Abstract
Annexin 7 (ANX7) acts as a tumor suppressor gene in prostate cancer, where loss of heterozygosity and reduction of ANX7 protein expression is associated with aggressive metastatic tumors. To investigate the mechanism by which this gene controls tumor development, we have developed an Anx7(+/-) knockout mouse. As hypothesized, the Anx7(+/-) mouse has a cancer-prone phenotype. The emerging tumors express low levels of Anx7 protein. Nonetheless, the wild-type Anx7 allele is detectable in laser-capture microdissection-derived tumor tissue cells. Genome array analysis of hepatocellular carcinoma tissue indicates that the Anx7(+/-) genotype is accompanied by profound reductions of expression of several other tumor suppressor genes, DNA repair genes, and apoptosis-related genes. In situ analysis by tissue imprinting from chromosomes in the primary tumor and spectral karyotyping analysis of derived cell lines identify chromosomal instability and clonal chromosomal aberrations. Furthermore, whereas 23% of the mutant mice develop spontaneous neoplasms, all mice exhibit growth anomalies, including gender-specific gigantism and organomegaly. We conclude that haploinsufficiency of Anx7 expression appears to drive disease progression to cancer because of genomic instability through a discrete signaling pathway involving other tumor suppressor genes, DNA-repair genes, and apoptosis-related genes.
Keywords: hepatocellular carcinoma, lymphosarcoma of the thymus, SKY analysis, cDNA microarray
Annexin 7 (ANX7) is a newly described tumor suppressor gene (TSG) for prostate cancer in men, featuring loss of heterozygosity and reduced ANX7§ protein expression in a large fraction of archived metastatic tumors (1). ANX7 maps to chromosome 10q21, where unknown TSGs had been postulated but not yet identified (2-4). In a previous work, we established an Anx7(+/-) knockout mouse, in which Ca2+-dependent endocrine secretory defects were identified (5). We also noted substantial reductions of Anx7 expression in the pancreas, heart, and other tissues. The nullizygous Anx7(-/-) knockout could not be studied. The embryo failed to develop beyond embryonic day 10 in utero. However, as the Anx7(+/-) colony aged, we began to observe an increasing frequency of disparate tumors (6). The mechanism of tumorigenesis in humans and mice often differ (7). Therefore, given the apparent TSG-like ANX7 parallel between tumors in mice and humans, we felt compelled to investigate the mechanisms of tumor transformation in the Anx7(+/-) mouse model.
Mechanistically, we reasoned that we might be able to distinguish between a Knudson two-hit model, as might be considered possible based on the human prostate tumor data, and a haploinsufficiency model, based on low Anx7 protein levels in otherwise normal knockout-mouse tissues. We find that the data clearly support a haploinsufficiency model for tumor development in the knockout mouse. In addition, the tumors demonstrate an increased level of genetic instability, based on spectral karyotyping (SKY) analysis of primary tumors and an established cell line derived from a hepatocellular carcinoma in an Anx7(+/-) mouse. Finally, gene-expression profiling shows that Anx7(+/-) tumors express not only low levels of Anx7 protein but also reveal down-regulation of several specific TSGs and DNA-repair genes. Taken together these data indicate that the haploinsufficiency of Anx7 has global consequences and that this cancer-prone phenotype of the Anx7(+/-) mouse model may yield important insights into the mechanism by which this gene exerts its control of prostate and other cancers in humans.
Materials and Methods
Production of Anx7(+/-) Mice. The Anx7 targeting vector, the transfection and selection of ES cells, the preparation of chimeras, and the breeding protocols were as described in Srivastava et al. (5). All animals described in this article were the progeny of at least six successive backcrosses into the wild-type C57BL6/J strain.
Anatomical, Histological, and Pathological Studies. For the growth study, 30 F2 generation hybrids from Anx7(+/-) and Anx7 (+/+) mice were weighed at regular intervals. Nine-month-old Anx7(+/-) and Anx7(+/+) mice were killed, and their internal organs were weighed. For pathological studies, the tissues were embedded in paraffin, cut in 5-μm sections, and stained with hematoxylin/eosin. Attempts were made to put Anx7(+/-) tumors in culture. One example, which was particularly successful, was from a hepatocellular carcinoma.
cDNA Microarray Analysis of mRNAs from Tumor and Adjacent Normal Tissues. Total RNA was prepared from tumor tissues and adjacent normal tissues (8), and cDNA copies were labeled with 32P as described by Srivastava et al. (9). Mouse cDNA microarrays on nylon membranes were obtained from Clontech, were prehybridized by using Express Hyb (Clontech) at 68°C for 1 h, and were then incubated overnight in Express Hyb with the denatured and neutralized labeled probes. After this incubation, the membranes were washed three times and exposed for 6 h on a Storm PhosphorImager plate (Molecular Dynamics). Data were collected in duplicate.
Imaging and Quantitation of cDNA Microarrays. Imaging data from the Storm PhosphorImager were downloaded into a Microsoft excel spreadsheet. Duplicate data points were normalized either to the ubiquitin standard or to the average expression of the entire array. Data were then analyzed by using the Stanford University SCANALYZE software. These data were also evaluated in parallel with the PSCAN program for point identification and with the JMP program for graphical organization.
Statistical Data Mining from cDNA Arrays by GRASP. The initial strategy we used is embodied in the GRASP algorithm (Gene Ratio AnalysiS Paradigm; ref. 9). The GRASP algorithm allows us to specify the changes in specific intensities of given genes that are greater or less than one or more SDs from the average changes of all genes in the entire array.
Spectral Karyotyping. Metaphase chromosome spreads were prepared from the hepatocellular carcinoma cell line LC01 derived from an Anx7(+/-) heterozygote, from passage 4 after incubation in 0.1 μg/ml Colcemid (GIBCO/BRL) for 1 h. Cells were harvested according to standard cytogenetic methods, and SKY was performed as described (10, 11). Chromosome aberrations were defined according to the nomenclature rules from the Committee on Standardized Genetic Nomenclature for Mice (12). Twenty metaphase spreads were analyzed. Comparative genomic hybridization analysis was performed as described (10, 13). RP23-7J10 and RP23-206N14 bacterial artificial chromosomes for fluorescence in situ hybridization (FISH) mapping (Research Genetics, Huntsville, AL) were identified by using the National Center for Biotechnology Information map-viewer database. Genomic DNA was labeled with biotin-dUTP by nick translation, hybridized overnight to tumor metaphases, and detected by avidin-FITC antibody. Protocol details can be found at www.riedlab.nci.nih.gov/.
Results
Anx7(+/-) Mice Have a High Frequency of Spontaneous Tumors. During the course of ≈2 year's experience husbanding the Anx7(+/-) knockout mouse, we gradually became aware of the cumulative existence of a significant number of spontaneous tumors in these animals. Heterozygotes and normal littermate-control animals of approximately 1 year of age were systematically killed and assessed for visible tumors of ≈1-cm diameter or greater. Of these animals, 58 were males and 79 were females. Tissues were also reserved for microscopic examination by our veterinary pathologist. The average incidence of visible tumor occurrence is ≈23%. Of the 33 animals found to have visible tumors, 14 (42%) had tumors in multiple locations. No tumors were observed in the normal littermate controls. As shown in Table 1, the most frequent tumors are those affecting lymphoid tissue, including thymus and spleen. In males this class constitutes 50% of visible tumors, whereas in females they constitute 76%. The tissue diagnosis has routinely been lymphosarcoma of the thymus, both localized and metastatic. In addition, liver tumors have been noted (data not shown) in both male and female animals. The tissue diagnosis has routinely been hepatocellular carcinoma. One primary colon tumor has been noted in a male, and two primary lung tumors have been detected in females. Single instances of prostate carcinoma, ovarian carcinoma, and salivary gland tumor have been detected. Data in the table indicate that, although tumors occur in both males and females, a definite increased propensity exists for tumor development in the female mutants. By contrast, in normal littermate controls examined in parallel, we did not note a single tumor.
Table 1. Tumor registry for Anx7(+/-) mice.
| Tumors by gender, n
|
|||
|---|---|---|---|
| Tumor location | Tumor diagnosis | Male | Female |
| Lymphoid tissue (thymus, spleen) | Lymphosarcoma, localized or metastatic | 4 | 19 |
| Liver | Hepatocellular carcinoma | 2 | 3 |
| Lung | Bronchioalveolar tumor | 2 | |
| Salivary gland | Adenocarcinoma | 1 | |
| Prostate | Carcinoma | 1 | |
| Ovary | Neoplasia | 1 | |
| Total | 8 (13.8%) | 25 (31.6%) | |
Heterozygotes and normal littermate control animals of ≈ 1 year of age were systematically killed and assessed for tumors. Of these animals, 58 were males and 79 were females. Tissues were also reserved for microscopic examination by our veterinary pathologist. The incidence of tumor occurrence is ≈23%. Of the 32 animals found to have tumors, 14 (44%) had tumors in multiple locations. No tumors were observed in the normal littermate controls.
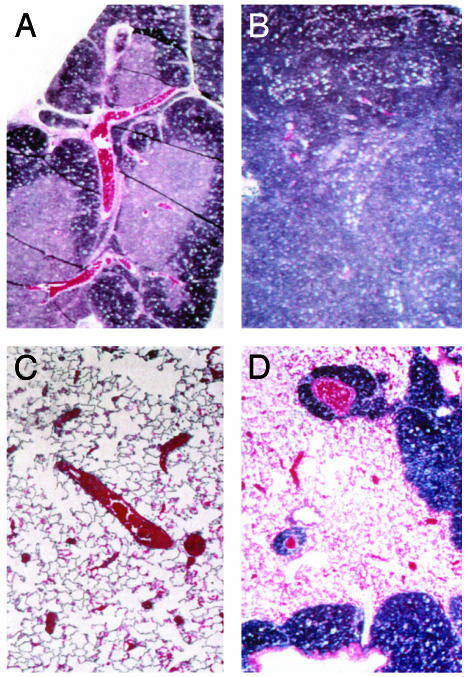
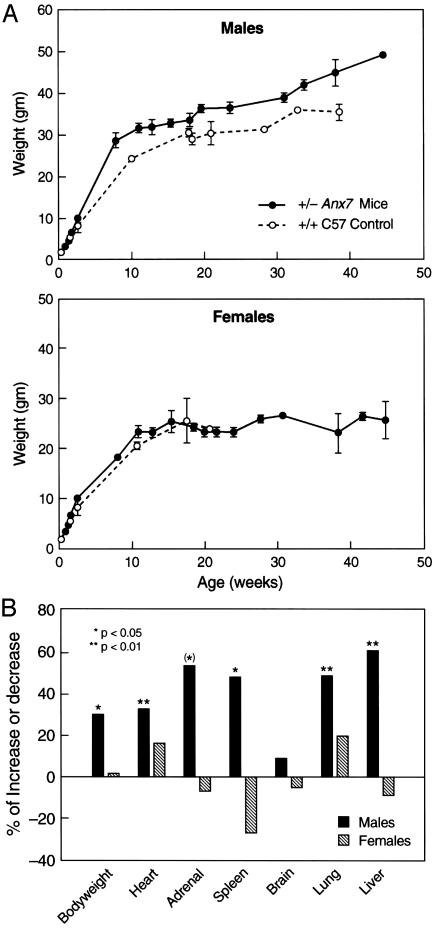
A section from a lymphosarcoma of the thymus, taken at ×50 magnification, is shown in Fig. 1B, with a sample of normal thymus shown in Fig. 1 A for comparison. The thymus architecture in the tumor had numerous large ”tingible body” macrophages scattered among the neoplastic cells. The mass is unencapsulated. Neoplastic cells infiltrate and expand the mediastinum, extend into the lung along branches of the pulmonary artery, efface the bronchial lymph nodes, and disseminate to the kidneys. Cell morphology is consistent with lymphosarcoma. A section is shown of tumor-cell infiltration into the lung in Fig. 1D, in which extensions along branches of the pulmonary artery are prominent. For comparison, control lung from an Anx7(+/+) mouse is shown in Fig. 1C. In many other examples of lymphosarcoma of the thymus, metastases to the pancreas have also been frequently noted. The mass is unencapsulated and neoplastic cells infiltrate adjacent to hepatic parenchyma. These results suggest that the effects of the Anx7(+/-) mutation on cancer occurrence are clearly not limited exclusively to any one type of tumor. Anx7(+/-) Mice Exhibit Gender Dimorphic Gigantism and Organomegaly. In addition to the tendency to develop tumors, the Anx7(+/-) heterozygote mice also exhibit profound gender-dimorphic growth anomalies and organomegaly. The male Anx7(+/-) mouse begins an extraordinarily enhanced growth spurt relative to growth of normal littermate controls after postpartum week 4 (Fig. 2A). The data in Fig. 2 show growth up to 10 months of age. Growth continues uninterrupted for at least 12 months, leading to 40- to 60-g mice. By contrast, the normal littermate controls had long since equilibrated at lower weights. Many internal organs also increase in weight proportionately (Fig. 2B). This relative weight increment for males of ≈25% is statistically significant and not due to obesity. Postmortem examination has systematically shown that the animals are not fat, merely large. By contrast, no changes are noted in the relative weight gains with age for females (Fig. 2 A).
Fig. 1.
Metastatic lymphosarcoma of the thymus. Thymus was taken from mice and fixed in buffered formalin. Sections were stained with hematoxylin/ eosin. (A) Thymus from a control littermate of the mouse shown in B. (B) Lymphosarcoma in anx7(+/-) mouse. (Magnification, ×200.) (C) Lung from a control littermate for the mouse shown in D.(D) Metastatic lymphosarcoma of the thymus to the lung in an anx7(+/-) mouse.
Fig. 2.
Increased growth of anx7(+/-) mouse compared with control (+/+) mouse. (A) Representative growth curve of 30 Anx7(+/+) and Anx7(+/-) littermates as a function of age. The size of mice is a function of Anx7 gene copy number. Anx7(+/+) is represented by a dotted line, and Anx7(+/-) is represented by a solid line. (B) Mean and 95% confidence interval of organ weights of 30 control and anx7(+/-) mice. The percent increase in knockout mice compared with control mice is plotted. In the graph, the hatched bar represents female Anx7(+/-) mice, and the filled bar represents male Anx7(+/+) mice.
Finally, to determine whether sexually dimorphic anomalies of growth hormone or other hormones of pituitary origin might explain the male gigantism, we examined the concentrations of insulin-like growth factor 1 (IGF-1) and corticosterone in plasma from wild-type and heterozygous animals. Neither IGF-1 nor corticosterone levels varied between wild-type or mutant, male or female (14). Moreover, the pituitaries of mutant males and mutant females were not found to be histologically different from wild-type littermate controls. In addition, no variations were noted among these groups for luteinizing hormone, growth hormone, thyrotropin, prolactin, α-melanocyte-stimulating hormone, β-endorphin, and corticotropin (n = 6, each; data not shown). Therefore, although tumors develop in only ≈23% of mice, male mice apparently have a more general tendency to exhibit growth anomalies in many organs and tissues that may not be realized as tumors later in life.
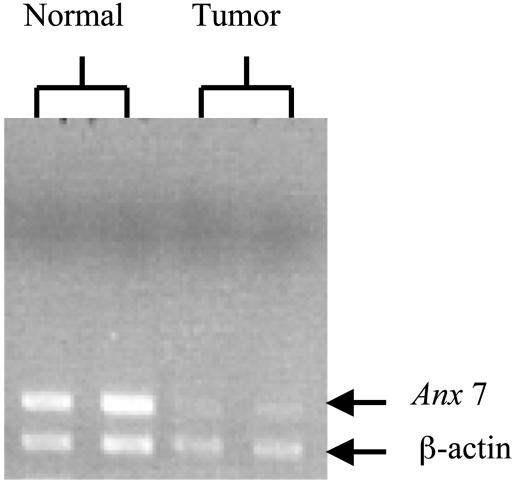
Mechanism of Tumor Development in Anx7(+/-) Mice. We have observed that heterozygous Anx7(+/-) mice have both low levels of ANX7 expression and a high incidence of tumors. It is possible that this observation may relate to predictions of the Knudson two-mutation hypothesis in which tumor occurrence depends on functional inactivation of both copies of the TSG by mutation or deletion (15, 16). In the Anx7(+/-) mouse, one of the Anx7 alleles is already inactivated by design. We therefore searched for deletions or global loss of the allele by using Anx7-specific primers and PCR. Although less frequently found, the hepatocellular carcinomas were remarkable for their size. We therefore concentrated on this tumor type for our analysis. Hepatocellular carcinoma and adjacent normal liver tissues from Anx7(+/-) mice were immediately embedded in OCT (Miles) and frozen at -70°C. We used laser capture microdissection to microdissect tumors from 1-μm frozen sections. We stained initial sections by hematoxylin/eosin and used these stained sections as optical templates for identification and isolation of tumor and normal cells from serial unstained sections from the same block. DNA was extracted from both tumor cells and adjacent histologically normal tissue domains that did not contain identifiable tumor cells. As shown in Fig. 3, DNA from laser capture microdissection-derived tumor samples also revealed the presence of normal Anx7 allele. We interpret these data to indicate that the mechanism of tumor induction does not involve loss of the normal allele and suggest that haploinsufficiency could be the mechanism by which the hepatocellular carcinoma develops in the Anx7(+/-) mice.
Fig. 3.
Detection of Anx7 allele in hepatocellular carcinoma specimens. Laser capture microdissection was used to microdissect tumors from 1-μm frozen sections of hepatocellular carcinoma. DNA was extracted from tumor cells and adjacent histologically normal tissue domains that did not contain identifiable tumor cells. Samples are run in 1% agarose gel by using Anx7-specific primers. β-Actin primers were used as reference.
Differential Expression of Genes Isolated from Tumor Tissue and Nearby Normal Area of the Tissue from Anx7(+/-) Mice. To assess the potential downstream targets and signaling pathway(s) of Anx7 that might be involved in tumorigenesis, we compared mRNAs from liver tumor and neighboring normal liver tissue from the same animal by using the ATLAS Mouse cDNA expression array. By using the GRASP algorithm for analysis (9), we have been able to detect those genes whose expression levels change specifically and significantly in tumor specimens in comparison with the cognate normal tissue. As shown in Table 2, 18 genes were identified which are expressed at ≥2 SDs more or less than the average of all genes in the tumor. Remarkably, these include seven known TSGs. TSGs down-regulated at 3 SDs greater than average include BRCA1, BRCA2, WT1 (Wilms tumor), and DCC (deleted in colorectal carcinoma). TSGs expressed at ≥2 SDs greater than average include adenomatous polyposis coli (APC), TSG101, and Von Hippel-Lindau (VHL). We conjecture that the altered level of expression of these TSGs might indicate that they could contribute to ANX7-regulated signaling pathways toward tumor development.
Table 2. Effect of reduced Anx7 expression on global gene expression.
| Gene assignment | Ratio | SD* | Tumor | Normal |
|---|---|---|---|---|
| Glutathione S-transferase (microsomal) | 5.9 | –4.87 | 0.11 | 0.38 |
| BRCA2, breast cancer susceptibility locus 2 product | 5.3 | –4.60 | 0.27 | 0.83 |
| WT1, Wilms tumor protein, tumor suppressor | 5.0 | –4.42 | 0.04 | 0.12 |
| BRCA1, breast/ovarian cancer susceptibility locus 1 product | 4.7 | –4.25 | 0.05 | 0.13 |
| GST Pi1, glutathione 5-transferase π1, preadipocyte growth factor | 4.1 | –3.88 | 0.07 | 0.17 |
| RNA polymerase/termination factor TTF-1 | 3.5 | –3.41 | 0.26 | 0.52 |
| DCC, netrin receptor, immunoglobulin gene superfamily member, former tumor suppressor protein candidate | 3.4 | –3.33 | 0.06 | 0.11 |
| APC, adenomatous polyposis coli protein | 3.0 | –2.99 | 0.03 | 0.05 |
| TSG 101, tumor susceptibility protein | 3.0 | –2.98 | 0.14 | 0.24 |
| Interleukin 12 (p40) β-chain | 2.9 | –2.94 | 0.09 | 0.15 |
| Cathepsin B | 2.9 | –2.94 | 0.18 | 0.30 |
| MHR23B, Rad23 UV excision repair protein homologue, xeroderina pigmentosuin group C (XPC) repair complementing protein | 2.8 | –2.87 | 0.06 | 0.09 |
| Bone morphogenetic protein 4 (BMP4) | 2.5 | 2.57 | 0.89 | 0.20 |
| VHL, Von Hippel-Lindau tumor suppressor protein | 2.5 | –2.51 | 0.04 | 0.05 |
| Brain-specific transcription factor NURR | 2.5 | 2.51 | 0.08 | 0.02 |
| Transcription factor Ci | 2.5 | 2.46 | 0.11 | 0.02 |
Effect of the Anx7 mutation on significantly and differentially affected expression of genes associated with tumor development in the Anx7(+/–) knockout mouse.
Number of standard deviations away from average log ratio
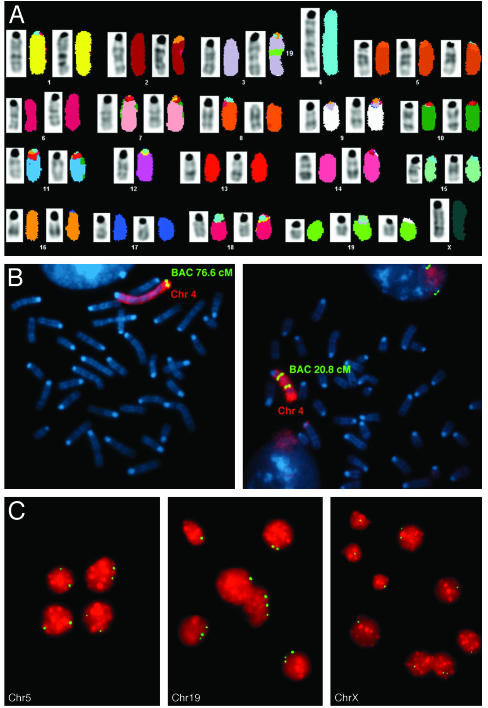
SKY and Comparative Genomic Hybridization Analyses Provide Evidence for Chromosome Instability in Anx7(+/-) Hepatocellular Carcinoma. To determine whether changes in gene expression might be linked to specific rearrangements or chromosome gain or loss, we analyzed the hepatocellular carcinoma cell line (AHCC) derived from the Anx7(+/-) mouse model by comparative genomic hybridization and SKY analyses. These cells had very low amounts of Anx7 protein (data not shown). Eighteen of the 20 cells analyzed by SKY revealed a hypodiploid genome (modal number of 39), and two cells were hypotetraploid (<4n = 73-76). We could detect a recurrent structural rearrangement, consisting of an insertion Is (3, 19) in 15 of 20 cells (Fig. 4A). Of 18 cells, 17 revealed monosomy for chromosome 12; and 15 of 18 cells displayed a trisomy of chromosome 19. Both numerical aberrations were also present in the near-tetraploid clone. All cells in the near-diploid clone showed a duplication of chromosome 4, Dp4 (Fig. 4B). FISH with bacterial artificial chromosomes specific for chromosome 4 allowed us to map the region of duplication between 48 Mb (band B3) and 148.99 Mb (band E2), resulting in the deletion of a region of 3 Mb toward the telomere. Additional clonal, yet less consistent changes were observed on chromosome X (loss, 11 of 20 cases), chromosome 5 (trisomy, 10 of 20 cases), and chromosome 16 (loss, 4 of 20 cases). The comparative genomic hybridization analysis therefore confirmed the SKY results. However, an additional chromosome 19 insertion Is(19A1;3) was detected. The complete karyotypes can be found at www.ncbi.nlm.nih.gov/sky/.
Fig. 4.
SKY analysis of a metaphase from a hepatocellular carcinomas in the ANX7-deficient mouse. (A) SKY analysis of a representative metaphase from hepatocellular carcinoma in the ANX7-deficient mouse. The inverted 4′,6-diamidino-2-phenylindole banding is shown next to the classification colors for the SKY analysis. The karyotype is 39, X, Is (3, 19), -4,+5, -12,+19. (B) FISH mapping hybridization with locus-specific bacterial artificial chromosomes for mouse chromosome 4 mapping at 76.6 and 20.8 cM. As shown on the left, the signal located at 76.6 cM is present in one copy, whereas the clone at 20.8 cM on the right shows two signals. This finding indicates that the tandem fusion of chromosome 4 results in an unbalanced translocation 4,4. (C) FISH hybridization on touch preparations from a liver tumor by using locus-specific probes for chromosomes 5, 19, and X (green signal).
The presence of chromosomal aberration in the original hepatocellular carcinoma was established with FISH on touch preparations from the tumor tissue by using locus-specific probes for chromosomes 5, 19, and X. Of the cells, 62% carried three copies of chromosome 19 and 82% contained only one copy of chromosome X (Fig. 4C). These results indicate that gain of chromosome 19 and loss of chromosome X are cytogenetic abnormalities not only in established cell lines but also in the primary tumor tissue. We interpret these results to support the hypothesis that hepatocellular carcinoma in the Anx7(+/-) heterozygotes could have arisen because of genomic instability.
Discussion
These data show that the Anx7 gene joins a small but select group of TSGs for whom inactivation of just one allele is sufficient to permit the formation of tumors (17). Others in this group include CBFA2 (18), Cdh1 (19), Dmp1 (20), Lkb1 (21), NF1 (22), p27kip1 (23), Ptch (24, 25), PTEN/MMAC (26), Atm (27), Blm (28), Fen1 (29), and p53 (30). Such genes have been termed haploinsufficient because the mutation or loss of one allele may be sufficient to exert a cellular phenotype that leads to tumorigenesis without inactivation of the wild-type allele. By contrast, Knudson's classical two-hit hypothesis suggests that mutation of both alleles of a TSG are needed to trigger tumor formation (15, 16). Tumors influenced by haploinsufficiency usually have a later age of onset when compared with those caused by inactivation of the second allele (31). Indeed, the haploinsufficient Anx7(+/-) heterozygous mice tend to develop observable spontaneous neoplasms at older ages in multiple organs, including the liver, prostate, endometrium, salivery gland adenocarcinoma, and lymphosarcoma of the thymus. For example, Pten/ Mmac1+/- (32) and p27kip1 (23) heterozygous mice develop tumors in multiple organs, some of which overlap with the tissue tropism of Anx7(+/-) mice.
Downstream Members of ANX7 Signaling Pathway Involved in Tumorigenesis Are Identified by cDNA Microarray Analysis. The signaling pathways regulated by the Anx7 TSG are not known. Our cDNA microarray analysis using the hepatocellular carcinoma tumor specimens and the adjacent normal tissues from the Anx7(+/-) heterozygous mice reveal that Anx7 modulates the expression of TSGs such as BRAC1, BRAC2, Wilms tumor protein, TSG101 tumor susceptibility protein, and tumor suppressor protein VHL. Other categories of affected genes include DNA-repair genes and apoptotic related genes, such as Akt and Flice. We conjecture that the altered levels of expression of these other TSGs and DNA-repair genes might indicate that ANX7 haploinsufficiency may result in defective DNA repair and increased genetic instability, leading to somatic mutations and alterations in other TSGs and oncogene expression. The multiplicity of procarcinogenic genes down-regulated by the Anx7(+/-) genotype emphasizes the possible ”gatekeeper” character of Anx7, much like many other TSGs. We have described such global gatekeeper-like properties of the Anx7(+/-) genotype for islets of Langerhans (14).
Gigantism Phenotype of the Anx7 (+/-) Mouse. Gigantism is part of the phenotype for a limited number of mouse knockout or transgenic models, some of which are for bona fide TSGs. In the Anx7(+/-) genotype, male Anx7(+/-) mice begin to grow at a greater rate than normal littermate controls by about the 4th week after birth. By contrast, female Anx7(+/-) mice do not vary from their controls. The growth phenotype of gender-dimorphic gigantism and organomegaly of the Anx7(+/-) mouse is therefore fundamentally different from that of other reported mouse knockouts of which we are aware. For example, with the exception of generalized, gender-independent organomegaly reported for the p27kip1(-/-) mouse (33, 34), the reported instances of mutation-based gigantism are mostly endocrine in origin and are due to increases of either growth hormone (35), IGF-1 (36), or IGF-2 (37). However, the levels of serum IGF-1 and growth hormone in the Anx7(+/-) mouse are within normal limits (14). One qualitative parallel between the growth kinetics of male Anx7(+/-) mice and those mice transgenic for growth hormone or IGF-1 is a postpartum delay in the onset of enhanced growth. The Anx7(+/-) male mice and mice transgenic for growth hormone begin to grow at 3-4 weeks, whereas those transgenic for IGF-1 begin to grow only after 6-8 weeks. Mice overproducing IGF-2 are heavier than control mice at birth but do not sustain the increase in weight into adulthood. Finally, pituitary gland histology in male and female Anx7(+/-) mutants cannot be distinguished from wild-type histology (data not shown). By contrast, the p27kip1(-/-) mouse (33, 34) is characterized by pituitary hyperplasia and by an increased frequency of pituitary tumors. Together, these data thus further validate the conclusion that the documented growth anomalies in the Anx7(+/-) mouse are probably not related to pituitary hyperfunction. The fact that unique growth anomalies in the Anx7(+/-) mouse are gender-specific constitutes a distinct internal genetic control for the Anx7(+/-) mouse mutation. Thus, independent of the involvement of the Anx7 gene in tumorigenesis, the gigantism phenotype of the Anx7(+/-) mutant indicates of a critical role for this gene in control of growth.
Role of anx7 Gene in Proliferative Phase of Dictyostelium. In retrospect, experimental suggestions have been made regarding the possible involvement of the anx7 gene in proliferation, particularly in the slime mold Dictyostelium discoidum. This primitive eukaryotic organism can switch from a growth phase, in which the cells are proliferating, to a differentiated phase, in which the cells form multicellular aggregates and fruiting bodies. The first anx7 gene disruption mutants in this organism were noted to have growth defects (38). More recent studies have shown that these anx7 knockout mutants lose many properties related to growth, differentiation, motility, and chemotaxis, especially in Ca2+-limiting conditions (39, 40). Bonfils et al. (40) have also shown that, compared with the differentiated form of Dictyostelium, the proliferating form possesses only 20% of anx7-mRNA and only 1.6% of anx7 protein. The mechanism of this transition involves synthesis by the organism of anx7 antisense mRNA from the complementary strand (41). In summary, the anx7 gene seems to control Dictyostelium differentiation by a mechanism in which a relative decrease in anx7 protein enhances growth and proliferation at the expense of Ca2+-dependent differentiated functions. This finding sounds very familiar in terms of the data for mouse and tumors presented in this article and human tumors (1).
Reduced Anx7 Gene Dosage Results in Genomic Instability. The genetic events that characterize hepatocellular carcinoma in Anx7(+/-) knockout mice are identified by SKY analysis. We attribute these changes to genomic instability from the low haplotype mechanism. The genomic changes accompanying the Anx7(+/-) tumor formation occurred on chromosomes 3, 4, 5, 8, 9, 12, 14, and 19. The loss of chromosome 12 and gain of chromosome 19 have been reported in other mouse models for hepatocellular carcinomas (42-45). As observed in tumors of epithelial origin in humans, SKY analysis of chromosomal aberrations in this hepatocellular mouse tumor clearly show that balanced translocations are rare and that virtually all changes result in genomic imbalances. This insight is important, because, until now, the mammalian ANX7 gene has not been considered from this point of view to play a role in tumorigenesis.
Conclusion
Taken together, these results strongly suggest that ANX7 acts as a tumor suppressor gene, not only in prostate cancer cell lines and in clinical prostate cancer, but also in the model systems of the Anx7(+/-) knockout mouse. In the mouse, haploinsufficiency of Anx7 expression appears to drive progression to cancer because of genomic instability through a discrete signaling pathway involving other tumor suppressor genes, DNA-repair genes, and apoptosis-related genes.
Acknowledgments
We thank Drs. Tom Darling and Eli Heldman for helpful discussions and other support and Ms. Ling Li for technical support. This work was supported by the Cystic Fibrosis Foundation, the Juvenile Diabetes Foundation International, and Department of Defense Grant PC 020228.
Abbreviations: ANX7, annexin 7; SKY, spectral karyotyping; FISH, fluorescence in situ hybridization; TSG, tumor suppressor gene; IGF-1, insulin-like growth factor 1.
Footnotes
Nomenclature: Anx7, protein, mouse; Anx7, nucleic acid, mouse; ANX7, protein, human; ANX7, nucleic acid, human; anx7, protein, Dictyostelium; anx7, nucleic acid, Dictyostelium.
References
- 1.Srivastava, M., Bubendorf, L., Nolan, L., Glasman, M., Leighton, X., Koivisto, P., Willi, N., Gasser, T., Kononen, J., Sauter, G., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 4575-4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lacombe, L., Orlow, I., Reuter, V. E., Fair, W. R., Dalbagni, G., Zhang, Z. F. & Cordon-Cardo, C. (1996) Int. J. Cancer 69, 110-113. [DOI] [PubMed] [Google Scholar]
- 3.Li, J., Yen, C., Liaw, D., Podsypanina, K., Bose, S., Wang, S. I., Puc, J., Miliaresis, C., Rodgers, L., McCombie, R., et al. (1997) Science 275, 1943-1947. [DOI] [PubMed] [Google Scholar]
- 4.Maier, D., Zhang, Z., Taylor, E., Hamou, M. F., Gratzl, O., Van Meir, E. G., Scott, R. J. & Merlo, A. (1998) Oncogene 16, 3331-3335. [DOI] [PubMed] [Google Scholar]
- 5.Srivastava, M., Atwater, I., Glasman, M., Leighton, X., Goping, G., Caohuy, H., Mears, D., Rojas, E., Westfal, H., Pichil, J. & Pollard, H. B. (1999) Proc. Natl. Acad. Sci. USA 96, 13783-13788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Srivastava, M., Bubendorf, L., Nolan, L., Glasman, M., Leighton, X., Miller, G., Fehrle., W., Raffeld, M., Eidelman, O., Kallioniemi, O. P., et al. (2001) Dis. Markers 17, 115-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hahn, W. C. & Weinberg, R. A. (2002) N. Engl. J. Med. 347, 1593-1603. [DOI] [PubMed] [Google Scholar]
- 8.Chirgwin, J. M., Przybyla, A. E., MacDonald, R. J. & Rutter, W. J. (1979) Biochemistry 18, 5294-5299. [DOI] [PubMed] [Google Scholar]
- 9.Srivastava, M., Eidelman, O. & Pollard, H. B. (2000) Mol. Med. 5, 753-767. [PMC free article] [PubMed] [Google Scholar]
- 10.Liyanage, M., Coleman, A., du Manoir, S., Veldman, T., McCormack, S., Dickson, R. B., Barlow, C., Wynshaw-Boris, A., Janz, S., Wienberg, J., et al. (1996) Nat. Genet. 14, 312-315. [DOI] [PubMed] [Google Scholar]
- 11.Weaver, Z. A., McCormack, S. J., Liyanage, M., du Manoir, S., Coleman, A., Schrock, E., Dickson, R. B. & Ried, T. (1999) Genes Chromosomes Cancer 25, 251-260. [PubMed] [Google Scholar]
- 12.Davisson, M. T. (1996) in Genetic Variants and Strains of the Laboratory Mouse, eds. Lyon, M. F., Rastan, S. & Brown, S. D. M. (Oxford Univ. Press, New York) 3rd Ed., pp. 1443-1445.
- 13.Montagna, C., Andrechek, E. R., Padilla-Nash, H., Muller, W. J. & Ried, T. (2002) Oncogene 21, 890-898. [DOI] [PubMed] [Google Scholar]
- 14.Srivastava, M., Eidelman, O., Leighton, X., Glassman, M., Goping, G. & Pollard, H. B. (2002) Mol. Med. 8, 781-797. [PMC free article] [PubMed] [Google Scholar]
- 15.Knudson, A. G. (1971) Proc. Natl. Acad. Sci. USA 68, 820-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knudson, A. G. (1993) Proc. Natl. Acad. Sci. USA 90, 10914-10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balmin, A. (2002) Nature 417, 235-236.12015584 [Google Scholar]
- 18.Song, W. J., Sullivan, M. G., Legare, R. D., Hutchings, S., Tan, X., Kufrin, D., Ratajczak, J., Resende, I. C., Haworth, C., Hock, R., et al. (1999) Nat. Genet. 23, 166-175. [DOI] [PubMed] [Google Scholar]
- 19.Smits, R., Ruiz, P., Diaz-Cano, S., Luz, A., Jagmohan-Changur, S., Breukel, C., Birchmeier, C., Birchmeier, W. & Fodde, R. (2000) Gastroenterology 119, 1045-1053. [DOI] [PubMed] [Google Scholar]
- 20.Inoue, K., Zindy, F., Randle, D. H., Rehg, J. E. & Sherr, C. J. (2001) Genes Dev. 15, 2934-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyoshi, H., Nakau, M., Ishikawa, T. O., Seldin, M. F., Oshima, M. & Taketo, M. M. (2002) Cancer Res. 62, 2261-2266. [PubMed] [Google Scholar]
- 22.Gutmann, D. H., Loehr, A., Zhang, Y., Kim, J., Henkemeyer, M. & Cashen, A. (1999) Oncogene 18, 4450-4459. [DOI] [PubMed] [Google Scholar]
- 23.Fero, M. L., Randel, E., Gurley, K. E., Roberts, J. M. & Kemp, C. J. (1998) Nature 396, 177-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wetmore, C., Eberhart, D. E. & Curran, T. (2000) Cancer Res. 60, 2239-2246. [PubMed] [Google Scholar]
- 25.Zurawel, R. H., Allen, C., Wechsler-Reya, R., Scott, M. P. & Raffel, C. (2000) Genes Chromosomes Cancer 28, 77-81. [PubMed] [Google Scholar]
- 26.Kwabi-Addo, B., Giri, D., Schmidt, K., Podsypanina, K., Parsons, R., Greenberg, N. & Ittmann, M. (2001) Proc. Natl. Acad. Sci. USA 98, 11563-11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spring, K., Ahangari, F., Scot, T. S. P., Waring, P., Purdie, D. M., Chen, P. C., Hourigan, K., Ramsay, J., McKinnon, P. J., Swift, M. & Lavin, M. F. (2002) Nat. Genet. 32, 185-190. [DOI] [PubMed] [Google Scholar]
- 28.Goss, K. H., Risinger, M. A., Kordich, J. J., Sanz, M. M., Straughen, J. E., Slovek, L. E., Capobianco, A. J., German, J., Boivin, G. P. & Groden, J. (2002) Science 297, 2051-2053. [DOI] [PubMed] [Google Scholar]
- 29.Kucherlapati, M., Yang, K., Kuraguchi, M., Zhao, J., Lia, M., Heyer, J., Kane, M. F., Fan, K., Russell, R., Brown, A. M., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 9924-9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venkatachalam, S., Shi, Y. P., Jones, S. N., Vogel, H., Bradley, A., Pinkel, D. & Donehower, L. A. (1998) EMBO J. 17, 4657-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fodde, R. & Smits, R. (2002) Science 298, 761-763. [DOI] [PubMed] [Google Scholar]
- 32.Podsypanina, K., Ellenson, L. H., Nemes, A., Gu, J., Tamura, M., Yamada, K. M., Cordon-Cardo, C., Catoretti, G., Fisher, P. E. & Parsons, R. (1999) Proc. Natl. Acad. Sci. USA 96, 1563-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fero, M. L., Rivkin, M., Tasch, M., Porter, P., Carow, C. E., Firpo, E., Polyak, K., Tsai, L. H., Broudy, V., Perlmutter, R. M., et al. (1996) Cell 85, 733-744. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama, K., Ishida, N., Shirane, M., Inomata, A., Inoue, T., Shishido, N., Horii, I., Loh, D. Y. & Nakayama, K. (1996) Cell 85, 707-720. [DOI] [PubMed] [Google Scholar]
- 35.Palmiter, R., Brinster, R., Hammer, R., Trumbauer, M., Rosenfeld, M., Birnberg, N. & Evans, R. (1982) Nature 300, 611-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quaife, C., Mathews, L., Pinkert, C., Hanner, R., Brinster, R. & Palmiter, R. (1989) Endocrinology 114, 40-48. [DOI] [PubMed] [Google Scholar]
- 37.Wolf, E., Kramer, R., Blum, W., Foll, J. & Brem, G. (1994) Endocrinology 135, 1877-1886. [DOI] [PubMed] [Google Scholar]
- 38.Doring, V., Schleicher, M. & Noegel, A. A. (1991) J. Biol. Chem. 266, 17509-17515. [PubMed] [Google Scholar]
- 39.Doring, V., Veretout, F., Albrecht, R., Muhlbauer, B., Schlatterer, C., Schleicher, M. & Noegel, A. A. (1995) J. Cell Sci. 108, 2065-2076. [DOI] [PubMed] [Google Scholar]
- 40.Bonfils, C., Greenwood, M. & Tsang, A. (1994) Mol. Cell. Biochem. 139, 159-166. [DOI] [PubMed] [Google Scholar]
- 41.Okafuji, T., Abe, F. & Maeda, Y. (1997) Gene 189, 49-56. [DOI] [PubMed] [Google Scholar]
- 42.Danielsen, H. E., Brogger, A. & Reith, A. (1991) Carcinogenesis 10, 1777-1780. [DOI] [PubMed] [Google Scholar]
- 43.Dragani, T. A., Manenti, G., Gariboldi, M., De Gregorio, L. & Pierotti, M. A. (1995) Toxicol. Lett. 82, 613-619. [DOI] [PubMed] [Google Scholar]
- 44.Sargent, L. M., Zhou, X., Keck, C. L., Sanderson, N. D., Zimonjic, D. B., Popescu, N. C. & Thorgeirsson, S. S. (1999) Am. J. Pathol. 154, 1047-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zimonjic, D. B., Keck, C. L., Thorgeirsson, S. S. & Popescu, N. C. (1999) Hepatology 29, 1208-1214. [DOI] [PubMed] [Google Scholar]