Abstract
Pathogenetic processes that facilitate the entry, replication, and persistence of Mycobacterium tuberculosis (MTB) in the mammalian host likely include the regulated expression of specific sets of genes at different stages of infection. Identification of genes that are differentially expressed in vivo would provide insights into host-pathogen interactions in tuberculosis (TB); this approach might be particularly valuable for the study of human TB, where experimental opportunities are limited. In this study, the levels of selected MTB mRNAs were quantified in vitro in axenic culture, in vivo in the lungs of mice, and in lung specimens obtained from TB patients with active disease. We report the differential expression of MTB mRNAs associated with iron limitation, alternative carbon metabolism, and cellular hypoxia, conditions that are thought to exist within the granulomatous lesions of TB, in the lungs of wild-type C57BL/6 mice as compared with bacteria grown in vitro. Analysis of the same set of mRNAs in lung specimens obtained from TB patients revealed differences in MTB gene expression in humans as compared with mice.
The clinical course of tuberculosis (TB) comprises a sequence of different stages (1). Airborne bacilli that implant in the lung alveoli are phagocytosed by alveolar macrophages (2). Intracellular replication of Mycobacterium tuberculosis (MTB) results in a primary lesion, followed by lymphohematogenous dissemination and secondary lesions in the lungs and other organs. In most individuals, disease progression is arrested at this stage by the acquired immune response, and a clinically latent state ensues. Postprimary disease arises from the subsequent reactivation of dormant tuberculous foci (3, 4). For the nearly 2 billion individuals worldwide who have been infected with MTB, the lifetime risk of developing TB is ≈10% (5). In the absence of effective treatment, TB case fatality rates can exceed 50%, as evidenced by longitudinal clinical studies in the preantimicrobial era (6).
Despite the global importance of TB as a leading cause of morbidity and mortality, little is known about the host–pathogen interactions at each stage of infection in humans. This information has been difficult to obtain because medical ethics demand that anti-TB therapy be initiated immediately on diagnosis. The uninterrupted course of disease can be studied in TB patients who are unresponsive to therapy, typically due to multidrug-resistant MTB. In such cases, palliative resection of diseased tissue is sometimes undertaken (7). Resected human tissues, which would otherwise be discarded, can be used to analyze host–pathogen interactions in situ (8, 9).
Adaptation of MTB to environmental changes in the course of infection is likely mediated by differential gene expression. An important role for transcriptional regulation in the life cycle of MTB is suggested by genes encoding 13 RNA polymerase σ-factors and >100 putative regulatory proteins in the genome (10). Although the cellular processes controlled by these transcription factors are largely unknown, recent studies (11–16) confirm the importance of gene regulatory networks in MTB pathogenesis. To date, gene expression studies in MTB have been limited to tissue culture systems and animal models. These models do not reproduce several key features of TB in humans, and their relevance to human TB is unclear (17). We have begun to address these issues by using quantitative real-time RT-PCR (QRT-PCR) (18) to study MTB gene expression in lung specimens from infected mice and patients with chronic active TB.
Here, we present evidence that adaptation of MTB to the in vivo environment involves the differential expression of genes responsive to iron limitation, alternative carbon metabolism, and cellular hypoxia; conditions that are thought to occur within the granulomatous lesions of TB. Different temporal patterns of gene expression in vivo were observed at early and late stages of murine infection. Differences were also observed between bacterial gene expression in mouse lungs as compared with lung specimens obtained from patients with chronic active TB.
Materials and Methods
Bacterial Strains and Media. MTB H37Rv and clinical strains were grown in plastic roller bottles at 37°C in Middlebrook 7H9 broth (Difco) containing 10% oleic acid-albumin-dextrose-catalase (OADC) (Difco), 0.5% glycerol, and 0.05% Tween 80. For RNA preparation, bacterial cultures were snap-chilled by the addition of ice and centrifuged at 3,000 × g for 10 min at 4°C. Bacterial pellets were snap-frozen and stored at -80°C. Bacterial stocks for mouse infections were prepared as described (19).
Mouse Infections. Female C57BL/6 mice (The Jackson Laboratory) were infected by using a Lovelace nebulizer (Intox, Albuquerque, NM) to deliver ≈ 500 colony forming units (cfu) per mouse. At this dose, aerosol-infected C57BL/6 mice survive for ≈5–6 mo (20). Mice were killed at selected time points and bacterial cfu were enumerated by homogenizing the left lung in PBS containing 0.05% Tween 80, plating lung homogenates on Middlebrook 7H10 agar (Difco) containing 10% OADC (Difco) and 0.5% glycerol, and incubating at 37°C for 3–4 wk before counting colonies.
Human Lung Specimens. Tissues were obtained from patients undergoing pulmonary resection at Groote Schuur Hospital (7). Tissues were resected by the thoracic surgeon (G.B.W.), immediately dissected by the pathologist (H.C.W.), and snap-frozen in liquid nitrogen. Here, we focus on four HIV-negative patients with chronic active TB, who were unresponsive to therapy. The patients' clinical histories are available in Supporting Text, which is published as supporting information on the PNAS web site.
RNA Preparation from Tissues. Human and murine (right lung) tissues were homogenized in TRI Reagent (Molecular Research Center, Cincinnati) with a Polytron (PRO Scientific, Oxford, CT) and centrifuged at 3,000 × g for 5 min at 4°C. Bacterial pellets were resuspended in 0.5 ml TRI Reagent containing 1% polyacryl carrier (Molecular Research Center), transferred to screwcap tubes with 0.25 ml of zirconia/silica beads, and broken in a bead beater (Biospec Products, Bartlesville, OK). RNA was isolated by using TRI Reagent per the manufacturer's instructions and treated with DNase I (DNA-free kit, Ambion).
Primers and Molecular Beacons. The sequences of primers and molecular beacons are available on Tables 1 and 2, which are published as supporting information on the PNAS web site. Primers with similar melting temperatures (60–66°C) were designed by using PRIMER 3 software (21). All primers were tested in PCRs with 100 H37Rv genome equivalents as template and the amplification products were evaluated by gel electrophoresis. Molecular beacons (Eurogentec, Brussels) were designed as described (www.molecular-beacons.org); standard curves were generated by using serial dilutions of H37Rv genomic DNA as template.
Reverse Transcription (RT) and QRT-PCR. QRT-PCRs were as described (18). Mock reactions (no RT) were done on each RNA sample to rule out DNA contamination. Control reactions with uninfected murine and human tissues were performed to rule out crosshybridization. RT reactions were performed with Klenow DNA polymerase from Carboxydothermus hydrogenoformans (Roche Applied Science): annealing, 95°C (1 min), 65°C (3 min), and 57°C (3 min); polymerization, 60°C (30 min); RT inactivation, 95°C (5 min). Real-time PCRs were run and were analyzed on an ABI7900 real-time PCR machine. PCR conditions: AmpliTaq Gold activation, 95°C (10 min); 15 cycles of 95°C (30 sec), 65°C (0.5° stepdown each cycle, 30 sec), and 72°C (30 sec); 25 cycles of 95°C (30 sec), 57°C (30 sec), and 72°C (30 sec).
Normalization of Transcripts. For each RNA sample, the control transcripts (16S rRNA or sigA mRNA) and target mRNAs were reverse-transcribed together in one reaction, and the resulting cDNAs were quantified by real-time PCR. The target cDNA was normalized internally to the sigA cDNA or 16S cDNA levels in the same sample and expressed as [Target mRNA]/[sigA mRNA] or [Target mRNA]/[16S rRNA].
Results
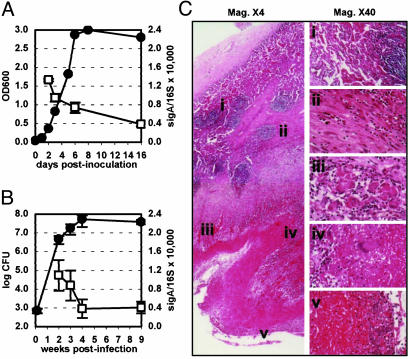
Expression of MTB mRNAs in aerosol-infected mice (Fig. 1B) was analyzed by QRT-PCR (18), with 16S rRNA (22) and sigA mRNA (23–25) as internal normalization standards. The sigA/ 16S varied little during the bacterial growth cycle in mice (Fig. 1B) or in vitro (Fig. 1 A); other σ-factor mRNA levels were more variable (Fig. 9, which is published as supporting information on the PNAS web site). A small decline in the sigA/16S ratio occurred at later stages of infection, when bacterial growth was inhibited by the host community (Fig. 1B). A similar decline in the sigA/16S ratio occurred in vitro when cultures entered stationary phase (Fig. 1 A). These observations are consistent with evidence that MTB might adopt a nonreplicating state in persistently infected mice (22, 26, 27).
Fig. 1.
Sources of RNA samples. (A) MTB H37Rv was grown in Middlebrook 7H9 medium. Samples were withdrawn for OD600 measurements and RNA extraction; sigA mRNA and 16S rRNA were quantified by QRT-PCR. •, OD600; □, sigA/16S ratio × 10,000 ± SD (n ≥ 3). (B) C57BL/6 mice were aerosol-infected with ≈500 cfu of MTB H37Rv. Lungs were collected for cfu enumeration and RNA extraction; sigA mRNA and 16S rRNA were quantified by QRT-PCR. •, cfu ± SD (n ≥ 4); □, sigA/16S ratio × 10,000 ± SD (n ≥ 3). (C) Multilayered organization of granulomatous tissue bordering a 4-cm-diameter cavity in the left upper lung lobe of patient 2 (hematoxylin/eosin-stained). At the periphery were residual airspace and lymphocytic aggregates (i). A capsule of fibroblasts and fibrin with scattered epithelioid macrophages (ii) surrounded a cellular zone containing clusters of multinucleated giant cells (iii). Giant cells were also prominent at the border between the cellular zone and the underlying zone of caseation necrosis (iv). The surface of the cavity wall bordering on the necrotic zone showed extensive leukocytic infiltration and numerous acid-fast bacilli, not revealed by hematoxylin/eosin staining (v). Magnification: ×4(Left), ×40 (Right). Similar pathology was observed in lung specimens from all four patients analyzed in this study.
Here, we focus on seven MTB genes involved in adaptation to iron limitation (mbtB, bfrA), fatty acid catabolism (icl1, mls), glucose starvation (pckA, pca), and cellular hypoxia (hspX). These environmental stresses are thought to occur within the granulomatous lesions of TB. Target mRNAs were quantified by QRT-PCR in RNA samples extracted from MTB grown in vitro (Fig. 1 A) or in the lungs of mice (Fig. 1B). Time points for in vitro-grown bacteria represent log phase (2 and 3 d), stationary phase (16 d), and the transition phase (6 d; Fig. 1 A). Time points in mice represent the acute (2 wk), chronic (9 wk), and transition (4 wk) phases of infection (Fig. 1B); earlier and later time points were queried in separate experiments, with similar results (data not shown).
In parallel, the same transcripts were quantified in seven lung specimens obtained from four HIV-negative patients with chronic active TB (see Materials and Methods). These individuals all had documented histories of long-standing disease with extensive pulmonary destruction and fibrosis necessitating lobectomy. Histopathologic analysis revealed that all of the lung specimens comprised organized multilayered granulomas with extensive caseation necrosis and, in some cases, central cavitation (Fig. 1C). Thus, although it is impossible to ascertain the precise age of individual lesions in TB patients, the human lung specimens analyzed in this study are presumed to represent a chronic, rather than an acute, tuberculous process.
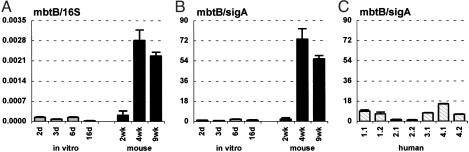
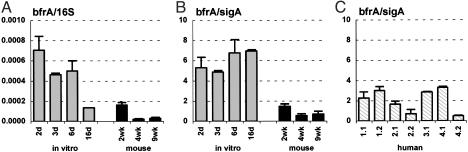
Iron Limitation. Iron is an essential nutrient for most pathogens, and iron limitation at sites of infection is an important mechanism of host defense. A number of MTB genes are known to be regulated in response to iron limitation (28–31). Iron-responsive genes include some that are induced such as mbtB, encoding an enzyme involved in siderophore biosynthesis, and others that are repressed such as bfrA, encoding an iron-storing bacterioferritin.
When bacteria were cultured in vitro in iron-replete medium (Materials and Methods), the 16S (Figs. 2A and 3A)- and sigA (Figs. 2B and 3B)-normalized levels of the mbtB (Fig. 2 A and B) and bfrA (Fig. 3 A and B) transcripts showed little fluctuation throughout the bacterial growth cycle (Fig. 1 A). Between 6 and 16 d postinoculation, when the culture entered stationary phase (Fig. 1 A), there was a small drop in the 16S-normalized levels of the mbtB and bfrA transcripts (Figs. 2 A and 3A), which was not observed when normalization was to sigA (Figs. 2B and 3B). This result likely reflects the slower turnover of 16S rRNA compared with mRNAs, because we have observed a similar drop in the 16S-normalized levels of many transcripts as cells enter stationary phase (e.g., Figs. 2 A, 3A, 5A, and 7A).
Fig. 2.
MbtB peptide synthetase (mbtB) mRNA levels in MTB grown in vitro (A and B; gray bars), in aerosol-infected mice (A and B; black bars), and in lung specimens from TB patients (C). Ratios of mbtB/16S (A) or mbtB/sigA (B and C) are means ± SD (n ≥ 3).
Fig. 3.
Bacterioferritin A (bfrA) mRNA levels in MTB grown in vitro (A and B; gray bars), in aerosol-infected mice (A and B; black bars), and in lung specimens from TB patients (C). Ratios of bfrA/16S (A) or bfrA/sigA (B and C) are means ± SD (n ≥ 3).
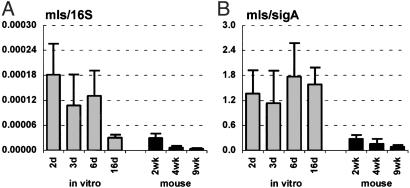
Fig. 5.
Malate synthase (mls) mRNA levels in MTB grown in vitro (A and B; gray bars) and in aerosol-infected mice (A and B; black bars). Ratios of mls/16S (A) or mls/sigA (B) are means ± SD (n ≥ 3).
Fig. 7.
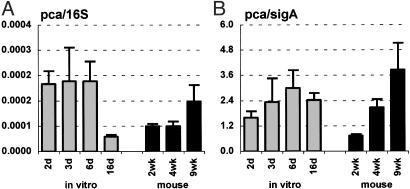
Pyruvate carboxylase (pca) mRNA levels in MTB grown in vitro (A and B; gray bars) and in aerosol-infected mice (A and B; black bars). Ratios of pca/16S (A) or pca/sigA (B) are means ± SD (n ≥ 3).
Consistent with the idea that iron availability might be restricted in vivo, the 16S (Fig. 2 A)- and sigA (Fig. 2B)-normalized levels of mbtB mRNA were markedly increased at 4 and 9 wk postinfection in mice, as compared with bacteria grown in vitro. The increase in normalized levels of mbtB mRNA roughly coincided with the control of bacterial replication by the immune response (Fig. 2 A and B). Normalized levels of mbtB mRNA were not increased at 2 wk postinfection (Fig. 2 A and B), when bacteria were still growing exponentially in the lungs (Fig. 1B). In contrast to mbtB, 16S (Fig. 3A)- or sigA (Fig. 3B)-normalized levels of bfrA mRNA were lower in vivo, as compared with bacteria grown in vitro, particularly at later stages of infection.
The levels of sigA-normalized mbtB mRNA in lung specimens obtained from TB patients (Fig. 2C) were variable, but, in all cases, were lower than in late-stage infected mice (Fig. 2B). In some of the human specimens, normalized mbtB mRNA levels were comparable to levels in bacteria grown in vitro under iron-replete conditions (Fig. 2B). Levels of sigA-normalized bfrA mRNA in the human samples were also variable (Fig. 3C); although there was a trend toward slightly higher levels in the human samples as compared with late-stage infected mice, this finding was not consistent in all of the specimens. Also, there was no clear correlation between the levels of mbtB and bfrA mRNAs in individual lung specimens. We considered the possibility that the patients might have been suffering from iron overload, which is a common clinical condition in subSaharan Africa. However, iron overload was ruled out on clinical grounds and by analysis of ferritin levels in plasma and lung tissues (data not shown).
Fatty Acid Metabolism. Pathogenic bacteria must compete successfully for nutrients, including carbon, to replicate and survive in host tissues. Previously (15), we reported that isocitrate lyase (ICL), the first enzyme of the glyoxylate cycle, is required for long-term persistence of MTB in mice. This finding suggests that mycobacteria might switch to a diet of C2 carbon sources such as fatty acids in vivo, thus engaging the glyoxylate cycle for carbon anaplerosis.
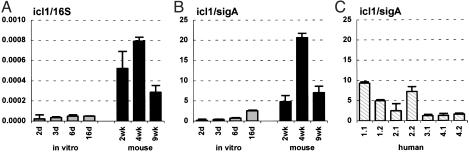
Consistent with this idea, we found that the 16S (Fig. 4A)- and sigA- (Fig. 4B)-normalized levels of icl1 mRNA were markedly increased in mice, as compared with bacteria grown in vitro with glucose and glycerol as carbon sources, conditions where the glyoxylate shunt is nonessential. In contrast, we found that the levels of 16S (Fig. 5A)- and sigA (Fig. 5B)-normalized mls mRNA, encoding the second enzyme of the glyoxylate shunt (malate synthase), were not increased in vivo. These results are in agreement with in vitro studies demonstrating that icl1 expression is induced by growth of MTB on C2 carbon sources (acetate or palmitate), whereas mls expression is not (32). The lack of coordinated regulation of icl1 and mls sets MTB apart from other bacteria, including closely related Actinomycetes such as Corynebacterium glutamicum (33); in these species, coordinate regulation of the icl and mls genes is facilitated by their contiguity on the chromosome, whereas in mycobacteria these genes are unlinked.
Fig. 4.
Isocitrate lyase (icl1) mRNA levels in MTB grown in vitro (A and B; gray bars), in aerosol-infected mice (A and B; black bars), and in lung specimens from TB patients (C). Ratios of icl1/16S (A) or icl1/sigA (B and C) are means ± SD (n ≥ 3).
The levels of sigA-normalized icl1 mRNA in lung specimens from TB patients (Fig. 4C) were highly variable: in some specimens, normalized icl1 mRNA levels were similar to levels in bacteria grown in vitro; in others, icl1 mRNA levels were more similar to bacteria grown in mice. One potential source of variability might be differences in tissue pathology between specimens; in a recent study (8), it was shown by RNA-RNA in situ hybridization, that icl1 mRNA was detectable in MTB in cellular regions of human lesions, whereas bacteria in necrotic regions were negative.
Gluconeogenesis. The observations that ICL is essential for MTB persistence in vivo (15), and that normalized levels of icl1 mRNA increase during infection, suggest that glucose might be unavailable in vivo. Glucose 6-phosphate is an essential intermediate in a number of metabolic pathways; if an external source of glucose is unavailable, glucose 6-phosphate is synthesized de novo by gluconeogenesis (34). Carbon derived from β-oxidation of fatty acids is diverted into gluconeogenesis through phosphoenolpyruvate carboxykinase (PckA), which converts oxaloacetate to phosphoenolpyruvate. Expression of pckA is induced when mycobacteria are grown in vitro on fatty acid substrates (35). The PckA-catalyzed reaction bypasses an alternative entry point for carbon into gluconeogenesis, namely, conversion of pyruvate to oxaloacetate by pyruvate carboxylase (Pca).
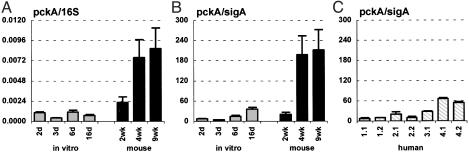
Consistent with the hypothesis that glucose might not be available to MTB in vivo, the levels of 16S (Fig. 6A)- and sigA (Fig. 6B)-normalized pckA mRNA were increased at 4 and 9 wk postinfection in mice, as compared with bacteria grown in vitro in the presence of glucose. Similar to the temporal pattern of mbtB expression, normalized levels of pckA mRNA were lower during acute infection (2 wk) than at later stages. In contrast, the levels of 16S (Fig. 7A)- and sigA (Fig. 7B)-normalized pca mRNA were similar in bacteria grown in vivo or in vitro, which is consistent with the hypothesis that fatty acids are the dominant carbon source used by MTB during infection. An essential role for gluconeogenesis in mycobacterial metabolism in vivo has been confirmed by recent reports that disruption of pckA in virulent Mycobacterium bovis (36), or the vaccine strain M. bovis bacillus Calmette–Guérin (35), resulted in attenuated virulence in animal models of infection.
Fig. 6.
pckA mRNA levels in MTB grown in vitro (A and B; gray bars), in aerosol-infected mice (A and B; black bars), and in lung specimens from TB patients (C). Ratios of pckA/16S (A) or pckA/sigA (B and C) are means ± SD (n ≥ 3).
The levels of sigA-normalized pckA mRNA in lung specimens obtained from TB patients (Fig. 6C) were variable, but, in all cases, were lower than in late-stage infected mice (Fig. 6B). Both icl1 and pckA are required for sustained metabolism on fatty acid substrates; however, there was no obvious parallel between the normalized levels of icl1 (Fig. 4C) and pckA (Fig. 6C) mRNAs in the human lung specimens.
Cellular Hypoxia. Expression of the MTB hspX gene encoding α-crystallin, a chaperonin, was previously shown to respond to hypoxia (37) and nitric oxide (NO) (38). Induction of hspX expression by these disparate stimuli can be explained by the inhibition of cytochrome oxidase by NO (39). It is thought that mycobacteria within granulomatous lesions are exposed to NO (produced by inducible nitric oxide synthase) and derivative reactive nitrogen species, as well as reduced oxygen tension due to inflammation and impaired vascularization.
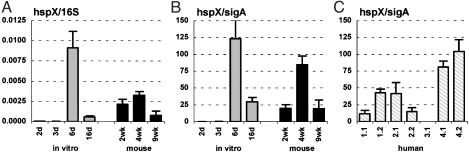
When bacteria were grown in vitro, the levels of 16S (Fig. 8A)and sigA (Fig. 8B)-normalized hspX mRNA were low in the exponential phase of growth (2 and 3 d postinoculation) but increased dramatically at the transition between exponential and stationary phase (6 d) before falling to an intermediate level in stationary phase (16 d; Fig. 1 A). These observations are consistent with previous reports (37) that hspX expression is induced in stationary phase.
Fig. 8.
α-crystallin (hspX) mRNA levels in MTB grown in vitro (A and B; gray bars), in aerosol-infected mice (A and B; black bars), and in lung specimens from TB patients (C). Ratios of hspX/16S (A) or hspX/sigA (B and C) are means ± SD (n ≥ 3). hspX mRNA could not be quantified in lung specimen 3.1 from patient 3 (C) because of a polymorphism in hspX in this patient's strain that interfered with QRT-PCR.
The levels of 16S (Fig. 8A)- and sigA (Fig. 8B)-normalized hspX mRNA in mice were relatively high, and were comparable to the levels in stationary-phase bacteria grown in vitro. Entry into stationary phase per se is unlikely to be the sole factor contributing to increased expression of hspX in vivo, because normalized levels of hspX mRNA were similar in acutely infected (2 wk) and chronically infected (9 wk) mice (Fig. 1B).
The sigA-normalized levels of hspX mRNA were also high in the lung specimens from TB patients; however, the level of expression varied markedly between patients (Fig. 8C). Although less dramatic than the variation between patients, significant quantitative variation in normalized levels of hspX mRNA was also observed in specimens obtained from different regions of the lung in the same patient (Fig. 8C).
Discussion
Strategies to identify genes that are differentially expressed in bacteria grown in vivo vs. in vitro have contributed to the current understanding of pathogenetic processes and adaptations that promote bacterial survival in host tissues (40). However, these strategies typically require genetic manipulation of the pathogen before infection, and cannot be applied to the evaluation of bacterial gene expression in humans. Consequently, with few exceptions (41), little is known about changes in bacterial gene expression in the context of human infection. Here, we use QRT-PCR for direct quantification of MTB mRNAs in bacteria grown in vitro and in tissues obtained from infected mice and TB patients. We report the differential expression of MTB genes involved in the adaptive responses to iron limitation, glucose starvation, fatty acid substrates, and cellular hypoxia, conditions that are thought to occur within the granulomatous lesions of TB, when bacteria were grown in vivo (lungs of mice or TB patients) as compared with being grown in vitro (liquid cultures). Quantitative differences were also observed between the levels of target mRNAs in mice as compared with TB patients; the significance of these observations is unknown, however, they do tend to support the idea that the physiology of MTB in vivo might vary with the host species, and underscore the importance of studying host–pathogen interactions in the natural host.
The results of our murine studies were highly consistent between experiments; in contrast, there was considerable variability in results obtained with lung specimens from different patients. We believe that these differences are real and are likely to be biologically significant, although we do not understand the underlying causes. A number of factors could plausibly influence MTB gene expression in humans: age, nutritional status, immune status, concurrent infections, stage of disease, region of the lung affected, cellular composition of individual lesions, etc. The correlation of specific patient characteristics and specific patterns of MTB gene expression in patients is an important goal; however, the identification of correlates will require the analysis of large numbers of patients and specimens.
Our observation that the in vivo induction (Fig. 2 A and B) and repression (Fig. 3 A and B) of iron-responsive MTB genes was most pronounced in late-stage infected mice suggests the possibility of a link between growth arrest and iron limitation at sites of infections. Consistent with this hypothesis is clinical and experimental evidence that susceptibility to TB is increased in humans and mice suffering from iron overload (42–44), and that MTB virulence is attenuated by mutations that interfere with adaptation to iron-restricted environments (28, 45, 46). The host mechanism(s) that might restrict iron availability to MTB within macrophage phagosomes is unknown; however, cytokine activation of macrophages has been shown to result in down-regulation of transferrin receptor expression and reduced iron uptake (47, 48), as well as maturation of mycobacteria-containing phagosomes to a stage where they no longer intersect with endosomal vesicles carrying iron-loaded transferrin (49, 50).
Our results are also consistent with the hypothesis that MTB switches its metabolism to use of fatty acid substrates in late-stage infected mice (15). Sustained metabolism of fatty acid substrates depends on carbon anaplerosis through the glyoxylate shunt and de novo synthesis of glucose-6-phosphate through gluconeogenesis. We showed previously (15) that deletion of icl1 encoding isocitrate lyase, an enzyme of the glyoxylate shunt, had no effect on early growth in vivo, but impaired MTB persistence at later stages of infection. Similarly, deletion of pckA in M. bovis bacillus Calmette–Guérin had little effect on the bacterial load in mice during the first 5 wk of infection, but survival of the pckA mutant thereafter was impaired (35). The factors responsible for triggering the putative switch in MTB metabolism in vivo are unknown, although previous studies (15) have suggested a role for the host immune response.
There are a number of possible explanations for the observed differences in MTB gene expression in human vs. murine lung specimens. There could be host-specific differences in the pulmonary tissue environment in humans and mice, or MTB might deploy host-specific mechanisms for acquisition of key nutrients such as iron (51). The growth state of MTB might be different in humans with chronic active TB as compared with persistently infected mice; differences in the human and murine immune responses at the site of infection might also influence bacterial gene expression. With respect to the latter points, it is important to stress that in humans TB comprises a spectrum, ranging from latent TB infection (LTBI) with no clinical signs or symptoms, to progressive and life-threatening disease. The human lung specimens analyzed here represent one end of the spectrum, and it is not clear whether the persistently infected mouse is an accurate model of this state of disease, or should, instead, be regarded as a model of LTBI (3). It might be possible, in the future, to clarify these issues through quantitative analysis of MTB mRNAs in tissues obtained from individuals with LTBI, where the host response has successfully contained the pathogen. This approach will be technically challenging, due to the paucity of mycobacterial RNA obtainable from individuals with LTBI (our unpublished results). If feasible, comparative analysis of MTB gene expression in individuals with LTBI vs. individuals with active TB could shed new light on an important and unresolved issue, namely, why some infected individuals develop progressive and potentially fatal disease, whereas others harbor the organism for a lifetime without ever becoming ill.
Supplementary Material
Acknowledgments
This work was supported by grants from GlaxoSmithKline and the Sequella Global Tuberculosis Foundation (J.D.M.) and by National Institutes of Health Grants AI051702 (to J.D.M.), AI054338 (to G.K.), and HL64544 (to I.S.). L.-G.B. and F.A.P. were supported by Fogarty Fellowship AITRP TW00231.
Abbreviations: TB, tuberculosis; MTB, Mycobacterium tuberculosis; LTBI, latent TB infection; QRT-PCR, quantitative real-time RT-PCR; cfu, colony-forming units; PckA, phosphoenolpyruvate carboxykinase; Pca, pyruvate carboxylase.
References
- 1.Iseman, M. D. (2000) in A Clinician's Guide to Tuberculosis (Lippincott Williams & Wilkins, Philadelphia), pp. 129-144.
- 2.Nyka, W. (1962) Am. Rev. Respir. Dis. 85, 33-39. [DOI] [PubMed] [Google Scholar]
- 3.Flynn, J. L. & Chan, J. (2001) Infect. Immun. 69, 4195-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lillebaek, T., Dirksen, A., Baess, I., Strunge, B., Thomsen, V. O. & Andersen, A. B. (2002) J. Infect. Dis. 185, 401-404. [DOI] [PubMed] [Google Scholar]
- 5.Dye, C., Scheele, S., Dolin, P., Pathania, V. & Raviglione, M. C. (1999) J. Am. Med. Assoc. 282, 677-686. [DOI] [PubMed] [Google Scholar]
- 6.Barnes, H. L. & Barnes, L. R. P. (1928) Am. Rev. Tuberc. 18, 412-424. [Google Scholar]
- 7.van Leuven, M., De Groot, M., Shean, K. P., von Oppell, U. O. & Willcox, P. A. (1997) Ann. Thorac. Surg. 63, 1368-1372. [PubMed] [Google Scholar]
- 8.Fenhalls, G., Stevens, L., Moses, L., Bezuidenhout, J., Betts, J. C., van Helden, P., Lukey, P. T. & Duncan, K. (2002) Infect. Immun. 70, 6330-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenhalls, G., Stevens, L., Bezuidenhout, J., Amphlett, G. E., Duncan, K., Bardin, P. & Lukey, P. T. (2002) Immunology 105, 325-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S., Barry, C. E., III, et al. (1998) Nature 393, 537-543. [DOI] [PubMed] [Google Scholar]
- 11.Chen, P., Ruiz, R. E., Li, Q., Silver, R. F. & Bishai, W. R. (2000) Infect. Immun. 68, 5575-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zahrt, T. C. & Deretic, V. (2000) J. Bacteriol. 182, 3832-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manganelli, R., Voskuil, M. I., Schoolnik, G. K. & Smith, I. (2001) Mol. Microbiol. 41, 423-437. [DOI] [PubMed] [Google Scholar]
- 14.Graham, J. E. & Clark-Curtiss, J. E. (1999) Proc. Natl. Acad. Sci. USA 96, 11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKinney, J. D., Höner zu Bentrup, K., Muñoz-Elías, E. J., Miczak, A., Chen, B., Chan, W. T., Swenson, D., Sacchettini, J. C., Jacobs, W. R., Jr., & Russell, D. G. (2000) Nature 406, 735-738. [DOI] [PubMed] [Google Scholar]
- 16.Wilkinson, R. J., DesJardin, L. E., Islam, N., Gibson, B. M., Kanost, R. A., Wilkinson, K. A., Poelman, D., Eisenach, K. D. & Toossi, Z. (2001) Mol. Microbiol. 39, 813-821. [DOI] [PubMed] [Google Scholar]
- 17.McKinney, J. D., Bloom, B. R. & Modlin, R. L. (2001) in Samter's Immunologic Diseases, eds. Austen, K. F., Frank, M. M., Atkinson, J. P. & Cantor, H. (Lippincott Williams & Wilkins, Baltimore), 6th Ed., pp. 995-1012.
- 18.Manganelli, R., Tyagi, S. & Smith, I. (2001) in Mycobacterium tuberculosis Protocols, eds. Parish, T. & Stoker, N. (Humana, Totowa, NJ), pp. 295-310.
- 19.Moreira, A. L., Wang, J., Tsenova-Berkova, L., Hellmann, W., Freedman, V. H. & Kaplan, G. (1997) Infect. Immun. 65, 305-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.North, R. J. (1995) J. Infect. Dis. 172, 1550-1553. [DOI] [PubMed] [Google Scholar]
- 21.Rozen, S. & Skaletsky, H. (2000) Methods Mol. Biol. 132, 365-386. [DOI] [PubMed] [Google Scholar]
- 22.Shi, L., Jung, Y. J., Tyagi, S., Gennaro, M. L. & North, R. J. (2003) Proc. Natl. Acad. Sci. USA 100, 241-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manganelli, R., Dubnau, E., Tyagi, S., Kramer, F. R. & Smith, I. (1999) Mol. Microbiol. 31, 715-724. [DOI] [PubMed] [Google Scholar]
- 24.Boshoff, H. I., Reed, M. B., Barry, C. E., III, & Mizrahi, V. (2003) Cell 113, 183-193. [DOI] [PubMed] [Google Scholar]
- 25.Dawes, S. S., Warner, D. F., Tsenova, L., Timm, J., McKinney, J. D., Kaplan, G., Rubin, H. & Mizrahi, V. (2003) Infect. Immun. 71, 6124-6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rees, R. J. W. & Hart, P. D. (1961) Br. J. Exp. Pathol. 42, 83-88. [PMC free article] [PubMed] [Google Scholar]
- 27.Wallace, J. G. (1961) Am. Rev. Respir. Dis. 83, 866-871. [DOI] [PubMed] [Google Scholar]
- 28.De Voss, J. J., Rutter, K., Schroeder, B. G. & Barry, C. E., III (1999) J. Bacteriol. 181, 4443-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez, G. M., Gold, B., Gomez, B. M., Dussurget, O. & Smith, I. (1999) Tuber. Lung Dis. 79, 287-298. [DOI] [PubMed] [Google Scholar]
- 30.Wong, D. K., Lee, B. Y., Horwitz, M. A. & Gibson, B. W. (1999) Infect. Immun. 67, 327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gold, B., Rodriguez, M., Marras, S., Pentecost, M. & Smith, I. (2001) Mol. Microbiol. 42, 851-865. [DOI] [PubMed] [Google Scholar]
- 32.Smith, C. V., Huang, C.-C., Miczak, A., Russell, D. G., Sacchettini, J. C. & Höner zu Bentrup, K. (2003) J. Biol. Chem. 278, 1735-1743. [DOI] [PubMed] [Google Scholar]
- 33.Wendisch, V. F., Spies, M., Reinsheid, D. J., Schnicke, S., Sahm, H. & Eikmanns, B. J. (1997) Arch. Microbiol. 168, 262-269. [DOI] [PubMed] [Google Scholar]
- 34.Tuckman, D., Donnelly, R. J., Zhao, F. X., Jacobs, W. R., Jr., & Connell, N. D. (1997) J. Bacteriol. 179, 2724-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, J., Yu, J. & Russell, D. G. (2003) Microbiology 149, 1829-1835. [DOI] [PubMed] [Google Scholar]
- 36.Collins, D. M., Wilson, T., Campbell, S., Buddle, B. M., Wards, B. J., Hotter, G., De Lisle & G. W. (2002) Microbiology 148, 3019-3027. [DOI] [PubMed] [Google Scholar]
- 37.Yuan, Y., Crane, F. D. & Barry, C. E., III (1996) J. Bacteriol. 178, 4484-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garbe, T. R., Hibler, N. S. & Deretic, V. (1999) Infect. Immun. 67, 460-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cooper, C. E. & Davies, N. A. (2000) Biochim. Biophys. Acta 1459, 390-396. [DOI] [PubMed] [Google Scholar]
- 40.Angelichio, M. J. & Camilli, A. (2002) Infect. Immun. 70, 6518-6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merrell, D. S., Butler, S. M., Qadri, F., Dolganov, N. A., Alam, A., Cohen, M. B., Calderwood, S. B., Schoolnik, G. K. & Camilli, A. (2002) Nature 417, 642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moyo, V. M., Gangaidzo, I. T., Gordeuk, V. R., Kiire, C. F. & Macphail, A. P. (1997) Cent. Afr. J. Med. 43, 334-339. [PubMed] [Google Scholar]
- 43.Lounis, N., Truffot-Pernot, C., Grosset, J., Gordeuk, V. R. & Boelaert, J. R. (2001) J. Clin. Virol. 20, 123-126. [DOI] [PubMed] [Google Scholar]
- 44.Schaible, U. E., Collins, H. L., Priem, F. & Kaufmann, S. (2002) J. Exp. Med. 196, 1507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manabe, Y. C., Saviola, B. J., Sun, L., Murphy, J. R. & Bishai, W. R. (1999) Proc. Natl. Acad. Sci. USA 96, 12844-12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Voss, J. J., Rutter, K., Schroeder, B. G., Su, H., Zhu, Y. & Barry, C. E., III (2000) Proc. Natl. Acad. Sci. USA 97, 1252-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mulero, V. & Brock, J. H. (1999) Blood 94, 2383-2389. [PubMed] [Google Scholar]
- 48.Kim, S. & Ponka, P. (2000) J. Biol. Chem. 275, 6220-6226. [DOI] [PubMed] [Google Scholar]
- 49.Schaible, U. E., Sturgill-Koszycki, S., Schlesinger, P. H. & Russell, D. G. (1998) J. Immunol. 160, 1290-1296. [PubMed] [Google Scholar]
- 50.MacMicking, J. D., Taylor, G. A. & McKinney, J. D. (2003) Science 302, 654-659. [DOI] [PubMed] [Google Scholar]
- 51.Schryvers, A. B. & Stojiljkovic, I. (1999) Mol. Microbiol. 32, 1117-1123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.