum stub and serially imaged with Gatan 3View™ mounted on a FEI Quanta 600 FEG scanning EM (raw images were 2k × 2k, acquired at 5KeV, spot 3, 2000×).
Whole-mount TUNEL
Animals were placed in glass vials and incubated at room temperature (RT), with rocking, for 5 min. in 10% n-acetyl cysteine (diluted in PBS). This step simultaneously kills planarians and removes their thick coating of mucous (Pearson et al., 2009). Aniimals were then fixed and permeabilized, respectively, in 4% formaldehyde [diluted in PBST (PBS with 0.3% triton X−100)] and 1% SDS (diluted in PBS) for 20 min. each (both steps at RT with rocking). Fixed animals were bleached overnight at RT, without rocking, in 6% H2O2 (diluted in PBST). Bleached animals were rinsed in PBST, transferred to microfuge tubes (2–4 animals per tube), and stored at 4°C in PBST. For staining, animals were rinsed in PBS and incubated for 4 hrs. at 37°C in terminal transferase enzyme (Chemicon, Cat. No. 90418) diluted in reaction buffer (Chemicon, Cat. No. 90417). Enzyme-treated animals were then rinsed in stop/wash buffer (Chemicon, Cat. No. 90419), rinsed in PBSTB (PBST with 0.25% BSA), and incubated for 4 hrs. at RT, on a platform shaker, in anti-digoxigenin-rhodamine (Chemicon, Cat. No. 90429) diluted in blocking solution (Chemicon, Cat. No. 90425). Stained animals were rinsed on a platform shaker at RT in PBSTB for 4 × 10 min. and then overnight, mounted under cover slips on glass slides in Vectashield (Vector, Cat. No. H-1000), and imaged as described below. For all TUNEL figures, “3 Days” = fixed 70–74 hours post-amputation (post-irradiation for Supplementary Fig. 1F). For Supplementary Figure 1C, “ −TdT” = reaction buffer alone substituted for terminal transferase; “ −Antibody” = blocking solution alone substituted for anti-digoxigenin antibody; “+HCl” = treated with 2N HCl for 40 minutes after fixation to generate DNA nicks (Tourigny et al., 1997); “+DNase I” = treated with DNase I (5 U/ml) for 1 hour after fixation.
TUNEL image acquisition and analysis
All TUNEL-stained animals were photographed on a Zeiss Lumar V12 stereomicroscope and photographed with an Axiocam HRc camera. This enabled visualization of each animal in a single focal plane. Image acquisition parameters and brightness, contrast, and gamma (BCG) adjustments (Adobe Photoshop) were optimized for each individual image shown in the Figures. For all quantitative TUNEL analyses, every image was acquired using identical parameters and linear BCG settings. Tagged Image Files (TIFs) were then analyzed using Kalaimoscope (Rink et al., 2005) (Transinsight GmbH) to generate a count of the total number of TUNEL-positive nuclei (TPN) and the specimen area for each image. This analysis was restricted to the specimen area within 100 µm of the amputation site for 4-hour timepoints in Figure 1C, Figure 4B, and Supplementary Figure 2E. Identical Kalaimoscope settings were used within each experiment. Between experiments, minor adjustments to Kalaimoscope settings were required to contend with qualitative variability in staining results (e.g., background). Supplementary Figure 1B shows a comparison of manual and automated TPN counts for a representative sample of experimental images.
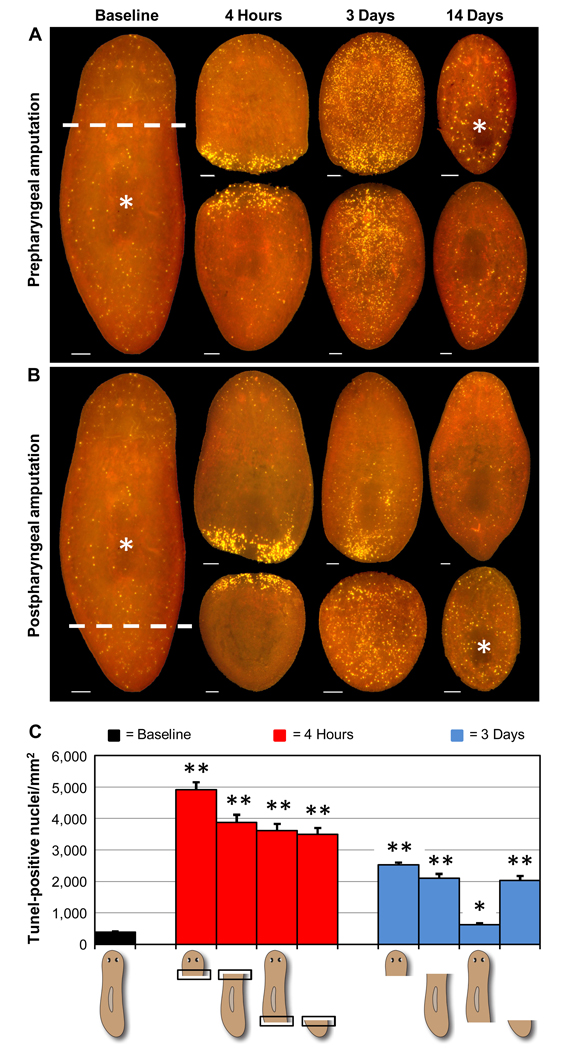
Fig. 1.
Apoptosis in regenerating planarians. Animals were amputated as indicated (dashed white lines) at Time 0, prior to visualization of apoptosis by TUNEL. These and all subsequent TUNEL pictures show whole-mounted animals imaged in a single focal plane. (A, B) Representative results for prepharyngeal (A) or postpharyngeal (B) amputation. Asterisks denote the original position of the pharynx (left) and the newly regenerated pharynges in head and tail fragments (right). Scale bars = 100 µm. (C) Average TUNEL levels for 5 independent experiments (n ≥ 21 for each condition). Values for the 4-hour timepoint are restricted to the area within 100 µm of the amputation site (boxed areas in schematic diagrams). Error bars = +/− s.e.m. * = p value < 0.01 for two-tailed Student’s t-Test comparing regenerating fragments with intact controls. ** = p value < 1 × 10−10. See Supplementary Figure 2A for extended regeneration timecourses.
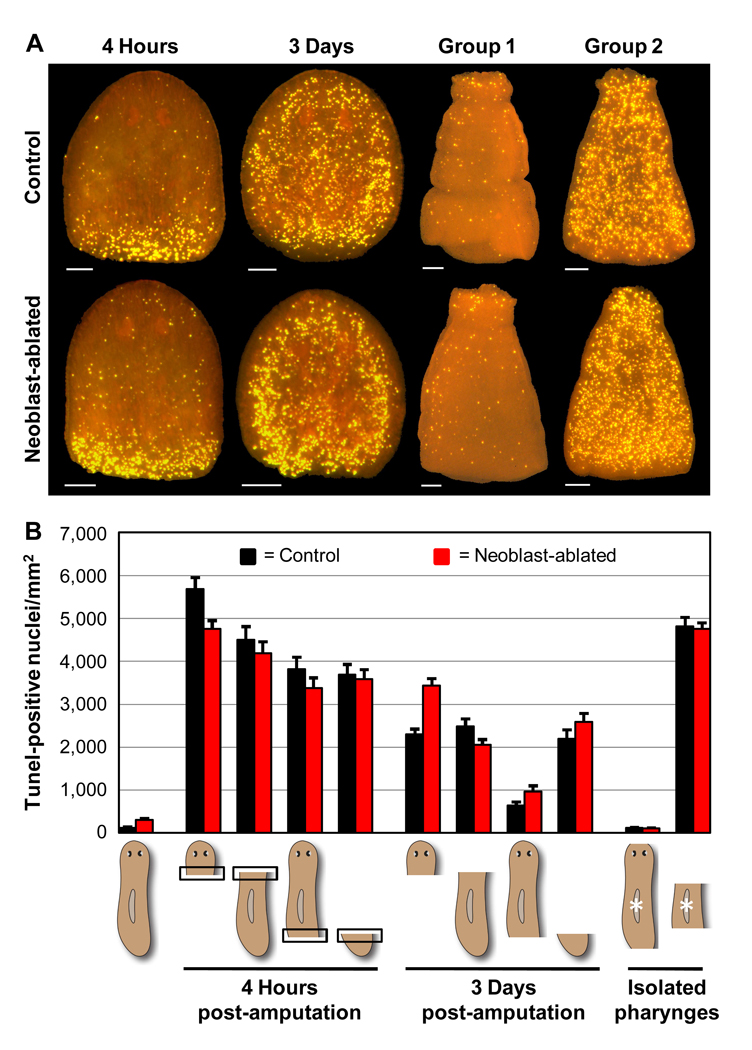
Fig. 4.
Amputation-induced apoptosis is stem cell-independent. Control and neoblast-ablated (irradiated) animals (Materials and methods) were amputated as described in Figure 1 and Figure 2. (A) Representative TUNEL patterns in head fragments (left) or pharynges isolated from regenerating trunk fragments (right). Head fragments were fixed at 4 hours or 3 days post-amputation. All pharynges were isolated and fixed at 3 days post-amputation. Scale bars = 100 µm. (B) Average TUNEL levels for 3 independent experiments (n ≥ 17 for each condition). Values for the 4-hour timepoint are restricted to the area within 100 µm of the amputation site (boxed areas in schematic diagrams). Error bars = +/− s.e.m. p values for two-tailed Student’s t-Tests comparing paired control and irradiated regenerating animals were all > 0.05, except for prepharyngeal anterior fragments. p values for comparisons of regenerating animals with their respective intact controls (or Group 1 vs. Group 2 for isolated pharynges) were all < 1 × 10−3.
Apoptosis induction
Apoptosis was induced by incubating live animals in planarian water with a final concentration of 20 nM staurosporine (BioVision, Cat. No. 1048) or 100 µM cycloheximide (Chemicon, Cat. No. APT800) for 12 or 72 hrs., respectively. Equivalent dilutions of straight DMSO (vehicle) were added to negative control dishes.
Neoblast ablation
Live animals subjected to a 10,000 Rad dose of gamma irradiation using a J.L. Shepherd and Associates model 30, 6000 Ci Cs137 instrument exhibit an increase in TUNEL staining that peaks approximately 24 hrs. later, but returns to near baseline levels from 3 through 7 days post-irradiation (Supplementary Fig. 1F). This presents a window of opportunity in which the TUNEL staining pattern in irradiated animals devoid of neoblasts and their division progeny (Eisenhoffer et al., 2008; Reddien et al., 2005a; Reddien et al., 2005b), is very close to that of unirradiated controls. Accordingly, all experiments with neoblast-ablated animals involved performing amputations at 72 hrs. post-irradiation and fixing fragments prior to the time when a secondary increase in TUNEL staining arises between 7 and 12 days (Supplementary Fig. 1F), in association with the onset of a characteristic ‘no-neoblast’ phenotype (head regression and ventral curling).
RNA-mediated interference
The full-length Smed-bcl2-1 cDNA (GenBank accession number FJ807655) was cloned according to the protocol in (Gurley et al., 2008). dsRNA corresponding to Smed-bcl2-1 was synthesized by in vitro transcription and column purified using the MEGAscript RNAi Kit (Ambion, Cat. No. 1626). A 500bp nonplanarian control sequence (included in the Ambion MEGAscript RNAi Kit) bioinformatically designed to avoid homology to previously identified genes was used to generate negative control dsRNA. Purified dsRNA samples were mixed with homogenized calf liver at a final concentration of approximately 0.25 mg/mL. Animals were then administered a single RNAi feeding and fixed for TUNEL just prior to the appearance of gross anatomical phenotypes (Fig. 6E) at approximately 5 days post-feeding.
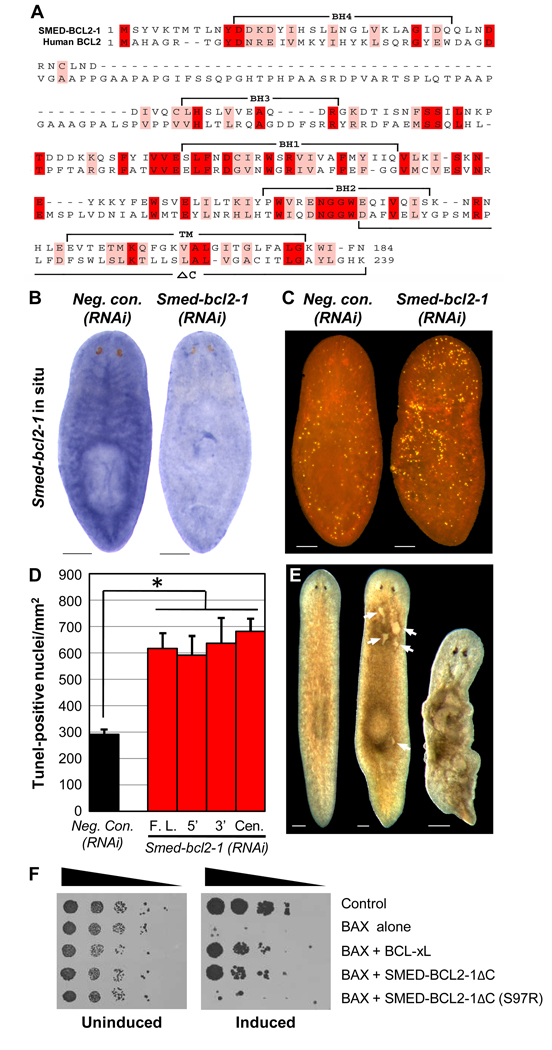
Fig. 6.
A S. mediterranea homolog of BCL2. (A) Alignment of SMED-BCL2-1 predicted protein sequence (GenBank accession no. FJ807655) with human BCL2 protein sequence (GenBank accession no. NM_000633) generated with Expresso (Armougom et al., 2006). Identical residues are shaded red and conserved residues are shaded pink. The 4 BCL2-Homology (BH) domains, transmembrane region (TM), and C-terminal region deleted in SMED-BCL2-1ΔC (ΔC) are indicated. (B) Smed-bcl2-1 expression pattern visualized by in situ hybridization. Representative animals from 2 independent experiments (n ≥ 12 for each condition) are shown. Scale bars = 200 µm. (C) Smed-bcl2-1(RNAi) animals exhibited elevated TUNEL staining. Scale bars = 100 µm. (D) Average TUNEL levels for 3 independent experiments (n ≥ 21 for each condition). RNAi constructs generated dsRNA corresponding to the full-length ORF (F.L.) or nonoverlapping regions at the 5’, 3’, and central parts of the ORF. Error bars = +/− s.e.m. * = p value for two-tailed Student’s t-Test < 1 × 10−3. (E) > 90% of Smed-bcl2-1(RNAi) animals (n > 100) developed tissue lesions on their dorsal surface (middle, arrows) and went on to exhibit atrophy and lysis (right), as compared to 0% of Negative control(RNAi) animals (n > 100). Scale bars = 200 µm. Results in (C, D) are for animals analyzed prior to the appearance of lesions. (F) S. cerevisiae was transformed with the indicated constructs (empty vector used for controls) and plated in serial dilution (left to right in each panel). Induced expression of murine BAX (right) is lethal. This effect was rescued by human BCL-xL or SMED-BCL2-1ΔC, and was abrogated by mutation of SMED-BCL2-1ΔC Ser97 to Arg (S97R).
In situ hybridization
Animals were stained as described (Pearson et al., 2009) using a riboprobe targeting nucleotides 140–555 of the Smed-bcl2-1 ORF. A full-length riboprobe gave identical results (data not shown). The Smed-bcl2-1(RNAi) animal in Figure 6B was fixed and stained 4 days after a single feeding with dsRNA targeting nucleotides 1–141 of the Smed-bcl2-1 ORF. Smed-bcl2-1(RNAi) animals exhibited variable staining intensity at this timepoint, but all showed lower levels of staining than Negative control(RNAi) animals (n ≥ 12 animals per condition, tested in 2 independent experiments). This is likely to reflect modest differences in the extent of gene knockdown at this timepoint, but we cannot exclude differences in staining efficiency from one animal to the next (which may be exaggerated in animals with very low levels of gene expression) as a source of this variability.
Yeast lethality assay
S. cerevisiae was cotransformed with a pGilda plasmid expressing murine BAX under the control of a galactose-inducible promoter (a gift from Shigemi Matsuyama, Case Western Reserve University) and a pADH plasmid (a gift from Mike Forte, Oregon Health and Science University) expressing human BCL-xL or Smed-bcl2-1 under the control of a constitutive alcohol dehydrogenase promoter. Transformed cells were pooled and replica plated on selective media in serial dilution prior to induction of BAX expression. SMED-BCL2-1ΔC, in which the 42 C-terminal amino acids were replaced by the sequence VSLSLFLDH, exhibited consistent protection from BAX-induced lethality (full-length SMED-BCL2-1 yielded inconsistent results).
Results and discussion
A whole-mount TUNEL assay for detection of apoptotic cells in adult planarians
We characterized ultrastructural features of dying cells in S. mediterranea by electron microscopy (EM) and observed chromatin condensation, a hallmark of apoptosis, in regenerating animals (Supplementary Fig. 1A). Chromatin condensation is accompanied by DNA fragmentation in apoptotic cells and this event can be detected by TUNEL (Gavrieli et al., 1992), an assay using terminal transferase to add modified nucleotides to double-strand DNA breaks. TUNEL has previously been used to label dissociated cells and tissue sections in planarians (Bueno et al., 2002; Gonzalez-Estevez et al., 2007; Hwang et al., 2004; Inoue et al., 2007) and we succeeded in developing a protocol that extends this capacity to whole-mounted animals (Materials and methods). One advantage to our assay is that it allows for an automated quantitative analysis of apoptotic cells throughout the entire planarian anatomy (Materials and methods; Supplementary Fig. 1B). Control experiments showed that TUNEL staining depends on both terminal transferase and the rhodamine-conjugated antibody recognizing the modified nucleotide used in our assay; additionally, fixed animals treated with DNase I or hydrochloric acid, reagents known to induce DNA strand breaks, exhibited labeling in all cells (Supplementary Fig. 1C). Live animals exposed to known apoptosis inducers, including irradiation, cycloheximide, and staurosporine exhibited a significant increase in TUNEL staining (Supplementary Figs. 1D–F). These findings, together with results reported below, argue that apoptosis is a prominent form of cell death in S. mediterranea that contributes to ongoing cell turnover during adult tissue homeostasis. Intriguingly, TUNEL levels in intact adult animals exhibited some variability from one experiment to the next. This observation may indicate that the rate of cell turnover is influenced by environmental factors, such as population density, and/or physiological factors, such as time since most recent fission event during asexual reproduction.
Spatial and temporal changes in cell death during regeneration
We used whole-mount TUNEL staining to characterize changes in apoptosis following amputation (Fig. 1; Supplementary Fig. 2A). Adult planarians were initially transected approximately midway between the photoreceptors and the pharynx (prepharyngeal amputation), as depicted in Figure 1A. Regenerating fragments were then fixed at multiple timepoints through 2 weeks post-amputation and stained by TUNEL. From approximately 1 hour (data not shown) through 4 hours post-amputation (Figs. 1A, C), we detected a significant increase in TUNEL-positive cells within an area extending approximately 100 µm from the wound site in both fragments. Small incisions were also sufficient to trigger this response (Supplementary Fig. 2B). Following the initial localized increase in apoptosis, we detected a second, systemic increase in TUNEL staining that peaked at approximately 3 days post-amputation, followed by a gradual return to baseline (Figs. 1A, C; Supplementary Fig. 2A). Fragments from animals amputated approximately midway between the tip of tail and the pharynx (postpharyngeal amputation) also exhibited an initial localized increase in TUNEL staining, but only posterior fragments displayed a strong systemic response at 3 days post-amputation (Figs. 1B, C). In summary, we find that amputation triggers two successive waves of apoptosis – an initial localized response near the wound site that peaks approximately 1 to 4 hours after any type of injury, and a second systemic response that peaks approximately 3 days after amputation and varies in magnitude depending on the type of fragment examined. The latter response was particularly noteworthy in that it was most pronounced in fragments requiring functional integration of new and preexisting organs (e.g., a new pharynx in head and tail fragments and new cephalic ganglia and photoreceptors in decapitated animals).
Cell death increases in association with remodeling of uninjured organs
One possible explanation for the systemic increase in cell death is that apoptosis promotes remodeling of preexisting tissues. This process is essential for restoration of proper scale and proportion, particularly in cases where very small fragments are transformed into whole new organisms. For example, a head fragment generated by prepharyngeal amputation (Fig. 1A) forms a new planarian not only by replacing posterior structures via blastema formation, but also through elongation of the preexisting tissue, which must integrate new trunk structures such as the pharynx (Fig. 1A, right asterisk), and reduce the size of anterior organs such as the cephalic ganglia and photoreceptors (Reddien and Sánchez Alvarado, 2004).
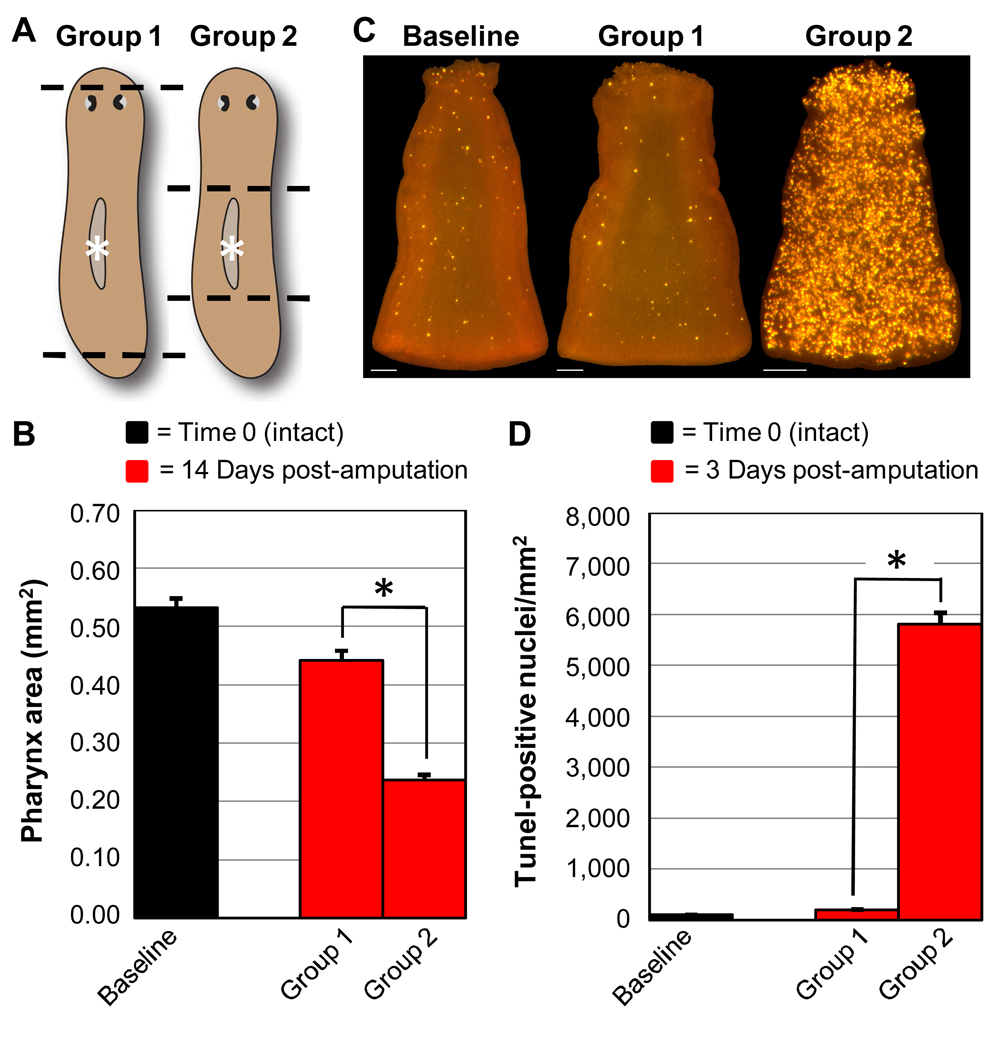
In the above experiments, amputation fragments differed in more than just the degree of tissue remodeling that was required for restoration of scale and proportion. Thus, the differences in TUNEL staining at 3 days post-amputation could be attributed to other variables, such as the absolute size of the regenerating fragments. To address this possibility, we tested whether different head and tail amputation schemes can affect the rate of apoptosis in an organ that is not directly injured by amputation – the pharynx. Specifically, size-matched adult animals were divided into two groups. Only the anterior and posterior tips of animals in Group 1 were resected, whereas entire head and tail structures were removed from animals in Group 2 (Fig. 2A). At 14 days post-amputation, pharynx size had decreased more than 2-fold in Group 2 trunk fragments, as anatomical scale and proportion was restored through tissue remodeling, whereas Group 1 trunk fragments exhibited only a modest reduction in pharynx size (Fig. 2B). We reasoned that if the systemic increase in cell death promotes tissue remodeling, then the level of apoptosis in the pharynx at 3 days post-amputation should accordingly show a greater increase in Group 2 than in Group 1. This is what we observed, using both TUNEL (Figs. 2C, D) and serial block-face scanning EM (Denk and Horstmann, 2004), which allowed us to conduct a semiquantitative analysis of cells exhibiting chromatin condensation (Materials and methods; Supplementary Movie 1, Supplementary Movie 2). Control experiments showed that trunk fragments from both Groups exhibited a similar localized increase in apoptosis near the wound sites at 4 hours post-amputation (Supplementary Figs. 2C, E), arguing that the above results are not due to a difference in the extent of the initial injury response. Additionally, there was no elevation in pharyngeal TUNEL staining for either Group at 4 hours post-amputation, confirming that the pharynx was not directly injured in either amputation scheme (Supplementary Figs. 2D, F). We conclude that the systemic increase in apoptosis at 3 days post-amputation correlates with tissue remodeling, and is not an artifact associated with differences in the size or anatomy of the regenerating fragments examined. Furthermore, this response can be induced in organs that have not sustained direct physical injury.
Fig. 2.
Induction of apoptosis during remodeling of an uninjured organ. (A) Animals were amputated as indicated (dashed lines) at Time 0 and anterior and posterior fragments were discarded. Pharynges (grey ovals marked by asterisks) were then isolated from the regenerating trunk fragments as well as intact control animals. (B) Average size of isolated pharynges at Time 0 (Baseline) and 14 days post-amputation (Group 1 and Group 2) for 3 independent experiments (n ≥ 35 for each condition). (C) Representative TUNEL staining patterns in isolated pharynges at Time 0 (Baseline) and 3 days post-amputation (Group 1 and Group 2). Scale bars = 100 µm. (D) Average TUNEL levels for 3 independent experiments (n ≥ 26 for each condition). In (B, D), error bars = +/− s.e.m. * = p value for two-tailed Student’s t-Test < 1 × 10−15.
Disproportionate induction of cell death by cephalic amputation
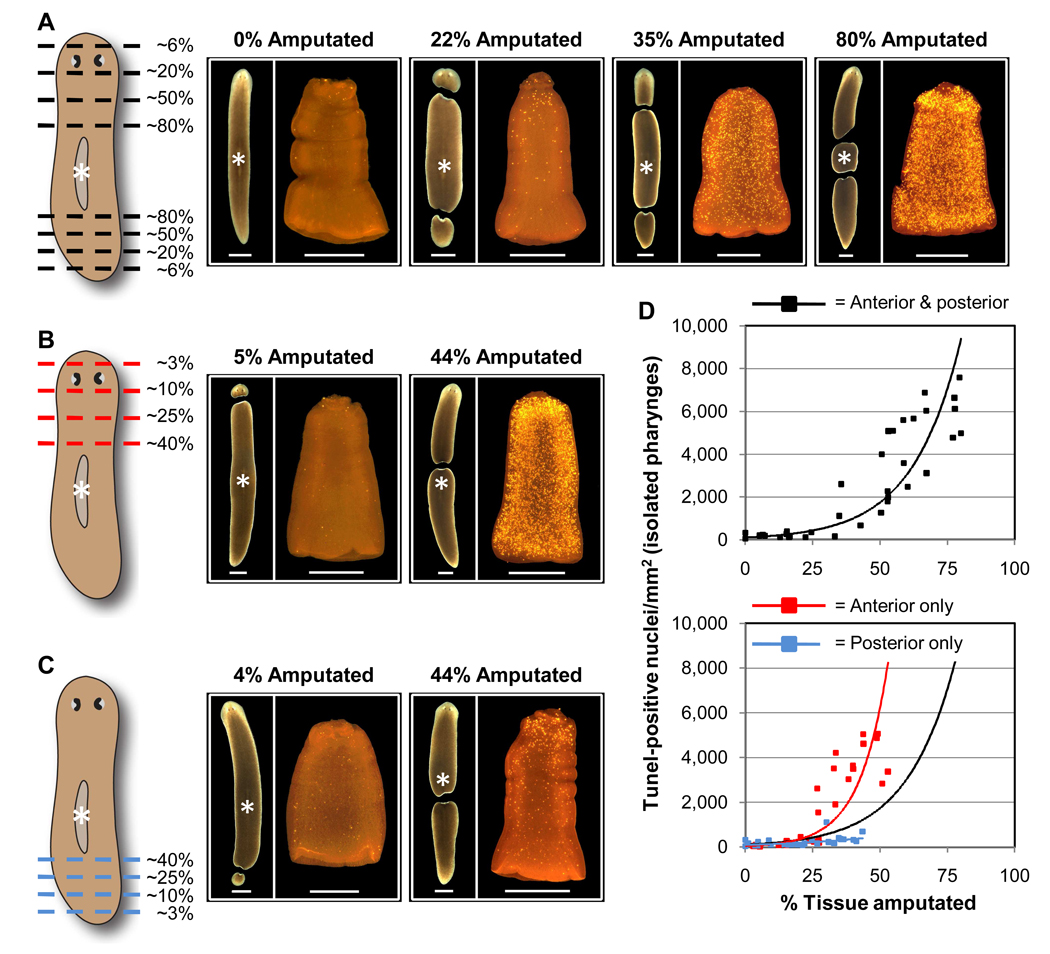
We next performed amputations at multiple different positions along the anterior/posterior axis, removing approximately equal amounts of both head and tail tissue (Fig. 3A). The total percentage of tissue removed was then plotted against the resulting apoptotic response in the pharynx at 3 days post-amputation (Fig. 3D). There was a clear positive correlation between these variables, supporting our hypothesis that apoptosis helps to restore scale and proportion to regenerating animals. However, this relationship was nonlinear, with removal of less than ~25% of total body mass resulting in relatively minor increases in pharyngeal TUNEL staining (Fig. 3D). Furthermore, selective amputation of anterior structures had a much stronger effect than selective amputation of posterior structures (Figs. 3B–D). These results may indicate that stem cell proliferation and blastema formation are largely sufficient to regenerate a functional anatomy following removal of postpharyngeal tissue, whereas fragments that must replace complex organs in the anterior and/or trunk of the animal (e.g., the photoreceptors, brain, and pharynx) rely upon more extensive remodeling of preexisting tissues. Our data point to the existence of regulatory mechanism(s) that integrate information about both the size and type of tissue lost to injury during initiation of regenerative tissue remodeling.
Fig. 3.
Disproportionate induction of apoptosis by cephalic amputation. (A–C) Animals were amputated as indicated (dashed lines) at Time 0, with approximately equal amounts of both anterior and posterior tissue removed (A) or selective amputation of anterior (B) or posterior (C) tissues. Each fragment was photographed (left of each panel; scale bars = 1 mm) and the total amount of tissue removed was determined from measurements of fragment areas. At 3 days post-amputation, pharynges (marked by white asterisks) were isolated from the regenerating trunk fragments and stained by TUNEL (right of each panel; scale bars = 0.5 mm). (D) The total percentage of tissue removed was plotted against TUNEL levels in the pharynx. Scatter plots show results for animals amputated both anterior and posterior to the pharynx (top) vs. only anterior or posterior to the pharynx (bottom). Each data point shows the value for a single animal tested in 1 of 3 independent experiments (n ≥ 26 for each condition). Exponential trendlines (color coded to match the schematic diagrams) are included. The trendline for anterior plus posterior amputation (black) is included in the lower graph for comparison.
Regeneration-induced cell death is stem cell-independent
Because proliferating neoblasts are absent from the pharynx (Baguñà, 1976a; Newmark and Sánchez Alvarado, 2000), the above results may indicate that the systemic cell death response occurs primarily in differentiated cells. However, we sought to test whether some fraction of the neoblast population might undergo apoptosis as well in regenerating head and tail fragments. Given that neoblasts increase their rate of division within a few hours of amputation (Baguñà, 1976b; Saló and Baguñà, 1984), we also sought to determine whether these rapidly dividing cells might trigger the death of their postmitotic neighbors, in analogy to cell competition in the Drosophila imaginal disc (Senoo-Matsuda and Johnston, 2007). To test these possibilities, we compared spatial and temporal changes in apoptosis induced by amputation in irradiated animals devoid of neoblasts and unirradiated controls (Materials and methods). We found that the prior elimination of neoblasts by irradiation had little to no qualitative effect on either the localized increase in apoptosis at 4 hours post-amputation or the systemic increase in apoptosis at 3 days post-amputation (Fig. 4A). Moreover, a quantitative analysis showed that the relative magnitude of different amputation-induced cell death responses followed the same trend in both neoblast-ablated animals and unirradiated controls (Fig. 4B). Neoblast ablation also had no detectable influence on the induction of apoptosis in the pharynx after head and tail amputations (Figs. 4A, B). We conclude that both the localized and systemic regenerative cell death responses occur predominantly, if not exclusively, in differentiated cells, and that initiation of these responses does not require planarian stem cells.
The converse scenario – that apoptosis triggers neoblast division – has yet to be tested. There is compelling evidence that dead or dying cells can generate mitogens in developing tissues (Huh et al., 2004; Ryoo et al., 2004) and it is interesting to note that neoblasts exhibit a biphasic mitotic increase during regeneration, with successive peaks at approximately 4–12 hours and 2–4 days post-amputation (Baguñà, 1976b; Saló and Baguñà, 1984). Thus, the roughly coincident increases in apoptosis that we observed (Fig. 1; Supplementary Fig. 2A) represent a candidate source for the signal that triggers neoblast division and blastema formation. A direct test of this hypothesis will require development of strategies for the inhibition of cell death in vivo.
Cell death increases during starvation-induced tissue remodeling
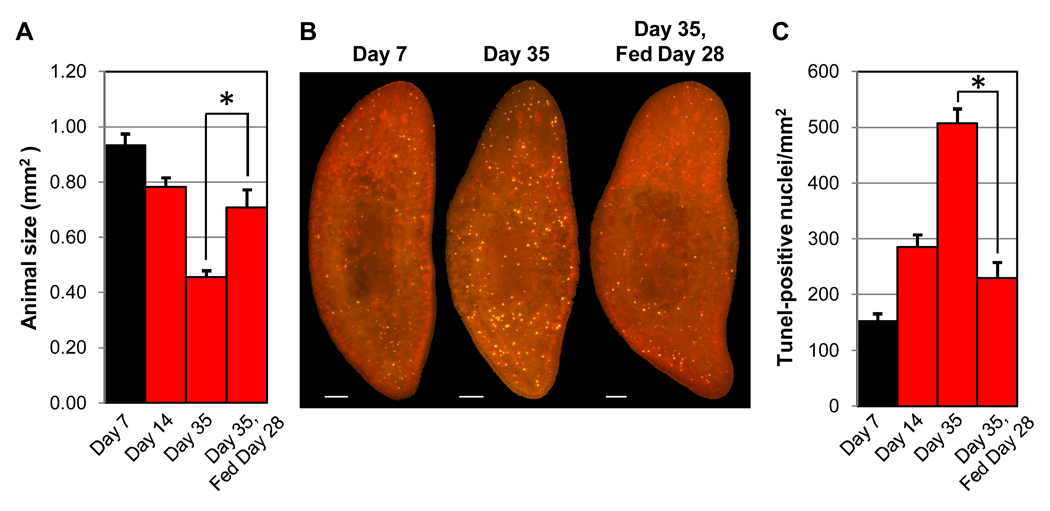
Prolonged starvation also triggers tissue remodeling in planarians. ‘Degrowth’ is rapidly reversed upon feeding and results primarily from a reduction in cell number, rather than a reduction in cell size (Baguñà and Romero, 1981; Oviedo et al., 2003; Romero and Baguñà, 1991). Remarkably, anatomical proportion is preserved as the overall size of starving animals decreases (Oviedo et al., 2003). In principle, this process could occur through a decrease in the rate of neoblast division, an increase in the rate of cell death, or both. There are conflicting reports about the former variable (Baguñà, 1976a; Bowen et al., 1976), though it is clear that at least some neoblast division persists during starvation. Autophagy has been proposed as a means of starvation-induced tissue remodeling in a related planarian species (Gonzalez-Estevez et al., 2007), but it is not yet clear whether this is a cell survival or cell death mechanism. In order to evaluate a possible role for apoptosis in degrowth, we measured changes in the number of dying cells over time in starved animals by TUNEL. Between 1 and 5 weeks post-feeding, animals displayed a significant increase in TUNEL-positive cells, in correlation with a significant decrease in overall size (Fiig. 5). These changes were rapidly reversed when animals were fed again (Fig. 5). Thus, planarians are able to regulate the rate of apoptosis to facilitate morphological changes to fully developed tissues in response to diverse environmental cues (e.g., injury or nutrient availability). How the rate of cell death is altered by these cues is presently unknown, but the ability to quantify apoptotic cells in whole-mounted animals by TUNEL should facilitate future attempts to address this issue.
Fig. 5.
Apoptosis increases during degrowth. Size-matched adult animals were fed at Time 0, prior to fixation at 7, 14, or 35 days post-feeding. After 28 days of starvation, a subset of the remaining animals was administered a single feeding and fixed on Day 35. (A) Average animal size for 3 independent experiments (n ≥ 31 for each timepoint). (B) Representative TUNEL results. Scale bars = 100 µm. (C) Average TUNEL levels. In (A, C), error bars = +/− s.e.m. * = p value for two-tailed Student’s t-Test < 1 × 10−3.
A S. mediterranea homolog of BCL2 is required for cell survival in adult tissues
To begin addressing the molecular mechanisms of planarian apoptosis, we identified S. mediterranea homologs of known cell death genes in other organisms and used RNA-mediated interference (RNAi) to assess their loss-of-function phenotypes. This approach identified a planarian homolog of the antiapoptotic gene BCL2 required for cell survival in adult animals (Fig. 6). The predicted protein for this gene, which we named Smed-bcl2-1, shows high overall sequence conservation with human BCL2 and includes 4 apparent BCL2-Homology (BH) domains (Fig. 6A). Smed-bcl2-1 is expressed throughout the planarian body plan, with enriched expression in the gastrovascular system (Fig. 6B). Animals fed Smed-bcl2-1 dsRNA (Materials and methods) displayed two notable phenotypes. First, they exhibited a significant increase in TUNEL-positive cells, relative to animals fed negative control dsRNA (Figs. 6C, D). Second, they developed tissue lesions over their dorsal surface, followed by overall atrophy and lysis, or disintegration of the entire animal (Fig. 6E). Because the increase in TUNEL staining preceded the appearance of any gross morphological phenotypes, lesions and lysis are likely caused, at least in part, by increased apoptosis, rather than the other way around.
To further evaluate a potential antiapoptotic function for Smed-bcl2-1, we used a heterologous system for testing interactions between cell death genes. Enforced expression of murine BAX, a proapoptotic BCL2 family member, is sufficient to kill S. cerevisiae and this lethality can be blocked by antiapoptotic proteins such as human BCL-xL (Sato et al., 1994) (Fig. 6F). We tested full-length SMED-BCL2-1 in this assay (Materials and methods), as well as a truncated version lacking 42 C-terminal amino acids (SMED-BCL2-1ΔC; Fig. 6A). The latter construct was included because the C-termini of some BCL2 family proteins can disrupt heterotypic interactions with other family members (Denisov et al., 2003; Suzuki et al., 2000). SMED-BCL2-1ΔC conferred significant protection from BAX-induced lethality and this effect was abolished by mutation of a residue corresponding to one in human BCL-xL implicated in BAX binding (Yin et al., 1994) (S97R; Fig. 6F). Full-length SMED-BCL2-1 failed to provide consistent protection in this assay (data not shown). Taken together, the SMED-BCL2-1 predicted protein sequence, RNAi phenotypes, and protective effects against BAX-induced lethality suggest that this is an important regulator of cell death in planarians that protects against inappropriate apoptosis in adult tissues. In other animals, BCL2 family members ultimately control the activity of caspases, the core effectors of apoptosis (Danial and Korsmeyer, 2004). Caspase-related genes have previously been identified in the planarian Dugesia japonica (Hwang et al., 2004) and we are presently cloning and functionally characterizing S. mediterranea caspases, as well as other candidate cell death genes, to characterize molecular mechanisms of apoptosis downstream of Smed-bcl2-1.
A model for cell death and regenerative tissue remodeling
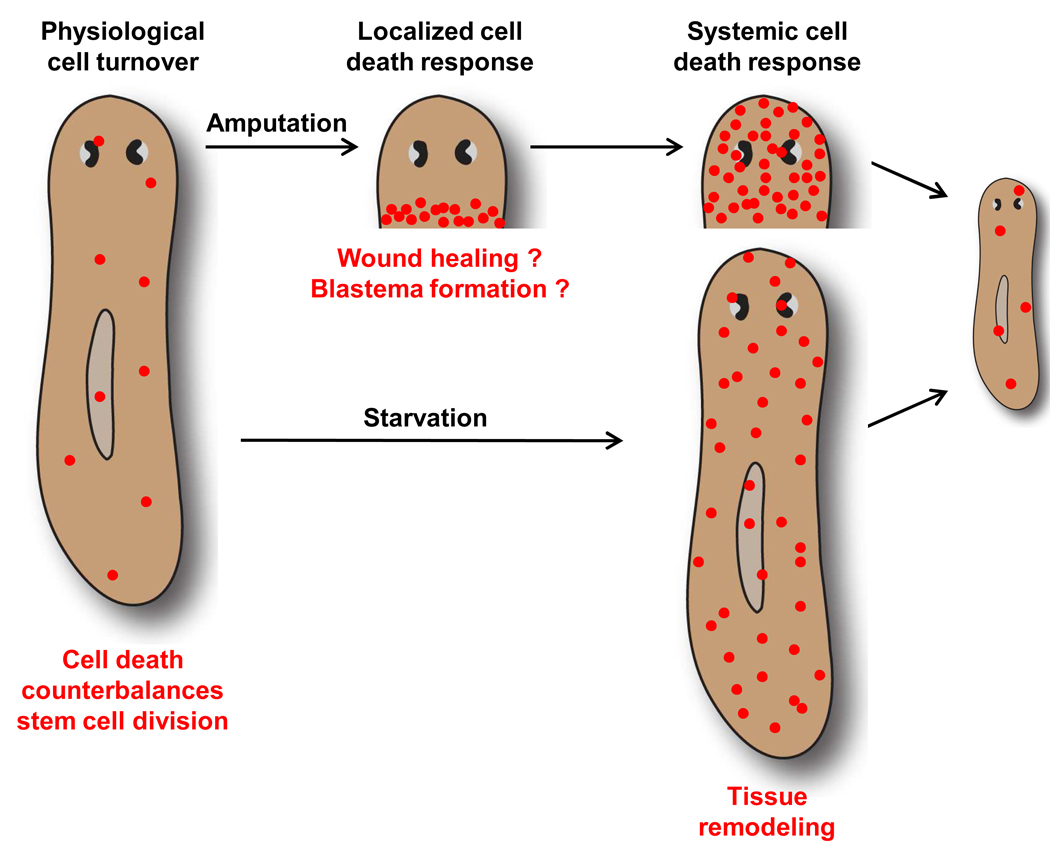
In conclusion, we propose that apoptosis complements the stem cell-mediated addition of new tissue during planarian regeneration by remodeling preexisting structures (Fig. 7). A determination of whether cell death is required for this process will necessitate future studies involving apoptosis inhibiton in vivo. One likely outcome of an increased rate of cell death is a reduction in the size of organs that are too large for the ultimate dimensions of the regenerated animal, such as a pharynx in a small trunk fragment (Fig. 2, Group 2) or the photoreceptors in a small head fragment (Fig. 7). This reproportioning of existing structures is necessary because regenerating animals do not, or in fragments lacking a pharynx, cannot ingest enough food to replace the entirety of lost tissues de novo via stem cell proliferation. A similar, but more gradual process occurs as overall animal size decreases during degrowth. The selective deletion of a portion of cells scattered throughout an organ like the pharynx (Fig. 2C) would not, in and of itself, change the overall dimensions of that organ; however, this could be accomplished by a subsequent reorganization of the surviving cells through transdifferentiation and/or cell migration. Alternatively, an organ’s size could be reduced by the eventual elimination of every differentiated cell with selective replacement of only a fraction of those cells by neoblast division progeny.
Fig. 7.
Model of apoptosis functions in planarian tissue remodeling. Amputation leads to 2 distinct waves of apoptosis (red circles) in regenerating fragments. The initial localized increase in cell death at 1 to 4 hours post-amputation might promote wound healing and/or blastema formation. We propose that the later systemic increase in cell death represents a key component of the tissue remodeling process that restores scale and proportion and integrates new and preexisting tissues. In this case, remodeling results in a reduction in the size of the photoreceptors, lengthening and narrowing of the entire animal, and integration of the newly formed pharynx. The systemic cell death response is also induced in intact animals by prolonged starvation, contributing to a reduction in the animal’s overall size.
Regenerative cell death responses have also been observed in a variety of other organisms. In the freshwater polyp Hydra, for example, midgastric amputation triggers a localized increase in apoptosis reminiscent of what we have observed in planarians; the apoptotic cells then secrete Wnt3, which induces cell proliferation and the formation of a blastema-like structure (Chera et al., 2009). In Xenopus laevis, formation of the regeneration blastema following tail amputation requires a similar cell death response (Tseng et al., 2007). Apoptosis has further been implicated in cycles of zooid resorbtion and regeneration in the colonial ascidian Botryllus schlosseri (Ballarin et al., 2008), the replacement of hepatic tissue with pharyngeal tissue during anterior regeneration of the hemichordate Ptychodera flava (Rychel and Swalla, 2008), and the conversion of cartilage to bone during forelimb regeneration in the adult newt (Vlaskalin et al., 2004). Finally, apoptosis plays an important role in localized tissue remodeling during cutaneous wound healing in humans and dysregulated apoptosis is likely one important cause of excessive scarring (Huang et al., 2003). Thus, apoptosis, and perhaps also nonapoptotic forms of cell death (Bowen and Ryder, 1974; Gonzalez-Estevez et al., 2007), appears to play a widespread role in promoting morphological changes during regeneration. Our results emphasize the importance of investigating not only how new tissue is created during such phenomena, but also how changes to preexisting structures restore a functional and properly proportioned anatomy. Together with our extensive knowledge of neoblast biology (Baguñà, 1976a; Baguñà, 1976b; Baguñà and Romero, 1981; Dubois, 1949; Eisenhoffer et al., 2008; Newmark and Sánchez Alvarado, 2000; Reddien et al., 2005a; Reddien et al., 2005b; Romero and Baguñà, 1991; Saló and Baguñà, 1984), the analysis reported here positions S. mediterranea as an ideal model system for future studies investigating how stem cell division and apoptosis are coordinated to drive adult tissue renewal and repair.
Supplementary Material
Supplementary Movie 1. Serial block-face scanning EM of a Group 1 pharynx. Serial block-face scanning electron microscopy was used to conduct a semi-quantitative analysis of cells with chromatin condensation in a Group 1 pharynx (see Fig. 2) isolated at 3 days post-amputation.
Supplementary Movie 2. Serial block-face scanning EM of a Group 2 pharynx. Serial block-face scanning electron microscopy was used to conduct a semi-quantitative analysis of cells with chromatin condensation in a Group 2 pharynx (see Fig. 2) isolated at 3 days post-amputation. Some of the cells exhibiting chromatin condensation are marked by white arrows.
Supplementary Fig. 1. TUNEL labels apoptotic planarian cells. (A) Transmission electron micrograph of a dying planarian cell exhibiting chromatin condensation (white arrow), a hallmark of apoptosis (compare to nuclei in healthy cells marked by black arrows). Section is from a regenerating head fragment at 3 days post-amputation. Scale bar = 2 µm. (B) Evaluation of automated TUNEL quantification results. The total number of TUNEL-positive nuclei (TPN) in each of 40 images were counted manually and plotted against the corresponding automated TPN counts (Materials and methods). At least 1 image per experiment was analyzed in this manner. (C) TUNEL assay controls. Animals were stained by TUNEL as described, with elimination of terminal transferase (−TdT) or anti-digoxigenin antibody (−Antibody), or following pretreatment with 2N HCl (+HCl) or DNase I (+DNase I). Images show representative animals from 2 independent experiments (n ≥ 14 for each condition). Insets = magnified views (5×) revealing staining in all nuclei. (D, E) Live animals were exposed to gamma irradiation, staurosporine, or cycloheximide prior to visualization of apoptosis by TUNEL (D). Irradiated = 24 hrs. after a 10,000 Rad dose. In (C, D), scale bars = 100 µm. Average TUNEL levels for 3 independent experiments (n ≥ 23 for each condition) are shown (E). * = p value < 1 × 10−5 for two-tailed Student’s t-Test comparing experimental conditions with matched controls. (F) Extended timecourses showing TUNEL staining following gamma irradiation at Time 0. Each data point shows the average TUNEL level for 2 independent experiments (n ≥ 15 per timepoint). The lack of a significant increase at 10 minutes indicates that the later increases are a biological response to irradiation, rather than a direct result of irradiation-induced DNA strand breaks. In (E, F), error bars = +/− s.e.m.
Supplementary Fig. 2. Amputation-induced apoptosis responses. (A) Animals were amputated as indicated in Figure 1 at Time 0, prior to visualization of apoptosis by TUNEL. Each data point shows the average TUNEL level for 2 independent experiments (n ≥ 11 per timepoint). 4-hour TUNEL levels in this graph are normalized for the area of the entire fragment. (B) Localized apoptosis induced by incision wounds (arrowheads) made 4 hours prior to fixation. A representative animal from 2 independent experiments with a total of 10 animals is shown. (C–F) Pharynx TUNEL controls. Animals were amputated as indicated in Figure 2 at Time 0. (C) TUNEL staining in representative Group 1 and Group 2 trunk fragments at 4 hours post-amputation. (D) TUNEL staining in representative pharynges isolated from Group 1 and Group 2 trunk fragments at 4 hours post-amputation. In (B–D), scale bars = 100 µm. (E, F) Average TUNEL levels at 4 hours post-amputation for trunk fragments (E) and isolated pharynges (F) from 3 independent experiments (n ≥ 25 for each condition). Values for the trunk fragments are restricted to the areas within 100 µm of the amputation sites (boxed areas in schematic diagrams). p values for two-tailed Student’s t-Tests comparing conditions were all > 0.05. In (A, E, F), error bars = +/− s.e.m.
Acknowledgements
We thank Mark Yandell and Eric Ross for assistance with automated image analysis and Carrie Adler, Kyle Gurley, Bret Pearson, and Jochen Rink for helpful comments on this manuscript. J.P. was supported by a fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. A.S.A. is an investigator of the Howard Hughes Medical Institute. D.G. is supported by NIH grants AI40646 and AI47891 and the American Lebanese Syrian Associated Charities.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armougom F, Moretti S, Poirot O, Audic S, Dumas P, Schaeli B, Keduas V, Notredame C. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 2006;34:W604–W608. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguñà J. Mitosis in the intact and regenerating planarian Dugesia mediterranea n.sp. I. Mitotic studies during growth, feeding and starvation. J. Exp. Zool. 1976a;195:53–64. [Google Scholar]
- Baguñà J. Mitosis in the intact and regenerating planarian Dugesia mediterranea n.sp. II. Mitotic studies during regeneration, and a possible mechanism of blastema formation. J. Exp. Zool. 1976b;195:65–80. [Google Scholar]
- Baguñà J, Romero R. Quantitative analysis of cell types during growth, degrowth and regeneration in the planarians Dugesia mediterranea and Dugesia tigrina. Hydrobiologia. 1981;84:181–194. [Google Scholar]
- Ballarin L, Burighel P, Cima F. A tale of death and life: natural apoptosis in the colonial ascidian Botryllus schlosseri (Urochordata, Ascidiacea) Curr. Pharm. Des. 2008;14:138–147. doi: 10.2174/138161208783378798. [DOI] [PubMed] [Google Scholar]
- Bowen E, Ryder T, Dark C. The effects of starvation on the planarian worm Polycelis tenuis Iijima. Cell Tissue Res. 1976;169:193–209. doi: 10.1007/BF00214208. [DOI] [PubMed] [Google Scholar]
- Bowen ID, Ryder TA. Cell autolysis and deletion in the planarian Polycelis tenuis Iijima. Cell Tissue Res. 1974;154:265–274. doi: 10.1007/BF00223168. [DOI] [PubMed] [Google Scholar]
- Bueno D, Fernandez-Rodriguez J, Cardona A, Hernandez-Hernandez V, Romero R. A novel invertebrate trophic factor related to invertebrate neurotrophins is involved in planarian body regional survival and asexual reproduction. Dev. Biol. 2002;252:188–201. doi: 10.1006/dbio.2002.0851. [DOI] [PubMed] [Google Scholar]
- Cebria F, Newmark PA. Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development. 2005;132:3691–3703. doi: 10.1242/dev.01941. [DOI] [PubMed] [Google Scholar]
- Chera S, Ghila L, Dobretz K, Wenger Y, Bauer C, Buzgariu W, Martinou JC, Galliot B. Apoptotic cells provide an unexpected source of Wnt3 signaling to drive hydra head regeneration. Dev. Cell. 2009;17:279–289. doi: 10.1016/j.devcel.2009.07.014. [DOI] [PubMed] [Google Scholar]
- Cotter TG. Apoptosis and cancer: the genesis of a research field. Nat. Rev. Cancer. 2009;9:501–507. doi: 10.1038/nrc2663. [DOI] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Denisov AY, Madiraju MS, Chen G, Khadir A, Beauparlant P, Attardo G, Shore GC, Gehring K. Solution structure of human BCL-w: modulation of ligand binding by the C-terminal helix. J. Biol. Chem. 2003;278:21124–21128. doi: 10.1074/jbc.M301798200. [DOI] [PubMed] [Google Scholar]
- Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- Dubois F. Contribution á l ’ètude de la migration des cellules de règènèration chez les Planaires dulcicoles. Bull. Biol. Fr. Belg. 1949;83:213–283. [Google Scholar]
- Eisenhoffer GT, Kang H, Alvarado AS. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell. 2008;3:327–339. doi: 10.1016/j.stem.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Estevez C, Felix DA, Aboobaker AA, Salo E. Gtdap-1 promotes autophagy and is required for planarian remodeling during regeneration and starvation. Proc. Natl. Acad. Sci. U.S.A. 2007;104:13373–13378. doi: 10.1073/pnas.0703588104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley KA, Rink JC, Sánchez Alvarado A. Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 2008;319:323–327. doi: 10.1126/science.1150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley KA, Sánchez Alvarado A. Stem cells in animal models of regeneration. In: StemBook, editor. The Stem Cell Research Community. StemBook; 2008. doi/10.3824/stembook.1.32.1. [PubMed] [Google Scholar]
- Hammarlund M, Palfreyman MT, Watanabe S, Olsen S, Jorgensen EM. Open syntaxin docks synaptic vesicles. PLoS Biol. 2007;5:e198. doi: 10.1371/journal.pbio.0050198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang NF, Zac-Varghese S, Luke S. Apoptosis in skin wound healing. Wounds. 2003 [Google Scholar]
- Huh JR, Guo M, Hay BA. Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr. Biol. 2004;14:1262–1266. doi: 10.1016/j.cub.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Hwang JS, Kobayashi C, Agata K, Ikeo K, Gojobori T. Detection of apoptosis during planarian regeneration by the expression of apoptosis-related genes and TUNEL assay. Gene. 2004;333:15–25. doi: 10.1016/j.gene.2004.02.034. [DOI] [PubMed] [Google Scholar]
- Inoue T, Hayashi T, Takechi K, Agata K. Clathrin-mediated endocytic signals are required for the regeneration of, as well as homeostasis in, the planarian CNS. Development. 2007;134:1679–1689. doi: 10.1242/dev.02835. [DOI] [PubMed] [Google Scholar]
- Morgan TH. Regeneration. New York: The Macmillan Company; 1901. [Google Scholar]
- Newmark P, Sánchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev. Biol. 2000;220:142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- Oviedo NJ, Newmark PA, Sánchez Alvarado A. Allometric scaling and proportion regulation in the freshwater planarian Schmidtea mediterranea. Dev. Dyn. 2003;226:326–333. doi: 10.1002/dvdy.10228. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sánchez Alvarado A. Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev. Dyn. 2009;238:443–450. doi: 10.1002/dvdy.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J, Sánchez Alvarado A. Cell turnover and adult tissue homeostasis: from humans to planarians. Annu. Rev. Genet. 2007;41:83–105. doi: 10.1146/annurev.genet.41.110306.130244. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sánchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev. Cell. 2005a;8:635–649. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005b;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Ann. Rev. Cell Dev. Bio. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Romero R, Baguñà J. Quantitative cellular analysis of growth and reproduction in freshwater planarians(Turbellaria; Tricladida). I. A cellular description of the intact organism. Invert. Reprod. Dev. 1991;19:157–165. [Google Scholar]
- Rychel AL, Swalla BJ. Anterior regeneration in the hemichordate Ptychodera flava. Dev. Dyn. 2008;237:3222–3232. doi: 10.1002/dvdy.21747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo HD, Gorenc T, Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev. Cell. 2004;7:491–501. doi: 10.1016/j.devcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Saló E, Baguñà J. Regeneration and pattern formation in planarians. I. The pattern of mitosis in anterior and posterior regeneration in Dugesia (G) tigrina, and a new proposal for blastema formation. J. Embryol. Exp. Morphol. 1984;83:63–80. [PubMed] [Google Scholar]
- Sato T, Hanada M, Bodrug S, Irie S, Iwama N, Boise LH, Thompson CB, Golemis E, Fong L, Wang H, Reed JC. Interactions among members of the Bcl-2 protein family analyzed with a yeast two-hybrid system. Proc. Natl. Acad. Sci. U.S.A. 1994;91:9238–9242. doi: 10.1073/pnas.91.20.9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo-Matsuda N, Johnston LA. Soluble factors mediate competitive and cooperative interactions between cells expressing different levels of Drosophila Myc. Proc. Natl. Acad. Sci. U.S.A. 2007;104:18543–18548. doi: 10.1073/pnas.0709021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Youle RJ, Tjandra N. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell. 2000;103:645–654. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- Tourigny MR, Mazel S, Burtrum DB, Petrie HT. T cell receptor (TCR)-beta gene recombination: dissociation from cell cycle regulation and developmental progression during T cell ontogeny. J. Exp. Med. 1997;185:1549–1556. doi: 10.1084/jem.185.9.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng AS, Adams DS, Qiu D, Koustubhan P, Levin M. Apoptosis is required during early stages of tail regeneration in Xenopus laevis. Dev. Biol. 2007;301:62–69. doi: 10.1016/j.ydbio.2006.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlaskalin T, Wong CJ, Tsilfidis C. Growth and apoptosis during larval forelimb development and adult forelimb regeneration in the newt (Notophthalmus viridescens) Dev. Genes Evol. 2004;214:423–431. doi: 10.1007/s00427-004-0417-1. [DOI] [PubMed] [Google Scholar]
- Yin XM, Oltvai ZN, Korsmeyer SJ. BH1 and BH2 domains of Bcl-2 are required for inhibition of apoptosis and heterodimerization with Bax. Nature. 1994;369:321–323. doi: 10.1038/369321a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1. Serial block-face scanning EM of a Group 1 pharynx. Serial block-face scanning electron microscopy was used to conduct a semi-quantitative analysis of cells with chromatin condensation in a Group 1 pharynx (see Fig. 2) isolated at 3 days post-amputation.
Supplementary Movie 2. Serial block-face scanning EM of a Group 2 pharynx. Serial block-face scanning electron microscopy was used to conduct a semi-quantitative analysis of cells with chromatin condensation in a Group 2 pharynx (see Fig. 2) isolated at 3 days post-amputation. Some of the cells exhibiting chromatin condensation are marked by white arrows.
Supplementary Fig. 1. TUNEL labels apoptotic planarian cells. (A) Transmission electron micrograph of a dying planarian cell exhibiting chromatin condensation (white arrow), a hallmark of apoptosis (compare to nuclei in healthy cells marked by black arrows). Section is from a regenerating head fragment at 3 days post-amputation. Scale bar = 2 µm. (B) Evaluation of automated TUNEL quantification results. The total number of TUNEL-positive nuclei (TPN) in each of 40 images were counted manually and plotted against the corresponding automated TPN counts (Materials and methods). At least 1 image per experiment was analyzed in this manner. (C) TUNEL assay controls. Animals were stained by TUNEL as described, with elimination of terminal transferase (−TdT) or anti-digoxigenin antibody (−Antibody), or following pretreatment with 2N HCl (+HCl) or DNase I (+DNase I). Images show representative animals from 2 independent experiments (n ≥ 14 for each condition). Insets = magnified views (5×) revealing staining in all nuclei. (D, E) Live animals were exposed to gamma irradiation, staurosporine, or cycloheximide prior to visualization of apoptosis by TUNEL (D). Irradiated = 24 hrs. after a 10,000 Rad dose. In (C, D), scale bars = 100 µm. Average TUNEL levels for 3 independent experiments (n ≥ 23 for each condition) are shown (E). * = p value < 1 × 10−5 for two-tailed Student’s t-Test comparing experimental conditions with matched controls. (F) Extended timecourses showing TUNEL staining following gamma irradiation at Time 0. Each data point shows the average TUNEL level for 2 independent experiments (n ≥ 15 per timepoint). The lack of a significant increase at 10 minutes indicates that the later increases are a biological response to irradiation, rather than a direct result of irradiation-induced DNA strand breaks. In (E, F), error bars = +/− s.e.m.
Supplementary Fig. 2. Amputation-induced apoptosis responses. (A) Animals were amputated as indicated in Figure 1 at Time 0, prior to visualization of apoptosis by TUNEL. Each data point shows the average TUNEL level for 2 independent experiments (n ≥ 11 per timepoint). 4-hour TUNEL levels in this graph are normalized for the area of the entire fragment. (B) Localized apoptosis induced by incision wounds (arrowheads) made 4 hours prior to fixation. A representative animal from 2 independent experiments with a total of 10 animals is shown. (C–F) Pharynx TUNEL controls. Animals were amputated as indicated in Figure 2 at Time 0. (C) TUNEL staining in representative Group 1 and Group 2 trunk fragments at 4 hours post-amputation. (D) TUNEL staining in representative pharynges isolated from Group 1 and Group 2 trunk fragments at 4 hours post-amputation. In (B–D), scale bars = 100 µm. (E, F) Average TUNEL levels at 4 hours post-amputation for trunk fragments (E) and isolated pharynges (F) from 3 independent experiments (n ≥ 25 for each condition). Values for the trunk fragments are restricted to the areas within 100 µm of the amputation sites (boxed areas in schematic diagrams). p values for two-tailed Student’s t-Tests comparing conditions were all > 0.05. In (A, E, F), error bars = +/− s.e.m.