Abstract
As a means to study surface proteins involved in the yeast to hypha transition, human monoclonal antibody fragments (single chain variable fragments, scFv) have been generated that bind to antigens expressed on the surface of Candida albicans yeast and/or hyphae. A cDNA expression library was constructed from hyphae, and screened for immunoreactivity with scFv5 as a means to identify its cognate antigen. A reactive clone contained the 3′ end of the C. albicans gene, orf 19.7136, designated SPT6 based on homology to S. cerevisiae, where its product functions as a transcription elongation factor. A mutant containing a homozygous deletion of SPT6 was isolated, demonstrating that unlike S. cerevisiae, deletion of this gene in C. albicans is not lethal. Growth of this strain was severely impaired, however, as was its capacity to undergo filamentous growth. Reactivity with scFv5 was not detected in the mutant strain, although its impaired growth complicates the interpretation of this finding. To assess C. albicans SPT6 function, expression of the C. albicans gene was induced in a defined S. cerevisiae spt6 mutant. Partial complementation was seen, confirming that the C. albicans and S. cerevisiae genes are functionally related in these species.
Index descriptors: Candida albicans, Saccharomyces cerevisiae, single-chain variable fragment, hyphae
Introduction
Candida albicans is a well recognized human pathogen that causes both mucocutaneous and systemic infections primarily in immunocompromised hosts (Calderone, 2002). Systemic infections caused by this organism have increased in frequency and carry a high mortality despite antifungal therapy (Benjamin, et al., 2006; Viudes, et al., 2002). The capacity of this organism to shift its morphology from yeast to hyphal form is important for its virulence and has been the subject of intensive study (Calderone, 2002; San-Blas, et al., 2000). The shift to hyphal growth is marked by significant changes in gene expression and expression of novel surface antigens, and some of these have been implicated in interaction with the host and virulence (Kumamoto and Vinces, 2005).
Because of its importance in disease states, several approaches have been used to probe specifics of the yeast to hypha transition. Traditional genetic approaches have been hampered by the diploid nature of C. albicans, and unlike the related yeast, Saccharomyces cerevisiae, the lack of a well defined sexual cycle. S. cerevisiae can be induced to grow in a pseudohyphal form, and C. albicans homologues to the genes involved in pseudohyphal growth have been studied (Leberer, et al., 1996; Liu, et al., 1994). Screening C. albicans gene libraries for their capacity to elicit pseudohyphal growth in S. cerevisiae has also met with some success (Feng, et al., 1999; Kadosh and Johnson, 2001; Stoldt, et al., 1997). Another fruitful approach involved large-scale transposon mutagenesis of C. albicans with selection of clones that had altered hyphal phenotypes (Uhl, et al., 2003). The yeast to hypha transition is also amenable to study via genomic microarray. Such an approach has identified 61 genes induced and 25 genes repressed in response to exposure to serum at 37°C (Kadosh and Johnson, 2005).
As the outermost structure, the cell wall is in closest contact with host defense mechanisms during infection and modulates the host-pathogen interaction. As such, defining immunogenic cell wall components and the capacity of antibody specific to these components to be protective has received much study. Screening of sera from both human and animals infected with C. albicans for specific antibodies has defined gene products from the cell wall as well as cytoplasmic and secreted proteins that elicit an antibody response. Antibodies against some of these proteins are well documented to have protective properties (Lopez-Ribot, et al., 2004). More recently, sophisticated proteomic and bioinformatic approaches have also been applied to identify gene products of the organism that elicit potentially protective antibody responses from the host. Studies comparing substantive collections of sera from patients with systemic candidiasis compared to controls have demonstrated unique signatures between the commensal and disease state that have both diagnostic and therapeutic implications (Pitarch, et al., 2006). Components of an effective cell wall extract vaccine that were associated with protective responses have also been identified using a proteomic approach (Thomas, et al., 2006). Advances in technology have also allowed systematic genomic analyses to be applied to determine gene products of the organism that are preferentially expressed under in vivo conditions. Potential virulence factors have been identified by methods including differential display, signature-tagged mutagenesis, transcriptional profiling by microarray, and antibody based screening strategies (Nguyen, et al., 2004). This approach has identified novel virulence factors and allows additional insights into the organism's pathogenesis and the impact of varied host environments (Cheng, et al., 2005).
As a means to obtain additional reagents to explore the antigenic milieu of the hyphal surface and potentially identify novel proteins that may have a role in the organism's virulence, we used phage display technology to isolate human antibody fragments (single-chain variable fragments, scFv) that are reactive with both the yeast and/or hyphal form of C. albicans (Bliss, et al., 2003; Haidaris, et al., 2001). To identify clones specific for C. albicans surface antigens expressed under native conditions, the human scFv phage display library was panned against live, whole cells growing in either the yeast or germ tube morphology. These scFv have been shown to facilitate interaction between the fungus and host immune cells (Wellington, et al., 2003). Additionally, one of these scFv (scFv3) recognizes the well-characterized fungal adhesin, Als3p, on the hyphal surface and interferes with Als3p mediated adhesion to human epithelial and endothelial cells in vitro (Laforce-Nesbitt, et al., 2008). The cognate antigen of another hyphal-specific scFv (scFv5) has remained elusive. To identify candidate gene products that may be the target of scFv5, a cDNA expression library from C. albicans hyphae was constructed and screened. We report the identification of the as yet poorly characterized C. albicans gene product, Spt6p, as a potential target for scFv5 and the characterization of a strain harboring a homozygous SPT6 deletion.
Materials and Methods
Strains and media
Strains used in this study are listed in Table 1. Media used include YPD (1% yeast extract, 2% peptone, 2% dextrose), Agar + serum (1% agar, 4% fetal calf serum), and spider medium (1% nutrient broth, 1% mannitol, 0.4% potassium phosphate, 1.3% agar, pH 7.2). Yeast dropout media for strains with auxotrophic markers were prepared by standard methods (Sambrook and Russell, 2001).
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| C. albicans | ||
| SC5314 | Wild-type clinical isolate | (Gillum, et al., 1984) |
| SN87 | leu2Δ/leu2Δ his1Δ/his1Δ URA3/ura3Δ∷imm434 IRO1/iro1Δ∷imm434 | (Noble and Johnson, 2005) |
| JMB220 | spt6Δ∷C.d HIS1/SPT6 leu2Δ/leu2Δhis1Δ/his1Δ URA3/ura3Δ∷imm434 IRO1/iro1Δ∷imm434 | This study |
| JMB31 | spt6Δ∷C.d HIS1/spt6Δ∷C.m.LEU2 leu2Δ/leu2Δ his1Δ/his1Δ URA3/ura3Δ∷imm434 IRO1/iro1Δ∷imm434 | This study |
| JMB222 | spt6Δ∷C.d HIS1/spt6*-SPT6 leu2Δ/leu2Δ his1Δ/his1Δ URA3/ura3Δ∷imm434 IRO1/iro1Δ∷imm434 | This study |
| S. cerevisiae | ||
| FY137 | MATa ura3, his4-912δ, lys2-1288spt6-140 | (Hartzog, et al., 1998) |
spt6* represents a truncated gene, containing the first 615 bp of the 4206 bp ORF
Construction and immunoscreening of C. albicans hyphae cDNA expression library
Starter cultures of C. albicans strain SC5314 were grown 16 h at 37°C with vigorous agitation in YPD medium. Cultures were predominantly (>99%) yeast forms following this incubation. Cells were washed and resuspended at 5 × 105 cells/ml in Medium 199 and incubated at 37°C for 24 h to induce hyphal growth. A cDNA expression library in λTriplEx2 (Clontech, Mountain View, CA) was constructed from hyphae by Bio S&T (Montreal, Quebec). Approximately 1.5 × 106 primary recombinant clones were amplified in 15 pools. Each pool was grown as plaques in an E. coli (XL-1 Blue) top agar lawn on 10 – 150 mm petri dishes, with approximately 1000 plaques per dish according to the manufacturer's instructions. Expression of cDNA insert was achieved by induction with isopropyl-β-D-thiogalactopyranoside (IPTG). Plaques were transferred to nitrocellulose and filters were blocked with 3% skim milk. Filters were probed with scFv5 prepared as described previously (Laforce-Nesbitt, et al., 2008), washed, and scFv5 binding was detected with mouse anti-FLAG antibody (Sigma) followed by alkaline phosphatase conjugated secondary antibody and chromogenic substrate. Reactive plaques were picked from the original top agar overlay and amplified. Successive rounds of immunostaining were done to confirm continued reactivity with scFv5 and to assure clonality. A positive phage clone (designated clone 3-7) was converted to plasmid (designated pBMJ9) following manufacturer's instructions and the insert sequenced using manufacturer's sequencing primers.
Construction of homozygous SPT6 deletion mutant in C. albicans
A homozygous deletion in SPT6 was derived from C. albicans strain SN87 (Noble and Johnson, 2005). Sequences for oligonucleotides used in this study are listed in Table 2. PCR products for targeting the SPT6 open reading frame (ORF) were generated using oligonucleotides 1 and 2 to amplify the 5′ flank of SPT6 and oligonucleotides 3 and 4 to amplify the 3′ flank of SPT6. Selectable marker sequences (Candida dubliniensis HIS1 and Candida maltosa LEU2) were also amplified from plasmids pSN52 and pSN40, respectively, as previously described (Noble and Johnson, 2005). Fusion PCR products were generated by using oligonucleotides 1 and 4. These PCR primers amplify the flanking sequences of SPT6 together with either the HIS1 or LEU2 marker PCR product (Noble and Johnson, 2005). The first allele of SPT6 was replaced using the HIS1 marker in strain SN87 (designated JMB220). The second allele of SPT6 was replaced with the LEU2 marker to generate a homozygous knockout of this gene (designated JMB31). Correct integration of the PCR products was verified by PCR across the 5′ disruption junction using oligonucleotides 5 and 6 (HIS1 marker) or 5 and 7 (LEU2 marker), and across the 3′ disruption junction using oligonucleotides 8 and 9 (HIS1 marker) or 8 and 10 (LEU2 marker). Loss of the ORF was confirmed using PCR primers internal to the SPT6 ORF (oligonucleotides 11 and 12).
TABLE 2.
Oligonucleotides used in this study
| Name | Sequencea |
|---|---|
| 1 | GCCCACGATTATCTCAACTTTATCC |
| 2 | cacggcgcgcctagcagcggACACCTCTTTCTCTCTTGTTGGGGTT |
| 3 | gtcagcggccgcatccctgcTTGTTGTCGTAGTGGAAGGTGATTG |
| 4 | TTATGGGACAAACAAGAACTCGAAA |
| 5 | GACATACAAATCCTTCCAATGGTCA |
| 6 | ATTAGATACGTTGGTGGTTC |
| 7 | AGAATTCCCAACTTTGTCTG |
| 8 | CCTCATTGAAAATCCAATGGAGCAA |
| 9 | AACACAACTGCACAATCTGG |
| 10 | AAACTTTGAACCCGGCTGCG |
| 11 | CATTTCGAGGTTGTGTGCTG |
| 12 | ACATCTTGACCGCCTGCTT |
| 13 | AGAGGGTGGTCTCTGGGA |
| 14 | GGGGGGGCCCGGGGCAATAAGGTTGAGTGA |
| 15 | GGGGCTCGAGACGGCGACAGATGGTTAATG |
| 16 | GGGGATCCGCGCAATACACGACCTATATGATG |
| 17 | GGCCACTATTTTCCAATAAGAGCTCC |
| 18 | GGAGCTCTTATTGGAAAATAGTGGCC |
| 19 | GGCTGCAGCGACGTATCAATTCGGGGACCTTGC |
| 20 | CAATCAAAGGTGGTCCTGCAG |
| 21 | GTAAATACCCTCCCCGGATCC |
| 22 | CAAACTGAACAACTTGTCCC |
| 23 | GGGACTAGTATGATGAAAAAAAAAATCTCTGCC |
| 24 | GGGCTCGAGCTAATAATACCCCGTGTTATATGACT |
Lowercase letters indicate exogenous sequences used for fusion PCR reactions as described previously (Noble and Johnson, 2005). Underlined sequences are restriction sites described in the text.
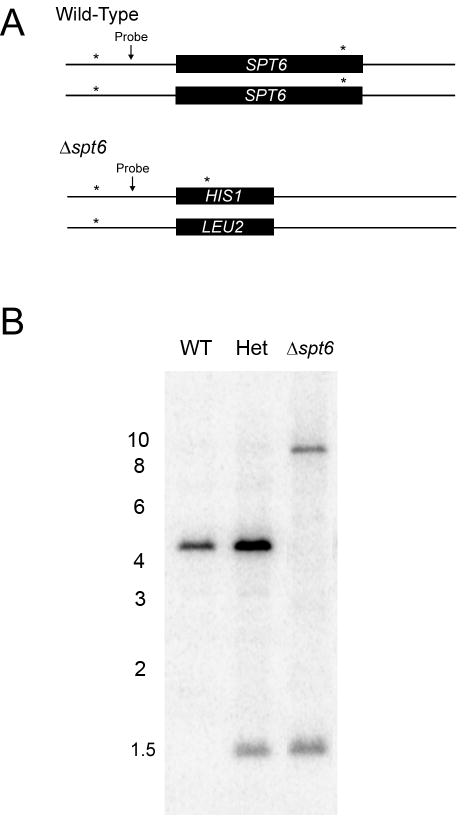
Southern analysis was also performed to confirm loss of SPT6 in JMB31. Genomic DNA was prepared from SN87 (parent), JMB220 (heterozygote) and JMB31 (Δspt6/Δspt6) using a yeast DNA purification kit (Epicentre Biotechnologies) according to maufacturer's instructions and digested with ScaI. DNA fragments were separated by agarose gel electrophoresis and transferred to nylon membrane by standard methods (Sambrook and Russell, 2001). A 197 nucleotide probe was generated by PCR using oligonucleotides 13 and 2, gel purified, and labeled with 32P-dCTP using the random primers method. The blot was hybridized overnight at 65°C, washed, and bound probe was detected by digital phosphorimaging.
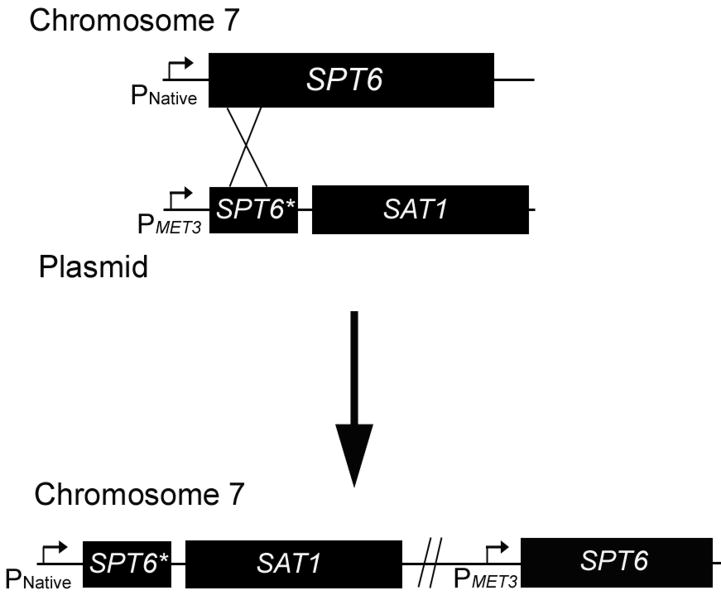
To construct a strain containing a reintegrated copy of SPT6, the gene was amplified by PCR using oligonucleotides 14 and 15, which incorporate an ApaI and XhoI site, respectively. The full length gene was then cloned into the corresponding sites of pSFS2a (Reuβ, et al., 2004). This construct contained the dominant nourseothricin resistance marker, SAT1, which allowed positive selection of transformants. The construct was linearized with EcoNI and transformations were conducted with the knockout strain, JMB31, and plated on YPD + nourseothricin. No transformants were obtained despite multiple attempts, suggesting that the impaired growth of JMB31 may adversely affect its capacity for transformation. To circumvent this issue, a strategy was devised to allow replacement of the native SPT6 with a truncated form in the heterozygote, JMB220, while simultaneously placing the gene under control of the inducible MET3 promoter (Fig. 1). First, to allow positive selection with nourseothricin, the SAT1 gene was excised from pSFS2a using EcoRI and PstI and cloned into the corresponding sites of pCaEXP (Care, et al., 1999), designated pBMJ40. Next, a fragment containing the 5′ end of SPT6 (615 bp) was amplified by PCR in two steps to incorporate a unique SacI restriction site in the middle of the fragment. Creation of the SacI site required 2 nucleotide changes relative to the native SPT6 ORF, but both were translationally silent. Oligonucleotides 16 and 17, or 18 and 19, were used to amplify the 5′ and 3′ ends of the 615 bp fragment, respectively. Oligonucleotides 17 and 18 contain a SacI site and are complementary, allowing these fragments to be combined in a second PCR with primers 16 and 19 to generate the 615 bp fragment with flanking BamHI and PstI sites. This fragment was cloned into the corresponding sites of pBMJ40, and designated pBMJ41. This plasmid was then linearized with SacI, transformed into the SPT6 heterozygote, JMB220, and plated on YPD + nourseothricin. Correct integration of the construct was verified by PCR across the 5′ integration junction using oligonucleotides 13 and 20 and across the 3′ disruption junction using oligonucleotides 21 and 22. These PCR products confirmed that the native SPT6 promoter was adjacent to the truncated SPT6 gene and the MET3 promoter was adjacent to the full length SPT6 gene. The reintegrant strain was designated JMB222.
Fig.1. Strategy for construction of SPT6 reintegrant strain.
Because transformation of the Δspt6/Δspt6 mutant was inefficient, the heterozygous strain, JMB220, was transformed with the construct depicted. Homologous recombination (depicted by “X”) resulted in a truncated SPT6 gene (SPT6*) adjacent to the native promoter (PNative), and a full length gene adjacent to the MET3 promoter (PMET3). The SAT1 gene was included in the construct to allow for positive selection of transformants. See Materials and Methods for details.
Indirect immunofluorescence assay (IFA) of C. albicans strains with scFv5
IFAs of SN87 (parent), JMB31 (Δspt6/Δspt6), and JMB222 (Δspt6/spt6*-SPT6) were conducted on glass coverslips using scFv as described previously (Bliss, et al., 2003).
Complementation of S. cerevisiae SPT6 mutant
C. albicans SPT6 was amplified by PCR from wild-type strain SC5314 using oligonucleotides 23 and 24, which incorporate a SpeI and XhoI site, respectively. The 4.2 kb fragment was cloned in the corresponding sites of the S. cerevisiae expression vector, p416-GAL1 (Mumberg, et al., 1994) to create plasmid pBMJ10. This plasmid was transformed into S. cerevisiae strain FY137 (Hartzog, et al., 1998), which contains a mutation in SPT6 that leads to temperature sensitivity and suppression of auxotrophic markers for histidine and leucine. The parent vector, p416-GAL1 was transformed as a negative control, and a positive control plasmid containing S. cerevisiae SPT6 under control of the GAL1 promoter, pBY011 (generously provided by Fred Winston) was also included. Growth phenotypes for FY137 containing each of the 3 plasmids were compared at 30°C on drop-out solid media lacking uracil (selectable marker for the plasmid) and either leucine or histidine, and on drop-out solid media lacking uracil at 37°C to test temperature sensitivity.
Results
Screening of a C. albicans hyphae expression library for immunoreactivity with scFv5
Messenger RNA from C. albicans hyphae was used to construct a cDNA expression library in bacteriophage λ. The library contained 1.5 × 106 independent clones which carried an average insert size of >1.2 kb. The library was grown as plaques on an E. coli lawn under conditions leading to expression of insert, and plaque lifts were performed. Filters were then probed for reactivity with scFv5, and positive plaques further purified. A reactive clone was purified to homogeneity and the insert sequenced. This clone (designated 3-7) contained 1951 bp homologous to the 3′ end of the gene identified as orf 19.7136 (SPT6) in the Candida genome database (Arnaud, et al., 2007). The 1951 bp insert represents 46% of this 4206 bp gene. Comparison of clone 3-7 sequence with the database sequence demonstrated 99% sequence identity. There were a total of 9 nucleotide and 6 amino acid discrepancies with the published sequence. DNA sequence data for clone 3-7 was deposited with GenBank (accession number GQ337860).
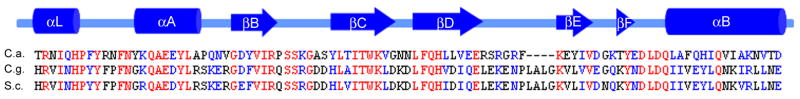
The published sequences of C. albicans SPT6 and S. cerevisiae SPT6 were compared by Basic Local Alignment Search Tool (BLAST, http://blast.ncbi.nlm.nih.gov/Blast.cgi). At the protein level, these sequences are 34% identical and 55% similar. Additionally, C. albicans Spt6p was found to contain the Src Homology 2 (SH2) domain that has been noted in Spt6 proteins of other organisms (Fig. 2) and is the only known SH2 domain in the yeast genome (Maclennan and Shaw, 1993).
Fig. 2. Comparison of Spt6p SH2 domain among species.
The alignment of the amino acid sequences of the Spt6p SH2 domains of C. albicans (C.a.), C. glabrata (C.g.), and S. cerevisiae (S.c.) is depicted. The SH2 domain represents residues 1203 - 1297, or 95 amino acids of the predicted 1402 in the entire protein (7%). Secondary structure elements are indicated above the alignment (cylinders for α helices, arrows for β strands) (Dengl, et al., 2009). Invariant residues are in red and conserved residues are in blue.
Deletion of C. albicans SPT6
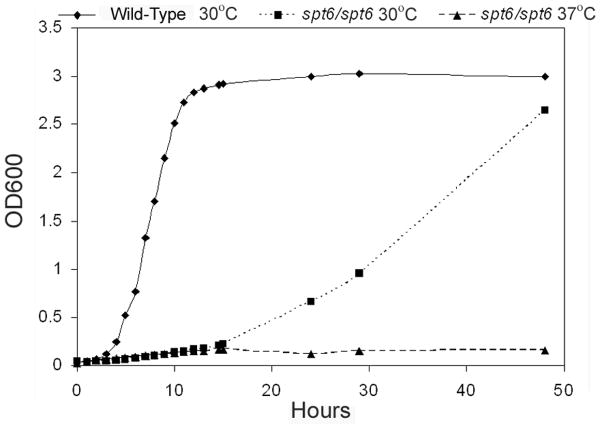
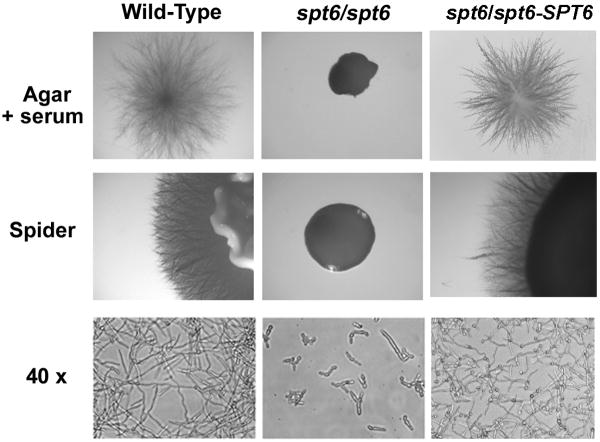
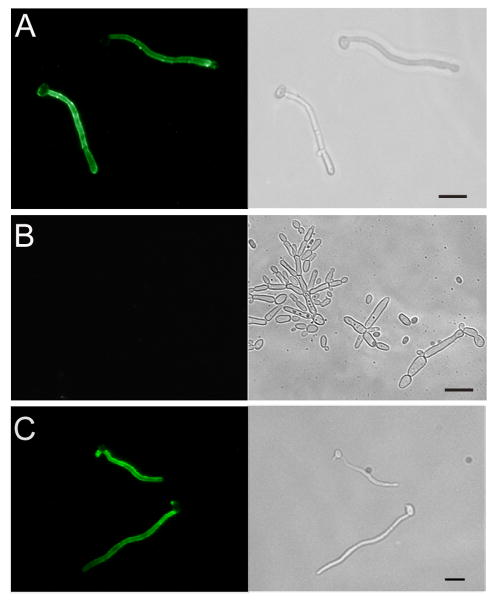
To establish whether the cognate antigen for scFv5 was the SPT6 gene product, a homozygous deletion mutant was constructed, designated JMB31. PCR with primers spanning both the 5′ and 3′ disruption junctions confirmed integration of the disruption constructs at the predicted locus and loss of the SPT6 open reading frame as described in Materials and Methods (data not shown). Deletion of SPT6 in this strain was also confirmed by Southern analysis (Fig. 3). Unlike S. cerevisiae, deletion of this gene in C. albicans was not lethal. However, the mutant's growth was markedly abnormal, and it was temperature sensitive. No growth occurred at 37°C, and growth at 30°C was very slow (Fig. 4). Additionally, the mutant had a severe defect in hyphal growth (Fig. 5). No hyphae were detected on solid media containing serum or on spider media. Microscopically, after a 6 hour incubation in Medium 199, the defective hyphal growth was readily apparent. To test reactivity with scFv5, immunofluorescence assays were conducted on JMB31 and its wild-type parent. No reactivity with scFv5 was detected on the mutant strain (Fig. 6). However, because the morphology of the mutant was drastically altered, the lack of binding may or may not reflect deletion of the target antigen. Disordered growth of the structure on which the target antigen is normally displayed is a reasonable alternative conclusion.
Fig. 3. Southern analysis to confirm homozygous deletion of SPT6 in JMB31.
Genomic DNA was prepared from the wild-type (WT) parent, from the heterozygous strain resulting from replacement of SPT6 with HIS1 (JMB220, Het), and from the strain with both copies of SPT6 replaced by HIS1 and LEU2 respectively (JMB31, Δspt6). Genomic DNA was digested with ScaI and analyzed by Southern blot. Recognition sites for this restriction enzyme exist within the SPT6 ORF, as well as the neighboring upstream and downstream sequences. The gene deletion strategy was predicted to eliminate the ScaI site both within and downstream to the ORF. Panel A shows the predicted ScaI recognition sites (*) and the binding location of the DNA probe. Digestion of wild-type DNA was predicted to yield a 4.2 kb fragment and digestion following replacement with HIS1 predicted a 1.5 kb fragment. SPT6 is adjacent to the telomere of chromosome 7. Because the LEU2 marker does not contain a ScaI site, and the only ScaI site in the region 3′ to the gene was deleted with SPT6, the fragment containing the LEU2 marker was predicted to extend to the telomeric region of the chromosome; ∼ 10 kb. Panel B demonstrates the predicted banding pattern, confirming homozygous deletion of SPT6. Positions of the relevant molecular weight markers (in kb) are depicted.
Fig. 4. Growth of Δspt6/Δspt6 mutant.
The Δspt6/Δspt6 mutant and its wild-type parent were grown in YPD media at 30°C and 37°C and growth was measured over time by optical density at 600 nm (OD600). The mutant demonstrated slower growth than the wild-type at 30°C and no growth at 37°C.
Fig. 5. Hyphal phenotype of Δspt6/Δspt6 mutant.
Wild-type and mutant cells were grown on the indicated media. In addition to markedly smaller colony size, the mutant generated no detectable hyphae under any conditions. Using phase contrast microscopy (40x), stunted filamentous forms were seen with no true hyphae. The SPT6 reintegrant strain (Δspt6/spt6-SPT6) resembled the wild-type phenotype.
Fig. 6. Indirect immunofluorescence assay (IFA) of wild-type and Δspt6/Δspt6 mutant probed with scFv5.
Wild-type (Panel A), Δspt6/Δspt6 mutant (Panel B), and SPT6 reintegrant cells (Panel C) were induced to form germ tubes and incubated with scFv5 followed by an appropriate fluorochrome labeled secondary antibody. Fluorescence and phase contrast photomicrographs of the same representative microscopic fields are depicted. The typical hyphae-specific binding of scFv5 to wild-type is seen, whereas no binding is detected to the mutant. The reintegrant strain showed binding with a wild-type pattern. Bar – 10 μm.
To establish a complete analysis of the mutant phenotype, reintegration of the wild-type SPT6 gene was attempted in the knockout strain JMB31. The gene was cloned into pSFS2A (Reuβ, et al., 2004) and DNA sequencing was used to confirm that the construct was appropriate. However, despite multiple attempts to transform the construct into JMB31, no transformants containing the reintegrated gene were obtained, while control transformations proceeded with expected efficiency. The significantly impaired growth phenotype of JMB31 likely compromises its capacity to undergo transformation. To circumvent this issue, a strategy was devised (Fig. 1) to replace the 2nd copy of SPT6 in the heterozygote with a truncated gene and simultaneously place the full length SPT6 gene under control of the MET3 promoter (Care, et al., 1999). PCR using primers that spanned the predicted integration site confirmed that the native SPT6 had been replaced with the truncated version and that full length SPT6 was adjacent to the MET3 promoter. Although transcription from the MET3 promoter is repressed in the presence of methionine or cysteine in the culture media (Care, et al., 1999), the spt6 mutant phenotype could not be recapitulated in the reintegrant strain regardless of growth conditions. Presumably, the MET3 promoter or other factors within the locus allow enough transcription of SPT6 to correct for the severe growth defect in the spt6 deletion mutant. Nevertheless, because truncation of native SPT6 was confirmed, the phenotype of the reintegrant strain was analyzed. This strain had a growth rate comparable to wild-type (data not shown) and had no defect in capacity for hyphal growth (Fig. 5). It is likewise reactive with scFv 5 by IFA (Fig. 6).
Complementation of a S. cerevisiae SPT6 mutant
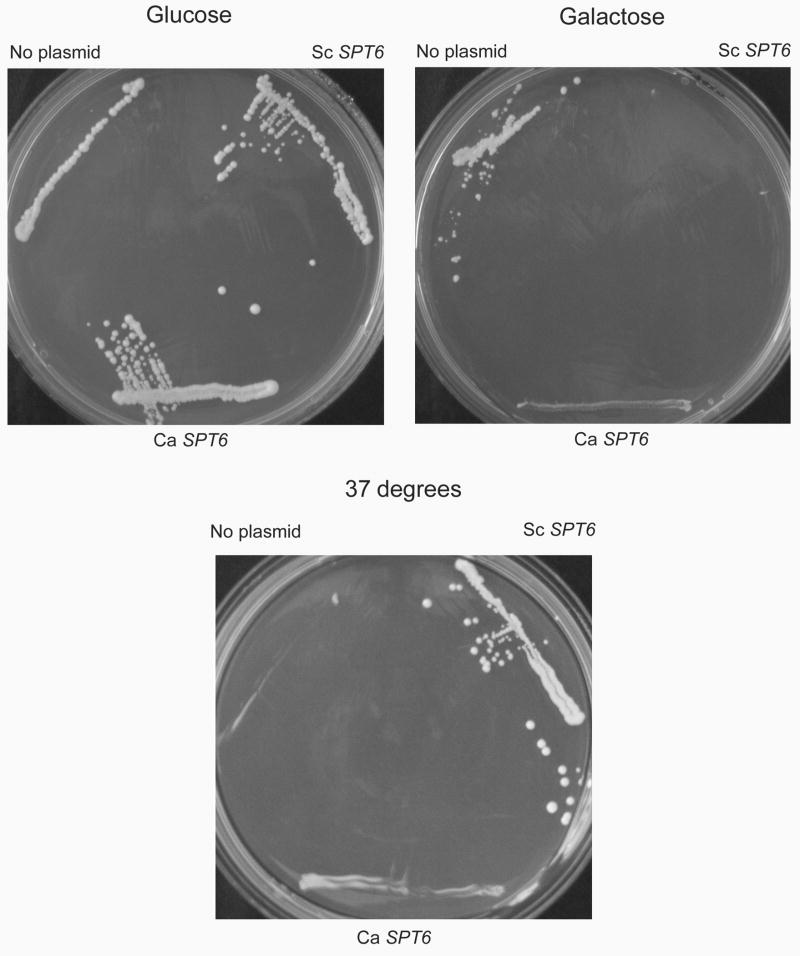
The S. cerevisiae SPT6 gene product functions as a transcription elongation factor, acting to modulate chromatin structure and prevent transcription from promoters within coding regions (Bortvin and Winston, 1996; Kaplan, et al., 2003). Deletion of SPT6 is lethal in S. cerevisiae (Clark-Adams and Winston, 1987), although mutants have been described that suppress defects in HIS4 and LEU2 (Winston, et al., 1984). FY137 contains such a mutation, spt6-140, that results in temperature sensitivity (ts) and suppresses his4 and leu2 mutations of the parent strain. Thus, its phenotype is ts, his+, leu+. Because spt6-140 is recessive, expression of wild-type S. cerevisiae SPT6 in trans restores the temperature resistant (tr), his-, leu- phenotype of the parent (Fig. 7). To test whether C. albicans SPT6 is capable of suppressing the S. cerevisiae mutant, the full length gene was cloned into a S. cerevisiae expression plasmid under the control of the GAL1 promoter. Proper in frame insertion and orientation of the insert was confirmed by DNA sequencing. Growth phenotypes on histidine or leucine drop-out media containing galactose to induce SPT6 expression, and growth at 37°C were determined (Fig. 7). Expression of the C. albicans gene partially suppressed the mutant phenotype, based on inhibition of growth on histidine drop-out media and restoration of growth at 37°C. However, the growth phenotypes were intermediate between the conditional mutant strain and complementation by S. cerevisiae SPT6.
Fig. 7. Complementation of spt6 in S. cerevisiae.
FY137 is a S. cerevisiae strain containing mutations in HIS4 and LYS2 that normally would make the strain auxotrophic for histidine and lysine. However, due to a recessive suppressor mutation in SPT6 (spt6-140), the phenotype of this strain is His+, Lys+. The strain also does not grow at 37°C. In the upper portion of the figure, growth of FY137 containing empty vector, S. cerevisiae (Sc) SPT6 or C. albicans (Ca) SPT6 under control of a galactose inducible promoter is depicted on His drop out media containing glucose or galactose at 30°C. Because wild-type SPT6 is not expressed on glucose, the strain grows normally. Induction of SPT6 expression by galactose leads to return to the His- phenotype, and the C. albicans gene has a similar effect to that seen with S. cerevisiae SPT6. In the lower portion of the figure, growth of the same strains on galactose at 37°C is shown. The temperature sensitive phenotype of FY137 is partially corrected by expression of C. albicans SPT6.
Discussion
The yeast to hypha transition in C. albicans is a complex, carefully orchestrated process in which the physiological state of the organism is significantly altered by the programmed activation of a subset of genes. We have examined scFv specific for the hyphal form of the organism as a means to identify potentially novel antigens expressed on the cell surface as part of this transition. Our screen of a hyphal cDNA expression library for clones reactive with hypha-specific human antibody fragments has identified the SPT6 gene product as the possible cognate antigen for scFv5. The altered physiology of a strain deleted for this gene has made definitive conclusions about its product's reactivity with scFv5 difficult to obtain. ScFv5 does not bind to the mutant, but this observation may reflect the defective hyphal growth in the strain rather than the specific lack of the cognate antigen.
We confirmed deletion of SPT6 in the mutant by PCR as well as Southern blot. Although Western analysis would also be helpful to confirm deletion at the protein level, this experiment was not possible due to technical limitations. Previous attempts have been made to extract the antigen reactive with scFv5 from wild-type C. albicans using multiple biochemical methods. These studies have never yielded antigen reactive with scFv5 in a soluble form that is amenable to Western blotting, either under denaturing or native conditions. We concluded from these studies that the protein either exhibits poor solubility, or that scFv5 recognizes a conformational epitope that is difficult to maintain following extraction.
The C. albicans gene, orf 19.7136, has been designated SPT6 based on homology to the S. cerevisiae gene (www.candidagenome.org). In S. cerevisiae, SPT6 codes for a transcription elongation factor involved in direct interaction with histones and mediation of chromatin structure (Bortvin and Winston, 1996). Although overall homology between the C. albicans and S. cerevisiae Spt6 proteins is somewhat limited (34% identical and 55% similar), they do bear some additional similarities. The predicted proteins are of similar size; C. albicans 1402 amino acids, S. cerevisiae 1452 amino acids. Both proteins have an acidic amino terminus, characteristic of proteins that interact with chromatin. In S. cerevisiae, the first 70 residues have a net charge of -30 and the amino terminal third of the protein has a net charge of -81 (Swanson, et al., 1990), whereas in C. albicans the net charge of these regions is -15 and -57, respectively. Additionally, the C. albicans Spt6p possesses a SH2 domain. This domain is important for the protein's interaction with RNA polymerase II (Pol II), as it associates with phosphoserine on the C-terminal repeat domain of the largest Pol II subunit (Andrulis, et al., 2000; Kaplan, et al., 2000). This observation provides strong evidence that the C. albicans gene is truly homologous to the gene in other organisms, as the SH2 domain in Spt6p is the only known SH2 domain encoded in the yeast genome. We also found evidence of similarity at a functional level. Inducing expression of the C. albicans gene in a well defined mutant of S. cerevisiae resulted in partial complementation of the mutant phenotype. Although the phenotype was less robust than that achieved with expression of S. cerevisiae SPT6, such a result is not surprising given the loose homology between the two organisms. Based on these observations, the most likely interpretation is that the C. albicans gene has a function homologous to that of S. cerevisiae. To our knowledge, however, this is the first report of an organism in which deletion of SPT6 is not lethal.
Direct experimental evidence for the function of Spt6p in C. albicans is generally unavailable. Several studies using genomic screening approaches to study an aspect of C. albicans biology or virulence have identified SPT6 in their screen. A large scale screen of Tn-7 transposon insertion mutants for those that had an alteration in filamentation phenotype identified 146 genes that affect the yeast to hypha transition, based on altered colony morphology relative to wild-type when grown on YEPD containing 1% serum and/or spider medium (Uhl, et al., 2003). Four individual clones with transposon insertions in SPT6 were identified in the screen. Three were either hyperfilamentous or had a textured colony morphology on serum agar, while the fourth was less filamentous than wild-type on spider medium. The authors noted that among the 60% of identified genes that had homologues in S. cerevisiae, the largest group were involved in transcription and its regulation. Genome wide transcript profiling of C. albicans in both a reconstituted human epithelium (RHE) model as well as in smear samples collected from 11 HIV-positive patients with oral thrush identified upregulation of SPT6 transcription, which persisted at all time points taken over 24 hours in the RHE model (Zakikhany, et al., 2007). This finding supports a role for this gene in the setting of infection, perhaps related to its role in filamentation. Finally, SPT6 was identified as one of 66 C. albicans genes that are preferentially expressed during oral candidiasis in humans using an antibody-based screening approach (Nguyen, et al., 2004). In this approach, pooled sera from 24 HIV-positive patients with active oral thrush were extensively adsorbed against whole cells and varied extracts of a C. albicans clinical isolate grown in vitro. The adsorbed sera were then used to screen a genomic DNA expression library for reactive clones. The approach was validated in a subset of these genes by showing preferential expression in an HIV-positive patient relative to growth in vitro, and localization of gene products to C. albicans cells within pseudomembranes using polyclonal antibodies raised against purified antigens (Cheng, et al., 2003). Again, among 58 genes with known or putative function, the largest group (23 genes) was comprised of genes involved in transcription or regulation. In the present study, we have demonstrated that lack of SPT6 expression resulted in disordered hyphal growth; a finding consistent with a role for this gene product in virulence.
If Spt6p truly has a role analogous to that of S. cerevisiae as suggested by the complementation data, its exposure on the hyphal surface and reactivity with scFv5 is difficult to reconcile. One possibility is that the actual cognate antigen happens to share an epitope with Spt6p that was recapitulated when the cDNA was expressed. Certainly expression of the library in E. coli introduces variability in epitope expression relative to the in vivo situation. However, the possibility of exposure of Spt6p on the cell surface cannot be excluded, as other cytoplasmic proteins are well documented to be surface exposed for at least part of the cell's life cycle. The glycolytic enzymes, enolase, 3-phosphoglycerate kinase (PGK) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), have been shown to be present in the cell wall as well as the cytoplasm, despite the absence of classical secretion signal sequences. Although enolase is not exposed at the cell surface, surface exposure of PGK and GAPDH has been demonstrated (Alloush, et al., 1997; Angiolella, et al., 1996; Gil-Navarro, et al., 1997; Sundstrom and Aliaga, 1994). A more recent study using a proteomic approach with biotinylation to identify surface exposed antigens also identified PGK and GAPDH (Urban, et al., 2003). Another protein, thiol-specific antioxidant-like protein 1 (Tsa1p), was localized to the cell surface in hyphae whereas only nuclear localization was detected in yeast forms. Thus, the localization of this protein varies with growth condition of the cell. Therefore, although not definitive, a dual localization of Spt6p cannot be excluded and will be the focus of ongoing study.
Acknowledgments
We thank Bethany McGonnigal for technical assistance, Fred Winston for providing strains and plasmids, and Richard Bennett for valuable discussions and for critically reviewing the manuscript. This work was supported in part by Basil O'Connor Starter Scholar Research Award Grant No. 5-FY05-1211 from the March of Dimes Foundation, a National Institute of Health grant (K08 AI064919), and a NIH COBRE grant (P20 RR018728).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alloush HM, Lopez-Ribot JL, Masten BJ, Chaffin WL. 3-phosphoglycerate kinase: a glycolytic enzyme protein present in the cell wall of Candida albicans. Microbiology. 1997;143(Pt 2):321–330. doi: 10.1099/00221287-143-2-321. [DOI] [PubMed] [Google Scholar]
- Andrulis ED, Guzman E, Doring P, Werner J, Lis JT. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angiolella L, Facchin M, Stringaro A, Maras B, Simonetti N, Cassone A. Identification of a glucan-associated enolase as a main cell wall protein of Candida albicans and an indirect target of lipopeptide antimycotics. J Infect Dis. 1996;173:684–690. doi: 10.1093/infdis/173.3.684. [DOI] [PubMed] [Google Scholar]
- Arnaud MB, Costanzo MC, Skrzypek MS, Shah P, Binkley G, Lane C, Miyasato SR, Sherlock G. Sequence resources at the Candida Genome Database. Nucleic Acids Res. 2007;35:D452–456. doi: 10.1093/nar/gkl899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin DK, Jr, Stoll BJ, Fanaroff AA, McDonald SA, Oh W, Higgins RD, Duara S, Poole K, Laptook A, Goldberg R. Neonatal candidiasis among extremely low birth weight infants: risk factors, mortality rates, and neurodevelopmental outcomes at 18 to 22 months. Pediatrics. 2006;117:84–92. doi: 10.1542/peds.2004-2292. [DOI] [PubMed] [Google Scholar]
- Bliss JM, Sullivan MA, Malone J, Haidaris CG. Differentiation of Candida albicans and Candida dubliniensis by using recombinant human antibody single-chain variable fragments specific for hyphae. J Clin Microbiol. 2003;41:1152–1160. doi: 10.1128/JCM.41.3.1152-1160.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- Calderone RA. Candida and Candidiasis. ASM Press; Washington D.C.: 2002. [Google Scholar]
- Care RS, Trevethick J, Binley KM, Sudbery PE. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol Microbiol. 1999;34:792–798. doi: 10.1046/j.1365-2958.1999.01641.x. [DOI] [PubMed] [Google Scholar]
- Cheng S, Clancy CJ, Checkley MA, Handfield M, Hillman JD, Progulske-Fox A, Lewin AS, Fidel PL, Nguyen MH. Identification of Candida albicans genes induced during thrush offers insight into pathogenesis. Mol Microbiol. 2003;48:1275–1288. doi: 10.1046/j.1365-2958.2003.03521.x. [DOI] [PubMed] [Google Scholar]
- Cheng S, Clancy CJ, Checkley MA, Zhang Z, Wozniak KL, Seshan KR, Jia HY, Fidel P, Jr, Cole G, Nguyen MH. The role of Candida albicans NOT5 in virulence depends upon diverse host factors in vivo. Infect Immun. 2005;73:7190–7197. doi: 10.1128/IAI.73.11.7190-7197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Adams CD, Winston F. The SPT6 gene is essential for growth and is required for delta-mediated transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:679–686. doi: 10.1128/mcb.7.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengl S, Mayer A, Sun M, Cramer P. Structure and in vivo requirement of the yeast Spt6 SH2 domain. J Mol Biol. 2009;389:211–225. doi: 10.1016/j.jmb.2009.04.016. [DOI] [PubMed] [Google Scholar]
- Feng Q, Summers E, Guo B, Fink G. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J Bacteriol. 1999;181:6339–6346. doi: 10.1128/jb.181.20.6339-6346.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- Gil-Navarro I, Gil ML, Casanova M, O'Connor JE, Martinez JP, Gozalbo D. The glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is a surface antigen. Journal of Bacteriology. 1997;179:4992–4999. doi: 10.1128/jb.179.16.4992-4999.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidaris CG, Malone J, Sherrill LA, Bliss JM, Gaspari AA, Insel RA, Sullivan MA. Recombinant human antibody single chain variable fragments reactive with Candida albicans surface antigens. J Immunol Methods. 2001;257:185–202. doi: 10.1016/s0022-1759(01)00463-x. [DOI] [PubMed] [Google Scholar]
- Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D, Johnson AD. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol Cell Biol. 2001;21:2496–2505. doi: 10.1128/MCB.21.7.2496-2505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D, Johnson AD. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol Biol Cell. 2005;16:2903–2912. doi: 10.1091/mbc.E05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Morris JR, Wu C, Winston F. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 2000;14:2623–2634. doi: 10.1101/gad.831900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto CA, Vinces MD. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol. 2005;7:1546–1554. doi: 10.1111/j.1462-5822.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- Laforce-Nesbitt SS, Sullivan MA, Hoyer LL, Bliss JM. Inhibition of Candida albicans adhesion by recombinant human antibody single-chain variable fragment specific for Als3p. FEMS Immunol Med Microbiol. 2008 doi: 10.1111/j.1574-695X.2008.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E, Harcus D, Broadbent ID, Clark KL, Dignard D, Ziegelbauer K, Schmidt A, Gow NA, Brown AJ, Thomas DY. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci U S A. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Kohler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- Lopez-Ribot JL, Casanova M, Murgui A, Martinez JP. Antibody response to Candida albicans cell wall antigens. FEMS Immunol Med Microbiol. 2004;41:187–196. doi: 10.1016/j.femsim.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Maclennan AJ, Shaw G. A yeast SH2 domain. Trends Biochem Sci. 1993;18:464–465. doi: 10.1016/0968-0004(93)90006-9. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MH, Cheng S, Clancy CJ. Assessment of Candida albicans genes expressed during infections as a tool to understand pathogenesis. Med Mycol. 2004;42:293–304. doi: 10.1080/13693780410001722485. [DOI] [PubMed] [Google Scholar]
- Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitarch A, Jimenez A, Nombela C, Gil C. Decoding serological response to Candida cell wall immunome into novel diagnostic, prognostic, and therapeutic candidates for systemic candidiasis by proteomic and bioinformatic analyses. Mol Cell Proteomics. 2006;5:79–96. doi: 10.1074/mcp.M500243-MCP200. [DOI] [PubMed] [Google Scholar]
- Reuβ O, Vik A, Kolter R, Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Woodbury, NY: 2001. [Google Scholar]
- San-Blas G, Travassos LR, Fries BC, Goldman DL, Casadevall A, Carmona AK, Barros TF, Puccia R, Hostetter MK, Shanks SG, Copping VM, Knox Y, Gow NA. Fungal morphogenesis and virulence. Med Mycol. 2000;38 1:79–86. [PubMed] [Google Scholar]
- Stoldt VR, Sonneborn A, Leuker CE, Ernst JF. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundstrom P, Aliaga GR. A subset of proteins found in culture supernatants of Candida albicans includes the abundant, immunodominant, glycolytic enzyme enolase. J Infect Dis. 1994;169:452–456. doi: 10.1093/infdis/169.2.452. [DOI] [PubMed] [Google Scholar]
- Swanson MS, Carlson M, Winston F. SPT6, an essential gene that affects transcription in Saccharomyces cerevisiae, encodes a nuclear protein with an extremely acidic amino terminus. Mol Cell Biol. 1990;10:4935–4941. doi: 10.1128/mcb.10.9.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DP, Viudes A, Monteagudo C, Lazzell AL, Saville SP, Lopez-Ribot JL. A proteomic-based approach for the identification of Candida albicans protein components present in a subunit vaccine that protects against disseminated candidiasis. Proteomics. 2006;6:6033–6041. doi: 10.1002/pmic.200600321. [DOI] [PubMed] [Google Scholar]
- Uhl MA, Biery M, Craig N, Johnson AD. Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C. albicans. EMBO J. 2003;22:2668–2678. doi: 10.1093/emboj/cdg256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban C, Sohn K, Lottspeich F, Brunner H, Rupp S. Identification of cell surface determinants in Candida albicans reveals Tsa1p, a protein differentially localized in the cell. FEBS Lett. 2003;544:228–235. doi: 10.1016/s0014-5793(03)00455-1. [DOI] [PubMed] [Google Scholar]
- Viudes A, Peman J, Canton E, Ubeda P, Lopez-Ribot JL, Gobernado M. Candidemia at a tertiary-care hospital: epidemiology, treatment, clinical outcome and risk factors for death. Eur J Clin Microbiol Infect Dis. 2002;21:767–774. doi: 10.1007/s10096-002-0822-1. [DOI] [PubMed] [Google Scholar]
- Wellington M, Bliss JM, Haidaris CG. Enhanced phagocytosis of Candida species mediated by opsonization with a recombinant human antibody single-chain variable fragment. Infect Immun. 2003;71:7228–7231. doi: 10.1128/IAI.71.12.7228-7231.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Chaleff DT, Valent B, Fink GR. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics. 1984;107:179–197. doi: 10.1093/genetics/107.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakikhany K, Naglik JR, Schmidt-Westhausen A, Holland G, Schaller M, Hube B. In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. Cell Microbiol. 2007;9:2938–2954. doi: 10.1111/j.1462-5822.2007.01009.x. [DOI] [PubMed] [Google Scholar]