Abstract
Dengue virus is a worldwide-distributed mosquito-borne flavivirus with a positive strand RNA genome. Its transcribed polyprotein is cleaved by host- and virus-encoded peptidases into 10 proteins, some of which are of unknown function. Although dengue virus-infected cells seem to be resistant to the antiviral action of IFN, the viral products that mediate this resistance are unknown. Therefore, we have analyzed the ability of the 10 dengue virus-encoded proteins to antagonize the IFN response. We found that expression in human A549 cells of the dengue virus nonstructural proteins NS2A, NS4A, or NS4B enhances replication of an IFN-sensitive virus. Moreover, expression of NS4B and, to a lesser extent, of NS2A and NS4A proteins results in down-regulation of IFN-β-stimulated gene expression. Cells expressing NS4B or infected with dengue virus do not exhibit nuclear signal transducer and activator of transcription (STAT) 1 on treatment with IFN-β or IFN-γ, indicating that NS4B might be involved in blocking IFN signaling during dengue virus infections. This protein, encoded by a positive strand RNA virus, is implicated as an IFN-signaling inhibitor.
Dengue virus (DEN) classifies in the family Flaviviridae (genus Flavivirus) of which >50 other members including West Nile (WN) and yellow fever (YF) viruses have been identified. Transmitted by the mosquito Aedes aegypti, DEN is the most prevalent arthropod-borne virus affecting humans, with >50 million new cases per year. The dengue fever is a relatively mild febrile illness with rash whereas the dengue hemorrhagic fever is a more severe, sometimes lethal disease.
The DEN virion contains a positive strand RNA molecule with an ≈10-kb-long ORF flanked by 5′ and 3′ nontranslated regions. After endocytosis and release of the viral nucleocapsid into the cytosol, a 3,391-aa-long polyprotein is translated from the viral RNA at the surface of the endoplasmic reticulum (ER). The combined activity of host and virus peptidases results in the cleavage of three structural (C, prM, and E) and seven nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) proteins. During viral RNA replication, catalyzed by NS5/NS3 proteins, a negative strand RNA intermediate is generated that is found in association with the genomic positive strand RNA (1, 2).
The onset of the IFN-α/β response in virus-infected cells presumably occurs on viral entry and release/synthesis of viral components, including double-stranded RNA (dsRNA) intermediates. The transcription factors IFN regulatory factor (IRF)-3, IRF-7, NF-κB, and activating transcription factor 2 (ATF2)/c-Jun are activated by some of these viral components and trigger the expression of IFN-α/β (3-7). Secreted IFN-α/β binds to the IFN alpha receptor (IFNAR) on the surface of infected and neighboring cells, resulting in activation of the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway and transcription of numerous genes from promoters containing IFN-stimulated regulatory elements (ISRE). About 100 genes are induced by IFN-α/β, resulting in the induction of an antiviral state (8). The pivotal role of IFN-α/β in protecting against viral infections has been demonstrated by the finding that STAT1-/- mice are extremely sensitive to viral infections (9-11).
The antiviral IFN-mediated response is often hindered by counteracting mechanisms developed by viruses. Experimentally, pretreatment with IFN-α/β or IFN-γ protects human cells against DEN infection (12). However, IFN treatment after DEN infection does not inhibit viral replication, indicating that DEN infection circumvents the action of IFN (12). In addition, the high pathogenicity of DEN in patients exhibiting high titers of IFN suggests that DEN is capable of antagonizing the IFN response (13). Interestingly, multiple anti-IFN mechanisms have been documented for the hepatitis C virus (HCV), a virus related to DEN. HCV NS5A inhibits the eukaryotic translation initiation factor 2α (eIF-2α) kinase PKR (protein kinase R), an IFN-induced protein with antiviral activity (14). The HCV envelope protein E2 also inhibits PKR by competing with eIF-2α as a PKR substrate (15). In addition, HCV NS3/4A protease inhibits activation of IRF-3 and IFN-α/β synthesis (16). Moreover, HCV seems to block the IFN-induced JAK/STAT pathway by mechanisms not yet elucidated (17). Pestiviruses, also related to DEN, express a protein, Npro, that inhibits IFN-α/β synthesis (18). In contrast, the mechanism(s) used by arthropod-born human flaviviruses, including DEN virus, remain unknown. Although HCV and DEN have common structural features, they do not share enough sequence similarity to allow predictions about the possible function of many of their genes. In addition, the Npro gene of pestivirus is not present in DEN and other members of the genus Flavivirus.
We have analyzed the ability of the 10 proteins encoded by DEN type 2 (DEN-2) to block the IFN system. Our results show that NS4B strongly blocks the IFN-induced signal-transduction cascade by interfering with STAT1 function. In addition, NS4A and, to a lesser extent, NS2A also block IFN signaling, and the cumulative effect of the three proteins inhibits IFN signaling completely. This inhibition is also observed in DEN-infected cells, suggesting a role for NS4B in DEN pathogenicity.
Materials and Methods
Plasmid Constructs, Virus Strains, and Cell Lines. The DEN-2 infectious cDNA clone pD2/IC-30P-A (19) served as template for the cloning of the 10 DEN-2 genes and to derive infectious virus used in this manuscript. This clone was kindly provided by R. Kinney (Arbovirus Disease Branch, Centers for Disease Control and Prevention, Fort Collins, CO). Each DEN-2 gene was cloned in the mammalian expression vector pCAGGS containing the β-actin promoter (20). Where appropriate, 5′ end PCR primers were targeted to upstream gene regions to include the signal peptide necessary for ER translocation and signal peptidase cleavage of the encoded protein (Table 1, which is published as supporting information on the PNAS web site). Hemagglutinin (HA) tags were placed at the 3′ end of each DEN-2 gene to generate C-terminal HA-tagged DEN-2 proteins. Two reporter plasmids containing ISRE promoters driving the expression of chloramphenicol acetyl transferase (CAT) were used: pHISG-54-CAT (21) and pISRE4-9-27-CAT (4), the latter kindly provided by M. Whatelet (Department of Molecular and Cellular Physiology, University of Cincinnati). pCAGGS-NS1(SAM) and pCAGGS-NiPV(-C) express the influenza A/PR8/34 virus NS1 and the Nipah virus V proteins, respectively (22, 23). The firefly luciferase (FL) control plasmid is also a pCAGGS-based plasmid. For the tumor necrosis factor (TNF) signal transduction assays, the plasmid pκB-luc encoding the FL gene under the control of NF-κB responsive promoter was used (24). The pRL-TK-luc (Promega) encoding Renilla luciferase (RL) under the control of herpes simplex virus (HSV) thymidine kinase promoter was used as an internal control of transfection efficiency in the TNF-signaling assays. The pEAK-8 plasmid was kindly provided by A. Ting (Department of Immunobiology, Mount Sinai School of Medicine). This plasmid expresses IκBαS32/36A, a dominant-negative mutant of IκBα (IκBαDN) containing serine-alanine substitutions at positions 32 and 36, and it was used as a control of TNF-signaling inhibition. The construction and growth of the GFP-tagged Newcastle disease virus (NDVGFP) is described elsewhere (23). Sendai virus (SeV), Cantell strain, was propagated at 37°C in 10-day-old embryonated chicken eggs. 293T, Vero, LLCMK2, and A549 cells were obtained from American Type Culture Collection (ATCC) and maintained in DMEM containing 10% FBS. Chicken embryo fibroblasts (CEF) where obtained from 10-day-old chicken embryos and maintained in MEM with 10% FBS.
Transfection and NDV-GFP Infection of CEF and A549 Cells. CEF and A549 cells were seeded onto 24-well plates. In transfection experiments, media were replaced by DMEM supplemented with 5% FBS for both CEF and A549 cells. To transfect CEF, 2 μg of plasmid DNA diluted in 50 μl of OptiMEM (GIBCO) and 6 μl of FuGENE 6 (Roche) diluted in 50 μl of OptiMEM were mixed. Transfection mixtures were incubated at room temperature for 20 min and added to the cells. A549 cells were transfected by using Lipofectamine 2000 (Invitrogen) as specified by the manufacturer. After 20 h of incubation at 37°C, transfected cells were washed with PBS and infected with NDV-GFP at a multiplicity of infection (moi) of 1-2 at room temperature for 60 min. The inoculum was then aspirated, and cells were maintained in MEM-10% FBS (CEF) or DMEM-10%FBS (A549). The infected cells were incubated at 37°C before detection of GFP expression by fluorescence microscopy.
Reporter Gene Assays. 293T cells were transfected by using calcium phosphate (25). Vero cells were transfected by using Lipofectamine 2000 (Invitrogen). Each transfection of 5 × 106 cells contained three plasmids: 1.2 μg of either pHISG-54-CAT or pISRE4-9-27-CAT, 0.5 μg of pCAGGS-FL, and 5 μg of a pCAGGS construct of interest. Twenty-four hours posttransfection, cells were mock-treated or treated [293T cells were infected with SeV, moi of ≈2, and Vero cells were incubated with 1,000 units of human IFN-β (Calbiochem)]. Cells were maintained in DMEM/1% FBS. Twenty-four hours posttreatment, cells were harvested and lysed. CAT assays were performed as described (26). Luciferase assays were performed by using a luciferase assay system (Promega). In the NF-κB-FL assays, Vero cells were transfected with empty plasmid, pCAGGS-NS4B-HA, or pEAK-8-IκBαDN. FL and RL constructs were cotransfected. Twenty-four hours after transfection, cells were incubated for 30 min in the presence of TNF-α (10 ng/ml; R & D Systems), and FL and RL activities were measured by using a dual luciferase assay (Promega).
Immunofluorescence. Cells were grown on microscope coverslips and fixed/permeabilized in cold acetone:methanol (1:1). Cell nuclei were permeabilized for 5 min in 0.5% Nonidet P-40 (Sigma). Dengue mouse antibody (Biogenesis, Bournemouth, U.K.), HA-TAG mouse antibodies (Sigma), phosphorylated STAT1 (Tyr-701) rabbit polyclonal antibody (Cell Signaling Technology, Beverly, MA), and rabbit polyclonal anti-calnexin (N and C terminus) (BD Biosciences) were used as primary antibodies. Texas-red- and FITC-conjugated anti-rabbit or anti-mouse (Jackson Immunochemicals) were used as secondary antibodies. Nuclear chromatin staining was performed by incubation in a PBS solution containing 0.5 mg/ml 4′,6-diamidino-2-phenylindole (DAPI, Sigma). Human IFN-β and IFN-γ were used at 1,000 units/ml in STAT1 induction studies.
Results
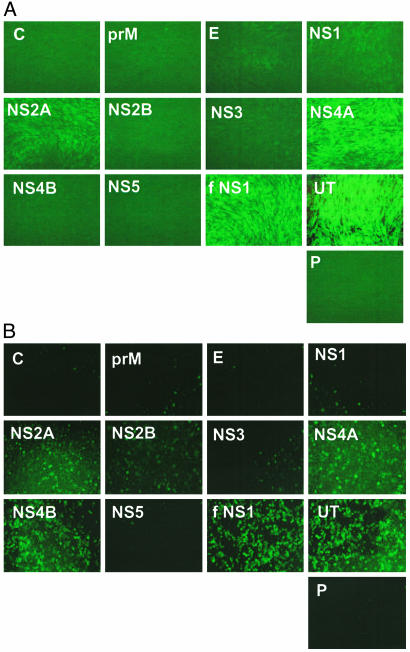
Detection of DEN-2 IFN Antagonist Proteins. The 10.7-kb RNA genome of DEN-2 encodes 10 polypeptides: C, prM, E, NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 proteins. The corresponding genes were amplified by PCR from pD2/IC-30P-A and cloned in pCAGGS(3′HA) to generate plasmids expressing C terminus HA-tagged DEN-2 proteins. Western blots indicated that all HA-tagged DEN proteins were expressed to comparable levels in transfected cells (data not shown). We and others have reported that IFN is produced after lipid-mediated transfection of cells with plasmid DNA (23, 27). As a consequence, replication of an IFN-sensitive virus, such as NDV, is greatly inhibited. Transfection of plasmids expressing known IFN antagonists, such as influenza A virus NS1 (fNS1), Ebola virus VP35, and Nipah virus V, W, and C proteins, enhances replication of a NDV-expressing GFP (NDVGFP) in plasmid-transfected CEF (23). Therefore, we attempted to identify potential DEN-encoded IFN antagonists based on their ability to facilitate NDV-GFP replication in transfected CEF. In addition, we have adapted this screening assay to human cells (A549) to detect antagonists of the human IFN response. CEF (Fig. 1A) and A549 cells (Fig. 1B) were transfected with each of the 10 DEN-2-derived plasmids. Twenty-four hours posttransfection, cells were infected with NDV-GFP, and GFP expression was analyzed the following day by fluorescence microscopy. The results revealed that NDV-GFP replication was enhanced in CEF transfected with DEN-2 NS2A and NS4A expression plasmids. We found that NDV-GFP replication was enhanced in A549 expressing NS2A and NS4A, but an additional DEN-2 protein, NS4B, also promoted viral replication. As expected, NDV-GFP replication was compromised in empty plasmid transfected cells and enhanced in cells expressing fNS1 (Fig. 1). These results pointed to the identification of three potential DEN-2 anti-IFN proteins, NS2A, NS4A, and NS4B. We considered particularly relevant the results obtained in A549 cells because these are human cells.
Fig. 1.
Identification of DEN-2 proteins that facilitate NDV-GFP replication in the presence of an IFN response. CEF (A) and A549 (B) cells were transfected with pCAGGS plasmids expressing HA-tagged DEN-2 proteins (C, prM, E, NS1, NS2A, NS2B, NS3, NS4A, NS4B, or NS5), influenza A virus NS1 (fNS1), empty pCAGGS (P), or untransfected (UT). Twenty-four hours later, cells were infected with NDV-GFP, and green fluorescence was visualized 24 h after infection under the fluorescence microscope.
Although the ability of DEN-2 NS2A, NS4A, and NS4B proteins to enhance replication of NDV-GFP suggests that they inhibit the IFN system, additional experiments are needed to demonstrate that this is the case. We then analyzed the ability of DEN-2 proteins to block either IFN induction by viral infection (Fig. 2A) or IFN-stimulated signal transduction (Fig. 2B) by using a CAT reporter gene under the control of an ISRE promoter. Virus-induced activation of the ISRE-54 promoter is known to be mediated by activation of IRF-3, a transcription factor also involved in IFN-β promoter activation (28). 293T cells have been demonstrated to be a suitable model to study the effects that a foreign IFN antagonist protein has in virus-induced activation of ISRE (29). High transfection efficiency of plasmids is obtained by using calcium phosphate, and unpublished results in our lab indicate that direct stimulation of ISRE-54 by endogenous IFN does not occur in 293T cells. To determine whether DEN-2 proteins inhibit virus-mediated activation of the ISRE-54 promoter, 293T cells were calcium phosphate transfected with an ISRE-54-CAT plasmid and pCAGGS plasmids expressing DEN-2 proteins. A plasmid expressing FL was used to control for transfection efficiency. Empty pCAGGS plasmid provided a control for basal expression of the ISRE reporter gene. After transfection, only very low levels of CAT activity were detected (data not shown). CAT activity, indicative of activation of the ISRE-54 promoter, was readily detected 24 h after infection with SeV (Fig. 2 A, penultimate column). None of the DEN-2 proteins significantly inhibited CAT expression, indicating that these proteins do not inhibit virus-mediated activation of the ISRE-54 promoter in 293T cells. By contrast, expression of fNS1, a known inhibitor of IRF-3 activation (22), resulted in strong inhibition of CAT activity. FL values in all transfections were comparable (data not shown), indicating similar transfection efficiencies and a lack of general inhibition of gene expression by individual DEN-2 proteins.
Fig. 2.
Inhibition of virus- and IFN-β-mediated induction of ISRE by DEN-2 proteins. (A) Fold induction of ISRE-54-CAT reporter gene on infection with SeV. 293T cells were transfected with pCAGGS plasmids expressing the indicated DEN-2 proteins, pCAGGS-fNS1, or empty pCAGGS (P), as well as with ISRE-54-CAT and pCAGGS-FL reporter plasmids. Twenty-four hours posttransfection, cells were infected with SeV (moi of ≈2) (+SeV) or mock-infected (-SeV), and CAT and luciferase activities were measured 24 h later. The CAT activities were normalized to the corresponding FL activities to determine the fold induction. (B) Fold induction of ISRE-54-CAT reporter gene on treatment with IFN-β. Vero cells were transfected with each of the indicated plasmids as in A. As positive control of IFN-signaling inhibition, pCAGGS-NipV was used. Twenty-four hours posttransfection, cells were treated with 1,000 units of human IFN-β (+IFN-β) or mock-treated (-IFN-β). CAT and luciferase activities were measured 24 h posttreatment. Average values and SD from three independent experiments are shown, except in the case of the NipV, which was used as positive control of inhibition of IFN signaling in only one of the experiments. Asterisks indicate significant differences (P < 0.05) as compared with empty plasmid-transfected cells.
In addition to directly responding to IRF-3 activation, the ISRE-54 promoter is stimulated by IFN-α/β through the activation of the STAT1/STAT2/IRF-9 transcription factor (ISGF3). To investigate the ability of the 10 DEN-2 proteins to inhibit IFN-mediated signal transduction, we studied ISRE-54 promoter activation in transfected Vero cells after stimulation with exogenously added IFN-β. Vero cells do not produce IFN (30), and therefore the IFN signaling detected is due only to stimulation by exogenous IFN and not by IFN that might be produced due to transfection. Each DEN-2 protein expression plasmid was transfected in Vero cells in combination with the ISRE-54-CAT reporter plasmid. Cells were incubated with IFN-β 24 h later, and cell lysates were prepared and assayed for CAT activities (Fig. 2B). We found that expression of NS4B (P = 0.04) and, to a lesser extent, NS2A (P = 0.06) and NS4A (P = 0.03) resulted in reduced CAT expression, suggesting that these DEN-2 proteins interfere with the IFN-α/β-signaling pathway. As expected, expression of Nipah virus V protein, a known inhibitor of IFN signaling (23, 31), also resulted in inhibition of ISRE-54-CAT activation by IFN-β (Fig. 2B).
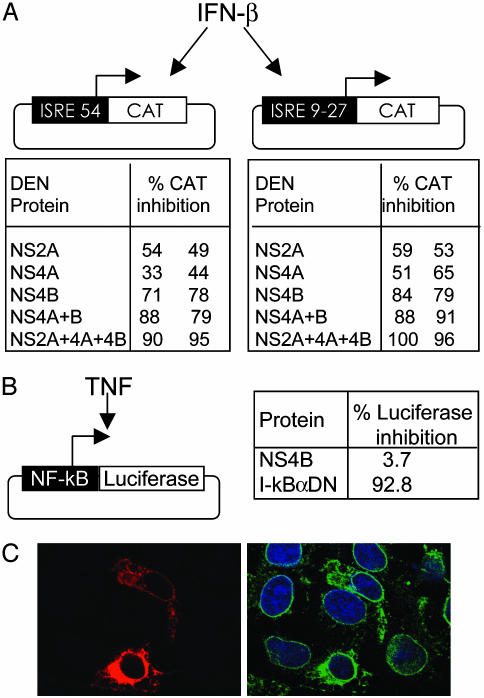
DEN-2 NS4B Inhibits IFN-β Signaling. Our reporter gene assays show that expression of NS2A, NS4A, or NS4B proteins of DEN-2 inhibits IFN-β stimulation of ISRE-54 to different extents. To further assess the specificity of this inhibition, Vero cells were transfected with pCAGGS-NS2A-HA, NS4A-HA, or NS4B-HA in combination with a plasmid containing the ISRE-9-27 promoter driving CAT expression. In addition, we tested the combined effects of these three DEN proteins. The ISRE-9-27 promoter is stimulated by IFN and not by virus-mediated activation of IRFs (4, 32). After IFN treatment, CAT activity was measured and normalized as in previous experiments, and the percentage of CAT inhibition was obtained for NS2A, NS4A, and NS4B. The results with each of the three individual proteins are comparable to the corresponding values obtained with the ISRE-54 promoter. In addition, we found that the antagonistic effects of these proteins in the IFN pathway is stronger when these proteins are coexpressed (Fig. 3A).
Fig. 3.
DEN-2 NS4B blocks IFN-β signaling and localizes to the ER. (A) Percentage of inhibition of CAT activity due to expression of the indicated DEN-2 proteins in Vero cells transfected with ISRE-54-CAT or ISRE4-9-27-CAT reporter constructs and incubated with human IFN-β. Values from two independent experiments are shown. (B) Vero cells were transfected with plasmids expressing DEN-2 NS4B or IκBαDN and a reporter plasmid expressing FL from an NF-κB-responsive promoter. A plasmid expressing RL under the control of the herpes simplex virus TK promoter was cotransfected to normalize FL values. Twenty-four hours posttransfection, cells were incubated with TNF for 30 min, and FL and RL activities were measured. (C) Detection of NS4B in pCAGGS-NS4B-HA-transfected Vero cells by immunofluorescence using an anti-HA mouse MoAb (Left, red). (Right) The same field stained in green with polyclonal antibodies specific for calnexin (an ER resident protein); DAPI (4′,6-diamidino-2-phenylindole) staining (blue) indicates nuclear chromatin.
In the next set of experiments, we focused our studies on NS4B because this protein showed the strongest induction of IFN-mediated gene expression. To determine the specificity of NS4B for IFN signaling, we tested the potential of this protein to inhibit a different signal transduction pathway, namely the activation of NF-κB by TNF. Vero cells were transfected with a plasmid containing an NF-κB responsive promoter driving the expression of FL. This reporter plasmid was cotransfected with a plasmid constitutively expressing RL to control for transfection efficiency, and with pCAGGS-NS4B-HA. A plasmid expressing IκBαDN provided a positive control for repression of NF-κB. The transfected Vero cells were incubated with TNF, and FL and RL activities were determined. We found that FL values were not altered by expression of NS4B, indicating that TNF-signaling pathway is not inhibited by this protein (Fig. 3B). Therefore, NS4B does not cause a general inhibition of cell-signaling pathways. We also confirmed that in these experiments NS4B is properly located in association with the ER (33, 34) by double immunofluorescence analysis of NS4B-HA and of the endogenous ER protein calnexin (Fig. 3C). NS2A and NS4A are also ER-associated (data not shown).
Inhibition of IFN-Mediated STAT1 Phosphorylation by DEN-2 NS4B. IFN-α/β binds to the cell surface receptor IFN-α receptor, resulting in activation of JAK1 and TYK2 kinases, which then tyrosine phosphorylate STAT1 and STAT2, causing their heterodimerization and nuclear translocation. To further analyze the inhibition of IFN signaling by NS4B, we tested the effects of NS4B in IFN-mediated phosphorylation (activation) of STAT1. LLCMK2 cells were transfected with pCAGGS or with pCAGGS-NS4B-HA and stimulated with IFN-β. NS4B-HA and tyrosine phosphorylated STAT1 were then visualized by immunofluorescence (Fig. 4A). Phosphorylated STAT1 was undetectable in cells not treated with IFN-β (data not shown). By contrast, all cells transfected with empty plasmid exhibited intense nuclear staining indicative of phosphorylated STAT1 on IFN-β treatment (not shown). Strikingly, phosphorylated STAT1 was undetectable in the majority (77%) of IFN-β-treated cells expressing NS4B (Fig. 4A). Immunofluorescence with a STAT1-specific antibody indicated that NS4B-expressing cells had levels of cytoplasmic STAT1 similar to those in nonexpressing cells (not shown), indicating that the lack of phosphorylated STAT1 is probably not due to STAT1 degradation.
Fig. 4.
Expression of NS4B and DEN-2 infection inhibit IFN-mediated STAT1 phosphorylation. (A) LLCMK2 cells were transfected with pCAGGS-NS4B-HA. Twenty-four hours posttransfection, cells were stimulated with IFN-β or IFN-γ for 30 min and processed for immunofluorescence. NS4B-HA was detected by using a monoclonal antibody against HA and donkey anti-mouse FITC-conjugated antibody (green). Phosphorylated STAT1 was visualized by using a polyclonal antibody against phosphorylated STAT1 and a donkey anti-rabbit Texas red-conjugated antibody (red). (B) LLCMK2 cells infected with DEN-2 at an moi of 1-2. After 6 days, cells were starved for 2 h and incubated in the presence of human IFN-β or IFN-γ for 30 min at 37°C. Cells were permeabilized and immunostained by using monoclonal antibody for DEN-2 E protein and Texas red-conjugated donkey anti-mouse (red). Phosphorylated STAT1 was visualized by using a polyclonal antibody against phosphorylated STAT1 and FITC-conjugated donkey anti-rabbit (green). Arrows show examples of cells with no or low levels of nuclear staining of phosphorylated STAT1 and high expression of NS4B or DEN-2 epitopes.
Tyrosine phosphorylation and nuclear translocation of STAT1 is also induced by IFN-γ. Stimulation of IFN-γ receptor causes phosphorylation of the JAK1 and JAK2 kinases, which in turn phosphorylate STAT1. STAT1 homodimers translocate to the nucleus and stimulate transcription of IFN-γ responsive genes. To determine whether NS4B also prevents IFN-γ-mediated phosphorylation of STAT1, we performed similar immunofluorescence experiments in cells transfected with a pCAGGSNS4B-HA and treated with IFN-γ. The results (Fig. 4 Lower) indicate that expression of NS4B also blocks IFN-γ phosphorylation (activation) of STAT1 (83% of transfected cells).
Inhibition of IFN-Mediated STAT1 Phosphorylation by DEN-2 Infection. To determine whether the inhibition of the IFN-signaling pathway also occurs during DEN-2 infection, we investigated STAT1 phosphorylation in LLCMK2 cells infected with DEN-2 and stimulated with IFNs. LLCMK2 cells were infected with DEN-2 for 6 days, followed by treatment with IFN-β or IFN-γ (Fig. 4B) for 30 min. The presence of virus antigens and phosphorylated STAT1 was visualized by immunofluorescence. The results revealed that 89% and 93% of the infected cells did not exhibit phosphorylated STAT1 in response to IFN-β and IFN-γ, respectively, whereas almost 100% of uninfected cells exhibited phosphorylated nuclear STAT1 after IFN-β or IFN-γ stimulation. Therefore, the inhibition of IFN signaling found in cells expressing NS4B is also present in DEN-2-infected cells. Collectively, the data suggest that NS4B contributes to the inhibition of IFN signaling during DEN-2 infection.
Discussion
Our results show two lines of evidence indicating that NS2A, NS4A, and NS4B proteins of DEN-2 are IFN antagonists. The NDV-GFP-based assay allows the detection of viral-encoded IFN antagonists acting through different mechanisms (23); for example, the fNS1 blocks IFN production as well as the activation of the IFN-inducible enzyme protein kinase R (35) whereas the V protein of Nipah virus blocks IFN signaling (23, 36). We found that expression of DEN-2 NS2A, NS4A, and NS4B facilitates NDV-GFP replication in human A549 cells whereas only NS2A and NS4A increased replication of NDVGFP in CEF. The differences in NS4B action between human and chicken cells may be indicative of a species-specific mechanism of IFN inhibition. Species specificity has also been reported for the IFN antagonist V proteins of several paramyxoviruses (37, 38). The second line of evidence implicating NS2A, NS4A, and NS4B as IFN antagonists comes from reporter gene assays. Expression of DEN-2 NS4B, and to a lesser extent NS2A and NS4A, inhibited the activation of two different ISRE promoters in response to IFN-β (Fig. 3A), suggesting that these proteins inhibit IFN-β signaling. Interestingly, coexpression of NS2A, NS4A, and NS4B blocked ISRE-54- and ISRE-9-27-driven CAT activity in response to IFN-β almost completely. All these three proteins are located in association with the ER. Because interactions between ER-associated proteins of flaviviruses have been described (39, 40), it is tempting to speculate that these three proteins might interact during DEN infection, resulting in strong IFN inhibition by a mechanism not yet identified. Future work will focus on elucidating those possible interactions.
Our work has mostly focused on NS4B because this protein exhibited the highest inhibitory effects on ISRE promoter activation. We further defined NS4B anti-IFN function by investigating STAT1 tyrosine phosphorylation (activation) in response to IFN by immunofluorescence. In these assays, IFN-β treatment of cells expressing NS4B did not result in detectable levels of nuclear phosphorylated STAT1, suggesting that NS4B blocks IFN signaling by preventing phosphorylation of STAT1. Alternatively, NS4B may promote dephosphorylation or degradation of activated STAT1. NS4B also blocks IFN-γ-mediated phosphorylation of STAT1. Therefore, a possible mechanism by which NS4B could block IFN may involve interactions with common players implicated in both IFN-α/β and IFN-γ-signaling pathways, such as JAK1 or STAT1. Future work is aimed to characterize the mechanism of action of NS4B.
A role for NS4B in flavivirus infections had not been previously established. Interestingly, a mutation in NS4B has recently been found to impair replication of DEN-4 in mosquito cells, but to enhance replication in mammalian cells, indicating a dual role of this protein in viral pathogenesis (41). Whether those findings relate to an IFN-antagonistic function of NS4B remains to be determined. NS4B has a 78-85% amino acid sequence identity among the four DEN serotypes whereas the NS4B proteins of yellow fever, West Nile virus, and DEN exhibit ≈35% amino acid identity, and HCV NS4B bears a negligible resemblance. Despite this divergence, the predicted topology of NS4B, containing several ER and cytoplasmic domains separated by transmembrane regions, is strikingly similar among the Flaviviridae (34, 42). Future work will elucidate whether the anti-IFN function of NS4B is conserved in this family.
Uncleaved NS4A-NS4B of HCV virus causes reduced transport of cell surface proteins through the ER. This inhibitory effect is not seen when the two proteins are cleaved or expressed separately (43). Nevertheless, because HCV NS4B expression induces distinct ER morphological changes (43, 44), it might be possible that DEN-2 NS4B generally inhibits cell surface expression of proteins/receptors, resulting in unspecific inhibition of IFN signaling. However, TNF signaling was not affected by NS4B expression (Fig. 3B), and immunofluorescence experiments have not reveled significant differences in the surface expression of transferrin and IFN-α receptors between cells expressing and not expressing NS4B (data not shown). Our data then point to a specific inhibition of IFN signaling by DEN-2 NS4B.
Inhibitors of IFN signaling have been previously identified in other viral families with DNA or negative strand RNA genomes. Among the negative strand RNA viruses, paramyxovirus V proteins block IFN signaling by inducing STAT degradation or by blocking STAT phosphorylation, and C proteins bind to and inhibit STAT1 (for a recent review, see ref. 45). IFN signaling has also been shown to be inhibited by the adenovirus E1A protein (46, 47) and by the large T antigen of murine polyoma virus (48). By contrast, no examples precede the identification of an IFN-signaling inhibitor encoded by a positive strand RNA virus. Although previous work indicates that the HCV virus may encode IFN-signaling inhibitor(s) (17), no viral proteins have yet been found to be directly responsible for this effect. We have described a previously unrecognized function of NS4B and possibly NS2A and NS4A of DEN-2 as IFN-signaling inhibitors. By counteracting the IFN-mediated antiviral immune response, NS4B is likely to play a role in DEN pathogenicity, and therefore this protein might represent an attractive target for the development of new antiviral compounds against this emerging virus.
Supplementary Material
Acknowledgments
We thank Georg Kochs and members of the Palese, García-Sastre, and Basler laboratories for valuable critiques, comments, and reagents. E. Nistal, R. Cadagan, and N. Morales contributed technical support. R. Kinney provided the DEN-2 infectious clone and valuable protocols. M. Wathelet provided the ISRE4-9-27-CAT plasmid. A. Ting provided the pEAK-8-IFκBαDN. This work was supported by a National Institutes of Health grant fellowship (to J.L.M.-J.), a Fulbright fellowship (to G.G.S.-B.), and National Institutes of Health research grants (to A.G.-S).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DEN, dengue virus; ISRE, IFN-stimulated response element; FL, firefly luciferase; NDV, Newcastle disease virus; RL, Renilla luciferase; SeV, Sendai virus; STAT, signal transducer and activator of transcription; ER, endoplasmic reticulum; IRF, IFN regulatory factor; JAK, Janus kinase; HCV, hepatitis C virus; HA, hemagglutinin; CAT, chloramphenicol acetyl transferase; TNF, tumor necrosis factor; CEF, chicken embryo fibroblast.
References
- 1.Bartholomeusz, A. & Thompson, P. (1999) J. Viral. Hepat. 6, 261-270. [DOI] [PubMed] [Google Scholar]
- 2.Cleaves, G., Ryan, T. & Schlesinger, R. (1981) Virology 111, 73-83. [DOI] [PubMed] [Google Scholar]
- 3.Weaver, B., Kumar, K. & Reich, N. (1998) Mol. Cell. Biol. 18, 1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wathelet, M. G., Lin, C., Parekh, B. S., Ronco, L. V., Howley, P. M. & Maniatis, T. (1998) Mol. Cell 1, 507-518. [DOI] [PubMed] [Google Scholar]
- 5.Yoneyama, M., Suhara, W., Fukuhara, Y., Fukuda, M., Nishida, E. & Fujita, T. (1998) EMBO J. 17, 1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma, S., tenOever, B. R., Grandvaux, N., Zhou, G. P., Lin, R. & Hiscott, J. (2003) Science 300, 1148-1151. [DOI] [PubMed] [Google Scholar]
- 7.Fitzgerald, K. A., McWhirter, S. M., Faia, K. L., Rowe, D. C., Latz, E., Golenbock, D. T., Coyle, A. J., Liao, S. M. & Maniatis, T. (2003) Nat. Immunol. 4, 491-496. [DOI] [PubMed] [Google Scholar]
- 8.Der, S., Zhou, A., Williams, B. & Silverman, R. (1998) Proc. Natl. Acad. Sci. USA 95, 15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durbin, J., Hackenmiller, R., Simon, M. & Levy, D. (1996) Cell 9, 443-450. [DOI] [PubMed] [Google Scholar]
- 10.Meraz, M., White, J., Sheehan, K., Bach, E., Rodig, S., Dighe, A., Kaplan, D., Riley, J., Greenlund, A., Campbell, D., et al. (1996) Cell 84, 431-441. [DOI] [PubMed] [Google Scholar]
- 11.García-Sastre, A., Durbin, R., Zheng, H., Palese, P., Gertner, R., Levy, D. & Durbin, J. (1998) J. Virol. 72, 8550-8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diamond, M., Roberts, T., Edgil, D., Lu, B., Ernst, J. & Harris, E. (2000) J. Virol. 74, 4957-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurane, I., Innis, B., Nimmannitya, S., Nisalak, A., Meager, A. & Ennis, F. (1993) Am. J. Trop. Med. Hyg. 48, 222-229. [DOI] [PubMed] [Google Scholar]
- 14.Gale, M., Blakely, C., Kwieciszewski, B., Tan, S., Dossett, M., Tang, N., Korth, M., Polyak, S., Gretch, D. & Katze, M. (1998) Mol. Cell. Biol. 18, 5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor, D. R., Shi, S. T., Romano, P. R., Barber, G. N. & Lai, M. M. C. (1999) Science 285, 107-110. [DOI] [PubMed] [Google Scholar]
- 16.Foy, E., Li, K., Wang, C., Sumpter, R., Keda, M., Lemon, S. & Gale, M. (2003) Science 300, 1145-1148. [DOI] [PubMed] [Google Scholar]
- 17.Heim, M., Moradpour, D. & Blum, H. (1999) J. Virol. 73, 8469-8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruggli, N., Tratschin, J. D., Schweizer, M., McCullough, K. C., Hofmann, M. A. & Summerfield, A. (2003) J. Virol. 77, 7645-7654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinney, R., Butrapet, S., Chang, G., Tsuchiya, K., Roehring, J., Bharapravati, N. & Gubler, D. (1997) Virology 230, 300-308. [DOI] [PubMed] [Google Scholar]
- 20.Niwa, H., Yamamura, K. & Miyazaki, J. (1991) Gene 108, 193-199. [DOI] [PubMed] [Google Scholar]
- 21.Bluyssen, H., Vlietstra, R., van der Made, A. & Trapman, J. (1994) Eur. J. Biochem. 220, 395-402. [DOI] [PubMed] [Google Scholar]
- 22.Talon, J., Horvath, C., Polley, R., Basler, C., Muster, T., Palese, P. & García-Sastre, A. (2000) J. Virol. 74, 7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park, M., Shaw, M., Muñoz-Jordán, J., Cros, J., Nakaya, T., Bouvier, N., Palese, P., García-Sastre, A. & Basler, C. F. (2002) J. Virol. 77, 1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, X., Li, M., Zheng, H., Muster, T., Palese, P., Beg, A. & García-Sastre, A. (2000) J. Virol. 74, 11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 26.Percy, N., Barclay, W., García-Sastre, A. & Palese, P. (1994) J. Virol. 68, 4486-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bridge, A., Pebernard, S., Ducraux, A., Nicoulaz, A. & Iggo, R. (2003) Nat. Genet. 34, 263-264. [DOI] [PubMed] [Google Scholar]
- 28.Sato, M., Suemori, H., Hata, N., Asagiri, M., Ogasawara, K., Nakao, K., Nakaya, T., Katsuki, M., Noguchi, S., Tanaka, N. & Taniguchi, T. (2000) Immunity 13, 539-548. [DOI] [PubMed] [Google Scholar]
- 29.Basler, C., Mikulosova, A., Martinez-Sobrido, L., Paragas, J., Muhlberger, E., Bray, M., Klenk, H., Palese, P. & García-Sastre, A. (2003) J. Virol. 77, 7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diaz, M. O., Ziemin, S., Le Beau, M. M., Pitha, P., Smith, S. D., Chilcote, R. R. & Rowley, J. D. (1988) Proc. Natl. Acad. Sci. USA 85, 5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parisien, J., Lau, J., Rodriguez, J., Sullivan, B., Moscona, A., Parks, G., Lamb, R. & Horvath, C. (2001) Virology 283, 230-239. [DOI] [PubMed] [Google Scholar]
- 32.Deblandre, G. A., Marinx, O. P., Evans, S. S., Majjaj, S., Leo, O., Caput, D., Huez, G. A. & Wathelet, M. G. (1995) J. Biol. Chem. 270, 23860-23866. [DOI] [PubMed] [Google Scholar]
- 33.Preugschat, F. & Strauss, J. (1991) Virology 185, 689-697. [DOI] [PubMed] [Google Scholar]
- 34.Lundin, M., Monne, M., Widell, A., Von Heijne, G. & Persson, M. A. (2003) J. Virol. 77, 5428-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.García-Sastre, A. (2001) Virology 279, 375-384. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez, J., Parisien, J. & Horvath, C. (2002) J. Virol. 76, 11476-11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young, D. F., Chatziandreou, N., He, B., Goodbourn, S., Lamb, R. A. & Randall, R. E. (2001) J. Virol. 75, 3363-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park, M. S., García-Sastre, A., Cros, J. F., Basler, C. F. & Palese, P. (2003) J. Virol. 77, 9522-9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mackenzie, J., Khromykh, A., Jones, M. & Westaway, E. (1998) Virology 245, 203-215. [DOI] [PubMed] [Google Scholar]
- 40.Lindenbach, B. & Rice, C. (1999) J. Virol. 73, 4611-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanley, K. A., Manlucu, L. R., Gilmore, L. E., Blaney, J. E., Hanson, C. T., Murphy, B. R. & Whitehead, S. S. (2003) Virology 312, 222-232. [DOI] [PubMed] [Google Scholar]
- 42.Qu, L., McMullan, L. K. & Rice, C. (2001) J. Virol. 75, 10651-10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konan, K., Giddings, T., Ikeda, M., Li, K., Lemon, S. & Kirkegaard, K. (2003) J. Virol. 77, 7843-7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Egger, D., Wolk, B., Gosert, R., Bianchi, L., Blum, H., Moradpour, D. & Bienz, K. (2002) J. Virol. 76, 5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gotoh, B., Komatsu, T., Takeuchi, K. & Yokoo, J. (2002) Rev. Med. Virol. 12, 337-357. [DOI] [PubMed] [Google Scholar]
- 46.Leonard, G. & Sen, G. (1997) J. Virol. 71, 5095-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Look, D., Roswit, W., Frick, A., Gris-Alevy, Y., Dickhaus, D., Walter, M. & Holtzman, M. (1998) Immunity 9, 8771-8880. [DOI] [PubMed] [Google Scholar]
- 48.Weihua, X., Ramanujam, S., Lindner, D., Kudaravalli, R., Freund, R. & Kalvakolanu, D. (1998) Proc. Natl. Acad. Sci. USA 95, 1085-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.