Abstract
During sexual development the human fungal pathogen Cryptococcus neoformans undergoes a developmental transition from yeast-form growth to filamentous growth. This transition requires cellular restructuring to form a filamentous dikaryon. Dikaryotic growth also requires tightly controlled nuclear migration to ensure faithful replication and dissemination of genetic material to spore progeny. Although the gross morphological changes that take place during dikaryotic growth are largely known, the molecular underpinnings that control this process are uncharacterized. Here we identify and characterize a C. neoformans homolog of the Saccharomyces cerevisiae BIM1 gene, and establish the importance of BIM1 for proper filamentous growth of C. neoformans. Deletion of BIM1 leads to truncated sexual development filaments, a severe defect in diploid formation, and a block in monokaryotic fruiting. Our findings lead to a model consistent with a critical role for BIM1 in both filament integrity and nuclear congression that is mediated through the microtubule cytoskeleton.
Keywords: Cryptococcus neoformans, BIM1, filamentation, microtubule binding, nuclear migration, sexual development, congression, dikaryon
Introduction
Proper nuclear migration is critical for the faithful replication and propagation of genetic material in all eukaryotic cells (1). The importance of this process is underscored by the fact that cells in which nuclear migration is defective cannot survive, and many human disorders have been linked to ineffective nuclear migration, including various forms of cancer and neurodegenerative disorders (2-4). The molecular events that control nuclear migration have been most fully characterized in ascomycete fungi, including the budding yeast Saccharomyces cerevisiae and the filamentous fungus Aspergillus nidulans (5). In S. cerevisiae, screens to identify mutants with defects in nuclear migration resulted in the discovery of microtubule motor proteins and their associated binding factors. (6). Screens in A. nidulans to identify mutants with defects in nuclear distribution (NUD) also resulted in the discovery of microtubule structural components and microtubule-associated proteins (7). The results of both studies indicate a strong dependence of nuclear migration on the microtubule cytoskeleton and related proteins in both bud-form and filamentous growth in ascomycete fungi.
In other fungi, however, very little is known about how more complex nuclear movements are coordinated. In basidiomycete fungi, a process takes place during sexual development in which filaments are formed that contain two (and only two) nuclei of distinct mating types. While the significance of maintaining these dikaryons is unclear in terms of basidiomycete development, basidiomycetes provide a unique system in which to study coordinated nuclear migration in the context of eukaryotic development. One basidiomycete in which the dikaryotic phase can be manipulated in the laboratory is the human fungal pathogen Cryptococcus neoformans. C. neoformans causes meningoencephalitis in immunocompromised individuals, and studies of its pathogenic properties have led to the development of many genetic and molecular tools in the system (8). In particular, the entire process of sexual development from mate recognition through spore production can be followed in vitro. Sexual development in C. neoformans can be defined by five general morphological stages (9, 10). The first stage begins when haploid budding yeast (a or α) recognize a mating partner and fuse. Although the cells fuse after mate recognition, the nuclei of each previously haploid cell remain distinct. Upon cell fusion, a new developmental pathway is initiated that establishes filamentous growth. The resulting dikaryotic filaments are characterized by the maintenance and propagation of two independent haploid nuclei. This stage continues until unknown signals trigger the terminal filament cell to form a basidium, in which the two haploid nuclei fuse and undergo meiosis. The resulting meiotic products are then replicated and packaged into spores that bud in four chains on the surface of the basidium. Although these stages have been identified morphologically, the molecular mechanisms that govern the process of sexual development are not well defined, and virtually nothing is known about how the nuclei in dikaryotic filaments are managed during growth.
To begin to understand the molecular underpinnings of nuclear migration in basidiomycetes, we compared predicted protein sequences of C. neoformans with sequences of proteins known to be important for nuclear migration in S. cerevisiae and A. nidulans to identify potential homologs. Here, we present the identification and characterization of Bim1, a likely homolog of the S. cerevisiae Bim1 that was originally identified in a screen for microtubule binding proteins. In S. cerevisiae, Bim1 is required for proper nuclear movement, and deletion strains exhibit severe defects in nuclear congression during sexual development (11). To assess the role of Bim1 in C. neoformans, we constructed deletion strains and tested them for detectable phenotypes in vivo. We found that Bim1 is required for proper dikaryotic filament formation during sexual development and appears to participate in nuclear congression. Our data are consistent with a model in which Bim1 in C. neoformans functions to control microtubule stability to provide intracellular scaffolds needed for the structural integrity of filament cells and for efficient nuclear congression.
Materials and Methods
Sequence analysis
Sequence comparisons were performed using the BLASTp algorithm (12) and C. neoformans genome sequence (C. neoformans Genome Project, Stanford Genome Technology Center (http://www-sequence.stanford.edu/group/C.neoformans/). The Cn BIM1 sequence can be obtained using Genbank accession number CNB04790. The Cn KAR3 sequence can be obtained using Genbank accession number CNF02260. Protein motif comparisons were analyzed using the Pfam algorithm (http://pfam.sanger.ac.uk/).
Plasmid and strain construction
bim1Δ strains were constructed by replacing the BIM1 ORF with either a nourseothricin (NAT) or neomycin (NEO) resistance gene cassette. Knockout cassettes were made by overlap PCR using the oligos listed in Table S1 as described previously (13). The amplified bim1Δ∷NATR and bim1Δ∷NEOR deletion cassettes were introduced into the serotype D strains JEC20 (a) and JEC21 (α) using biolistic transformation (14). Positive clones were identified by PCR and confirmed by Southern blot analysis. Multiple independent deletion strains were created in both mating types (a and α) with both markers (NAT and NEO). Deletion strains were backcrossed by wild type mating partners, and mutant progeny were isolated. All experiments were conducted with at least three independent deletion strains. kar3Δ∷NATR strains were constructed by overlap PCR, and knockout strains were confirmed as described above. kar3Δ∷NATR strains were crossed with bim1Δ∷NEOR strains, and spores were isolated and grown on double selective media to recover kar3Δ∷NATR bim1Δ∷NEOR strains. Multiple independent a and α kar3Δ∷NATR bim1Δ∷NEOR strains were recovered and evaluated. Southern blot analysis: Genomic DNA was isolated from strains as described previously (15). Southern analysis was carried out according to (16). Briefly, 10 μg of genomic DNA for each sample were digested with either KpnI or XhoI and electrophoresed on agarose gels and transferred to Amersham Hybond N+ membrane blots. Radiolabeled DNA probes to the 5′ flanking regions of each construct were amplified from genomic DNA and labeled with 32P using a GE Amersham RediPrime II kit according to the manufacturer's protocol. Blots were hybridized at 65°C in Church hybridization buffer and washed in 0.5X SSC prior to autoradiography as described previously (17). Construction of the GFP-α-tubulin fusion was carried out by isolating 1 kb upstream sequence and the entire coding sequence of α-tubulin (Gene ID: CNB02810) via PCR using primers CHO2904/CHO2905 and CHO2906/CHO2907. PCR fragments were digested with BamHI, ligated and re-amplified to obtain a single fragment. The fragment was then cloned into pCR4blunt-TOPO (Invitrogen) and the sequence of the insert was confirmed (pCH1005). A GFP fragment (18) was amplified with primers CHO2908/CHO2909, digested with BamHI and cloned into the BamHI site of pCH1005 to create pCH1006. pCH1006 was then digested with EcoRI to release the GFP-α-tubulin construct with its native promoter, and the resulting fragment was cloned into pJAF7 harboring a URA5 gene to create pCH935. Strains expressing GFP-α-tubulin were created by transforming pCH935 into ura5 strains. Single GFP-α-tubulin integration events were confirmed by Southern blot analysis. Genomic DNA was digested with EcoRI and blots were probed with radiolabeled DNA probes to the URA5 ORF. The resulting strain, CHY2271, was crossed with the bim1Δ strain CHY1753, and bim1Δ progeny with a singly integrated reporter construct in the same genomic position as in the wild-type reporter strain were isolated (CHY2277).
Sexual development Assays
All strains were serotype D. Strain genotypes are given in Table 1. All strains were handled using standard techniques and media as described previously (19, 20). Sexual development assays were conducted on V8 medium at room temperature in the dark for 2-4 days and evaluated by observing the periphery of test spots. Fusion assays were carried out by mixing equal numbers of log phase mating partners on V8 agar plates and incubating at room temperature. After 24 hours the cells were plated under selection (200 μg/ml G418 and 200 μg/ml of nourseothricin) and incubated at 30°C for 2 days. Diploid analysis: Diploids were selected using an identical protocol except that selection was carried out at 37°C. Diploid cells were confirmed by DAPI staining and light microscopy to contain a single nucleus. Spore isolation and analysis: Spores were isolated as described previously with minor modifications (21). Briefly, crosses between differentially marked strains were incubated on V8 medium at room temperature in the dark for 4 days, resuspended in 65% Percoll (Fisher)/1% Triton/1X PBS, and centrifuged at 9,000 rpm for 20 minutes in a SW 40Ti Beckman-Coulter ultracentrifuge rotor. Spores were harvested from the bottom of the gradient and grown on YPD plates for 2 days at 30°C. Resulting colonies were grown on YPD+NAT and YPD+NEO media at 30°C and scored for resistance. Mating type was determined by crossing with a known a mating partner. Mating type information was collated with drug resistance to determine reassortment frequencies among markers in progeny from all crosses.
Table 1.
Strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| JEC20 | a | Kwon-Chung et al. 1992 |
| JEC21 | α | Kwon-Chung et al. 1992 |
| CHY1345 – 1346 | a NATR | this study |
| CHY1348 – 1349 | α NEOR | this study |
| CHY1750 – 1752 | a bim1Δ∷NEOR | this study |
| CHY1753 – 1755 | α bim1Δ∷NEOR | this study |
| CHY1744 – 1746 | a bim1Δ∷NATR | this study |
| CHY1747 – 1749 | α bim1Δ∷NATR | this study |
| CHY1675 – 1676 | a kar3Δ∷NATR | this study |
| CHY1679 – 1680 | α kar3Δ∷NATR | this study |
| CHY1859, 1861, 1863 | a bim1Δ∷NEOR kar3Δ∷NATR | this study |
| CHY1860, 1862, 1864 | α bim1Δ∷NEOR kar3Δ∷NEOR | this study |
| CHY1865 – 1867 | a bim1Δ∷NATR/α bim1Δ∷NEOR | this study |
| CHY1872 – 1873 | a bim1Δ∷NATR/α NEOR | this study |
| CHY1874 – 1875 | a NATR/α bim1Δ∷NEOR | this study |
| CHY1993, 1994 | a NATR/α NEOR | this study |
| CHY2271 – 2273 | a ura5 + GFP-α-tubulin-URA5 | this study |
| CHY1987 – 1989 & CHY2274 – 2276 |
α ura5 + GFP-α-tubulin-URA5 | this study |
| CHY1990 – 1992 & CHY2277 – 2279 |
α bim1Δ∷NEOR 5-FOAR + GFP-α-tubulin-URA5 | this study |
Phenotypic analyses
Growth Assay: Wild type and bim1Δ strains were grown in liquid culture to mid-log phase. Cell concentrations were equalized based on optical density, and 10-fold serial dilutions were made from a starting pool of ~5×105 cells. 5 μL of each serial dilution were spotted on YPD plates with or without 2.5 μg/mL benomyl. Growth at 15, 25, 30, 37, and 42°C were analyzed after 48 hours. Similar growth assays were used to analyze the effects of taxol (1.25 – 5 ug/mL). Capsule production was tested by growing strains in RPMI cell culture medium and staining with India ink to reveal capsule under light microscopy (22). Melanin production was assayed by growth on plates containing L-dopa and assessing pigment production macroscopically (23). Microscopy and staining: Light microscopy/photography was performed using a Zeiss Axioskop 2 microscope fitted with long working distance objectives and a Nikon Coolpix 5400 camera. Fluorescence microscopy was carried out using a Zeiss Axioskop 2 fluorescence microscope fitted with a MRM REV3 digital camera and corresponding AV4 software. For filament staining, crosses were incubated on a thin layer of V8 media on a microscope slide and incubated at room temperature for 24 hours. Cells were fixed with 2.5% gluteraldehyde/4% paraformaldehyde for 30 minutes, washed, and stained with 0.4 μg/mL calcofluor white and 1nM Sytox green for 20 minutes. Cells were mounted in 5% glycerol with 20% DABCO anti-fade (Sigma). For microtubule imaging, yeast were mounted on agarose pads and imaged on a swept field confocal (Nikon Eclipse Ti-E) microscope equipped with a Nikon 60X, 1.4 NA Planapo oil objective lens and a Roper Scientific CoolSNAP HQ2 CCD camera.
Results
Identification and deletion of Cn BIM1
To identify potential homologs of the S. cerevisiae Bim1 protein in C. neoformans, we analyzed the C. neoformans serotype D genome using both BLASTp and PFAM domain analyses. A significant sequence homolog of the Sc BIM1 gene was identified. This previously uncharacterized C. neoformans open reading frame (NCBI GeneID: CNB04790) contains 6 introns and 7 exons, and is predicted to encode a 253 amino acid protein. The predicted protein contains a Calponin Homology (CH) domain that is thought to bind actin, and an EB-1-like domain that is predicted to bind to microtubules. Overall, Cn Bim1 shares 37% identity and 49% similarity with Sc Bim1, with 77% and 56% similarity between the CH and the EB1 core domains, respectively (Figure S1). Based on sequence conservation and domain architecture we refer to this previously unidentified C. neoformans ORF as BIM1.
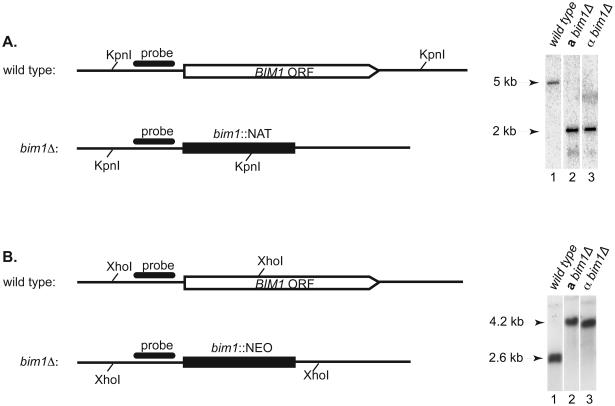
To assess the function of Bim1 in C. neoformans, strains were created in which the entire BIM1 ORF was deleted in both the a and the α mating types of a congenic strain pair (JEC20 and JEC21). The BIM1 ORF was replaced with two different dominant selectable markers (nourseothricin or neomycin resistance genes) to create the deletion strains. Multiple independent strains were identified using PCR genotyping and confirmed by Southern blot analysis (Figure 1). Northern analysis confirmed that no BIM1 transcript was present in deletion strains (Figure S2).
Figure 1. Deletion of the C. neoformans BIM1 ORF.
A. bim1Δ∷NAT strains. Schematic of BIM1 locus before and after deletion with the NAT construct are shown with KpnI restriction sites. Southern blot reveals a 5 kb wild type band in lane 1, and a 2 kb deletion band in both a and α strains in lanes 2 and 3 respectively. B. bim1Δ∷NEO strains. Schematic of BIM1 locus before and after deletion with the NEO construct are shown with XhoI restriction sites. Southern blot reveals a 4.2 kb wild type band in lane 1, and a 2.6 kb deletion band in both a and α strains in lanes 2 and 3 respectively. Representatives of multiple, independent deletion strains are shown.
bim1Δ strains exhibit phenotypes consistent with defects in microtubule stability
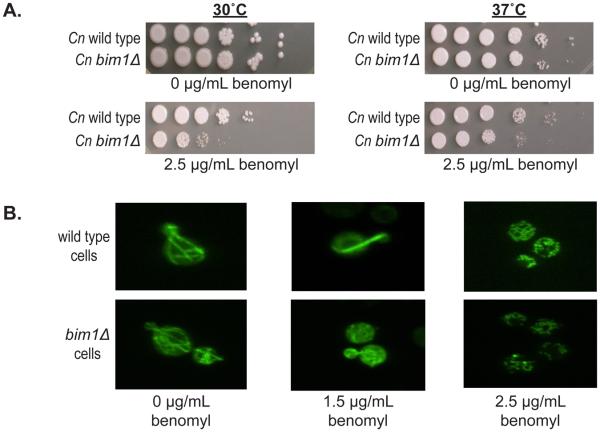
Sc BIM1 was originally identified in a screen for proteins that bind microtubules. Haploid Sc bim1Δ yeast cells were shown to be sensitive to traditional microtubule stressors such as growth at elevated temperature (37°C) and growth in the presence of the microtubule inhibitor benomyl (11). To characterize the phenotypes of Cn bim1Δ strains, cells were grown in the presence of benomyl and at 30°C and 37°C. Given the high level of conservation between the Sc and Cn Bim1 proteins, we predicted that Cn bim1Δ strains would exhibit defects similar to those seen in Sc bim1Δ strains. Indeed, Cn bim1Δ strains were more sensitive than wild type strains to 2.5 μg/mL benomyl (Figure 2A). Unlike what was observed in Sc bim1Δ strains, however, Cn bim1Δ strains showed no sensitivity to growth at 37°C relative to wild type strains, and showed no increased sensitivity to the combined stressors of growth at 37°C in the presence of benomyl (Figure 2A). Growth assays performed at other temperatures (14°C, 25°C, and 42°C) did not show differences between wild type and mutant strains (data not shown), consistent with the observations made at 30°C and 37°C. In each assay, Sc bim1Δ strains were used as controls and exhibited phenotypes reported previously by Schwartz, et al. (data not shown).
Figure 2. Phenotypes of bim1Δ strains.
A. bim1Δ haploid strains are sensitive to the microtubule inhibitor benomyl. Serial 10-fold dilutions of cells were spotted from left to right with an initial concentration of ~5 × 10−5 log phase cells on YPD plates in the presence and absence of 2.5 μg/mL of benomyl. Plates were incubated at 30°C and 37°C for 2 days. B. Microtubules are less stable in bim1Δ strains. Confocal fluorescence microscopy of wild type and bim1Δ strains expressing a GFP-α-tubulin construct in the presence of 0, 1.5, and 2.5 μg/mL of benomyl at 600X magnification.
Because Bim1 has been shown to bind microtubules and to affect microtubule stability in S. cerevisiae, we examined microtubule architecture in C. neoformans bim1Δ strains. To visualize microtubules, a GFP-α-tubulin fusion gene under the control of the α-tubulin promoter was transformed into wild type and bim1Δ strains. Multiple random integrants were recovered and evaluated for the ability to form intact microtubules. GFP-expressing strains were also constructed by crossing a wild type strain containing a single integrated cope of GFP-α-tubulin with a bim1Δ strain and recovering GFP-α-tubulin expressing bim1Δ progeny. Fluorescence microcopy revealed no differences in microtubule structure between wild type and bim1Δ strains in yeast cells grown at 30°C. However, in the presence of 1.5 μg/mL of benomyl, microtubules in the bim1Δ strains were visibly disrupted. That is, in the wild type strains, long microtubules transversing the cells were observed; whereas in bim1Δ strains, only punctate spots were detected. By 2.5 μg/mL of benomyl, both wild type and bim1Δ strains exhibited disrupted microtubules (Figure 2B). These data indicate that bim1Δ strains have a reduced ability to maintain proper microtubule architecture.
To test the possibility that Bim1 might play a in role C. neoformans pathways distinct from those in S. cerevisiae, we assessed capsule formation, melanin formation, and sexual development. Analyses of these processes revealed no defects in capsule or melanin formation (data not shown), but did reveal abnormal filament formation during sexual development.
bim1Δ strains produce abnormal filaments during sexual development
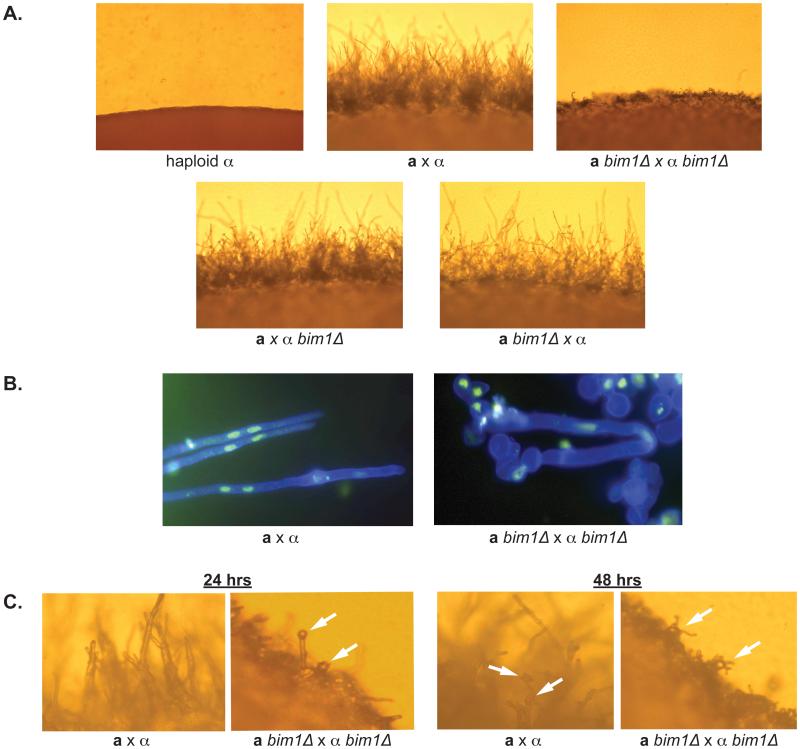
In sexual development assays, crosses between wild type and bim1Δ strains were incubated on V8 agar at room temperature for 2 days and assessed for filament formation. Crosses between a bim1Δ and α bim1Δ strains resulted in abnormally short filaments during sexual development as compared to wild type a and α strains (Figure 3A). Filaments in bim1Δ crosses did not extend beyond the cell mass and appeared to collapse upon themselves, creating a dense mat of filaments at the periphery of the cross. Crosses between wild type a and α bim1Δ or between a bim1Δ and wild type α strains resulted in no visible differences from wild type a by α crosses (Figure 3A), suggesting that Bim1 acts primarily after cell fusion and that a single copy of BIM1 is sufficient for wild type sexual development.
Figure 3.
A. bim1Δ strains show defects during sexual development. Panels show the edges of crosses after 48 hours on V8 medium at 200X magnification. B. Panels show filaments from wild type and mutant crosses stained with calcofluor white (cell wall stain - blue) and Sytox green (nuclear stain) at 1000X magnification. C. Basidia and spore formation occur earlier in bim1Δ strains. Left hand panels show the edge of wild type and bim1Δ crosses taken at 400X magnification after 24 hrs on V8 agar. Right hand panels show the same crosses after 48 hours on V8 agar. White arrows point to basidia and spores.
bim1Δ strains produce abnormal filaments during sexual development
Given the overall defect in sexual development observed in bim1Δ by bim1Δ crosses, experiments were conducted to more specifically address where in this process Bim1 acts. To confirm that Bim1 acts after cell fusion, we carried out fusion assays and compared the rates of cell fusion between wild type strains and bim1Δ strains. Differentially marked strains were mixed in equal numbers, incubated for 24 hours on V8 mating medium, and plated to double selective medium that would allow colony growth only if both markers were present. No significant differences in the rates of fusion were observed between wild type a by α crosses, bim1Δ by bim1Δ crosses, or crosses between bim1Δ strains and wild type mating partners (data not shown).
To determine if Bim1 is required for the formation of dikaryons, filaments from wild type and bim1Δ mutant crosses were fluorescently stained with calcofluor white (cell wall stain) and Sytox green (nuclear stain), and were examined by fluorescence microscopy (Figure 3B). As expected, wild type filaments appeared as straight, elongated filaments, with two nuclei characteristically positioned at the midline of each cell. Filaments from bim1Δ by bim1Δ crosses, however, were curved, folded back upon one another, and seemed to lack the structural integrity present in wild type filaments. In addition, the nuclei in bim1Δ filament cells were not consistently positioned at the midline of each cell.
Later stages of sexual development, including basidium formation and spore production were also examined. Bright field observations of basidia and spores from bim1Δ by bim1Δ crosses were indistinguishable from wild type crosses. Both wild type and deletion crosses formed robust basidia with long spore chains. It was noted, however, that basidia and spores were observed much earlier in bim1Δ by bim1Δ crosses than in wild type crosses (24 hours vs. 48 hours or later) (Figure 3C).
To assess the viability of spores from bim1Δ mutant strains, spores from bim1Δ by bim1Δ crosses were isolated and confirmed to germinate at wild type frequencies, demonstrating that bim1Δ strains produce viable spores. In addition, marker analysis of progeny from bim1Δ by bim1Δ crosses revealed a Mendelian distribution of markers, as was also the case for wild type a by α crosses and bim1Δ strains crossed with wild type mating partners (data not shown). While basidia and spores appeared to form at near wild type levels, it was noted that bim1Δ strains formed these structures earlier during development than wild type crosses. Overall, this assessment of each stage of sexual development reveals that Bim1 appears to act primarily after cell fusion and plays a role in the formation of filaments, suggesting that poor filament integrity may lead to premature development of later sexual stages.
bim1Δ strains show defects in diploid formation and diploid filamentation
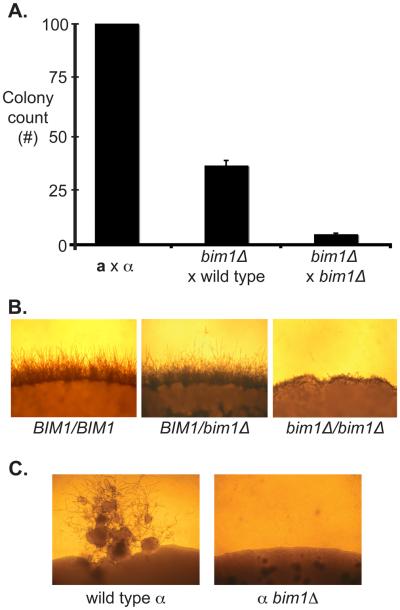
S. cerevisiae bim1Δ mutants have a severe defect in nuclear congression during mating, so we anticipated that C. neoformans bim1Δ strains might encounter similar difficulties when forming diploids (11). C. neoformans can be induced to form stable diploid cells by crossing differentially marked a and α cells on V8 at room temperature for 24 hrs and then outgrowing them under double selection at 37°C (24). This process requires the movement of nuclei toward one another (congression) and nuclear fusion to create mononucleate, bud-form diploid cells. To assess diploid formation in C. neoformans, we crossed differentially marked strains and compared the number of colonies that grew after selection between wild type and mutant strains. Compared to wild type diploid formation from an a × α cross, Cn bim1Δ strains displayed a 90% reduction in the ability to form diploid colonies from a bim1Δ × α bim1Δ crosses, and a 60% reduction in diploid formation from crosses between bim1Δ strains and wild type mating partners (Figure 4A). This result shows that Bim1 plays an important role in diploid formation, likely by facilitating nuclear congression.
Figure 4.
A. bim1Δ crosses show reduced diploid formation. Equal numbers of each mating type were crossed and selected for diploids. Colony counts are graphed as a percentage of wild type colony number. X-axis represents the crosses that were carried out. Y-axis represents normalized number of colonies recovered. B. bim1Δ diploids show defects in filament formation. The edges of wild type and mutant diploid colonies are shown after growth under sexual development conditions. 100X magnification. C. bim1Δ strains are unable to undergo monokaryotic fruiting. Panels show monokaryotic fruiting from a wild type α haploid (left) and a bim1Δ α haploid (right) grown on filament agar for 2 weeks.
Another feature of diploid cells of C. neoformans is that when they are placed under sexual development conditions, they form mononucleate filaments, basidia, and spores (24). When bim1Δ/bim1Δ diploids were grown at 25°C on V8 agar, the observed phenotype was similar to that seen in crosses between bim1Δ haploid strains: the filaments were short and lacked structural integrity in contrast to the long filaments seen with wild type BIM1/BIM1 strains (Figure 4B). This result indicates that the bim1Δ filament phenotype observed during sexual development is independent of the number of nuclei present within a filament cell. This suggests that Bim1 plays a role in the structural integrity of filaments.
bim1Δ strains show defects in α fruiting
Another form of sexual development in C. neoformans is monokaryotic fruiting in which α haploid yeast cells produce filaments and spores in response to severe nutrient limitation and desiccation. Monokaryotic fruiting is a rare event, occurs only in α cells, and appears to be the result of α-α mating or endoreplication (25, 26). To assess the effect of Bim1 on α fruiting, we placed wild type and bim1Δ strains on filament agar at 25°C for 10 days. We observed that although wild type α strains formed characteristic filaments, basidia, and spores, α bim1Δ strains were not able to undergo monokaryotic fruiting (Figure 4C). One possibility for this defect is that mononucleate bim1Δ strains cannot form filaments; however, mononucleate diploid bim1Δ/bim1Δ strains form filaments under sexual development conditions, suggesting that ploidy is not a general factor in the ability of bim1Δ strains to form filaments. Another possibility is that defects in α fruiting are due to defects in nuclear congression during the very early stages of fruiting. In this scenario a diploid nucleus must be formed prior to α filamentation, and this event is inefficient in bim1Δ strains (consistent with decreased diploid formation) such that no filaments are observed.
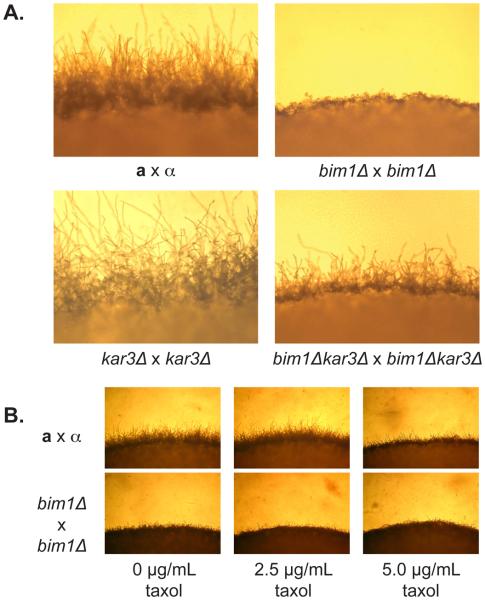
Deletion of Cn KAR3 partially suppresses the bim1Δ filament phenotype
Studies of nuclear congression in many fungi have identified an essential role for microtubules. In S. cerevisiae the loss of BIM1 causes shortened microtubules in vivo. Another microtubule-interacting protein, Kar3, acts to lengthen microtubules. Deletion of KAR3 in a bim1Δ background was shown to produce microtubules of intermediate length and to largely suppress bim1Δ phenotypes (27). To assess the role of KAR3 in C. neoformans, we identified a KAR3 sequence homolog in C. neoformans (NCBI GeneID: CNF02260), constructed kar3Δ and bim1Δkar3Δ knockout strains, and carried out sexual development assays. kar3Δ by kar3Δ crosses did not appear different than wild type crosses; however, bim1Δkar3Δ by bim1Δkar3Δ crosses developed filaments shorter than wild type filaments but longer than bim1Δ filaments (Figure 5A). This partial rescue of the bim1Δ filament phenotype is consistent with the observations in S. cerevisiae and supports the hypothesis that Bim1 acts on the microtubule cytoskeleton in C. neoformans. Crosses between bim1Δ strains were also carried out in the presence of the microtubule stabilizer taxol to determine whether chemical stabilization of microtubules could also restore full-length filaments in bim1Δ strains. While taxol had no apparent effect on the filaments in bim1Δ crosses, the presence of 5 μg/mL taxol caused short filaments in wild type crosses (Figure 5B). Taxol treatment creates hyperstable microtubules that cannot undergo normal growth/shrinkage (dynamic instability). Because taxol did not alleviate filament defects in bim1Δ crosses, it appears that microtubule dynamic instability, and not simply microtubule length, is vital to proper filament growth.
Figure 5.
A. Deletion of KAR3 partially suppresses the bim1Δ filament phenotype. Top panels show the edges of wild type and bim1Δ crosses, and bottom panels show the edges of kar3Δ and bim1Δkar3Δ crosses. All strains were photographed after 48 hours at room temperature in the dark on V8 under 200X magnification. B. Taxol does not suppress the truncated filament phenotype of bim1Δ strains, but wild type crosses show reduced filament length at high taxol concentrations. Light microscopy of wild type a by α and bim1Δ by bim1Δ crosses grown in 0, 2.5 and 5.0 μg/mL taxol. Pictures were taken at 100X magnification after 48 hours on V8 medium containing taxol.
Discussion
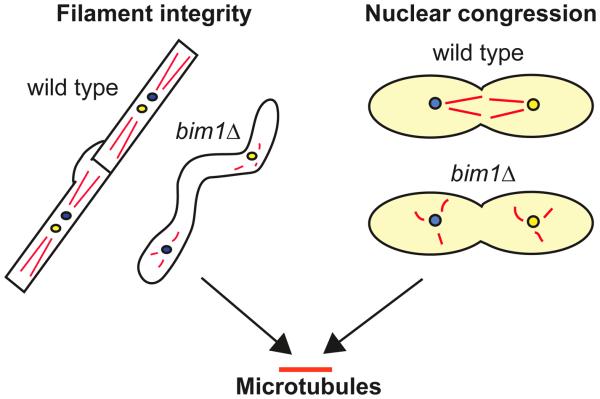
In this study we identified a previously uncharacterized C. neoformans BIM1 gene and showed that Bim1 is important for filamentous growth during sexual development, diploid formation and filamentation, and monokaryotic fruiting of C. neoformans. We also found that bim1Δ strains are sensitive to the microtubule inhibitor benomyl and that BIM1 interacts genetically with KAR3. From these data we have developed a model in which Bim1 is required for two distinct but related functions: maintaining the structural integrity of filaments and facilitating nuclear congression, both of which require proper microtubule dynamics (Figure 6).
Figure 6.
Model for the role of BIM1 in C. neoformans. Phenotypic analyses of bim1Δ strains revealed that the loss of Bim1 manifests itself in two primary ways: a loss of filament integrity, and a likely disruption of nuclear congression. Both of these phenotypes are consistent with Bim1 acting through the microtubule cytoskeleton. Left hand schematic represents sexual development filaments in wild type and bim1Δ cells. Right hand schematic shows yeast cells in wild type and bim1 crosses during diploid formation. Red lines represent microtubules. Yellow and blue circles represent a and α nuclei.
In the first part of this model, filament formation is dependent on microtubules for polar growth. The absence of directed filament growth in bim1Δ strains results from microtubule instability caused by the absence of Bim1. This part of the model is supported by our findings that sexual development filaments and those formed during diploid filamentation are abnormally short and appear relatively unstructured. Support for the role of microtubules in this process comes from findings in Schizosaccharomyces pombe in which the Bim1 homolog Mal3 has been shown to bind microtubules and suppress microtubule catastrophe (28). Deleting MAL3 shifts microtubule dynamics, causing shorter microtubule bundles that result in bent or “T” shaped cells. This phenotype in S. pombe is quite similar to the filament formation defects observed in C. neoformans bim1Δ crosses. The intermediate filament length observed in bim1Δkar3Δ double deletion stains also supports the importance of microtubule dynamics. The loss of Kar3 likely compensates for the loss of Bim1 by partially restoring the balance of microtubule dynamics. The importance of microtubule dynamics is further emphasized by the inability of the microtubule stabilizer taxol to rescue the bim1Δ phenotype. Although taxol may create longer microtubules in bim1Δ filaments, the hyperstability of these microtubules prevents normal filamentous growth. Finally, the reduction in wild type filament length when grown in the presence of taxol further supports the importance of microtubule dynamics for proper filaments.
Additional support for our model and the importance of microtubules for normal filamentous growth comes from work in Ustilago maydis. Chemical disruption of the microtubule cytoskeleton in U. maydis did not affect cell-cell recognition or cell-cell fusion of haploid yeast-like cells during early development. However, without an intact microtubule cytoskeleton, hyphae elongation rates and overall hyphae length were both severely reduced (29). These findings are very similar to what is observed in bim1Δ strains in C. neorfomans and add further support to the importance of microtubules for long range filamentous growth in basidiomycetes and the role Bim1 plays in this process.
In the second part of the model nuclear migration is dependent on intact microtubules for congression. Unstable or misdirected microtubules formed in the absence of Bim1 result in inefficient and/or misdirected nuclear movements. This part of the model is supported by our finding that diploid formation is severely affected in bim1Δ strains. Our findings are consistent with studies in S. cerevisiae in which genetic and biochemical analyses revealed defects in microtubule dynamics in bim1Δ mutant strains, leading to defects in nuclear congression during mating. In crosses between bim1Δ strains of S. cerevisiae, diploid formation was decreased by 99% (11). The loss of diploid formation in C. neoformans is over 90%, consistent with a model in which Bim1 is required for nuclear congression during diploidization.
An interesting finding in the context of this model was that α fruiting does not occur in bim1Δ strains. Our initial hypothesis was that the filaments from fruiting would be truncated; however, no filaments were formed. Fruiting appears to occur via rare α by α matings or endoreplication events (26). Although it is unclear how a congression defect would affect endoreplication, congression would likely be required for α-α mating. One possibility is that the inability of bim1Δ strains to undergo fruiting results from microtubule defects that prevent α nuclei from congressing efficiently after cell fusion. The result is that this already rare diploidization event does not occur, and no detectable filament formation occurs.
A paradox in our data is the finding that whereas the diploid formation and fruiting data are suggestive of a congression defect in bim1Δ strains, the distribution of markers observed in progeny from bim1Δ crosses is not. Although we observe short, unstructured filaments, each filament contains two nuclei, and analyses of spores from bim1Δ crosses reveal a Mendelian distribution of markers. This result indicates that 1) one nucleus from each parent strain was maintained throughout sexual development, 2) both nuclei entered the basidium, 3) the nuclei fused, 4) reassortment of markers occurred, and 5) viable spores were formed. We predicted that congression defects might occur during nuclear fusion in the basidium, but this does not appear to be the case based on marker analyses. One possibility is that nuclear congression in the basidium is mediated by a different set of molecules than during diploid formation, and perhaps, Bim1 plays no role in this process. Another possibility is that because the basidial head is much smaller than two fused haploid cells, the loss of Bim1 is mitigated because nuclear congression takes place over a much shorter distance. The role of Bim1 in this process, if any, remains to be determined.
Finally, we found that basidia and spores are formed much earlier in crosses between bim1Δ strains than wild type crosses. The signal that triggers a filament cell to form a basidium is not known. It is possible that the early emergence of basidia and spores observed in bim1Δ crosses represents a survival strategy in response to distress caused by the loss of BIM1 and the resulting loss of filament integrity. Further studies of molecules like Bim1 and how they contribute to nuclear migration and filament formation will continue to elucidate the molecular events required for sexual development.
Supplementary Material
A. PFAM analysis of S. cerevisiae and C. neoformans Bim1 proteins reveals similar domain structures between the two proteins with broad protein similarity throughout these domains. Specifically, the CH domains share 77% similarity, and the EB1 domains share 56% similarity. B. Complete sequence alignment of the Sc and Cn Bim1 proteins. Similarity across the entire protein is 41%.
No BIM1 transcript was present in bim1Δ strains. To determine whether BIM1 was regulated at the transcriptional level during dikaryon formation, northern analysis using a probe to the BIM1 transcript was carried out using standard procedures. RNA was isolated using hot-acid phenol from wild type and bim1Δ haploid cells, wild type a × α crosses, and bim1Δ × bim1Δ crosses after 24 hours under sexual development conditions. Previous observations indicated that by 24 hours substantial cell fusion has occurred and filament formation is just beginning. We found that BIM1 transcript levels remained largely unchanged through the initial stages of sexual development (lanes 1-3). As expected, no BIM1 transcript was detected in bim1Δ haploid strains or crosses (lanes 4-6). It was also noted that RNA from sxiΔ crosses appeared to contain slightly increased amounts of BIM1 transcript. The significance of this, if any, remains to be determined. GPD1 loading and hybridization controls are shown below each lane.
Acknowledgements
Thanks to J. Audyha for his expertise and assistance with confocal microscopy. Thanks to R. Borchardt, K. Klein, J. Webb, and A. Rubendall for laboratory support. Thanks to M. Botts, S. Giles, and B. Stanton for critical reading of and comments on the manuscript. Support for sequencing of the C. neoformans genome was provided by the NIAID/NIH under cooperative agreement U01 AI47087, and The Institute for Genomic Research, funded by the NIAID/NIH under cooperative agreement U01 AI48594. This work was supported by a March of Dimes Basil O'Connor Starter Scholar Award to C.M.H., a Burroughs Welcome Fund Career Award in the Biomedical Sciences to C.M.H., and grant # R01-AI064287 to C.M.H. from the NIH. E.K.K. was supported by the Molecular Biosciences Training Program, NIH grant # 5T32GMO3439. M.W.S. was supported by the Biotechnology Training Program, NIH grant # 5T32GMO3429.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Morris NR. Nuclear migration. From fungi to the mammalian brain. J Cell Biol. 2000;148:1097–1101. doi: 10.1083/jcb.148.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore JK, Sept D, Cooper JA. Neurodegeneration mutations in dynactin impair dynein-dependent nuclear migration. Proc Natl Acad Sci U S A. 2009;106:5147–5152. doi: 10.1073/pnas.0810828106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wynshaw-Boris A. Lissencephaly and LIS1: insights into the molecular mechanisms of neuronal migration and development. Clin Genet. 2007;72:296–304. doi: 10.1111/j.1399-0004.2007.00888.x. [DOI] [PubMed] [Google Scholar]
- 4.Schober JM, Cain JM, Komarova YA, Borisy GG. Migration and actin protrusion in melanoma cells are regulated by EB1 protein. Cancer Lett. 2009 doi: 10.1016/j.canlet.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Suelmann R, Fischer R. Nuclear migration in fungi--different motors at work. Res Microbiol. 2000;151:247–254. doi: 10.1016/s0923-2508(00)00151-0. [DOI] [PubMed] [Google Scholar]
- 6.Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiang X, Fischer R. Nuclear migration and positioning in filamentous fungi. Fungal Genet Biol. 2004;41:411–419. doi: 10.1016/j.fgb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Lin X. Cryptococcus neoformans: morphogenesis, infection, and evolution. Infect Genet Evol. 2009;9:401–416. doi: 10.1016/j.meegid.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 9.Alspaugh JA, Davidson RC, Heitman J. Morphogenesis of Cryptococcus neoformans. Contrib Microbiol. 2000;5:217–238. doi: 10.1159/000060352. [DOI] [PubMed] [Google Scholar]
- 10.Hull CM, Heitman J. Genetics of Cryptococcus neoformans. Annu. Rev. Genet. 2002;36:557–615. doi: 10.1146/annurev.genet.36.052402.152652. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz K, Richards K, Botstein D. BIM1 encodes a microtubule-binding protein in yeast. Mol Biol Cell. 1997;8:2677–2691. doi: 10.1091/mbc.8.12.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davidson RC, Blankenship JR, Kraus PR, de Jesus Berrios M, Hull CM, D'Souza C, Wang P, Heitman J. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology. 2002;148:2607–2615. doi: 10.1099/00221287-148-8-2607. [DOI] [PubMed] [Google Scholar]
- 14.Toffaletti DL, Cox GM, Rude TH, Perfect JR. A transformation system with a dominant selection marker for Cryptococcus neoformans; 95th ASM Meeting.1995. [Google Scholar]
- 15.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- 16.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Current Protocols in Molecular Biology. Vol. 2. John Wiley and Sons, Inc.; Boston, MA: 1997. p. 13. [Google Scholar]
- 17.Sambrook J, F. E, Maniatis T, editors. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1989. [Google Scholar]
- 18.Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- 19.Alspaugh JA, Perfect JR, Heitman J. Signal transduction pathways regulating differentiation and pathogenicity of Cryptococcus neoformans. Fungal Genet Biol. 1998;25:1–14. doi: 10.1006/fgbi.1998.1079. [DOI] [PubMed] [Google Scholar]
- 20.Sherman F, Fink GR, Hicks JB. Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 21.Botts MR, Giles SS, Gates MA, Kozel TR, Hull CM. Isolation and characterization of Cryptococcus neoformans spores reveal a critical role for capsule biosynthesis genes in spore biogenesis. Eukaryot Cell. 2009;8:595–605. doi: 10.1128/EC.00352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anna EJ. Rapid in vitro capsule production by cryptococci. Am J Med Technol. 1979;45:585–588. [PubMed] [Google Scholar]
- 23.Eisenman HC, Mues M, Weber SE, Frases S, Chaskes S, Gerfen G, Casadevall A. Cryptococcus neoformans laccase catalyses melanin synthesis from both D- and L-DOPA. Microbiology. 2007;153:3954–3962. doi: 10.1099/mic.0.2007/011049-0. [DOI] [PubMed] [Google Scholar]
- 24.Sia RA, Lengeler KB, Heitman J. Diploid strains of the pathogenic basidiomycete Cryptococcus neoformans are thermally dimorphic. Fungal Genet Biol. 2000;29:153–163. doi: 10.1006/fgbi.2000.1192. [DOI] [PubMed] [Google Scholar]
- 25.Lin X, Hull CM, Heitman J. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature. 2005;434:1017–1021. doi: 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- 26.Lin X, Patel S, Litvintseva AP, Floyd A, Mitchell TG, Heitman J. Diploids in the Cryptococcus neoformans serotype A population homozygous for the α mating type originate via unisexual mating. PLoS Pathog. 2009;5:e1000283. doi: 10.1371/journal.ppat.1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tirnauer JS, O'Toole E, Berrueta L, Bierer BE, Pellman D. Yeast Bim1p promotes the G1-specific dynamics of microtubules. J Cell Biol. 1999;145:993–1007. doi: 10.1083/jcb.145.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beinhauer JD, Hagan IM, Hegemann JH, Fleig U. Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J Cell Biol. 1997;139:717–728. doi: 10.1083/jcb.139.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuchs U, Manns I, Steinberg G. Microtubules are dispensable for the initial pathogenic development but required for long-distance hyphal growth in the corn smut fungus Ustilago maydis. Mol Biol Cell. 2005;16:2746–2758. doi: 10.1091/mbc.E05-03-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. PFAM analysis of S. cerevisiae and C. neoformans Bim1 proteins reveals similar domain structures between the two proteins with broad protein similarity throughout these domains. Specifically, the CH domains share 77% similarity, and the EB1 domains share 56% similarity. B. Complete sequence alignment of the Sc and Cn Bim1 proteins. Similarity across the entire protein is 41%.
No BIM1 transcript was present in bim1Δ strains. To determine whether BIM1 was regulated at the transcriptional level during dikaryon formation, northern analysis using a probe to the BIM1 transcript was carried out using standard procedures. RNA was isolated using hot-acid phenol from wild type and bim1Δ haploid cells, wild type a × α crosses, and bim1Δ × bim1Δ crosses after 24 hours under sexual development conditions. Previous observations indicated that by 24 hours substantial cell fusion has occurred and filament formation is just beginning. We found that BIM1 transcript levels remained largely unchanged through the initial stages of sexual development (lanes 1-3). As expected, no BIM1 transcript was detected in bim1Δ haploid strains or crosses (lanes 4-6). It was also noted that RNA from sxiΔ crosses appeared to contain slightly increased amounts of BIM1 transcript. The significance of this, if any, remains to be determined. GPD1 loading and hybridization controls are shown below each lane.