Abstract
Dengue virus (DENV) infection is a worsening global health problem. The plaque reduction neutralization test (PRNT) is currently considered to be the “gold standard” to characterize and quantify circulating levels of anti-DENV neutralizing antibody (NAb). Many variations of the PRNT are currently in use and neither the assay nor its performance conditions have been standardized or harmonized between laboratories. We used a well-characterized panel of acute and late convalescent follow-up sera samples from children experiencing primary and secondary DENV infections to evaluate the performance of the dengue PRNT under a variety of testing conditions. Investigators varied cell type, control virus passage, and the use of complement across multiple assay runs of the same sample panel. Our findings indicate wide variation in PRNT titer results in response to varied testing conditions.
Introduction
Dengue is a serious global health problem with an estimated 2.5 billion people at risk for infection and 50–100 million dengue virus (DENV) infections resulting in more than 25,000 deaths annually.1–3 The first DENV was isolated by Hotta during a 1942 epidemic in the Nagasaki-Sasebo area of Japan.4
Dengue viruses are characterized as small, enveloped viruses containing a single positive strand of ribonucleic acid (RNA) and are members of the family Flaviviridae, genus Flavivirus.5 There are four antigenically distinct DENV types (DENV-1, DENV-2, DENV-3, and DENV-4) that display a high degree of antigenic cross-reactivity with other mosquito and tick-borne flaviviruses such as Japanese encephalitis virus, yellow fever virus, and tick-borne encephalitis virus.6,7
In 1959, Henderson and Taylor8 first described a method to detect arbovirus plaques in an agar overlay stained with neutral red and described its ability to measure serum antibody neutralization titers. In 1964, Hammon and Sather9 wrote on the difficulties of typing DENV caused by serologic cross-reactivity and antigenic similarities. At that time, three assays were being used to distinguish DENV types: virus neutralization using a suckling mouse model, complement fixation, and hemagglutination inhibition. The latter two methods were plagued with a high degree of serologic cross-reactivity and the former by limitations associated with using suckling mice. The dengue virus neutralization assay using suckling mice in a challenge virus resistance test was developed by Halstead and Nisalak and adapted to BS-C-1 (African green monkey kidney), PS (porcine kidney), and LLC-MK2 (rhesus monkey kidney) cell lines.10 An in vitro assay using plaque reduction to measure DENV neutralizing antibody and DENV identification was developed in 1967 by Russell and Nisalak.11,12 The Russell and Nisalak assay became known as the plaque reduction neutralization test (PRNT) and used prototype dengue seed viruses, monkey anti-sera controls, LLC-MK2 cell lines, and an agar overlay media with neutral red staining. A probit analysis was used to determine the serum titer required to reduce dengue viral plaques by 50% (PRNT50) compared with control. The PRNT introduced a method of measuring DENV type-specific neutralizing antibodies and has remained the standard assay.
Variations of the Russell PRNT were subsequently introduced using a variety of cell lines and methods: 1) a micro-metabolic inhibition test and a microculture plaque-reduction test using BHK-21 (baby hamster kidney cells) and LLC-MK2 cells lines, respectively; 2) microplate cultures using BHK-21 cells, a focus reduction method using peroxidase-anti-peroxidase staining of BHK-21 cells; 3) a semi-micro method in LLC-MK2 cells using a 70% plaque reduction criteria; and 4) a simplified PRNT assay using BHK-21 cells.13–17
Today a wide variety of dengue PRNT assays are being used by dengue vaccine developers, academic research, and public health laboratories. The PRNT is being used to define the immunogenicity of dengue vaccine candidates, support dengue seroepidemiologic studies, and support pathogenesis studies of severe dengue illness.18–29 Despite its widespread use, neither the PRNT nor the required critical reagents (e.g., cell line, viral strains, passage, complement) have been standardized nor harmonized between laboratories. Guidelines on the conduct of the PRNT have recently been published by the World Health Organization (WHO) Initiative for Vaccine Research of the Department of Immunization, Vaccines and Biologicals with support from the Bill and Melinda Gates Foundation Pediatric Dengue Vaccine Initiative (http://whqlibdoc.who.int/hq/2007/WHO_IVB_07.07_eng.pdf).30
We conducted a series of experiments to define the variability in anti-dengue virus PRNT results using different cell lines, virus preparations, and the presence or absence of complement. Our study demonstrated that modification of these conditions had significant effects on the PRNT titers measured in a given serum sample. Significant associations were observed between certain testing conditions and increases and decreases in titers from different tests on the same serum sample. These findings underscore the need to harmonize assay methods, testing conditions, and key reagents if inter-laboratory comparison of PRNT results is desired.
Materials and Methods
Standardized sera panel
A standardized sera panel was used to test the performance of the PRNT under a variety of test conditions. The panel was assembled from blood samples collected as part of a hospital-based study evaluating children with suspected dengue admitted to the Queen Sirikit National Institute of Child Health (QSNICH) located in Bangkok, Thailand.31 The study was approved by the Thai Ministry of Health, QSNICH, University of Massachusetts Medical School, and U.S. Army ethical review committees. All volunteers were enrolled following an informed consent process with parent(s) and written documentation of the same.
Sera were characterized for the presence of dengue antibody by dengue enzyme immunoassay (EIA), hemagglutination inhibition (HAI), mosquito inoculation with viral isolation, and DENV identification by a typing enzyme immunoassay.32–35 A diagnosis of dengue and clinical characterization were guided by established criteria (WHO, monograph on Dengue/Dengue Hemorrhagic Fever [1997]) applied by a medical monitor, as previously described and outlined below.36
Paired sera from 18 patients were used in all neutralization assays (Table 1) testing all conditions (Figure 1). “Acute” samples were obtained between 8 and 11 days after hospital admission and “late convalescent” samples were obtained 354–380 days after admission; one convalescent sample was obtained 177 days after hospital admission. The panel consisted of 6 primary dengue infections (3 DENV-1, 1 DENV-2, and 2 DENV-3) and 12 secondary dengue infections (3 DENV-1, 3 DENV-2, 3 DENV-3, and 3 DENV-4).
Table 1.
Serum panel characteristics*
| Age (years) | Number of days after first hospital day sera was collected (acute and follow-up) | Peak acute EIA serology IgM/IgG (ratio) (OD units) | Acute dengue serologic diagnosis | Clinical diagnosis | Viral isolation |
|---|---|---|---|---|---|
| 4 | Day 11, Day 374 | 146/27 (5.4) | Primary dengue | DF | DENV-1 |
| 6 | Day 8, Day 368 | 82/13 (6.3) | Primary dengue | DF | DENV-1 |
| 11 | Day 9, Day 354 | 103/46 (2.2) | Primary dengue | DF | DENV-1 |
| 3 | Day 11, Day 368 | 130/65 (2.0) | Primary dengue | DF | DENV-2 |
| 9 | Day 9, Day 366 | 168/39 (4.3) | Primary dengue | DF | DENV-3 |
| 8 | Day 11, Day 367 | 221/0 (> 1.8) | Primary dengue | DF | DENV-3 |
| 10 | Day 9, Day 369 | 74 / 468 (0.16) | Secondary dengue | DF | DENV-1 |
| 7 | Day 11, Day 368 | 18 / 350 (0.05) | Secondary dengue | DHF Gr. I | DENV-1 |
| 10 | Day 9, Day 380 | 46/168 (0.27) | Secondary dengue | DHF Gr. II | DENV-1 |
| 10 | Day 11, Day 374 | 18/208 (0.087) | Secondary dengue | DHF Gr. II | DENV-2 |
| 10 | Day 10, Day 367 | 47/248 (0.19) | Secondary dengue | DHF Gr. II | DENV-2 |
| 3 | Day 8, Day 369 | 48/272 (0.18) | Secondary dengue | DF | DENV-2 |
| 5 | Day 9, Day 369 | 26/95 (0.27) | Secondary dengue | DHF Gr. III | DENV-3 |
| 6 | Day 10, Day 374 | 105/168 (0.63) | Secondary dengue | DF | DENV-3 |
| 7 | Day 9, Day 368 | 57/188 (0.30) | Secondary dengue | DHF Gr. II | DENV-3 |
| 13 | Day 9, Day 372 | 144/363 (0.40) | Secondary dengue | DF | DENV-4 |
| 9 | Day 8, Day 374 | 145/245 (0.59) | Secondary dengue | DHF II | DENV-4 |
| 9 | Day 10, Day 177 | 89/255 (0.35) | Secondary dengue | DF | DENV-4 |
EIA = enzyme immunoassay; OD = optical density; DF = dengue fever; DHF = dengue hemorrhagic fever; Gr. I–IV = grade I–IV; DENV = dengue virus.
Figure 1.
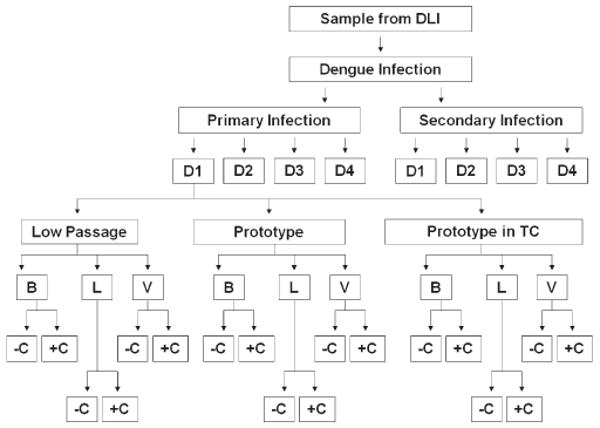
Schematic depicting the testing conditions throughout the experiment using a primary DENV-1 infection as an example. DLI = dengue-like illness; D1, 2, 3, 4 = DENV-1, -2, -3, -4; B = BHK-22; L = LLC-MK2; V = Vero cells; −C = without complement; +C = with complement.
Laboratory characterization of dengue virus infections
The following serologic definitions were used to characterize DENV infections. The collection of all acute and convalescent specimens was separated by more than 3 days.
Acute DENV infection
An acute DENV infection was defined as an acute febrile illness consistent with dengue fever (DF) or dengue hemorrhagic fever (DHF) confirmed by the isolation of a DENV from serum or plasma. Acute infection was defined serologically by a 4-fold rise in HAI antibody against any DENV type between the acute and convalescent specimen or a single specimen with a titer > 1:1280 by HAI. Dengue virus-specific IgM levels of greater than or equal to 40 units by IgM capture EIA was also considered diagnostic of an acute DENV infection.
Acute primary or secondary dengue
Primary DENV infection was defined as an acute dengue infection with an IgM:IgG ratio of ≥ 1.8 by IgM and IgG capture enzyme immunoassay in the acute or early follow-up specimen as previously described.32 Briefly, 96-well microtiter plates (Titertek, Flow Laboratories, McLean, VA) were sensitized with human mu (μ) or gamma (γ) chain antibody (Hyland Diagnostics, Garden Grove, CA) diluted 1:800 in 6 mM carbonate buffer, pH 9.0. Fifty (50) μL test serum diluted 1:100 in phosphate buffered saline (PBS) was then added to each of 2 wells and incubated overnight at 4°C. The plate was washed with PBS plus 0.05% Tween, 20 μL of tetravalent dengue antigen diluted in PBS plus 20% acetone extracted normal human serum (NHS). The dengue antigen was a mixture of 16 hemagglutination units each of DENV types 1–3 and 8 hemagglutination units of dengue type 4. After an overnight incubation at 4°C (or 2 hours at room temperature), plates were washed and 25 μL of an optimal dilution of horseradish peroxidase conjugated human anti-flavivirus IgG, prepared as previously described, was added to each well. The conjugate was diluted in PBS plus 20% acetone extracted NHS and incubated for 1 hour at 37°C. The dilution of conjugate was adjusted so that the positive standards yielded an absorbance at 492 nm (A492) of 0.40. After washing, 100 μL o-phenylene diamine, 0.5 mg/mL (Sigma, St. Louis, MO) in 3% H2O2 diluted 1:300 in 0.1 M citrate buffer, pH 5.0, was added to the wells for 30 min. Color development was stopped by the addition of 50 μL 4 M H2SO4/well. A492 were read on an automatic plate reader (Titertek). A ratio < 1.8 was defined as an acute secondary DENV infection.
Clinical grading of serologically confirmed acute dengue virus infection
All DENV infections were clinically classified according to the criteria described by the WHO monograph.
Determining infecting virus type
A dengue typing enzyme immunoassay was used for determining the DENV type isolated in C6/36 cell cultures following virus amplification in live mosquitoes.35 Briefly, Immulon “U” plates (Dynatech Laboratories, Inc, Chantilly, VA) are coated with goat anti-mouse IgG (Kirkegard and Perry Laboratories Inc., Gaithersburg, MD), incubated at room temperature for 2–3 hours or overnight for 16 to 24 hours at 4°C in a humidified box and washed 8 times with PBS-T (PBS with 0.05% Tween-20) and tap dried. After blocking with freshly prepared 1% bovine serum albumin (BSA), the plates are incubated at room temperature for 1 hour and washed with PBS. Specific mouse monoclonal antibodies (anti-flavivirus 4G2, anti-den-1 J93, anti-den-2 3H5, anti-den-3 10C10, anti-den-4 1H10, and anti-dengue group 2H2) are diluted at 1:1000 in 0.5% BSA and 50 μL of each monoclone is added to the appropriate wells. Control dengue antigens and test antigens are diluted at 1:2 in 0.5% BSA and 20% acetone extracted NHS in PBS and 50 μL added to the test wells and incubated overnight at 4°C. The conjugate step is performed using anti-flavivirus horseradish peroxidase diluted 1:500, 0.5% BSA in 20% acetone extracted NHS in PBS. Twenty-five (25) μL are added to each well and incubated for 2 hours at room temperature. A 100 μL freshly prepared substrate solution (5 mg o-phenylene diamine in 10 mL 0.1 M citrate phosphate buffer and 33 μL of 3% hydrogen peroxide) was added to each well, incubated at room temperature for 30 minutes in a humidified box protected from light. The reaction was stopped by adding 50 μL of 4 M sulfuric acid. Plates are read on a spectrophotometer at a wavelength of 492 nm, mode one, and filter four. Results are interpreted by comparing test OD with positive control where positive controls are greater than or equal to 0.20 and negative controls are less than 0.20.
PRNT Conditions
Control viruses and passage
Three viruses representing each DENV type with varied passage history (low passage wild type, prototype, and high passage virus [prototype in tissue culture]) were used in the PRNT assays. Prototype and prototype in tissue culture viruses represent the same viral strains with differing passage history, whereas low passage wild type viruses represent different viral strains from the prototype strains. Table 2 details the virus strains and passage history.
Table 2.
Viral strains used in the conduct of plaque reduction neutralization assays
| Passage history* | ||||||
|---|---|---|---|---|---|---|
| Virus label | DENV type | Strain | Suckling mouse | LLC-MK2 | C6/36 | VERO |
| Low passage wild type | DENV-1 | 16007 (Thailand) | 0 | 3 | 3 | 0 |
| DENV-2 | 16681 (Thailand) | 0 | 3 | 3 | 0 | |
| DENV-3 | 16562 (Philippines) | 0 | 3 | 2 | 0 | |
| DENV-4 | 1036 (Indonesia) | 0 | 3 | 2 | 0 | |
| Prototype | DENV-1 | Hawaii | 24 | 0 | 0 | 0 |
| DENV-2 | New Guinea-C | 40 | 0 | 0 | 0 | |
| DENV-3 | Hawaii 87 | 40 | 0 | 0 | 0 | |
| DENV-4 | Hawaii 241 | 38 | 0 | 0 | 0 | |
| Prototype in tissue culture | DENV-1 | Hawaii | 25 | 0 | 3 | 1 |
| DENV-2 | New Guinea-C | 41 | 0 | 4 | 2 | |
| DENV-3 | Hawaii 87 | 41 | 0 | 4 | 0 | |
| DENV-4 | Hawaii 241 | 39 | 0 | 4 | 0 | |
Passages occurred chronologically in order from left to right, i.e., passage in suckling mouse followed by passage in C6/36 (low passage) or passage in suckling mouse followed by passage in C6/36 followed by passage in Vero cells (prototype in tissue culture).
Complement
The PRNT assays were completed with and without the use of fresh-frozen guinea pig complement (GIBCO/BRL, Life Technologies, Carlsbad, CA).
Cell Lines
The PRNT assays were completed in three different cell lines: LLC-MK2 (Rhesus monkey kidney cells), Vero (African green monkey kidney), and BHK-21 (baby hamster kidney cells).
PRNT methods
All PRNT assays were completed by the same two technicians following the same standard assay procedures. Assay procedures were adopted from the published methods. Samples were uniquely coded; technicians were not aware of previous serologic or virologic characterizations. A single PRNT assay was performed for each sample under each condition.
LLC-MK2
The method of Russell and Nisalak 12 was used for determining the neutralizing antibodies against DENV-1 through 4, as previously described. Briefly, LLC-MK2 cells (ATCC) were propagated at 35°C without CO2 in an incubator in 162-cm2 flasks using a growth media consisting of 20% fetal bovine serum (heat-inactivated at 56°C for 30minutes), 1× L-glutamine, 100 units of penicillin-streptomycin, and M-199. Cell monolayers were prepared by removing medium and adding a trypsin-versine mixture (100 mL of 10× versine, 5 mL of 10% trypsin, 1.0 mL of 10,000 units/mL of penicillin/streptomycin mixture with distilled water added to a total volume of 500 mL) to cover the cell sheet and incubated at 35°C for 5 minutes. The trypsin-versine mixture was poured off and the flask containing cells was incubated at 35°C for another 10 minutes. Cells were re-suspended in 10 mL growth media (containing 10% heat-inactivated fetal bovine serum, 1× L-glutamine, 100 units of penicillin-streptomycin, 20 mM HEPES and 0.6% of 7.5% sodium bicarbonate). Cells were counted and diluted to 4 × 104 cells per mL in media. One mL was transferred to each well of a 24-well plate (COSTAR, Corning, NY). Plates were incubated at 35°C with 5% CO2 for 3 to 5 days. After incubation, media was removed from the cell monolayer and washed with Hank's balanced salt solution (HBSS). Well-characterized monkey sera were used as positive controls and repeated without and with complement. Positive control antibody titers with complement were 1:540 anti-DENV-1 (G246), 1:1280 anti-DENV-2 (G327), 1:1400 anti-DENV-3 (G189), and 1:370 anti-DENV-4 (G310). Test serum was heat inactivated at 56°C for 30 minutes. The complement used was fresh frozen guinea pig complement (GIBCO BRL, Life Technologies). In an ice bath, 4-fold dilutions of the test sera beginning with a 1:10 dilution (1:10; 1:40; 1:160; and 1:640) with reference virus (noted above) diluent (0.3 mL/dilution) were made and 5 units of guinea pig complement added to each dilution of serum. Virus-serum mixture was inoculated (0.1 mL per well) and absorbed for 1.5 hours at room temperature at which point the inoculums were removed. First overlay medium (containing low-melting point agarose (GIBCO BRL, Life Technologies) Hank's BSS, vitamins, amino acids, heat-inactivated calf serum, L-glutamine, and 7.5% sodium bicarbonate) were added, 0.5 mL per well and allowed to solidify for 15 minutes. Plates were incubated at 35°C in 5% CO2 for 7 days. Neutral red staining was performed by adding a neutral red second overlay (same as above without fetal calf serum and containing 4% Neutral Red [Sigma Chemical, St. Louis, MO]), 0.5 mL per well and allowed to solidify for 15 minutes. Plates were incubated overnight at 35°C 5% CO2 and plaques counted. Positive control wells (virus without sera) were established for each assay run to ensure infectivity of the cell monolayer (formation of > 25 plaques).
Vero cells
Dengue PRNT in Vero cells were performed by using monolayers of Vero cells in 24-well plates seeded with 1 mL of 3 × 104 Vero cell suspension per well in minimum essential media (MEM) (Gibco BRL) with 10% heat-inactivated fetal bovine serum, 1% glutamine, and 100 units of penicillin and streptomycin (Gibco BRL). Cells were incubated for 3 to 4 days at 35°C. Media for the first overlay consisted of 0.5 mL of 0.9% low-melting point agarose, MEM + 10% heat-inactivated fetal bovine serum with 0.5% vitamin, L-glutamine, 2.5% of 7.5% sodium bicarbonate solution, and 1,000 units of neomycin-streptomycin. The second overlay contained 0.9% low-melting point agarose in normal saline with neutral red. Preparation of virus-serum mixtures and cell monolayer inoculation was performed as previously described with LLC-MK2 cells. Cultures were incubated at 35°C in 5% CO2 and 0.5 mL of the second overlay was added on the 6th or 7th day depending on the control virus being used. Plaques were counted the following day. Positive control wells (virus without sera) were established for each assay run to ensure infectivity of the cell monolayer (formation of > 25 plaques)
BHK-21 cells
Dengue PRNT in BHK-21 cells was performed according to the method of Morens and others.17 BHK-21 cell clone 15 cells were a gift from Dr. Sukhathida Ubol, PhD, Department of Microbiology, Faculty of Sciences, Mahidol University, Bangkok, Thailand. Cells were maintained in MEM with 10% heat-inactivated fetal bovine serum with 1% glutamine and 100 units of penicillin-streptomycin with 2 × 104 BHK-21 cells used to seed each well of 24-well plates. The PRNT was performed by incubating serial dilution of serum-virus mixtures and taking 0.1 mL of the mixture and inoculating into each well. Plates were incubated at 35°C in 5% CO2 for 4 hours to let the cells form monolayers. The supernatant was then gently aspirated and overlaid with 1 mL per well of MEM with 5% heat-inactivated fetal bovine serum containing 0.75% carboxy-methyl-cellulose (Sigma). Plates are incubated at 35°C in 5% CO2 for 4 to 5 days or until cytopathic effect is observed. After incubation the media was removed and fixed and stained for 10 to 15 minutes with a solution of crystal violet-formaldehyde prepared by dissolving 1.3 gm of crystal violet in 50 mL of 95% isopropyl alcohol, 300 mL of 37% formaldehyde and distilled water to a final 1 L stock volume. Final solution is prepared with a 1:1 volume of PBS at a pH of 7.4. Fixing and staining solution was removed and plates rinsed and dried at room temperatures and plaques counted immediately. Positive control wells (virus without sera) were established for each assay run to ensure infectivity of the cell monolayer (formation of > 25 plaques).
Plaque reduction neutralization endpoints were calculated using probit analysis as described previously.37 Briefly, a reduction in plaque count of 50% (PRNT50) was used as the neutralizing end point. Plaques generated by test sera at varying dilutions and the control preparation were counted. The percentage of plaques counted in test sera were compared with the number of plaques from the control preparation. Log dilutions of test sera preparations (i.e., 1:10, 1:40, 1:160, 1:640) were plotted along the X axis, whereas percent reduction in plaque count was detailed along the Y axis. Percent count plaque reductions for each test sample at each dilution was plotted and a best-fit line drawn. The reciprocal of the lowest dilution of test sera to neutralize 50% of the control virus input represents the PRNT50 titer. Therefore, the PRNT50 titer is calculated by counting plaques and reporting the titer as the reciprocal of the last serum dilution to show 50% reduction of the control plaque count as based on the back-titration of control plaques.30
Statistical analysis
Multivariate mixed effects linear regression models were constructed using SAS's PROC MIXED procedure (SAS Institute, Inc., Cary, NC), including a random intercept to account for the effect of the individual and fixed effects for the conditions that were varied between assays. The random intercept was incorporated to account for the fact that multiple tests were run on a single sample, and that these results are therefore correlated. Fixed effects measures allowed testing of the influence of each of the conditions that were varied between assays, and the influence of important covariates (infecting DENV type, whether the infection was primary or secondary), controlling for all other factors.
Four models were constructed, with the outcomes of interest being the log titers to DENV-1–4 in late convalescent serum samples (i.e., samples that were collected 6–12 months after infection). The fixed effects included virus passage (prototype, prototype in tissue culture, and low passage wild type), cell line (LLC-MK2, Vero, and BHK-21), the infecting DENV type (as detected by viral isolation), whether the infection was primary or secondary, and the presence or absence of complement. The use of Vero cells, the absence of complement, and the use of a prototype virus were selected to be the referent categories for analysis, to facilitate comparison of assay results to those obtained using reagents recently recommended by the WHO (http://whqlibdoc.who.int/hq/2007/WHO_IVB_07.07_eng.pdf). For infecting DENV type, the referent category was assigned to match the DENV type-specific titer being measured (e.g., infection with DENV-1 was the referent category for the outcome of log titers to DENV-1).
A “main effects” model was constructed, which considered the influence of each of the fixed effects in the absence of interactions between variables. In interpreting the regression coefficients (betas) for each variable, positive values indicate an increase in the log antibody titer relative to the referent level for that variable; negative values indicate a decrease. As these are multivariate regressions, the coefficient indicates the relative change in the log antibody titer between levels of a given variable, controlling for the other conditions in the assay. These main effect regression parameters can be considered as averages across the range of possible interactions between variables.
In subsequent models, all possible two-way interactions were considered; backward elimination was conducted until only significant interaction terms remained in the model. These interactions terms are of interest because they reflect assay variability that is enhanced when two different parameters are in combination—for example, higher titers may be obtained using specific combinations of cell line and complement than may be expected by considering these variables independently.
Statistical analyses were performed using SAS analytic software (version 9.1, SAS Institute, Inc.).
Results
Main effect results for antibody titers to DENV1–4
Each of the four main effects models (with no interactions between variables) resulted in a significantly good fit to the data (P < 0.05, by the likelihood ratio test). Overall, the effects of virus passage, cell line, and the use of complement were found to be highly influential on the final PRNT titers determined for all DENV types (Table 3A–D). However, there was significant variability in the specific conditions that resulted in the highest and lowest titers for each DENV type and in the magnitude of the associations. The random intercepts resulted in a significantly improved model fit for each of the four models; the individual intercept values for each serum sample are omitted from the tables for simplicity.
Table 3.
Table 3A: Main effects models (no interaction terms). Outcome: log titers to DENV-1
| Parameter | Regression parameter (beta) | P value | Overall P value |
|---|---|---|---|
| Virus passage | |||
| Low passage wild type | −0.2714 | 0.0760 | |
| Prototype | 0 (ref) | – | < 0.001 |
| Prototype in tissue culture | −0.6222 | < 0.001 | |
| Cell line | |||
| BHK21 | −0.5212 | < 0.001 | < 0.001 |
| Vero | 0 (ref) | – | |
| LLC-MK2 | 0.04403 | 0.7729 | |
| Infecting serotype | |||
| DENV-1 | 0 (ref) | – | |
| DENV-2 | −0.2674 | 0.6473 | 0.6013 |
| DENV-3 | −0.7297 | 0.1762 | |
| DENV-4 | −0.2837 | 0.6723 | |
| Complement | |||
| Absent | 0 (ref) | – | < 0.001 |
| Present | 0.5137 | < 0.001 | |
| Primary or secondary | |||
| Primary | 0 (ref) | – | 0.4694 |
| Secondary | 0.3246 | 0.4694 | |
| Intercept | 5.0871 | < 0.001 | |
| Table 3B: Main effects models (no interaction terms). Outcome: log titers to DENV-2 | |||
| Parameter | Regression parameter (beta) | P value | Overall P value |
| Virus passage | |||
| Low passage wild type | −0.2706 | 0.0093 | 0.0232 |
| Prototype | 0 (ref) | – | |
| Prototype in tissue culture | −0.2140 | 0.0393 | |
| Cell line | |||
| BHK21 | −0.7510 | < 0.001 | < 0.001 |
| Vero | 0 (ref) | – | |
| LLC-MK2 | −0.02935 | 0.7767 | |
| Infecting serotype | |||
| DENV-1 | −0.9151 | 0.0862 | |
| DENV-2 | 0 (ref) | – | 0.1879 |
| DENV-3 | −0.8966 | 0.1008 | |
| DENV-4 | −0.00623 | 0.9921 | |
| Complement | |||
| Absent | 0 (ref) | – | < 0.001 |
| Present | 0.5263 | < 0.001 | |
| Primary or secondary | |||
| Primary | 0 (ref) | – | 0.0293 |
| Secondary | 0.9504 | 0.0293 | |
| Intercept | 4.3889 | < 0.001 | |
| Table 3C: Main effects models (no interaction terms). Outcome: log titers to DENV-3 | |||
| Parameter | Regression parameter (beta) | P value | Overall P value |
| Virus passage | |||
| Low passage wild type | 1.3049 | < 0.001 | < 0.001 |
| Prototype | 0 (ref) | – | |
| Prototype in tissue culture | 0.07537 | 0.4302 | |
| Cell line | |||
| BHK21 | −0.1354 | 0.1569 | < 0.001 |
| Vero | 0 (ref) | – | |
| LLC-MK2 | 0.4500 | < 0.001 | |
| Infecting serotype | |||
| DENV-1 | −0.3771 | 0.03870 | |
| DENV-2 | 0.5760 | 0.2348 | 0.2548 |
| DENV-3 | 0 (ref) | – | |
| DENV-4 | 0.1091 | 0.8415 | |
| Complement | |||
| Absent | 0 (ref) | – | < 0.001 |
| Present | 0.4544 | < 0.001 | |
| Primary or secondary | |||
| Primary | 0 (ref) | – | 0.1693 |
| Secondary | 0.5311 | 0.1693 | |
| Intercept | 2.8795 | 0.0012 | |
| Table 3D: Main effects models (no interaction terms. Outcome: log titers to DENV-4 | |||
| Parameter | Regression parameter (beta) | P value | Overall P value |
| Virus passage | |||
| Low passage wild type | −0.1620 | 0.0496 | 0.0017 |
| Prototype | 0 (ref) | – | |
| Prototype in tissue culture | 0.1961 | < 0.001 | |
| Cell line | |||
| BHK21 | −0.3623 | < 0.001 | 0.0014 |
| Vero | 0 (ref) | – | |
| LLC-MK2 | −0.2125 | 0.0334 | |
| Infecting serotype | |||
| DENV-1 | −1.2953 | 0.0015 | |
| DENV-2 | −0.4726 | 0.2582 | 0.0013 |
| DENV-3 | −1.3743 | < 0.001 | |
| DENV-4 | 0 (ref) | – | |
| Complement | |||
| Absent | 0 (ref) | – | < 0.001 |
| Present | 0.4219 | < 0.001 | |
| Primary or Secondary | |||
| Primary | 0 (ref) | – | 0.0950 |
| Secondary | 0.4820 | 0.0950 | |
| Intercept | 4.4952 | < 0.001 | |
Table 3A–D display the output from the four multivariate, linear, mixed-effects models for the log antibody titers to DENV-1–4. No interaction terms were included in the main effects models. These results are based on the titers observed for the “follow-up specimens” for each dengue patient (collected 6–12 months after presentation); the results from acute specimens are excluded. Multiple tests were run on each sample and therefore a random intercept for the effect of each sample was included in the model. The values of each intercept were of secondary interest and are excluded from the tables. P values shown in bold are those that were significant at an α-level of 0.05.
Virus passage
The effect of viral passage was significant in all four models (P < 0.05). Compared with prototype, mean PRNT50 titers were significantly lower in assays using low passage wild type virus for DENV types -2 and -4, and using prototype in tissue culture across DENV types -1 and -2 (P < 0.05 for each). In contrast, DENV-4 titers were significantly higher when using prototype in tissue culture and DENV-3 titers were significantly higher when using low passage wild type virus (P < 0.05 for each).
Cell line
The effect of cell line was significant overall in all four models (P < 0.01). DENV-1, -2, and -4 titers were significantly lower using BHK-21 cells compared with Vero cells (P < 0.05). Assays performed using LLC-MK2 cells increased titers to DENV-3 (0.45 log), but decreased titers for DENV-4 (−0.21 log, P < 0.05 for both).
Complement
The addition of complement significantly increased titers across all DENV types (range 0.42–0.53 log, P < 0.05 for all).
Infecting DENV type
The overall effect of infecting dengue serotype was significant only for titers to DENV-4, for which infection with DENV-1 and DENV-3 resulted in dramatically lower titers to DENV-4 (−1.29 log and −1.37 log, respectively). Antibody titers for the other DENV types tended to be non-significantly decreased if infection was caused by a heterologous serotype (e.g., antibody titers to DENV-2 were lower if the individual was infected with DENV-1, -3, or -4).
Primary or secondary infection
Secondary infection was associated with increased PRNT titers to the infecting DENV type relative to primary infection for all DENV type-specific titers (significant only for DENV-2, 0.95 log increase).
Main effect plus interaction results for antibody titers to DENV1–4
Significant two-way interaction effects were observed for three combinations of parameters when incorporated into the main effects models described previously. Interaction effects included: virus passage and the use of complement, cell line and the use of complement, and cell line and virus passage (Figure 2A–C). This implies that different virus passages and cell lines, for example, interact to influence antibody titers in a manner that is not captured by controlling for virus passage and cell line separately.
Figure 2.
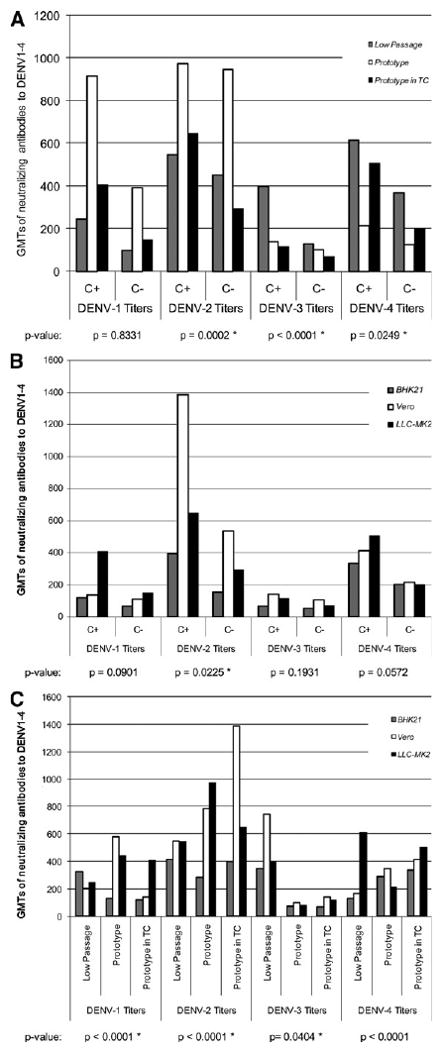
A, Interaction of viral strain passage and complement. C+ = with complement; C− = without complement; TC = tissue culture; GMT = geometric mean titer. B, Interaction of cell line and complement. TC = tissue culture; GMT = geometric mean titer. C, Interaction of cell type and viral strain passage. TC = tissue culture; GMT = geometric mean titer. A–C, display geometric mean neutralizing antibody titers (GMT) to DENV 1–4 for all two-way combinations of reference virus, cell type, and the presence or absence of complement. The GMTs for the interactions here are calculated from multivariate mixed-effects models like those presented in Table 3A–D, but with the inclusion of the two-way interaction term of interest (for example, cell line and complement). For convenience, the reference levels for variables other than the conditions being measured were used to estimate the expected antibody titer. As noted in Table 3A–D, referent levels include primary infection, the absence of complement, the use of Vero cells, and prototype virus. P values represent the overall significance of the interaction term in the model for that serotype, controlling for all other variables. Starred P values indicate those that were significant at a α-level of 0.05.
Virus passage and complement
The addition of complement increased antibody titers across all DENV types, cell lines and levels of viral passage. Significant increases with the addition of complement were observed for DENV-2 when using prototype virus in tissue culture (P < 0.001), DENV-3 titers when using the low passage wild type virus (P < 0.001), and DENV-4 titers using the prototype virus in tissue culture (P = 0.025).
Cell line and complement
The variation of antibody titers was less pronounced when analyzing the interactive effect of cell line and complement, as compared with the dramatic effects observed with some combinations of complement and virus passage. Only DENV-2 titers increased significantly for Vero cells and complement (P = 0.023).
Virus passage and cell line
Different combinations of cell line and virus passage resulted in significant differences between peak antibody titers for each DENV type. Vero cells produced the highest titers for DENV-1, -2, and -3, whereas LLC-MK2 produced the highest titers for DENV-4.
Discussion
Dengue is a worsening global health problem. The study of dengue epidemiology and pathogenesis of disease has increased considerably over the past decade. Numerous dengue vaccine candidates are now in pre-clinical and clinical development and two candidates have generated sufficient clinical data to support planning efficacy trials.38 The PRNT assay, or variations of the same, remains at the center of these efforts.
The PRNT has been used to document the development of neutralizing, DENV type-specific antibody to support basic science research and vaccine development efforts for nearly four decades. Understanding neutralizing antibody (NAb) “profiles” at the individual and population levels allow investigators to document the occurrence of a DENV infection, correlate immune status to clinical outcomes (i.e., asymptomatic versus symptomatic DENV infection), and functions as a marker of dengue vaccine immunogenicity. There is general consensus that if defining a surrogate marker of immunity/protection is possible for dengue, it will most likely be accomplished using quantitative NAb titers. Once vaccine efficacy trials are completed and clinical benefit of vaccination established, national regulatory authorities will likely evaluate vaccine candidate immunogenicity based on the ability to reproducibly generate NAbs. As such, many vaccine developers are transitioning to second generation neutralization assays (e.g., microneutralization assay) with greater potential for automation, higher throughput, and validation.
Despite the importance of NAb in understanding immune responses to DENV infection and vaccination and efforts by the WHO and Pediatric Dengue Vaccine Initiative (PDVI), there has been no standardization or harmonization of the PRNT. As a consequence, it has been difficult and potentially misleading to compare and contrast results and data across studies and institutions. The WHO recently offered its guidance and recommendations on PRNT assay performance (http://whqlibdoc.who.int/hq/2007/WHO_IVB_07.07_eng.pdf).30
Standardization of biologic assays, such as the PRNT, requires consistency among several assay conditions and harmonization between laboratories performing the assay. Standardization and thorough characterization of critical reagents (e.g., cell line, viral strains, use of complement), optimization of assay methods and sample preparation (e.g., incubation time), strict adherence to a single operating procedure (i.e., SOP), and the use of internal and external controls is key. Reproducibility of assay performance (e.g., lower bounds of reliable assay performance) within and between laboratories is also essential. Achieving these ends is typically accomplished using a well-characterized reference panel of natural (i.e., patient) or artificially generated (i.e., spiking serum with known virus and concentration) samples.
In our experiments, we attempted to examine the performance aspects of the PRNT using a well-characterized panel of patient sera representing acute and late convalescent, primary and secondary, and DENV type-specific DENV infections. All assays were performed by the same two technicians who were blinded to the previous serologic and virologic characterization of the samples and used the same assay procedure and method. The objective of the study was to reproduce the assay methods currently in use and identify how the differences in conditions (materials and methods) could impact assay results.
Our study showed significant variability in PRNT results depending on the assay conditions. There was considerable variation in the DENV NAb titers generated by the PRNT depending on the control viral passage and cell line used and the use of complement. The variation in PRNT titers was demonstrated not only to be impacted by manipulating a single condition but also by interaction between two or more conditions.
The effect of viral passage on NAb titers was significant but not uniform across all DENV types. There was varying effect on titer depending on the virus (wild type versus prototype) and passage (prototype versus prototype in tissue culture). The low passage wild type virus represents a different virus strain (genotype) compared with the prototype viruses and genetic diversity could impact NAb titer distinct from passage level. However, the differences in NAb titers between prototype and higher passaged prototype viruses appear to indicate passage level alone can impact assay results. Unfortunately, viral strains from the experiments are no longer available, and it is not possible to quantitate the impact of genetic diversity on NAb titer. A limited exploration of E gene sequences of viruses representative of those used in this study revealed significant divergence between low passage wild type and prototype viruses. There was no significant divergence between prototype viruses and prototype viruses serially passed in tissue culture. Therefore, although genetic variation could account for assay result differences observed between wild type and prototype viruses, it could not account for the differences observed between using prototype and prototype in tissue culture viruses.
The selection of control virus for any given assay is often dictated by the intended use of the assay. If the assay is being used to characterize regional epidemiology then representative strains (i.e., regional) may be chosen. Assays being used to define vaccine candidate immunogenicity use homologous viruses (i.e., vaccine parent strain). It is currently unknown whether vaccine candidates showing efficacy against DENV strains circulating in, for example, Asia will show similar efficacy against strains circulating in the Americas, Western Pacific, or Africa. Therefore, demonstration of vaccine immunogenicity against non-parental viruses (heterologous strains) may be considered important by vaccine developers seeking a global indication. Until large-scale efficacy trials are conducted the answer to this question will likely remain elusive.
The effect of cell line on NAb titers was significant but not uniform across all DENV types. BHK-21 cells appeared to exert a more uniform effect (lower titers for DENV-1, -2, and -4); a larger sample size may have revealed a significant reduction in NAb titer across all DENV types.
Choice of cell line is also likely impacted by intended assay use. Of the cell lines used in these experiments, a specific derivation of Vero cells are the only cells certified by the WHO for production of live virus dengue vaccine candidates and use in the PRNT.30 Because of this, dengue vaccine developers may be partial to using Vero cells.
The significant lowering effect of DENV-1 and -3 infections on DENV-4 NAb titers is interesting. The observation may be explained by the antigenic similarities between DENV-1 and -3 viruses and a shared reduced ability to cross-neutralize the DENV-4 control virus.39 A second point to consider is that the primary infecting DENV type was not known for the secondary infections. One would assume higher DENV-4 titers would have been observed if the primary infection were caused by a DENV-4 virus (i.e., DENV-4 primary infection followed by DENV-1 or -3 secondary infection).
The addition of complement resulted in a significant rise in titers across all DENV types. Neutralization of DENVs does not require complement and its use in PRNT assays is not recommended.30
Significant interaction effects were observed for virus passage and the use of complement, cell line and the use of complement, and cell line and virus passage. These results were not unexpected considering the significant effects each individual condition had on assay results. The interaction effects contribute to the high variability in titers generated by this assay, suggesting that not only is the choice of assay conditions important but also which conditions are used in combination.
One limitation in our data is the small sample size. Although statistical analyses were applied to the data, the results are considered descriptive and designed to highlight, but not quantify, differences in NAb data generated by PRNT assays under varied testing conditions. Accurately quantifying differences between different assay conditions and attempting to identify “optimal” assay methods would require repeating these experiments with expanded sample panel sizes. A second limitation is the absence of genetic characterizations of the viruses used in the assays. A comparison of virus sequences would have added clarity to understanding assay variation and the varied ability of patient sera to neutralize control viruses. Because of limited sera volume, assays could only be performed once for each condition. Inherent biologic inter-assay variability is a potential issue that would need to be addressed in subsequent experiments. A sera panel composed of more primary infections was desired but these samples are very difficult to acquire in Bangkok using hospital-based studies. An expanded analysis of PRNT assay conditions without the confounding of heterotypic antibody found in secondary infections would have been ideal.
The authors' intent was not to generate a consensus protocol or make recommendations on individual assay conditions. As mentioned, this effort has already been undertaken and the results published by the WHO. The intent of conducting these experiments was to explore how varying individual testing conditions impact assay results. The intra-assay variation experienced when conditions remain constant presupposed variation would be observed when key conditions were modified. The occurrence of intra- and interassay variation is expected with any biologic assay; conducting this series of experiments was an attempt to understand the magnitude of the variation. The data generated during this study supports the contention that significant variation in NAb titers occur when key assay conditions are varied. Furthermore, the data highlight the potential hazards of comparing NAb readouts between laboratories; these concerns are likely most relevant when comparing dengue vaccine candidate immunogenicity. Without the broad implementation of standardized assay reagents and harmonization of assay methods, comparison of NAb data across different vaccine candidates should be approached with caution. If inter-laboratory and manufacturer data comparisons are desired, efforts to standardize and harmonize the PRNT must continue.
Acknowledgments
We thank the volunteers, their parents, and the medical staff of the Queen Sirikit National Institute of Child Health (Bangkok, Thailand) for their valuable participation in this study. We thank the laboratory, project management, and clinical staff of the Department of Virology, Armed Forces Research Institute of Medical Sciences (Bangkok, Thailand) for performing laboratory assays to support the study and coordinating study activities. We also thank Suchitra Nimmannitya for providing expert consultation on the diagnosis and management of dengue cases.
Financial support: This work was funded by the United States Army Military Infectious Diseases Research Program (Fort Detrick, Frederick, MD) and NIH grant P01A134533.
Footnotes
Disclosure: One of the authors is employed by the GlaxoSmithKline Group or Companies. This statement is made in the interest of full disclosure and not because the authors consider this to be a conflict of interest.
Publisher's Disclaimer: Disclaimer: The opinions or assertions contained herein are the private views of the authors and are not to be construed as reflecting the official views of the United States Army, Royal Thai Army, or the United States Department of Defense.
Contributor Information
Stephen J. Thomas, Department of Virology, USAMC-AFRIMS, APO AP 96546, Tel: 662-644-5644, Fax: 662-644-4760.
Ananda Nisalak, Email: anandan@afrims.org, Department of Virology, Armed Forces Research Institute of Medical Sciences, 315/6 Rajvithi Road, Bangkok, 10400 Thailand, Tel: 662-644-5644, Fax: 662-644-4760.
Kathryn B. Anderson, Email: kbander@learnlink.emory.edu, Department of Epidemiology, Rollins School of Public Health, Emory University, Atlanta, GA 30030, Tel: 404-840-5004.
Daniel H. Libraty, Email: daniel.libraty@umassmed.edu, Rm S6-826, UMMS, 55 Lake Ave N., Worcester, MA 01655, Tel: 508-856-4182, Fax: 508-856-4890.
Siripen Kalayanarooj, Email: siripenk@gmail.com, Department of Pediatrics, Queen Sirikit National Institute of Child Health, Rajvithi Road, Bangkok 10400 Thailand.
David W. Vaughn, Email: david.w.vaughn@gsk.com, GlaxoSmithKline Biologicals, 2301 Renaissance Boulevard, RN0220, PO Box 61540, King of Prussia, PA 19406-2772, Tel: 610-787-3907, Fax: 610-787-7057.
Robert Putnak, Email: robert.putnak@us.army.mil, Division of Viral Diseases, Walter Reed Army Institute of Research, 503 Robert Grant Avenue, Silver Spring, MD 20910, Tel: 301-319-9425, Fax: 301-319-9661.
Robert V. Gibbons, Email: robert.gibbons@afrims.org, Department of Virology, USAMC-AFRIMS, APO AP 96546, Tel: 662-644-5644, Fax: 662-644-4760.
Richard Jarman, Email: richard.jarmnan@afrims.org, Department of Virology, USAMC-AFRIMS, APO AP 96546, Tel: 662-644-5644, Fax: 662-644-4760.
Timothy P. Endy, Email: endyt@upstate.edu, Infectious Disease Division, Department of Medicine, State University of New York, Upstate Medical University, 725 Irving Avenue, Suite 304, Syracuse, NY 13210, Tel: 315-464-5533, Fax: 315-464-5579.
References
- 1.Gibbons RV, Vaughn DW. Dengue: an escalating problem. BMJ. 2002;324:1563–1566. doi: 10.1136/bmj.324.7353.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gubler DJ. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 2002;10:100–103. doi: 10.1016/s0966-842x(01)02288-0. [DOI] [PubMed] [Google Scholar]
- 4.Hotta S. Experimental studies on dengue. I. Isolation, identification and modification of the virus. J Infect Dis. 1952;90:1–9. doi: 10.1093/infdis/90.1.1. [DOI] [PubMed] [Google Scholar]
- 5.Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 6.Calisher CH, Karabatsos N, Dalrymple JM, Shope RE, Porterfield JS, Westaway EG, Brandt WE. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J Gen Virol. 1989;70:37–43. doi: 10.1099/0022-1317-70-1-37. [DOI] [PubMed] [Google Scholar]
- 7.Wisseman CL, Jr, Sweet BH. Immunological studies with group B arthropod-borne viruses. III. Response of human subjects to revaccination with 17D strain yellow fever vaccine. Am J Trop Med Hyg. 1962;11:570–575. [PubMed] [Google Scholar]
- 8.Henderson JR, Taylor RM. Arthropod-borne virus plaques in agar overlaid tube cultures. Proc Soc Exp Biol Med. 1959;101:257–259. doi: 10.3181/00379727-101-24902. [DOI] [PubMed] [Google Scholar]
- 9.Hammon W, Sather G. Problems of typing dengue viruses. Mil Med. 1964;129:130–135. [PubMed] [Google Scholar]
- 10.Halstead SB, Sukhavachana P, Nisalak A. Assay of mouse adapted dengue viruses in mammalian cell cultures by an interference method. Proc Soc Exp Biol Med. 1964;115:1062–1068. doi: 10.3181/00379727-115-29117. [DOI] [PubMed] [Google Scholar]
- 11.Russell PK, Nisalak A. Dengue virus identification by the plaque reduction neutralization test. J Immunol. 1967;99:291–296. [PubMed] [Google Scholar]
- 12.Russell PK, Nisalak A, Sukhavachana P, Vivona S. A plaque reduction test for dengue virus neutralizing antibodies. J Immunol. 1967;99:285–290. [PubMed] [Google Scholar]
- 13.Sukhavachana P, Yuill TM, Russell PK. Assay of arbovirus neutralizing antibody by micro methods. Trans R Soc Trop Med Hyg. 1969;63:446–455. doi: 10.1016/0035-9203(69)90031-5. [DOI] [PubMed] [Google Scholar]
- 14.Fujita N, Tamura M, Hotta S. Dengue virus plaque formation on microplate cultures and its application to virus neutralization (38564) Proc Soc Exp Biol Med. 1975;148:472–475. doi: 10.3181/00379727-148-38564. [DOI] [PubMed] [Google Scholar]
- 15.Okuno Y, Igarashi A, Fukai K. Neutralization tests for dengue and Japanese encephalitis viruses by the focus reduction method using peroxidase-anti-peroxidase staining. Biken J. 1978;21:137–147. [PubMed] [Google Scholar]
- 16.Morens DM, Halstead SB, Larsen LK. Comparison of dengue virus plaque reduction neutralization by macro and “semi-micro” methods in LLC-MK2 cells. Microbiol Immunol. 1985;29:1197–1205. doi: 10.1111/j.1348-0421.1985.tb00909.x. [DOI] [PubMed] [Google Scholar]
- 17.Morens DM, Halstead SB, Repik PM, Putvatana R, Raybourne N. Simplified plaque reduction neutralization assay for dengue viruses by semimicro methods in BHK-21 cells: comparison of the BHK suspension test with standard plaque reduction neutralization. J Clin Microbiol. 1985;22:250–254. doi: 10.1128/jcm.22.2.250-254.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edelman R, Wasserman SS, Bodison SA, Putnak RJ, Eckels KH, Tang D, Kanesa-Thasan N, Vaughn DW, Innis BL, Sun W. Phase I trial of 16 formulations of a tetravalent live-attenuated dengue vaccine. Am J Trop Med Hyg. 2003;69:48–60. doi: 10.4269/ajtmh.2003.69.48. [DOI] [PubMed] [Google Scholar]
- 19.Sabchareon A, Lang J, Chanthavanich P, Yoksan S, Forrat R, Attanath P, Sirivichayakul C, Pengsaa K, Pojjaroen-Anant C, Chambonneau L, Saluzzo JF, Bhamarapravati N. Safety and immunogenicity of a three dose regimen of two tetravalent live-attenuated dengue vaccines in five- to twelve-year-old Thai children. Pediatr Infect Dis J. 2004;23:99–109. doi: 10.1097/01.inf.0000109289.55856.27. [DOI] [PubMed] [Google Scholar]
- 20.Sabchareon A, Lang J, Chanthavanich P, Yoksan S, Forrat R, Attanath P, Sirivichayakul C, Pengsaa K, Pojjaroen-Anant C, Chokejindachai W, Jagsudee A, Saluzzo JF, Bhamarapravati N. Safety and immunogenicity of tetravalent live-attenuated dengue vaccines in Thai adult volunteers: role of serotype concentration, ratio, and multiple doses. Am J Trop Med Hyg. 2002;66:264–272. doi: 10.4269/ajtmh.2002.66.264. [DOI] [PubMed] [Google Scholar]
- 21.Simasathien S, Thomas SJ, Watanaveeradej V, Nisalak A, Barberousse C, Innis BL, Sun W, Putnak JR, Eckels KH, Hutagalung Y, Gibbons RV, Zhang C, De La Barrera R, Jarman RG, Chawachalasai W, Mammen MP., Jr Safety and immunogenicity of a tetravalent live-attenuated dengue vaccine in Flavivirus naive children. Am J Trop Med Hyg. 2008;78:426–433. [PubMed] [Google Scholar]
- 22.Calisher CH, Nuti M, Lazuick JS, Ferrari JD, Kappus KD. Dengue in the Seychelles. Bull World Health Organ. 1981;59:619–622. [PMC free article] [PubMed] [Google Scholar]
- 23.Sangkawibha N, Rojanasuphot S, Ahandrik S, Viriyapongse S, Jatanasen S, Salitul V, Phanthumachinda B, Halstead SB. Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. Am J Epidemiol. 1984;120:653–669. doi: 10.1093/oxfordjournals.aje.a113932. [DOI] [PubMed] [Google Scholar]
- 24.Fukunaga T, Okuno Y, Tadano M, Fukai K. A retrospective serological study of Japanese who contracted dengue fever in Thailand. Biken J. 1983;26:67–74. [PubMed] [Google Scholar]
- 25.McBride WJ, Mullner H, LaBrooy JT, Wronski I. The 1993 dengue 2 epidemic in Charters Towers, north Queensland: clinical features and public health impact. Epidemiol Infect. 1998;121:151–156. doi: 10.1017/s0950268898001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papaevangelou G, Halstead SB. Infections with two dengue viruses in Greece in the 20th century. Did dengue hemorrhagic fever occur in the 1928 epidemic? J Trop Med Hyg. 1977;80:46–51. [PubMed] [Google Scholar]
- 27.Endy TP, Chunsuttiwat S, Nisalak A, Libraty DH, Green S, Rothman AL, Vaughn DW, Ennis FA. Epidemiology of inapparent and symptomatic acute dengue virus infection: a prospective study of primary school children in Kamphaeng Phet, Thailand. Am J Epidemiol. 2002;156:40–51. doi: 10.1093/aje/kwf005. [DOI] [PubMed] [Google Scholar]
- 28.Endy TP, Nisalak A, Chunsuttiwat S, Libraty DH, Green S, Rothman AL, Vaughn DW, Ennis FA. Spatial and temporal circulation of dengue virus serotypes: a prospective study of primary school children in Kamphaeng Phet, Thailand. Am J Epidemiol. 2002;156:52–59. doi: 10.1093/aje/kwf006. [DOI] [PubMed] [Google Scholar]
- 29.van Peenen PF, Sunoto, Sumarmo, Sulianti Saroso J, Sinto S, Joseph PL, See R. Dengue with haemorrhage and shock in Jakarta, Indonesia. Southeast Asian J Trop Med Public Health. 1978;9:25–32. [PubMed] [Google Scholar]
- 30.Roehrig JT, Hombach J, Barrett AD. Guidelines for plaque-reduction neutralization testing of human antibodies to dengue viruses. Viral Immunol. 2008;21:123–132. doi: 10.1089/vim.2008.0007. [DOI] [PubMed] [Google Scholar]
- 31.Vaughn DW, Green S, Kalayanarooj S, Innis BL, Nimmannitya S, Suntayakorn S, Endy TP, Raengsakulrach B, Rothman AL, Ennis FA, Nisalak A. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis. 2000;181:2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- 32.Innis BL, Nisalak A, Nimmannitya S, Kusalerdchariya S, Chongswasdi V, Suntayakorn S, Puttisri P, Hoke CH. An enzyme-linked immunosorbent assay to characterize dengue infections where dengue and Japanese encephalitis co-circulate. Am J Trop Med Hyg. 1989;40:418–427. doi: 10.4269/ajtmh.1989.40.418. [DOI] [PubMed] [Google Scholar]
- 33.Clarke DH, Casals J. Techniques for hemagglutination and hemagglutination inhibition with arthropod-borne viruses. Am J Trop Med Hyg. 1958;7:561–573. doi: 10.4269/ajtmh.1958.7.561. [DOI] [PubMed] [Google Scholar]
- 34.Rosen L, Gubler D. The use of mosquitoes to detect and propagate dengue viruses. Am J Trop Med Hyg. 1974;23:1153–1160. doi: 10.4269/ajtmh.1974.23.1153. [DOI] [PubMed] [Google Scholar]
- 35.Kuno G, Gubler DJ, Santiago de Weil NS. Antigen capture ELISA for the identification of dengue viruses. J Virol Methods. 1985;12:93–103. doi: 10.1016/0166-0934(85)90011-4. [DOI] [PubMed] [Google Scholar]
- 36.Kalayanarooj S, Vaughn DW, Nimmannitya S, Green S, Suntayakorn S, Kunentrasai N, Viramitrachai W, Ratanachu-eke S, Kiatpolpoj S, Innis BL, Rothman AL, Nisalak A, Ennis FA. Early clinical and laboratory indicators of acute dengue illness. J Infect Dis. 1997;176:313–321. doi: 10.1086/514047. [DOI] [PubMed] [Google Scholar]
- 37.Cutchins E, Warren J, Jones WP. The antibody response to smallpox vaccination as measured by a tissue culture plaque method. J Immunol. 1960;85:275–283. [PubMed] [Google Scholar]
- 38.Edelman R. Dengue vaccines approach the finish line. Clin Infect Dis. 2007;45(Suppl 1):S56–S60. doi: 10.1086/518148. [DOI] [PubMed] [Google Scholar]
- 39.Henchal EA, Gentry MK, McCown JM, Brandt WE. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am J Trop Med Hyg. 1982;31:830–836. doi: 10.4269/ajtmh.1982.31.830. [DOI] [PubMed] [Google Scholar]


