Kinetochore protein CENP-I is regulated by SUMO protease SENP6 and RNF4, a ubiquitin ligase which targets polysumoylated proteins for degradation during mitosis.
Abstract
We have analyzed the mitotic function of SENP6, a small ubiquitin-like modifier (SUMO) protease that disassembles conjugated SUMO-2/3 chains. Cells lacking SENP6 showed defects in spindle assembly and metaphase chromosome congression. Analysis of kinetochore composition in these cells revealed that a subset of proteins became undetectable on inner kinetochores after SENP6 depletion, particularly the CENP-H/I/K complex, whereas other changes in kinetochore composition mimicked defects previously reported to result from CENP-H/I/K depletion. We further found that CENP-I is degraded through the action of RNF4, a ubiquitin ligase which targets polysumoylated proteins for proteasomal degradation, and that SENP6 stabilizes CENP-I by antagonizing RNF4. Together, these findings reveal a novel mechanism whereby the finely balanced activities of SENP6 and RNF4 control vertebrate kinetochore assembly through SUMO-targeted destabilization of inner plate components.
Introduction
Small ubiquitin-like modifiers (SUMOs) are ubiquitin-related proteins (Johnson, 2004). All newly synthesized SUMOs are posttranslationally processed by ubiquitin-like protein–specific proteases (Ulps)/sentrin-specific proteases (SENPs; Hay, 2007; Mukhopadhyay and Dasso, 2007). Mature SUMOs become conjugated to lysine residues within target proteins in a manner similar to ubiquitin (Johnson, 2004). Ulps/SENPs also deconjugate sumoylated species, rendering this modification highly dynamic. Budding yeast expresses a single SUMO paralogue (Smt3p), whereas mammals express three major SUMO paralogues: SUMO-2 and -3 are ∼95% identical to each other, and both are ∼45% identical to SUMO-1. (SUMO-2 and -3 will be collectively called SUMO-2/3 in contexts in which they cannot be distinguished.) Smt3p and SUMO-2 and -3 can form chains, primarily through conjugation to a conserved acceptor lysine (Geoffroy and Hay, 2009).
SUMO chain formation is antagonized by a subset of Ulps/SENPs that are most closely related to the Ulp2p protease in budding yeast (Mukhopadhyay et al., 2006; Mukhopadhyay and Dasso, 2007). Ulp2p has been implicated in several aspects of chromosome segregation and cell cycle control (Mukhopadhyay and Dasso, 2007). Many phenotypes of ulp2Δ yeast are rescued by expression of smt3 mutants that are unable to form chains, confirming that these defects arise from the accumulation of Smt3p chains. The vertebrate Ulps/SENPs most closely related to Ulp2p are the SENP6 and -7 proteins, which preferentially act on substrates bearing SUMO-2/3 chains (Mukhopadhyay et al., 2006; Mukhopadhyay and Dasso, 2007; Lima and Reverter, 2008). Polysumoylated species are also substrates for a family of RING (really interesting new gene) finger ubiquitin ligases (Hunter and Sun, 2008; Geoffroy and Hay, 2009). The ubiquitination of polysumoylated target proteins leads directly to their proteasomal degradation. The single mammalian SUMO-targeted ubiquitin ligase that has been described is the RNF4 protein.
Vertebrate centromeres are specialized chromatin domains, maintained epigenetically by association with centromeric proteins (Vagnarelli et al., 2008). Centromeres are sites for the assembly of mitotic kinetochores, proteinaceous structures that bind the kinetochore microtubules (MTs [kMTs]) of the kinetochore fibers (k-fibers). Electron microscopic studies suggest that kinetochores have a trilaminar structure (Maiato et al., 2004; Chan et al., 2005): the electron-dense inner plate is located at the surface of the centromeric heterochromatin, and it is separated from the outer plate by a layer that appears translucent in fixed and stained samples. After attachment, the plus ends of kMTs terminate in the outer plate (Cheeseman and Desai, 2008). Kinetochores also play a key role in the spindle assembly checkpoint (SAC), which prevents mitotic exit until all kinetochores are attached and chromosomes are aligned on the metaphase plate (Musacchio and Salmon, 2007). The inner centromeric region is the chromatin domain between the sister kinetochores and the localization site for the chromosomal passenger complex (CPC), a kinase complex which is essential for the correction of kMT misattachments (Kelly and Funabiki, 2009). Multiple functions of sumoylation have been proposed within centromeres and kinetochores, and many proteins in these domains have been identified as potential sumoylation targets (for review see Dasso, 2008). In mammals, outer kinetochore proteins CENP-E, BubR1, and Nuf2 can be mitotically sumoylated, and CENP-E recruitment to kinetochores depends on noncovalent binding to SUMO-2/3 (Zhang et al., 2008).
To better understand the role of sumoylation in kinetochore function, we have analyzed the mitotic role of SENP6 in mammalian cells. In this study, we report that SENP6-depleted HeLa cells have defects in metaphase chromosome congression, and they show characteristic changes in spindle morphology. We examined kinetochore composition to find the molecular basis of these phenotypes: we found that a subset of the inner kinetochore proteins became undetectable at the kinetochores of SENP6-depleted cells, including components of the CENP-H/I/K and CENP-O complexes. At the same time, changes in outer kinetochore composition closely mimicked phenotypes observed after loss of CENP-H/I/K components (Cheeseman et al., 2008). Importantly, we found that the CENP-I protein was quantitatively degraded in the absence of SENP6 through a mechanism that requires both RNF4 and proteasome-mediated proteolysis. Together, these findings demonstrate a novel function of the SUMO pathway in the assembly of the inner kinetochore, which serves to finely balance the incorporation and degradation of structural components of the inner plate.
Results
SENP6 depletion causes defects in spindle assembly and mitotic progression
We depleted SENP6 from several human tissue culture cell lines by oligonucleotide-mediated RNAi, and we observed two distinct phenotypes: some cell lines such as U2OS cells showed persistent interphase arrest. This phenotype was typically found in cells with an intact p53 pathway (unpublished data), possibly reflecting activation of DNA damage or replication checkpoints (Stehmeier and Muller, 2009). Characteristics of such interphase arrest will be discussed in a future study. Other cell lines, such as HeLa cells, progressed through interphase but showed prolonged mitotic delays with one or more misaligned chromosomes. Similar mitotic defects were observed with a series of independent siRNAs directed against SENP6 (Fig. S1), substantiating the notion that this phenotype was directly caused by the absence of SENP6.
To quantitate mitotic delays, we performed RNAi using HeLa cells stably expressing GFP-tagged histone H2B (HeLaH2B-GFP; Arnaoutov et al., 2005). We imaged individual cells within these cultures as they progressed through mitosis and measured the timing between key events. Like untransfected HeLaH2B-GFP cells (Video 1), HeLaH2B-GFP cells transfected with a control oligonucleotide against lamin A formed a tight metaphase plate within 15 (±5) min of nuclear envelope breakdown (NEB) and underwent anaphase ∼97 (±36) min after NEB (n = 15; Video 2). These control cells did not show obvious chromosome segregation defects. SENP6-depleted HeLaH2B-GFP cells formed a loose metaphase plate with timing that was comparable with the control cells (15 ± 5 min after NEB; n = 15; Video 3). We followed several cells in the SENP6-depleted cultures for extended intervals (n = 13). Two cells showed entry and exit from mitosis with normal kinetics, suggesting either that they had some residual SENP6 or that they could bypass mitotic requirements for it. More interestingly, six cells arrested in mitosis for the duration of the experiment, >7 h in each case. Five depleted cells delayed in metaphase but eventually underwent anaphase with highly variable timing (224 ± 68 min after NEB); all of these anaphase cells showed either cell death upon mitotic exit (two cells) or obvious chromosome missegregation (three cells).
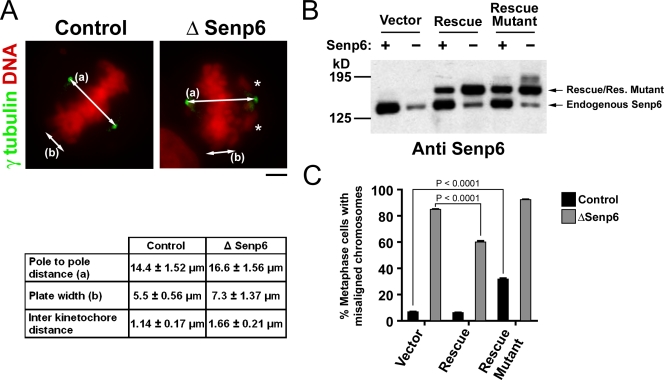
To understand these defects, we analyzed the spindles of SENP6-depleted HeLa cells. We synchronized control or SENP6-depleted cells using a double thymidine block (DTB) procedure. The use of a DTB allowed us to assure that all cells were fully depleted before we allowed them to enter mitosis and scored their mitotic phenotypes and also assured that none of the phenotypes were a secondary result of extended mitosis by itself. After release of the second thymidine block, the cells progressed into metaphase, followed by fixation and staining with Hoechst 33342 to visualize DNA and with anti–γ-tubulin antibodies to visualize spindle poles (Fig. 1 A). The spindles of SENP6-depleted cells were consistently bipolar (Figs. 1 and 2) but showed a series of other defects: the majority of SENP6-depleted spindles (83%) had one or more misaligned chromosomes, whereas misaligned chromosomes were rare in control cells (9%). The metaphase plates of SENP6-depleted cells were noticeably broader than control cells, and they showed an increased separation of their spindle poles. To measure the distance between sister kinetochore pairs, we stained similarly synchronized cells with antibodies against α-tubulin and CREST sera. CREST sera are autoimmune sera from scleroderma patients that recognize a group of centromeric proteins, including CENP-A, -B, and -C (Earnshaw and Rothfield, 1985). Strikingly, sister kinetochore separation in SENP6-depleted cells was 46% greater for chromosomes aligned on the metaphase plate than in control cells (Fig. 1 A). Sister kinetochore separation of unaligned chromosomes was equal in control and SENP6-depleted cells, as was the distance between sister pairs in both cultures after treatment with the MT poison nocodazole (unpublished data). Thus, the larger distance between sister kinetochore pairs of aligned chromosomes in SENP6-depleted cells reflects greater stretching after attachment, which is indicative either of increased tension being exerted by k-fibers or of changes in the structure of the kinetochores themselves.
Figure 1.
SENP6 depletion causes defects in chromosome congression and spindle assembly. (A) HeLa cells were treated with control or SENP6 siRNA and synchronized by DTB. Cells were fixed and stained for γ-tubulin and DNA. The asterisks indicate misaligned chromosomes. Distance between the poles (a) and the metaphase plate width (b) were measured and tabulated (mean ± SD; n = 40; P < 0.0001). Similarly treated cells were stained for α-tubulin and centromere (CREST; not depicted), and the distance between the sister kinetochore pairs on the metaphase plate were measured and tabulated (mean ± SD; n = 40; P < 0.0001). (B) HeLa cells were treated with anti-SENP6 (−) or control (+) siRNAs and DTB. Cells were transfected with the rescue constructs during the second thymidine block and harvested 48 h after siRNA treatment. The cells were lysed in sample buffer and immunoblotted using anti-SENP6 antibodies. (C) Cells treated as in B were fixed and stained with Hoechst 33342. The percentage of cells with one or more misaligned chromosomes was determined for each sample. The graph represents three independent experiments. Error bars represent the 95% confidence interval (n = 300). Bar, 5 µm.
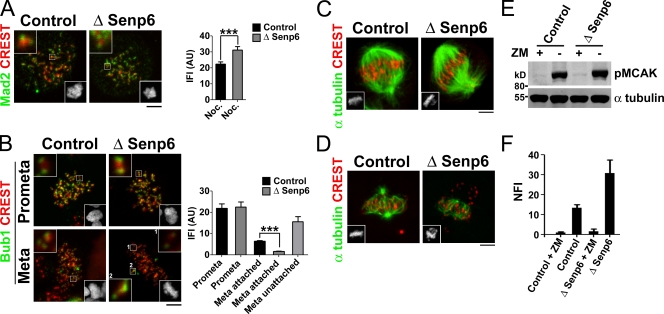
Figure 2.
SAC, spindle MTs, and Aurora B kinase activity in SENP6-depleted cells. (A–D) HeLa cells were treated with anti-SENP6 (−) or control (+) siRNAs and DTB for 48 h except in A, in which cells were treated with siRNA for 36 h and then with nocodazole (Noc.) for a further 12 h. Cells were fixed and stained for centromeres (CREST) and with antibodies against the indicated proteins, Mad2 (A) Bub1 (B), and α-tubulin (C and D). Bottom insets show Hoechst 33342 staining of the cells. (A and B) The boxed areas have been magnified and included as insets (top corners). Error bars represent the 95% confidence interval (***, P < 0.0001). (D) Cells were treated as in C but incubated at 0°C for 10 min before fixation. (E) HeLa cells were transfected with SENP6 or control siRNAs. 36 h after transfection, monastrol was added, and incubation was continued for 12 h. After monastrol wash out, the cells were allowed to recover in the presence or absence of ZM447439 (ZM) for 30 min, as indicated. The cells were lysed in sample buffer, and Western blots were performed with anti–phospho-MCAK (pMCAK; top) and α-tubulin (bottom) antibodies. (F) Phospho-MCAK signals from three separate experiments as shown in E were quantitated and normalized against the α-tubulin signal in the same samples. Mean normalized fluorescence intensity (NFI) signal is shown, and error bars indicate SEM. AU, arbitrary unit. Bars, 5 µm.
To confirm that the failure of chromosome alignment was caused by SENP6 depletion, we rescued this defect through the expression of an mRNA that was insensitive to the anti-SENP6 oligonucleotide (Fig. 1, B and C). We performed a DTB, transfecting oligonucleotides upon the first thymidine addition and rescuing plasmid upon the second thymidine addition. After the cells were released from the second block and allowed to progress into mitosis, they were fixed and stained with Hoechst 33342. Less than 8% of control cells failed to align all chromosomes onto the metaphase plate under these conditions. As expected, >80% of cells transfected with anti-SENP6 oligonucleotide showed congression defects if cotransfected with an empty vector, but this percentage was significantly reduced by cotransfection with the construct for expression of nondegradable SENP6 mRNA (60%). CENP-I sumoylation occurred most prominently during S phase (see Fig. 5 and not depicted), the interval during which it becomes incorporated into assembling inner kinetochore structures (Hemmerich et al., 2008). It appeared likely that the incomplete phenotypic restoration that we observed reflects an insufficient concentration of SENP6 protein expressed from the rescue mRNA in S phase. However, higher levels of SENP6 expression caused other toxic effects in HeLa cells during the later course of the experiment, preventing us from directly testing this assumption.
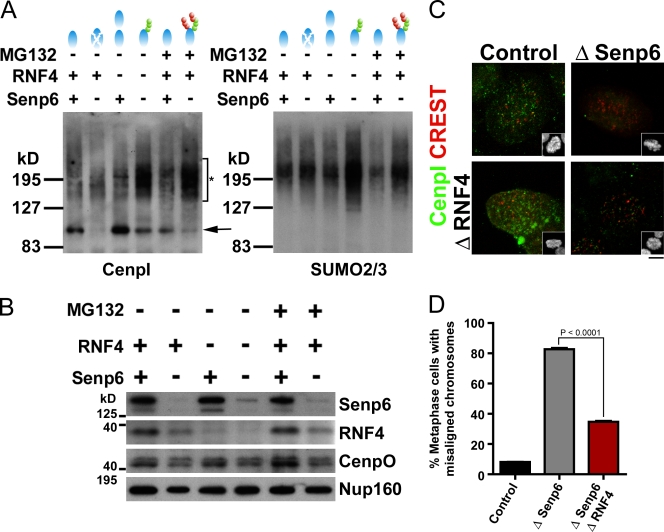
Figure 5.
CENP-I sumoylation and RNF4-dependent degradation after SENP6 depletion. (A) HeLa cells were treated with control or SENP6 siRNA for 36 h, followed by a 13-h thymidine block. Where indicated, RNF4 siRNA was transfected at 12 h after control or SENP6 siRNAs. Where indicated, MG132 was added during the final hour before cells were harvested. Cell lysates were subjected to immunoprecipitation using antibodies against CENP-I. The precipitates were subject to blotting with antibodies against CENP-I (left) or SUMO-2/3 (right). The arrow indicates unmodified CENP-I, and the asterisk indicates modified forms of CENP-I. Undepleted (+) or depleted (−) levels of SENP6 and RNF4 are indicated, along with a cartoon representation of SUMO (greens rounds) and ubiquitin (red rounds) modifications of CENP-I (light blue ovals) predicted after each treatment. (B) HeLa cells were treated as in A, and immunoblot analysis was conducted on whole cell extracts from each sample. Primary antibodies used for blotting are indicated to the right of each panel. (C) HeLa cells were treated with control or SENP6 siRNA and synchronized by DTB for 48 h. Where indicated, RNF4 siRNA was transfected at 12 h after control or SENP6 siRNAs. Cells were fixed and stained for CENP-I and centromeres (CREST) as indicated. Insets show the DNA. (D) Cells treated as in C were fixed and stained with Hoechst 33342. The percentage of cells with one or more misaligned chromosomes was determined for each sample. The graph represents three independent experiments. Error bars represent 95% confidence interval (n = 300). Bar, 5 µm.
Restoration required SENP6 enzymatic activity because a point mutant lacking a cysteine residue within SENP6's catalytic site failed to rescue, with <10% of cells showing full congression. Indeed, we found a modest but reproducible defect in chromosome congression in undepleted cells expressing the catalytically inactive form of SENP6 (Fig. 1 C). It is possible that this catalytically inactive form of SENP6 acted in a dominant-negative manner, perhaps by sequestering SENP6 substrates or binding partners, and thereby disrupted the function of the endogenous, wild-type SENP6 protein. Together, these results indicate that the misalignment phenotype is genuinely caused by the depletion of SENP6 function.
Spindle abnormalities observed after SENP6 depletion (Fig. 1 A) would be expected to provoke SAC activation, thereby causing the observed mitotic delays. As a measure of whether the SAC was working properly, we examined the behavior of Mad2 and Bub1, key SAC components which are abundantly recruited to unattached kinetochores but released or reduced upon attachment of kMTs (Musacchio and Salmon, 2007). After nocodazole treatment, Mad2 was recruited as well or better to the unattached kinetochores of SENP6-depleted cell (Fig. 2 A). In metaphase cells that had not been drug treated, Mad2 staining was lost from kinetochores of aligned chromatids in both depleted and control cells (unpublished data), suggesting that its redistribution in response to kMT–kinetochore attachment does not require SENP6. The levels of Bub1 were indistinguishable on the kinetochores of prometaphase control and SENP6-depleted cells before kMT–kinetochore attachment (Fig. 2 B). The kinetochores of aligned chromatids in control and SENP6-depleted metaphase cells showed decreased Bub1 staining in comparison with unaligned chromosomes or prometaphase cells; the residual level of Bub1 on aligned kinetochores of SENP6-depleted cells was somewhat lower than found in control cells. These findings are consistent with the notion that activation and inactivation of the SAC in response to unattached kinetochores were relatively normal in SENP6-depleted cells, so that their failure to fully align and correctly tension all kinetochores is likely to cause the observed mitotic delay.
MT assembly and Aurora B activation in SENP6-depleted cells
To examine the architecture of k-fibers in SENP6-depleted cells, we stained cells with antibodies against α-tubulin and CREST sera. Spindles in SENP6-depleted cells were bipolar and showed MT densities similar to control spindles (Fig. 2 C). In parallel, we subjected similar cultures to a 10-min cold treatment before staining to depolymerize MTs that were not stably attached to kinetochores. SENP6-depleted cells showed robust k-fibers for the aligned chromosomes (Fig. 2 D). Moreover, the SENP6-depleted cells showed normal loading of k-fiber components, such as HURP (hepatoma up-regulated protein; Fig. S2; Wilde, 2006), indicating that their composition was comparable with k-fibers of control cells. The lack of marked differences between the control and depleted cells argued against a gross change in k-fiber assembly or stability after loss of SENP6.
Attachment errors are frequent during the process of spindle assembly, and there are mechanisms to correct syntelic (both kinetochores attached to a single pole) and merotelic (a single kinetochore attached to both poles) configurations (Kelly and Funabiki, 2009). These mechanisms monitor tension across sister kinetochore pairs and rely on the Aurora B kinase, a component of the CPC. We wished to determine whether this tension-monitoring system was working properly after SENP6 depletion. To examine Aurora B activity, we treated control and SENP6-depleted cells for 12 h with monastrol, which inhibits the kinesin Eg5 (kinesin-5), causing the majority of cells to arrest in mitosis with syntelic attachments (Kapoor et al., 2000). 30 min after monastrol removal, the level of Aurora B activity was assayed by Western blotting with phospho-specific antibodies against mitotic centromere-associated kinesin (MCAK), a well-characterized Aurora B substrate (Kelly and Funabiki, 2009). We observed comparable levels of phospho-MCAK in control and SENP6-depleted cells, which were lost upon treatment with ZM447439, a specific inhibitor of Aurora kinases (Fig. 2, E and F). These results indicate that SENP6-depleted cells are capable of recognizing inappropriate MT attachments to kinetochores, of responding through Aurora B activation, and of thereby releasing such attachments. Additionally, the fact that unaligned chromosomes in SENP6-depleted cells lacked cold stable k-fibers (Fig. 2 D) indicates that syntelic and monotelic (only one kinetochore attached) configurations are appropriately unstable, further arguing against a role for SENP6 in the sensing of tension across kinetochores.
Collectively, these findings show that many aspects of k-fiber assembly and CPC activation are relatively normal in SENP6-depleted cells. Our results did not rule out subtle problems in either process, but such problems did not appear sufficient in themselves to account for mitotic defects observed after SENP6 depletion.
Defects in SENP6-depleted cells result from loss of the CENP-H/I/K complex
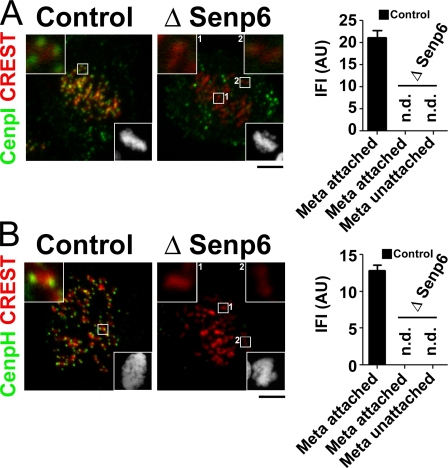
Because sumoylation has previously been implicated in kinetochore structure and function (for review see Dasso, 2008), we examined kinetochores of SENP6-depleted cells in detail by immunolocalization of outer and inner kinetochore components. We were particularly interested in the CENP-H/I/K complex: loss of CENP-H/I/K complex members causes mitotic arrest with multiple misaligned chromosomes, but a subset of the chromosomes still typically achieves alignment on the metaphase plate with cold stable kMTs (Cheeseman et al., 2008), a phenotype which is strikingly similar to our observations in SENP6-depleted cells (Figs. 1 and 2). We examined the status of CENP-H and -I by immunostaining. Although both proteins were clearly detected on kinetochores of control cells, they were undetectable at all kinetochores of SENP6-depleted cells (Fig. 3). Western blotting showed that total CENP-I levels were drastically reduced in SENP6-depleted cells (see Fig. 5 A), indicating that its absence from kinetochores reflected lower protein levels rather than simply mislocalization.
Figure 3.
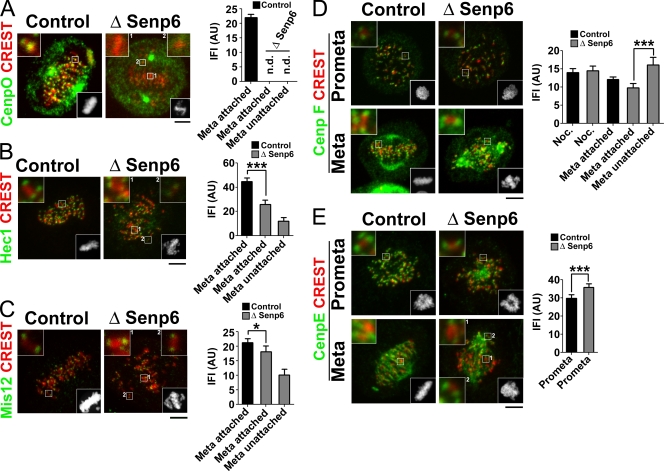
SENP6 depletion results in loss of the CENP-H/I/K complex from kinetochores. (A and B) HeLa cells were treated with control or SENP6 siRNA and synchronized by DTB. Metaphase cells were fixed and stained with CREST sera and with antibodies against the indicated centromeric proteins, CENP-I (A) and CENP-H (B). The bottom right inset in each panel shows Hoechst 33342 staining. Other insets show magnification of the indicated areas. IFIs of the centromeric proteins were measured. The graphs show mean ± 95% confidence interval for metaphase cells as indicated (n = 100 for bipolarly attached sister kinetochores; n. d. = measured value is not distinguishable from background signal). AU, arbitrary unit. Bars, 5 µm.
The CENP-H/I/K complex promotes the correct localization of some other kinetochore components. We reasoned that if the phenotype of SENP6-depleted cells results primarily from loss of the CENP-H/I/K complex, the behavior of these proteins should closely mimic oblation of CENP-H/I/K complex components by other means. For instance, the kinetochore loading of the CENP-O complex is completely dependent on it (Okada et al., 2006). We examined the status of CENP-O by immunostaining and found that it was entirely absent from kinetochores, as predicted (Fig. 4 A). However, unlike CENP-I, CENP-O was still clearly present within depleted cells (Fig. 5 B), indicating that it was subject to displacement but not degradation in the absence of SENP6.
Figure 4.
Changes in kinetochore composition after SENP6 depletion. (A–E) HeLa cells were treated with control or SENP6 siRNA and synchronized by DTB for 48 h. Cells were fixed and stained for kinetochore proteins as indicated: CENP-O (A), Hec1 (B), Mis12 (C), CENP-F (D), and CENP-E (E). Cells were simultaneously stained for centromeres (CREST). The bottom right inset in each panel shows Hoechst 33342 staining. Other insets show magnification of the indicated areas. IFIs of the proteins were measured on kinetochores of prometaphase cells as well as on attached (aligned) or unattached (nonaligned) kinetochores of metaphase cells, as indicated. The graphs show mean ± 95% confidence interval (n = 100 for bipolarly attached sister kinetochores; n = 40 for unattached misaligned kinetochores; ***, P < 0.0001; and *, P < 0.05, as indicated; n. d. = measured value is not distinguishable from background signal). AU, arbitrary unit. Bars, 5 µm.
The Hec1 complex contains Hec1, Nuf2, and Spc24 and -25, and it has a critical role in kMT–kinetochore attachment (Tanaka and Desai, 2008). Hec1 complex recruitment to outer kinetochores is partially dependent on the CENP-H/I/K complex (Cheeseman et al., 2008). The total level of Hec1 did not vary upon SENP6 depletion, nor did we observe prominently sumoylated forms of this protein (Fig. S3; Montpetit et al., 2006). However, analysis of Hec1 in metaphase cells revealed two interesting findings (Fig. 4 B). First, the level of Hec1 on kinetochores of aligned chromosomes of SENP6-depleted cells was roughly 45% less than in control cells. Second, unaligned chromatids near the poles of SENP6-depleted cells showed an even more substantial reduction of kinetochore-associated Hec1, roughly 75% less than the kinetochores of aligned chromosomes in control cells. The latter finding was particularly striking because Hec1 is stably recruited onto kinetochores during late G2 phase and because its levels do not normally vary in response to the MT attachment (Hori et al., 2003; DeLuca et al., 2005). The magnitude of these changes was consistent with the idea that inadequate Hec1 complex loading may contribute to kinetochore attachment defects, particularly for the unaligned kinetochores.
The Mis12 complex contains Dsn1, Nnf1, Nsl1, and Mis12; it acts in concert with the Hec1 complex to promote outer kinetochore assembly and MT capture (Cheeseman et al., 2006). Depletion of CENP-H/I/K components causes observable mislocalization of the Mis12 complex under some assay conditions but not others (Cheeseman et al., 2008). After SENP6 depletion, Mis12 showed a pattern of redistribution similar to Hec1, although the magnitude by which Mis12 association was reduced on attached (14%) and unattached (52%) kinetochores was less than for Hec1 (Fig. 4 C). Again, it is possible to speculate that erratic loading of the Mis12 complex occurs in the absence of SENP6 during late G2 phase, resulting in attachment failure for the underloaded kinetochores and thus spindle defects and mitotic arrest.
CENP-F is a large coiled-coil protein that localizes to outer kinetochores and has been implicated in several mitotic functions (Varis et al., 2006). CENP-F loading does not require CENP-H/I/K (Cheeseman et al., 2008). CENP-F staining was indistinguishable on kinetochores of control and SENP6-depleted cells after nocodazole treatment and similar on kinetochores of aligned sister chromatids of untreated metaphase cells. Unaligned sister pairs in SENP6-depleted metaphase cells showed slightly higher levels of CENP-F accumulation than aligned pairs (Fig. 4 D), which is consistent with the previous observation that CENP-F levels decrease after kinetochore attachment (Varis et al., 2006). These findings indicate that CENP-F loading is largely independent of SENP6. Thus, the profiles of CENP-O, Hec1, Mis12, and CENP-F redistribution after SENP6 depletion are all reminiscent of the situation after CENP-H/I/K complex depletion (Cheeseman et al., 2008). These findings are collectively consistent with the notion that defects caused by SENP6 depletion reflect CENP-H/I/K complex instability.
CENP-E distribution is not sensitive to SENP6 depletion
CENP-E is an MT motor found on outer kinetochores that has roles in MT attachment and spindle checkpoint control (Musacchio and Salmon, 2007). CENP-E is sumoylated in mitotic human cells, and its recruitment to kinetochores is facilitated by SUMO-binding motifs in its C-terminal domain (Zhang et al., 2008). We analyzed CENP-E distribution in control and SENP6-depleted prometaphase cells, which had undergone NEB but had not yet formed stable kMT attachments (Fig. 4 E). The recruitment of CENP-E was as good as or better in SENP6-depleted cells than it was in control cells. During metaphase, when the majority of aligned chromosomes had formed stable k-fibers, CENP-E association with kinetochores of aligned chromatids was similar in control and SENP6-depleted cells (Fig. 4 E), although it was not possible to quantitate the levels of CENP-E bound to kinetochores by itself because of staining along the kMTs. Notably, CENP-E loading was responsive to the attachment state of the kinetochores after of SENP6 depletion (Fig. 4 E, bottom right, compare inset 1 with inset 2), as has previously been observed (Hoffman et al., 2001).
In addition to CENP-E, human BubR1 and Nuf2 can be mitotically sumoylated (Zhang et al., 2008). We did not observe significant increases in sumoylated forms of CENP-E, BubR1, and Nuf2 after SENP6 depletion (unpublished data). Therefore, we suspect that other Ulps/SENPs may act as deconjugating enzymes for CENP-E, BubR1, and Nuf2 within mitosis. This observation implies that sumoylation acts in multiple distinct ways to control kinetochore assembly and function in mammalian cells.
Mechanism of CENP-I degradation after SENP6 depletion
SENP6 acts on conjugated species containing SUMO-2/3 chains (Mukhopadhyay et al., 2006). These species are also potential substrates for the SUMO-targeted ubiquitin ligase RNF4, which marks them for proteasome-mediated degradation (Geoffroy and Hay, 2009). We predicted that if SENP6 stabilizes CENP-I by removing SUMO-2/3 chains and thus preventing its ubiquitination by RNF4, depletion of RNF4 or inhibition of proteasomes should restore CENP-I stability in SENP6-depleted cells. Consistent with this prediction, the levels of CENP-I in SENP6-depleted cells increased with RNF4 depletion (Fig. 5 A). Moreover, higher molecular mass forms of CENP-I accumulated in the absence of both SENP6 and RNF4 that could be recognized by anti–SUMO-2/3 antibodies (Fig. 5 A, left and right, fourth lane). When proteasomal degradation was prevented by treatment with MG132, CENP-I was stabilized in the SENP6-depleted cells, primarily in SUMO-2/3–conjugated forms. Our findings indicate that sumoylated CENP-I is recognized by both SENP6 and RNF4 and that these proteins act antagonistically in regulating its abundance.
Notably, sumoylated forms of CENP-I were easily detected in S-phase cells but were less abundant in other cell cycle phases (unpublished data). S phase is the interval during which the CENP-H/I/K complex is deposited onto centromeres (Hemmerich et al., 2008), so it is possible that sumoylation of CENP-I is coordinated with this process. Interestingly, depletion of SENP6 lowered RNF4 levels, even in cells that were not transfected with anti-RNF4 oligonucleotides (Fig. 5 B). It is intriguing to speculate that RNF4 may be subject to autoregulation that is antagonized by SENP6, as this would imply that this control pathway is subject to elaborate cross talk between SENP6 and RNF4.
Finally, we examined the phenotypes of cells codepleted of both SENP6 and RNF4 (Fig. 5, C and D). As expected, CENP-I was present on metaphase kinetochores of cells that were not depleted of SENP6, regardless of whether RNF4 was depleted, and CENP-I was absent after SENP6 was depleted (Fig. 5 C). When SENP6 and RNF4 were codepleted, CENP-I was visible on kinetochores, suggesting that its accumulation is regulated by the balance of SENP6 and RNF4. Because CENP-H/I/K is constitutively present on centromeres, it was possible to see CENP-I colocalization to CREST staining foci in interphase nuclei. CENP-I foci were lost after SENP6 depletion but restored in the codepleted samples (unpublished data), arguing that SENP6 and RNF4 similarly control CENP-I during interphase. We also scored chromosome misalignment in codepleted cells (Fig. 5 D): the percentage of mitotic cells showing chromosome misalignment was slightly higher in RNF4-depleted cells (unpublished data) than in control cells (9%). In the same experiment, the vast majority of SENP6-depleted cells (83%) showed misalignment. The percentage of cells showing misalignment was markedly decreased (35%) when both proteins were depleted, arguing that RNF4 and SENP6 function antagonistically in this context.
Discussion
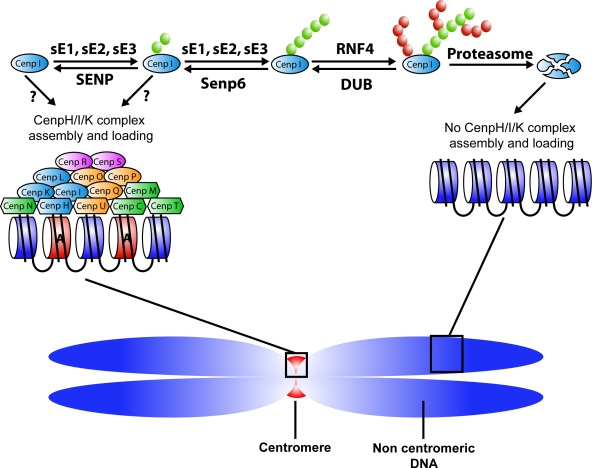
Genetic and cell biological studies have indicated that sumoylation plays a critical role at mitotic kinetochores (for review see Dasso, 2008). We have found that the inner kinetochore protein CENP-I is subject to conjugation with SUMO-2/3 and that its modification targets it for RNF4-dependent proteasomal degradation (Fig. 5). RNF4 was antagonized by the SENP6 SUMO protease, which acts specifically to shorten SUMO-2/3 chains. It seems likely that RNF4 and SENP6 both act directly on polysumoylated CENP-I, although this idea remains to be conclusively tested through biochemical studies. CENP-I degradation after SENP6 depletion leads not only to loss of the CENP-H/I/K complex from kinetochores but also to displacement of the stable CENP-O complex (CENP-O/P/Q/R/U; Figs. 4 and 5). Our findings show that one mechanism through which sumoylation controls vertebrate kinetochore function is through the assembly of constitutive components of the inner kinetochore and that RNF4 and SENP6 act as antagonistic regulators of this process. CENP-I is only the second substrate of RNF4 to be reported (Lallemand-Breitenbach et al., 2008; Tatham et al., 2008), and our findings provide the first demonstration that SENP6 acts in direct antagonism to RNF4.
Deposition of a histone H3 variant nucleosome, CENP-A, during late mitosis and early G1 phase specifies the centromeric chromatin domain (Jansen et al., 2007). CENP-A recruits six proteins (CENP-C, -H, -M, -N, -T, and -U(50)) that are collectively called the CENP-A nucleosome-associated complex (NAC; Foltz et al., 2006). The inner kinetochore also contains CENP-A distal (CAD) proteins (CENP-I, -K, -L, -O, -P, -Q, -R, and -S) that are believed to associate indirectly with CENP-A (Foltz et al., 2006). The amount of CENP-A on inner kinetochores was equivalent in the presence and absence of SENP6 (unpublished data). Although there is some evidence that CENP-C may be a sumoylation target (for review see Dasso, 2008), we did not observe any changes in its abundance, modification status or localization (unpublished data), arguing that it is likewise regulated in a SENP6-independent manner. In contrast, SENP6 depletion enhanced sumoylation and destruction of CENP-I and thereby blocked CENP-H/I/K accumulation on kinetochores (Figs. 3 and 5). Other members of the CENP-H/I/K complex may be similarly regulated by SENP6: the concentration of CENP-H decreased in SENP6-depleted cells, and CENP-H immunoprecipitates showed the accumulation polysumoylated species in the absence of both SENP6 and RNF4 (unpublished data). SUMO is frequently found to be conjugated to many members of large protein complexes (for example, Panse et al., 2006), so the modification of multiple CENP-H/I/K members would be broadly consistent with the action of SUMO in other pathways. To our knowledge, this is the first demonstration of a mechanism governing NAC/CAD assembly through posttranslational modification. Although CENP-O complex components were not similarly destabilized (Fig. 5 B), their localization was disrupted (Fig. 4), which is consistent with the dependence of their localization on CENP-H/I/K (Okada et al., 2006).
The similarity between the phenotypes of SENP6-depleted cells (Figs. 1–3) and cells lacking the CENP-H/I/K complex (Cheeseman et al., 2008) suggests that modulation of CENP-H/I/K complex stability may be a key aspect of SENP6 function for kinetochore assembly, although our findings certainly do not exclude the possibility that other aspects of mitotic physiology are changed through the altered sumoylation of additional targets. We speculate that this mechanism may limit CENP-H/I/K stability spatially or temporally, helping to assure that inner kinetochore structures are only assembled at the correct location and time (Fig. 6). The kinetochore loading of Hec1 was decreased but not completely lost after SENP6 depletion (Fig. 4), as in cells lacking the CENP-H/I/K complex (Cheeseman et al., 2008). Interestingly, Hec1 loading was not uniform in depleted cells; within an individual cell, unattached kinetochores had less Hec1 than attached kinetochores (Fig. 4). Because Hec1 levels do not change greatly upon kMT binding (Hori et al., 2003; DeLuca et al., 2005), these data may suggest that depleted cells enter mitosis with variable levels of Hec1 on their kinetochores, and that those kinetochores with the lowest level of Hec1 are destined to fail in forming stable kMT attachments. Mis12 showed a similar loading defect, although it was not underloaded to the same extent as Hec1 (Fig. 4). The variable Hec1 and Mis12 loading may either reflect random failure of individual centromeres to assemble functional kinetochores in the absence of SENP6 or some centromeres (for instance, those assembled last) may systematically fail in this situation. Although subtle changes in other outer kinetochore components (Bub1, Mad2, and CENP-E and -F; Fig. 4) did not appear sufficient in themselves to cause the mitotic defects observed after SENP6 depletion, such changes may have further contributed to the terminal phenotype of SENP6-depleted cells. These changes may have been consequences underlying defects in NAC-CAD or Hec1 and Mis12 complex loading, or they may have reflected other SENP6 functions.
Figure 6.
SENP6 and RNF4 act antagonistically to maintain CENP-I homeostasis. CENP-I conjugation to poly–SUMO-2/3 is mediated by SUMO activating (sE1) and conjugating (sE2) enzymes and perhaps one or more SUMO ligases (sE3). This allows it to be ubiquitinated by RNF4 and degraded by proteasomes. SENP6 deconjugates the SUMO-2/3 chains on CENP-I, preventing its degradation in this manner. By spatially or temporally restricting the activity of SENP6 in this pathway, it should be possible to restrict the stability of CENP-I and prevent its inappropriate accumulation at noncentromeric regions. NAC proteins are shown as hexagons, whereas CAD proteins are depicted as ovals (Foltz et al., 2006). Different colors indicate putative subcomplexes identified through biochemical studies (Okada et al., 2006; Cheeseman et al., 2008).
SENP6 appears uniquely important for control of NAC-CAD assembly through deconjugation of poly-SUMO chains. This is remarkable because a closely related enzyme, SENP7, shows similar in vitro activity in SUMO-2/3 chain editing (Lima and Reverter, 2008; Shen et al., 2009). It is possible that SENP7 activity is simply insufficient in the absence of SENP6. Consistent with this idea, SENP7 is a much less abundant protein in Xenopus laevis eggs and embryos (Wang et al., 2009) as well as in mammalian tissue culture cells (unpublished data). Alternatively, SENP7 and -6 may act on different subsets of poly-SUMO chains. Consistent with this idea, depletion of SENP7 from HeLa cells by RNAi causes a strikingly different phenotype from SENP6 depletion, with cells undergoing an interphase arrest followed by apoptotic cell death (unpublished data).
It is notable that sumoylation has other points of action at kinetochores that do not require SENP6. In particular, conjugation of SUMO-2/3 to outer kinetochore proteins has been described for CENP-E, Nuf2, and BubR1 (Zhang et al., 2008), and SUMO-1 conjugation of RanGAP1, the activating protein for the Ran GTPase, localizes it to outer kinetochores association (Joseph et al., 2002). Sumoylation of these substrates is believed to regulate their functions within mitosis (Joseph et al., 2002; Zhang et al., 2008), and we found no indication that sumoylation destabilizes any of these proteins. Staining of mitotic cells with anti-SENP6 antibodies did not show its accumulation at mitotic kinetochores (unpublished data). Although this localization pattern does not strictly exclude the possibility that SENP6 acts on mitotically sumoylated kinetochore proteins, it favors the notion that these targets are desumoylated by other SUMO proteases. In contrast, sumoylation of CENP-I occurs predominantly during S phase, perhaps corresponding to the interval when it is assembled onto nascent kinetochores (Hemmerich et al., 2008). SENP6 is nucleoplasmic throughout interphase (Mukhopadhyay et al., 2006), a localization which is consistent with its action on CENP-I in this context.
In summary, our results show that sumoylation regulates the construction of vertebrate inner kinetochores through the CENP-H/I/K complex. In this context, SENP6 promotes the deconjugation of multiply sumoylated CENP-I, whereas RNF4 promotes its proteasome-mediated proteolysis. In the future, it will be interesting to understand the circumstances that may tip the balance of this regulatory circuit in favor of CENP-I stability or destruction as well as how this mechanism is integrated with other events to spatially and temporally coordinate kinetochore assembly and function.
Materials and methods
Antibodies and reagents
Anti-SENP6 antibodies were raised in rabbit against the N-terminal 300 aa of SENP6 and affinity purified using an antigen column. CENP-I, CENP-O, Bub1, Mis12, phospho-MCAK, and anti-Nup160 antibodies were raised in rabbit against the recombinant proteins and affinity purified. Mcm21R (CENP-O) antibody was a gift from A. McAinsh (Marie Curie Research Institute, Surrey, England, UK). RNF4 antibody was a gift from J.J. Palvimo (University of Helsinki, Helsinki, Finland). CENP-H antibody was purchased from Bethyl Laboratories, Inc. Hec1 monoclonal antibody (9G3) was purchased from Abcam. Mad2 antibody was obtained from Covance. HA–SUMO-2–vinyl sulfone was a gift from K. Wilkinson (Emory University, Atlanta, GA). Alexa Fluor–labeled secondary antibodies, Lipofectamine RNAiMax, and RNF4 siRNA were purchased from Invitrogen, and HRP-conjugated rabbit secondary antibody was obtained from Jackson ImmunoResearch Laboratories, Inc. SuperSignal West Femto developer was purchased from Thermo Fisher Scientific. All other reagents were purchased from Sigma-Aldrich unless otherwise stated.
Cell culture, siRNA-mediated depletion, and drug treatments
HeLa cells and their derivative stable lines were maintained in DME supplemented with 10% fetal bovine serum at 37°C and 5% CO2. Plasmid transfections were performed using Effectine reagent (QIAGEN) according to the manufacturer's instructions. Duplex siRNAs with standard dTdT 3′ extension against SENP6 (5′-AAGAAAGTGAAGGAGATACAG-3′) or lamin A/C (5′-AACTGGACTTCCAGAAGAACA-3′) mRNAs were obtained from QIAGEN. siRNA experiments were performed in 6-well culture plates or 6- or 10-cm dishes. Cells were plated at a density of 3 × 103/cm2, grown for 24 h, and washed once with DME without antibiotics immediately before transfection. 2.5 µl of 20 µM siRNA and 7 µl of Lipofectamine RNAiMAX were suspended in separate tubes of 0.5 ml OptiMEM (Invitrogen). The tubes were kept at 23°C for 7 min, and the contents of the two tubes were mixed, kept at RT for a further 25 min, and added to a 6-cm dish containing 4 ml complete DME without antibiotics. The same procedure was followed for other dishes, but the reagents were scaled accordingly.
For synchronization by DTB, 2 mM thymidine was added after the addition of siRNA. After 12 h, the cells were washed once with thymidine-free DME and incubated for 14 h without thymidine. A second block was performed through incubation with 2 mM thymidine for 12 h and again released by washing in thymidine-free DME. Mitotic phenotypes were assayed 10 h after release, when ∼30% cells were mitotic.
In Fig. 5, HeLa cells were transfected with SENP6 or control siRNAs. 36 h after transfection, 2 mM thymidine was added to the medium, and incubation was continued for 13 h. Where indicated, 20 µM MG132 was added during the final hour of the incubation.
Aurora B activity assay
10-cm dishes of HeLa cells were transfected with SENP6 or control siRNAs. 36 h after transfection, 40 µM monastrol (EMD) was added to the medium, and incubation was continued for 12 h. Mitotic cells were collected by shake off, washed with ice-cold DME three times, and plated in 6-cm dishes with or without 2 µM ZM447439 (Tocris Bioscience). After 30 min, the cells were lysed in SDS sample buffer. The cell lysates were subjected to SDS-PAGE. Immunoblotting was performed using phospho-MCAK–specific antibodies, and the signals were quantitated using an infrared imaging system (Odyssey; LI-COR Biosciences).
Immunofluorescence, microscopy, and image analysis
For immunofluorescence, cells grown on poly-l-lysine–coated coverslips were either preextracted for 1 min with 0.5% Triton X-100 in BRB80 buffer (80 mM Pipes, pH 6.9, 1 mM EGTA, and 5 mM MgCl2) or directly fixed for 12 min at ambient temperature with 4% paraformaldehyde in PBS. The cells were permeabilized with 0.5% Triton X-100 in TBS (10 mM Tris-HCl, pH 7.5, and 150 mM NaCl) for 10 min. After washing with TBST (TBS + 0.1% [vol/vol] Tween 20), the cells were blocked for 10 min in 1% normal horse serum (Vector Laboratories) in TBST and incubated for 1 h with primary antibodies diluted 1:200 to 1:500 in blocking solution. The coverslips were washed and incubated for 45 min with secondary antibodies diluted 1:1,000 in blocking solution. Unbound antibodies were removed by washing, and the cells were briefly incubated in 100 ng/ml Hoechst 33258 DNA stain. Finally, the coverslips were mounted in Vectashield mounting medium (Vector Laboratories).
Fluorescence microscopy was performed at RT on a confocal microscope (LSM510 Meta; Carl Zeiss, Inc.) equipped with a 100× Plan-Apochromat objective. We used a 543-nm HeNe laser (5 mW output; detection LP560 nm) for detection of Alexa Fluor 568–labeled antibodies. The 488-nm line of an Argon laser (25 mW nominal output; detection BP 505–530 nm) was used for analysis of Alexa Fluor 488–labeled antibodies. Hoechst 33258 images were captured using the 364-nm line of an ion laser (Enterprise II ML UV; Coherent, Inc.; 80 mW nominal output; detection BP 385–470 nm). Confocal microscopy software (SP2 version 3.2; Carl Zeiss, Inc.) was used for the capture of images. In all figures, scale bars represent 5 µm.
The images were analyzed using ImageJ 4.1g software (National Institutes of Health), and the fluorescence signal of kinetochore proteins was quantitated as described previously (Howell et al., 2000). In brief, two concentric circular regions of interest (ROIs) were marked on the kinetochore fluorescent spot, making sure that the visible signal was inside the smaller circle. The integrated fluorescence intensities (IFIs) and area of both the ROIs were measured for all z slices. Then, the IFI minus the background was calculated using the formula: specific IFI = Fs – BG = [(FL – Fs)/(AL –As)] x As, where Fs = IFI of the small ROI, FL = IFI of the large ROI, As = area of the small ROI, and Fs = area of the large ROI. If the fluorescence signal of a particular kinetochore spot spanned multiple z slices, the specific IFIs for all of the slices were summed to generate the IFI for a single kinetochore spot. This process was repeated for 100 different kinetochore puncta, keeping the areas of the ROIs constant for a particular kinetochore protein. These values were then subjected to statistical analysis using Open Office Calc (Sun Microsystems), and graphs were generated.
Immunoprecipitation and immunoblotting of CENP-I
In Fig. 5 A, cells were washed once in ice-cold PBS, harvested by scraping in 200 µl of lysis buffer (50 mM K-Hepes, pH 7.5, 150 mM NaCl, 2% Empigen BB, 1 µg HA–SUMO-2–vinyl sulfone, 20 mM iodoacetamide, 2 µM AEBSF, and 10 µg/ml each of leupeptin, pepstatin, and chymostatin), and briefly sonicated on ice. Cell lysates were frozen in liquid nitrogen and stored at −80°C or used immediately. 100 µg of affinity-purified rabbit CENP-I antibody was bound to 1 ml of protein A–conjugated magnetic beads (Invitrogen) and conjugated with dimethyl pimelimidate and washed three times in ice-cold 100 mM glycine-HCl, pH 2.5. The beads were blocked for 30 min in 5% hydrolyzed gelatin in wash buffer (10 mM K-Hepes, pH 7.5, 150 mM NaCl, and 2% Empigen BB) at 23°C and washed twice in wash buffer. 200 µl of cell lysate was incubated with 50 µl of beads for 1 h at 4°C. The beads underwent three 5-min washes in ice-cold wash buffer. The bound proteins were eluted with 200 µl of 0.1 M glycine-HCl and immediately neutralized with 20 µl of 1 M Tris-HCl, pH 8.0. 5 µl of 1% Na deoxycholate was added to the eluates, and the proteins were precipitated overnight at 4°C with TCA at a final concentration of 20%. The TCA precipitates were centrifuged at 16,000 g for 30 min at 4°C, washed in −20°C acetone, and recentrifuged at 16,000 g for 30 min at 4°C. The pellets were dried at 37°C, solubilized in 15 µl of sample buffer, subjected to immunoblotting, and developed using SuperSignal West reagent (Thermo Fisher Scientific).
Online supplemental material
Fig. S1 shows the specificity of SENP6 knockdown by siRNA. Fig. S2 shows HURP/α-tubulin staining of a mitotic HeLa cell after SENP6 depletion. Fig. S3 shows that Hec1 abundance is unaffected by SENP6 depletion. Video 1 shows untreated HeLaH2B-GFP cells. Videos 2 and 3 show lamin siRNA– and SENP6 siRNA–treated HeLaH2B-GFP cells, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200909008/DC1.
Acknowledgments
We would like to thank Maia Ouspenskaia and Maiko Furuta for technical support and Dr. Michaela Serpe for her help with the Odyssey imaging system. We also thank Dr. Kara Lukasiewicz for critical reading of the manuscript.
This work was supported by National Institute of Child Health and Human Development intramural funds (NICHD project #Z01 HD001902-14 and # Z01 HD008740-07).
Footnotes
Abbreviations used in this paper:
- CAD
- CENP-A distal
- CPC
- chromosomal passenger complex
- DTB
- double thymidine block
- IFI
- integrated fluorescence intensity
- k-fiber
- kinetochore fiber
- kMT
- kinetochore MT
- MCAK
- mitotic centromere-associated kinesin
- MT
- microtubule
- NAC
- nucleosome-associated complex
- NEB
- nuclear envelope breakdown
- ROI
- region of interest
- SAC
- spindle assembly checkpoint
- SENP
- sentrin-specific protease
- SUMO
- small ubiquitin-like modifier
- Ulp
- ubiquitin-like protein–specific protease
References
- Arnaoutov A., Azuma Y., Ribbeck K., Joseph J., Boyarchuk Y., Karpova T., McNally J., Dasso M. 2005. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat. Cell Biol. 7:626–632 10.1038/ncb1263 [DOI] [PubMed] [Google Scholar]
- Chan G.K., Liu S.T., Yen T.J. 2005. Kinetochore structure and function. Trends Cell Biol. 15:589–598 10.1016/j.tcb.2005.09.010 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Hori T., Fukagawa T., Desai A. 2008. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell. 19:587–594 10.1091/mbc.E07-10-1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso M. 2008. Emerging roles of the SUMO pathway in mitosis. Cell Div. 3:5 10.1186/1747-1028-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Dong Y., Hergert P., Strauss J., Hickey J.M., Salmon E.D., McEwen B.F. 2005. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol. Biol. Cell. 16:519–531 10.1091/mbc.E04-09-0852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W.C., Rothfield N. 1985. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 91:313–321 10.1007/BF00328227 [DOI] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R., III, Cleveland D.W. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458–469 10.1038/ncb1397 [DOI] [PubMed] [Google Scholar]
- Geoffroy M.C., Hay R.T. 2009. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell Biol. 10:564–568 10.1038/nrm2707 [DOI] [PubMed] [Google Scholar]
- Hay R.T. 2007. SUMO-specific proteases: a twist in the tail. Trends Cell Biol. 17:370–376 10.1016/j.tcb.2007.08.002 [DOI] [PubMed] [Google Scholar]
- Hemmerich P., Weidtkamp-Peters S., Hoischen C., Schmiedeberg L., Erliandri I., Diekmann S. 2008. Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 180:1101–1114 10.1083/jcb.200710052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman D.B., Pearson C.G., Yen T.J., Howell B.J., Salmon E.D. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 12:1995–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Haraguchi T., Hiraoka Y., Kimura H., Fukagawa T. 2003. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 116:3347–3362 10.1242/jcs.00645 [DOI] [PubMed] [Google Scholar]
- Howell B.J., Hoffman D.B., Fang G., Murray A.W., Salmon E.D. 2000. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J. Cell Biol. 150:1233–1250 10.1083/jcb.150.6.1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T., Sun H. 2008. Crosstalk between the SUMO and ubiquitin pathways. Ernst Schering Found Symp Proc. 1:1–16 10.1007/2789_2008_098 [DOI] [PubMed] [Google Scholar]
- Jansen L.E., Black B.E., Foltz D.R., Cleveland D.W. 2007. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176:795–805 10.1083/jcb.200701066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73:355–382 10.1146/annurev.biochem.73.011303.074118 [DOI] [PubMed] [Google Scholar]
- Joseph J., Tan S.H., Karpova T.S., McNally J.G., Dasso M. 2002. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156:595–602 10.1083/jcb.200110109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor T.M., Mayer T.U., Coughlin M.L., Mitchison T.J. 2000. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 150:975–988 10.1083/jcb.150.5.975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.E., Funabiki H. 2009. Correcting aberrant kinetochore microtubule attachments: an Aurora B-centric view. Curr. Opin. Cell Biol. 21:51–58 10.1016/j.ceb.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V., Jeanne M., Benhenda S., Nasr R., Lei M., Peres L., Zhou J., Zhu J., Raught B., de Thé H. 2008. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 10:547–555 10.1038/ncb1717 [DOI] [PubMed] [Google Scholar]
- Lima C.D., Reverter D. 2008. Structure of the human SENP7 catalytic domain and poly-SUMO deconjugation activities for SENP6 and SENP7. J. Biol. Chem. 283:32045–32055 10.1074/jbc.M805655200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., DeLuca J., Salmon E.D., Earnshaw W.C. 2004. The dynamic kinetochore-microtubule interface. J. Cell Sci. 117:5461–5477 10.1242/jcs.01536 [DOI] [PubMed] [Google Scholar]
- Montpetit B., Hazbun T.R., Fields S., Hieter P. 2006. Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. J. Cell Biol. 174:653–663 10.1083/jcb.200605019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D., Dasso M. 2007. Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 32:286–295 10.1016/j.tibs.2007.05.002 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D., Ayaydin F., Kolli N., Tan S.H., Anan T., Kametaka A., Azuma Y., Wilkinson K.D., Dasso M. 2006. SUSP1 antagonizes formation of highly SUMO2/3-conjugated species. J. Cell Biol. 174:939–949 10.1083/jcb.200510103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Okada M., Cheeseman I.M., Hori T., Okawa K., McLeod I.X., Yates J.R., III, Desai A., Fukagawa T. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8:446–457 10.1038/ncb1396 [DOI] [PubMed] [Google Scholar]
- Panse V.G., Kressler D., Pauli A., Petfalski E., Gnädig M., Tollervey D., Hurt E. 2006. Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic. 7:1311–1321 10.1111/j.1600-0854.2006.00471.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L.N., Geoffroy M.C., Jaffray E.G., Hay R.T. 2009. Characterization of SENP7, a SUMO-2/3-specific isopeptidase. Biochem. J. 421:223–230 10.1042/BJ20090246 [DOI] [PubMed] [Google Scholar]
- Stehmeier P., Muller S. 2009. Regulation of p53 family members by the ubiquitin-like SUMO system. DNA Repair (Amst.). 8:491–498 10.1016/j.dnarep.2009.01.002 [DOI] [PubMed] [Google Scholar]
- Tanaka T.U., Desai A. 2008. Kinetochore-microtubule interactions: the means to the end. Curr. Opin. Cell Biol. 20:53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham M.H., Geoffroy M.C., Shen L., Plechanovova A., Hattersley N., Jaffray E.G., Palvimo J.J., Hay R.T. 2008. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10:538–546 10.1038/ncb1716 [DOI] [PubMed] [Google Scholar]
- Vagnarelli P., Ribeiro S.A., Earnshaw W.C. 2008. Centromeres: old tales and new tools. FEBS Lett. 582:1950–1959 10.1016/j.febslet.2008.04.014 [DOI] [PubMed] [Google Scholar]
- Varis A., Salmela A.L., Kallio M.J. 2006. Cenp-F (mitosin) is more than a mitotic marker. Chromosoma. 115:288–295 10.1007/s00412-005-0046-0 [DOI] [PubMed] [Google Scholar]
- Wang Y., Mukhopadhyay D., Mathew S., Hasebe T., Heimeier R.A., Azuma Y., Kolli N., Shi Y.-B., Wilkinson K.D., Dasso M. 2009. Identification and developmental expression of Xenopus laevis SUMO proteases. PLoS One. 4:e8462 10.1371/journal.pone.0008462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde A. 2006. “HURP on” we're off to the kinetochore! J. Cell Biol. 173:829–831 10.1083/jcb.200605150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.D., Goeres J., Zhang H., Yen T.J., Porter A.C., Matunis M.J. 2008. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol. Cell. 29:729–741 10.1016/j.molcel.2008.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]