Abstract
The mammalian Golgi apparatus is characterized by a ribbon-like organization adjacent to the centrosome during interphase and extensive fragmentation and dispersal away from the centrosome during mitosis. It is not clear whether this dynamic association between the Golgi and centrosome is of functional significance. We discuss recent findings indicating that the Golgi–centrosome relationship may be important for directional protein transport and centrosome positioning, which are both required for cell polarization. We also summarize our current knowledge of the link between Golgi organization and cell cycle progression.
Introduction
The Golgi apparatus plays a central role in the secretory pathway. Newly synthesized proteins are transported from the ER to the Golgi, where they are posttranslationally modified. They are sorted into carriers for delivery to the plasma membrane or the endosomal–lysosomal system. The basic structural unit of the Golgi apparatus is a stack of flattened cisternae that is morphologically conserved among most species. In mammalian cells, individual Golgi stacks are connected laterally to form a continuous membranous system called the Golgi ribbon, which is located in close physical proximity to the centrosome (Fig. 1, left).
Figure 1.
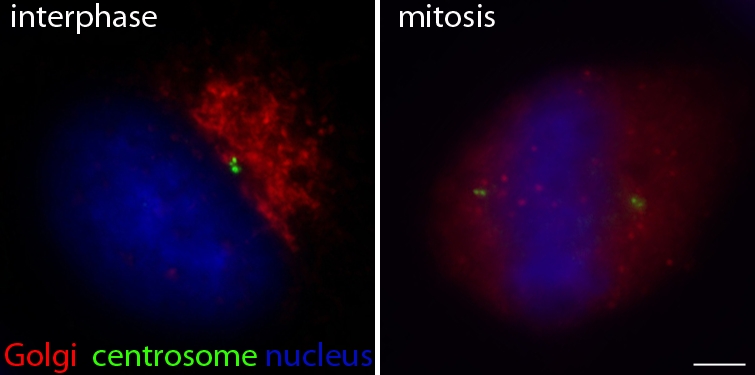
The spatial relationship between the Golgi and the centrosome during the mammalian cell cycle. Golgi (red, stained with antibodies to GM130) and centrosome (green, stained with antibodies to centrin) staining of nonsynchronized bone cancer cells (U2-OS) shows the physical proximity between these two organelles during interphase (left) and its temporary loss during mitosis (right). Bar, 10 µm.
The centrosome functions as the major microtubule-organizing center of the cell and plays an important role in cell polarization and ciliogenesis (Bettencourt-Dias and Glover, 2007). In a newly formed daughter cell, this nonmembrane-bound organelle is composed of a pair of centrioles that is surrounded by a cloud of electron-dense material called the pericentriolar matrix. γ-Tubulin ring complexes (γ-TuRCs) in the pericentriolar matrix allow the centrosome to nucleate the radial array of interphase microtubules whose minus ends are embedded in the centrosome and whose plus ends extend toward the cell periphery. After centrosome duplication in S phase, the two centrosomes move to opposite poles of the cell and become the spindle poles from which spindle microtubules grow. Centrosomes are generally located in the cell center close to the nucleus, although this central position is lost in response to a polarization stimulus, which prompts centrosomes to reorient toward the leading edge of the cell (Pouthas et al., 2008). In most cell types, centrosome reorientation is critical for the ability of cells to polarize and migrate (Yvon et al., 2002). The centrosome is also linked to ciliogenesis because one of its centrioles is converted into the basal body from which a primary cilium extends (D'Angelo and Franco, 2009).
The spatial relationship between the Golgi apparatus and the centrosome is altered by changes in Golgi organization that occur during the cell cycle (Fig. 1). These two organelles are only adjacent in interphase when the Golgi apparatus is arranged as a ribbon in the pericentriolar region (Colanzi et al., 2003). In contrast, Golgi membranes are fragmented and dispersed throughout the cytoplasm during mitosis. Intriguingly, the pericentriolar localization of the Golgi is a feature typical of some eukaryotic cells, ranging from mammalian and amphibian cells (Thyberg and Moskalewski, 1999; Reilein et al., 2003) to amoeba (Rehberg et al., 2005). However, other eukaryotes, including plants and flies, have isolated Golgi stacks (Stanley et al., 1997; Nebenführ and Staehelin, 2001) or isolated cisternae in the case of Saccharomyces cerevisiae that are scattered throughout the cytoplasm without an obvious connection with the centrosome (Preuss et al., 1992).
In this paper, we review recent findings indicating that the relationship between the Golgi and the centrosome in interphase is important for cell polarization. We also summarize the current understanding of how Golgi–centrosome interactions during mitosis affect cell division.
Are there functional interactions between the Golgi and the centrosome during interphase?
Golgi membranes are actively positioned in the pericentriolar position.
The localization of the mammalian Golgi ribbon next to the centrosome requires the microtubule and actin cytoskeleton (Brownhill et al., 2009). Microtubules have a dual role in organizing the pericentriolar Golgi ribbon. First, the subset of microtubules that is nucleated at the Golgi is necessary for the assembly of Golgi fragments into a connected ribbon in the cell periphery (Miller et al., 2009). Second, centrosomal microtubules provide the tracks along which Golgi membranes are transported to the cell center (Cole et al., 1996). Both steps depend on the minus end–directed motor complex dynein (Burkhardt et al., 1997; Miller et al., 2009). The actin cytoskeleton is also involved in localizing Golgi membranes. Actin fibers, which have been detected at the Golgi complex, are required for the maintenance of the pericentriolar position of this organelle by providing tracks for actin-based motors (Valderrama et al., 1998; Sahlender et al., 2005; Vicente-Manzanares et al., 2007). Actin fibers and microtubules are coordinated by proteins that associate with both cytoskeletal elements such as WHAMM, a Golgi-bound actin-nucleating factor, and MACF1, a microtubule–actin cross-linking protein (Lin et al., 2005; Campellone et al., 2008).
Golgi organization and localization in the pericentriolar region also depend on Golgi-associated proteins (Table I). These proteins include members of the GRASP and golgin families and provide structural support to the Golgi apparatus (Ramirez and Lowe, 2009). Their depletion produces defects in Golgi organization ranging from a disconnected Golgi ribbon in the pericentriolar region (Puthenveedu et al., 2006) to dispersed ministacks in the cytoplasm (Diao et al., 2003; Yadav et al., 2009).
Table I.
Golgi-associated proteins that control the pericentriolar position of the Golgi apparatus
Although the position of the Golgi next to the centrosome is actively maintained, it does not appear to be critical for basic Golgi functions. For example, membrane trafficking and the modification of secretory proteins are unaffected when the Golgi ribbon is severed into individual ministacks (Cole et al., 1996; Diao et al., 2003; Yadav et al., 2009). Furthermore, organisms such as S. cerevisiae secrete proteins with high efficiency, although their Golgi membranes are never pericentriolar (Preuss et al., 1992). Thus, the physiological role of the pericentrosomal positioning of the mammalian Golgi apparatus remains a major unanswered question.
An emerging role for Golgi–centrosome association in polarized secretion.
New studies indicate that the relationship between the Golgi and the centrosome may be important for specialized functions of mammalian cells. A prominent example is cell polarization, which is a prerequisite for cell migration (Li et al., 2005). Cell polarization depends on directional protein transport along Golgi-nucleated microtubules as well as centrosome reorientation toward the leading edge of the cell, which both appear to be affected by interactions between the Golgi and the centrosome.
In a recent study, Yadav et al. (2009) investigated the role of the pericentriolar Golgi ribbon in directional transport and cell polarization. Depletion of each of the two structural proteins of the golgin family, GMAP210 and Golgin-160, disrupted the ribbon-like structure of the Golgi and led to isolated ministacks in the cytoplasm. These dispersed stacks were competent of general protein transport to the cell surface. However, there were defects in directional protein transport, as shown by the failure to secrete vesicular stomatitis virus G protein in a directional manner toward the leading edge of a cell and the inability of these cells to migrate in a wound-healing assay. These results indicate that the pericentriolar Golgi ribbon is critical for directional protein transport, although it is not clear whether it is the ribbon-like organization or the position next to the centrosome that is important.
Golgi–centrosome interactions may also contribute to cell polarization through regulatory effects on centrosome positioning. Both the centrosome and the Golgi apparatus undergo reorientation toward the leading edge of a stimulated cell. Bisel et al. (2008) found that centrosome reorientation depends on phosphorylation of the Golgi protein GRASP65, which is proposed to promote Golgi stack disassembly (Wang et al., 2003; Yoshimura et al., 2005). In this study, expression of nonphosphorylatable forms of GRASP65 prevented Golgi and centrosome reorientation toward the leading edge and cell migration. Intriguingly, this block was overcome when Golgi membranes were artificially fragmented, indicating that Golgi membranes have to be remodeled to allow the coordinated reorientation of the centrosome and the Golgi. Thus, the ability of the Golgi to reorganize affects the positioning of the centrosome (Bisel et al., 2008).
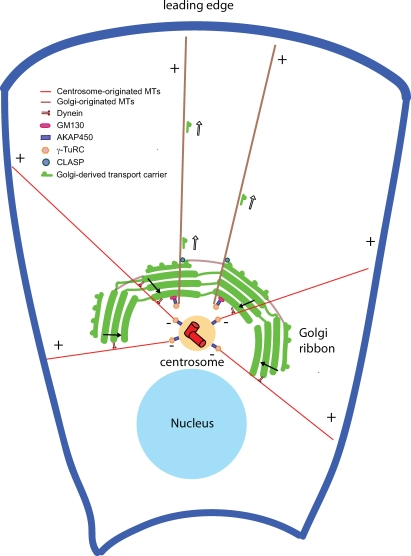
The peripheral Golgi protein, GM130, is an additional critical factor in the regulation of cell polarization (Preisinger et al., 2004; Kodani et al., 2009; Rivero et al., 2009). There are at least four reasons to explain why depletion of GM130 prevents cells from polarizing and migrating in wound-healing assays (Kodani et al., 2009). First, Kodani and Sütterlin (2008) showed that GM130 depletion altered the organization of the centrosome so that it was no longer able to nucleate microtubules or to reorient in response to a polarization stimulus. Second, GM130-dependent centrosome regulation involved the small GTPase Cdc42 (Kodani et al., 2009), a known regulator of cell polarization (Etienne-Manneville, 2006; Kodani et al., 2009). Third, Rivero et al. (2009) identified a novel role for GM130 in microtubule nucleation at the Golgi, which required GM130-dependent recruitment of the microtubule nucleation factor AKAP450 to the Golgi (Rivero et al., 2009). Golgi-nucleated microtubules, which were first identified in in vitro studies (Chabin-Brion et al., 2001), are preferentially oriented toward the leading edge of a motile cell and are necessary for directional protein transport (Fig. 2; Rivero et al., 2009). Fourth, GM130 binds and activates the protein kinase YSK1, which has a known role in cell migration (Preisinger et al., 2004). Thus, GM130 may affect cell polarization and migration through effects on centrosome organization, Cdc42 activation, microtubule nucleation at the Golgi, and YSK1 activation.
Figure 2.
Golgi- and centrosome-nucleated microtubules in cell migration. The centrosome nucleates a radial array of microtubules (red) whose minus ends (−) are anchored at the centrosome and whose plus ends (+) extend into the cell periphery. This population of microtubules depends on γ-TuRC complexes and the large scaffold protein AKAP450 for their nucleation and functions in maintaining the pericentriolar localization of the Golgi ribbon by a dynein-mediated mechanism (closed arrows). In contrast, the Golgi apparatus nucleates microtubules (brown) that extend asymmetrically toward the leading edge of a migrating cell. Microtubule nucleation at the Golgi requires the peripheral Golgi protein GM130, which recruits AKAP450 and γ-TuRC complexes to the Golgi apparatus. Golgi-nucleated microtubules are coated with CLASP proteins and are necessary for the formation of the Golgi ribbon from dispersed stacks. In addition, they are required for cell migration by facilitating polarized protein transport to the leading edge of a cell (open arrows).
The formation of a primary cilium is another process that involves interactions between the Golgi and the centrosome. During ciliogenesis, the centrosome moves to the plasma membrane, where one of its centrioles becomes the basal body from which the primary cilium extends. IFT20, a critical component of the intraflagellar transport machinery that is required for formation and extension of the cilium (Follit et al., 2006), localizes to the Golgi by binding to the structural Golgi protein GMAP210. Loss of either IFT20 or GMAP210 impairs ciliogenesis (Follit et al., 2006, 2008), which supports a role for Golgi-localized IFT20 in protein sorting at the Golgi to produce transport carriers involved in the formation of a primary cilium. A similar role in directing specific cargo molecules to the ciliary membrane has been proposed for the small GTPase Rab8, which also localizes to the Golgi and the basal body (Nachury et al., 2007). Collectively, these new findings are intriguing, as they provide support for a functional link between the Golgi and the centrosome.
Are there functional interactions between the Golgi and the centrosome during mitosis?
Regulation of mitotic Golgi reorganization from the centrosome.
The physical proximity of the Golgi apparatus and the centrosome is transiently lost during mitosis when Golgi membranes undergo extensive fragmentation. This dramatic change in Golgi structure is concomitant with a block in secretory trafficking and the reorganization of the microtubule cytoskeleton (Colanzi et al., 2003). Although the Golgi and the centrosome are physically separate at this stage of the cell cycle, there is evidence for functional interactions between these two organelles, which may control progression through mitosis.
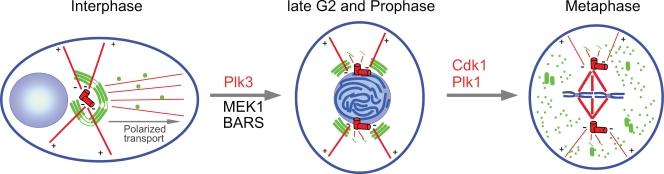
Many studies have identified possible links between mitotic Golgi fragmentation and the centrosome. For instance, breaking the Golgi ribbon into its constituent stacks during G2 requires the activity of the protein kinase Plk3 (Xie et al., 2004; López-Sánchez et al., 2009), which localizes to the centrosome and spindle poles (Xie et al., 2004; Jiang et al., 2006). The subsequent conversion of Golgi stacks into small, highly dispersed fragments (Jesch et al., 2001; Altan-Bonnet et al., 2006) and vesicular/tubular clusters next to astral spindle microtubules (Shima et al., 1998; Wei and Seemann, 2009) is regulated by two mitotic kinases, Cdk1 and Plk1, which are both associated with the centrosome (Fig. 3; Bailly et al., 1989; Dai and Cogswell, 2003). These findings suggest that components of the centrosome, spindle poles, or the spindle may initiate a signaling pathway that leads to the fragmentation of the Golgi and that may help coordinate Golgi dynamics with cell cycle progression. However, these regulatory factors also exist in the cytosol, and possible roles of cytosolic pools of Cdk1 and Plk3 in mitotic Golgi fragmentation have not been excluded.
Figure 3.
Golgi fragmentation during mitosis. The mammalian Golgi apparatus (green) forms an interconnected ribbon adjacent to the centrosome (red) and the nucleus (blue). It nucleates a population of microtubules that is necessary for polarized protein transport. Plus (+) and minus ends (−) are indicated. The activities of the protein kinases Plk3 and MEK1 and the fission protein BARS are required to convert the ribbon structure into isolated stacks in late G2 and prophase. In metaphase, the isolated stacks are further fragmented by a Plk1- and Cdk1-dependent mechanism, producing vesicular/tubular membranes that are dispersed throughout the cytoplasm. During this process, ribbon determinants, which are proteins required for postmitotic Golgi ribbon formation, remain associated with the mitotic spindle for their partitioning into daughter cells. Centrosome-associated regulators of mitotic Golgi fragmentation are labeled in red. Regulators of Golgi fragmentation that are not associated with the centrosome are labeled in black.
Further support for functional interactions between the Golgi and the centrosome during mitosis stems from a novel study on spindle-dependent reassembly of the Golgi ribbon after mitosis (Wei and Seemann, 2009). In a series of elegant experiments, Wei and Seemann (2009) demonstrated that the spindle is required for the postmitotic reformation of the Golgi ribbon. They induced asymmetric cell division so that the entire spindle segregated into only one daughter cell. Although Golgi membranes assembled into stacks in both daughter cells, they only formed a ribbon in the cell that inherited the spindle. Ribbon formation in the spindle-free cell required coinjection of Golgi extracts and tubulin or the addition of spindle-containing fractions. Collectively, these results suggest that Golgi ribbon formation occurs in two steps, with the initial assembly into stacks being mediated by factors that are partitioned by a spindle-independent mechanism. The subsequent formation of the Golgi ribbon from individual stacks, however, has an additional requirement for ribbon determinants, which are likely to be Golgi-associated proteins inherited with the spindle. Possible candidates include regulators of Golgi dynamics and the secretory pathway that have been identified in preparations of the spindle matrix (Ma et al., 2009).
Significance of the loss of Golgi–centrosome proximity during mitosis.
Several studies have identified an unexpected link between mitotic Golgi fragmentation and cell cycle progression (Sütterlin et al., 2002; Hidalgo Carcedo et al., 2004; Preisinger et al., 2005). For example, interfering with mitotic Golgi disassembly by blocking the function of the peripheral Golgi protein GRASP65 or the fission protein BARS resulted in cell cycle arrest in G2 (Sütterlin et al., 2002; Hidalgo Carcedo et al., 2004). Intriguingly, breaking the ribbon into isolated stacks, which occurs in G2, is sufficient to overcome this cell cycle arrest and allows cells to enter mitosis (Colanzi et al., 2007; Feinstein and Linstedt, 2007). It is not known how and why the presence of an intact pericentriolar Golgi ribbon prevents mitotic entry. The existence of a Golgi checkpoint, which monitors the correct inheritance of the Golgi complex, has been proposed because these inhibitory effects are not caused by activation of the DNA damage checkpoint (Sütterlin et al., 2002; Hidalgo Carcedo et al., 2004). It is conceivable that severing the Golgi ribbon in G2 separates ribbon determinants from the rest of the Golgi so that they can cosegregate with the spindle (Fig. 3). Such a mechanism for spindle-dependent Golgi inheritance would ensure that both daughter cells inherit the ability to form a Golgi ribbon and, thus, to transport proteins in a polarized manner. By analogy to the spindle checkpoint, which controls the exit from mitosis by monitoring the correct binding of spindle microtubules to kinetochores, this Golgi checkpoint may assess binding of spindle microtubules to these putative ribbon determinants to regulate entry into mitosis.
Golgi-dependent regulation of the spindle and mitotic progression.
A series of recent studies has identified a requirement for specific Golgi-associated proteins in the formation of a bipolar spindle (Table II). These Golgi factors are functionally diverse and include the poly-ADP ribosyl transferase, tankyrase-1 (Chang et al., 2005), the putative Golgi stacking factor, GRASP65 (Sütterlin et al., 2005), a regulator of the spindle checkpoint, RINT-1 (Lin et al., 2007), and the phosphatidylinositide phosphatase, Sac1 (Burakov et al., 2008). Depletion of any one of these proteins leads to multipolar spindles and mitotic cell death. For example, RNAi-mediated knockdown of Sac1 resulted in disorganization of the Golgi apparatus and mitotic defects characterized by multiple mechanically active spindles (Liu et al., 2008). Similarly, loss of GRASP65 led to the formation of multipolar spindles and mitotic arrest followed by cell death (Sütterlin et al., 2005). The molecular mechanisms by which Golgi-associated proteins regulate spindle formation are not known. Also, it is not known whether Golgi components control spindle formation when Golgi membranes are in the form of a pericentriolar ribbon, isolated stacks, or small fragments.
Table II.
Golgi-associated proteins with a role in regulating centrosome and spindle function
| Protein | Function | Depletion phenotype | Reference |
| Sac1 | Lipid phosphatase | Multiple mechanically active spindles | Liu et al., 2008 |
| GM130 | Golgin | Aberrant centrosome, multipolar spindles | Kodani and Sütterlin, 2008 |
| GRASP65 | Golgi matrix | Multipolar spindles, mitotic cell death | Sütterlin et al., 2005 |
| RINT-1 | Membrane traffic | Multipolar spindles, mitotic cell death | Sun et al., 2007 |
| Tankyrase-1 | ADP ribosyl transferase | Multipolar spindles, mitotic cell death | Chang et al., 2005 |
| Rab6′ | GTPase | Metaphase block, SAC activation | Miserey-Lenkei et al., 2006 |
| Clathrin | Vesicle coat | Defects in chromosome congression, SAC activation | Royle et al., 2005 |
SAC, spindle assembly checkpoint.
In addition to Golgi-dependent effects on spindle formation, other mitotic events are also regulated by disassembly of Golgi stacks during prophase and prometaphase. Indeed, this disassembly step correlates with the release of several peripheral proteins from Golgi membranes to carry out specific functions during mitosis. For instance, clathrin dissociates from the Golgi complex and from endocytic vesicles during mitosis and localizes to the spindle pole where it stabilizes mitotic spindle fibers involved in chromosome segregation (Royle et al., 2005). The small GTPase, Rab6A′, is also released from the Golgi during mitotic Golgi fragmentation (Miserey-Lenkei et al., 2006). If this dynamic behavior of Rab6A′ is inhibited, cells are no longer able to progress through mitosis and are blocked in metaphase through activation of the spindle checkpoint. Another example is the Golgi-associated protein ACBD3, whose release and cytoplasmic dispersal during mitotic Golgi breakdown is necessary for the activation of Numb in the regulation of asymmetric cell division (Zhou et al., 2007). Thus, in addition to facilitating the partitioning of Golgi membranes into the daughter cells, Golgi fragmentation may provide a unique mechanism for the regulation of signaling pathways that involve Golgi-associated components. In the case of ACBD3 and Rab6A′, Golgi fragmentation may relieve inhibitory effects that are either the result of proximity with the centrosome or the organization of the Golgi ribbon.
Conclusions
There is increasing evidence that the relationship between the Golgi apparatus and the centrosome in mammalian cells extends beyond physical proximity and involves functional interactions. Several features of this Golgi–centrosome relationship can be surmised from the recent studies reviewed. This relationship appears to be bidirectional because components of each organelle are able to influence the function of the other organelle. For example, Golgi proteins are necessary for centrosome organization and positioning (Chang et al., 2005; Sütterlin et al., 2005; Kodani and Sütterlin, 2008), whereas centrosome-nucleated microtubules are required for pericentriolar Golgi positioning (Corthésy-Theulaz et al., 1992; Cole et al., 1996). Importantly, these functional interactions affect fundamental cellular processes such as cell polarization and progression through mitosis (Sütterlin et al., 2002; Yadav et al., 2009). Intriguingly, there is evidence for functional interactions when the Golgi and the centrosome are in physical proximity during interphase but also during mitosis when they are physically separate.
What is the functional significance of the physical proximity between the Golgi and the centrosome? One possibility is that it may enhance the efficiency of signaling between the Golgi and centrosome and thereby facilitate directional protein transport. The Golgi apparatus is well known for its role in the exocytic pathway, and Golgi membranes, the intermediate compartment, and late endosomes are concentrated in the centrosomal area in mammalian cells (Marie et al., 2009). Thus, the centrosomal area may serve as a traffic hub, allowing integrated regulation of exocytic and endocytic transport routes for polarized delivery of cargo. In support of this idea, species in which Golgi membranes are not adjacent to the centrosome use alternative strategies for transporting proteins in a directional manner. For example, polarized secretion in Drosophila melanogaster is achieved by targeting mRNA to specific transitional ER–Golgi units in which the cargo is synthesized and secreted locally (Herpers and Rabouille, 2004).
Why has it taken so long to reveal functional Golgi–centrosome interactions during cell division? The phenomenon of a pericentriolar interphase Golgi ribbon, which is fragmented and dispersed during mitosis, is mainly seen in mammalian cells. Therefore, the significance of this dynamic spatial relationship cannot be studied in a more genetically tractable system such as yeast or Drosophila in which genome-wide screens can be readily performed. Furthermore, there has been a lack of tools to separate the Golgi and centrosome without affecting the functions of these organelles. Some recent studies have used new approaches such as severing the Golgi ribbon by depleting structural golgins, but there are still experimental limitations. For example, an intact Golgi ribbon cannot simply be displaced from the pericentriolar region, which makes it difficult to directly test the significance of Golgi localization versus organization. In addition, Golgi fragmentation, as induced by the depletion of structural Golgi proteins, is a multifactorial process that is marked by both the loss of the Golgi ribbon and dispersal from the pericentriolar position. The limited availability of experimental tools makes it difficult to separate these processes, which has hampered efforts to dissect their individual contributions to the Golgi–centrosome partnership. Also, until a recent study (Kodani et al., 2009), a molecular pathway linking the Golgi and the centrosome during interphase had not been described. For these reasons, it has been difficult to experimentally alter Golgi–centrosome proximity and assay the effects.
Although progress has been made, there are many unresolved questions about the Golgi–centrosome relationship during the cell cycle. For example, is there a single bidirectional regulatory pathway between the Golgi and the centrosome, or are there separate signaling pathways in each direction? Are there differences in signaling between these organelles during interphase when the organelles are adjacent and in mitosis when they are physically separate? There are also more specific unanswered questions. For example, how do Golgi proteins control spindle formation? Which factors on the mitotic spindle regulate postmitotic reassembly of the Golgi? How does the organization of the Golgi apparatus control progression through the cell cycle? Is there a Golgi organization checkpoint, and what does it monitor? The answers to these questions will help us better understand the significance of Golgi–centrosome interactions and could lead to the development of novel approaches for the treatment of several important diseases, including cancer.
Acknowledgments
We apologize to those colleagues whose work we were not able to discuss due to space limitation. We are grateful to Andrew Kodani for providing immunofluorescence images. We thank Dr. Ming Tan for insightful discussions and critical comments on the manuscript.
A. Colanzi acknowledges the Italian Association for Cancer Research (Milan, Italy; grant IG 6074) and Telethon (Italy) for financial support. C. Sütterlin is supported by a grant from the University of California Cancer Research Coordinating Committee.
Footnotes
Abbreviations used in this paper:
- γ-TuRC
- γ-tubulin ring complex
References
- Altan-Bonnet N., Sougrat R., Liu W., Snapp E.L., Ward T., Lippincott-Schwartz J. 2006. Golgi inheritance in mammalian cells is mediated through endoplasmic reticulum export activities. Mol. Biol. Cell. 17:990–1005 10.1091/mbc.E05-02-0155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly E., Dorée M., Nurse P., Bornens M. 1989. p34cdc2 is located in both nucleus and cytoplasm; part is centrosomally associated at G2/M and enters vesicles at anaphase. EMBO J. 8:3985–3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez I.B., Lowe M. 2009. Golgins and GRASPs: holding the Golgi together. Semin. Cell Dev. Biol. 20:770–779 10.1016/j.semcdb.2009.03.011 [DOI] [PubMed] [Google Scholar]
- Bejarano E., Cabrera M., Vega L., Hidalgo J., Velasco A. 2006. Golgi structural stability and biogenesis depend on associated PKA activity. J. Cell Sci. 119:3764–3775 10.1242/jcs.03146 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Glover D.M. 2007. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 8:451–463 10.1038/nrm2180 [DOI] [PubMed] [Google Scholar]
- Bisel B., Wang Y., Wei J.H., Xiang Y., Tang D., Miron-Mendoza M., Yoshimura S., Nakamura N., Seemann J. 2008. ERK regulates Golgi and centrosome orientation towards the leading edge through GRASP65. J. Cell Biol. 182:837–843 10.1083/jcb.200805045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownhill K., Wood L., Allan V. 2009. Molecular motors and the Golgi complex: staying put and moving through. Semin. Cell Dev. Biol. 20:784–792 10.1016/j.semcdb.2009.03.019 [DOI] [PubMed] [Google Scholar]
- Burakov A.V., Zhapparova O.N., Kovalenko O.V., Zinovkina L.A., Potekhina E.S., Shanina N.A., Weiss D.G., Kuznetsov S.A., Nadezhdina E.S. 2008. Ste20-related protein kinase LOSK (SLK) controls microtubule radial array in interphase. Mol. Biol. Cell. 19:1952–1961 10.1091/mbc.E06-12-1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt J.K., Echeverri C.J., Nilsson T., Vallee R.B. 1997. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J. Cell Biol. 139:469–484 10.1083/jcb.139.2.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone K.G., Webb N.J., Znameroski E.A., Welch M.D. 2008. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 134:148–161 10.1016/j.cell.2008.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabin-Brion K., Marceiller J., Perez F., Settegrana C., Drechou A., Durand G., Poüs C. 2001. The Golgi complex is a microtubule-organizing organelle. Mol. Biol. Cell. 12:2047–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P., Coughlin M., Mitchison T.J. 2005. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat. Cell Biol. 7:1133–1139 10.1038/ncb1322 [DOI] [PubMed] [Google Scholar]
- Colanzi A., Suetterlin C., Malhotra V. 2003. Cell-cycle-specific Golgi fragmentation: how and why? Curr. Opin. Cell Biol. 15:462–467 10.1016/S0955-0674(03)00067-X [DOI] [PubMed] [Google Scholar]
- Colanzi A., Hidalgo Carcedo C., Persico A., Cericola C., Turacchio G., Bonazzi M., Luini A., Corda D. 2007. The Golgi mitotic checkpoint is controlled by BARS-dependent fission of the Golgi ribbon into separate stacks in G2. EMBO J. 26:2465–2476 10.1038/sj.emboj.7601686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole N.B., Sciaky N., Marotta A., Song J., Lippincott-Schwartz J. 1996. Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol. Biol. Cell. 7:631–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corthésy-Theulaz I., Pauloin A., Pfeffer S.R. 1992. Cytoplasmic dynein participates in the centrosomal localization of the Golgi complex. J. Cell Biol. 118:1333–1345 10.1083/jcb.118.6.1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo A., Franco B. 2009. The dynamic cilium in human diseases. Pathogenetics. 2:3 10.1186/1755-8417-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W., Cogswell J.P. 2003. Polo-like kinases and the microtubule organization center: targets for cancer therapies. Prog. Cell Cycle Res. 5:327–334 [PubMed] [Google Scholar]
- Derby M.C., Lieu Z.Z., Brown D., Stow J.L., Goud B., Gleeson P.A. 2007. The trans-Golgi network golgin, GCC185, is required for endosome-to-Golgi transport and maintenance of Golgi structure. Traffic. 8:758–773 10.1111/j.1600-0854.2007.00563.x [DOI] [PubMed] [Google Scholar]
- Diao A., Rahman D., Pappin D.J., Lucocq J., Lowe M. 2003. The coiled-coil membrane protein golgin-84 is a novel rab effector required for Golgi ribbon formation. J. Cell Biol. 160:201–212 10.1083/jcb.200207045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz Añel A.M., Malhotra V. 2005. PKCeta is required for β1γ2/β3γ2- and PKD-mediated transport to the cell surface and the organization of the Golgi apparatus. J. Cell Biol. 169:83–91 10.1083/jcb.200412089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois T., Paléotti O., Mironov A.A., Fraisier V., Stradal T.E., De Matteis M.A., Franco M., Chavrier P. 2005. Golgi-localized GAP for Cdc42 functions downstream of ARF1 to control Arp2/3 complex and F-actin dynamics. Nat. Cell Biol. 7:353–364 10.1038/ncb1244 [DOI] [PubMed] [Google Scholar]
- Efimov A., Kharitonov A., Efimova N., Loncarek J., Miller P.M., Andreyeva N., Gleeson P., Galjart N., Maia A.R., McLeod I.X., et al. 2007. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell. 12:917–930 10.1016/j.devcel.2007.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S. 2006. In vitro assay of primary astrocyte migration as a tool to study Rho GTPase function in cell polarization. Methods Enzymol. 406:565–578 10.1016/S0076-6879(06)06044-7 [DOI] [PubMed] [Google Scholar]
- Feinstein T.N., Linstedt A.D. 2007. Mitogen-activated protein kinase kinase 1-dependent Golgi unlinking occurs in G2 phase and promotes the G2/M cell cycle transition. Mol. Biol. Cell. 18:594–604 10.1091/mbc.E06-06-0530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein T.N., Linstedt A.D. 2008. GRASP55 regulates Golgi ribbon formation. Mol. Biol. Cell. 19:2696–2707 10.1091/mbc.E07-11-1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit J.A., Tuft R.A., Fogarty K.E., Pazour G.J. 2006. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell. 17:3781–3792 10.1091/mbc.E06-02-0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit J.A., San Agustin J.T., Xu F., Jonassen J.A., Samtani R., Lo C.W., Pazour G.J. 2008. The Golgin GMAP210/TRIP11 anchors IFT20 to the Golgi complex. PLoS Genet. 4:e1000315 10.1371/journal.pgen.1000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Sztul E. 2001. A novel interaction of the Golgi complex with the vimentin intermediate filament cytoskeleton. J. Cell Biol. 152:877–894 10.1083/jcb.152.5.877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herpers B., Rabouille C. 2004. mRNA localization and ER-based protein sorting mechanisms dictate the use of transitional endoplasmic reticulum-golgi units involved in gurken transport in Drosophila oocytes. Mol. Biol. Cell. 15:5306–5317 10.1091/mbc.E04-05-0398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo Carcedo C., Bonazzi M., Spanò S., Turacchio G., Colanzi A., Luini A., Corda D. 2004. Mitotic Golgi partitioning is driven by the membrane-fissioning protein CtBP3/BARS. Science. 305:93–96 10.1126/science.1097775 [DOI] [PubMed] [Google Scholar]
- Jesch S.A., Mehta A.J., Velliste M., Murphy R.F., Linstedt A.D. 2001. Mitotic Golgi is in a dynamic equilibrium between clustered and free vesicles independent of the ER. Traffic. 2:873–884 10.1034/j.1600-0854.2001.21203.x [DOI] [PubMed] [Google Scholar]
- Jiang N., Wang X., Jhanwar-Uniyal M., Darzynkiewicz Z., Dai W. 2006. Polo box domain of Plk3 functions as a centrosome localization signal, overexpression of which causes mitotic arrest, cytokinesis defects, and apoptosis. J. Biol. Chem. 281:10577–10582 10.1074/jbc.M513156200 [DOI] [PubMed] [Google Scholar]
- Kodani A., Sütterlin C. 2008. The Golgi protein GM130 regulates centrosome morphology and function. Mol. Biol. Cell. 19:745–753 10.1091/mbc.E07-08-0847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani A., Kristensen I., Huang L., Sütterlin C. 2009. GM130-dependent control of Cdc42 activity at the Golgi regulates centrosome organization. Mol. Biol. Cell. 20:1192–1200 10.1091/mbc.E08-08-0834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Guan J.L., Chien S. 2005. Biochemistry and biomechanics of cell motility. Annu. Rev. Biomed. Eng. 7:105–150 10.1146/annurev.bioeng.7.060804.100340 [DOI] [PubMed] [Google Scholar]
- Lin C.M., Chen H.J., Leung C.L., Parry D.A., Liem R.K. 2005. Microtubule actin crosslinking factor 1b: a novel plakin that localizes to the Golgi complex. J. Cell Sci. 118:3727–3738 10.1242/jcs.02510 [DOI] [PubMed] [Google Scholar]
- Lin X., Liu C.C., Gao Q., Zhang X., Wu G., Lee W.H. 2007. RINT-1 serves as a tumor suppressor and maintains Golgi dynamics and centrosome integrity for cell survival. Mol. Cell. Biol. 27:4905–4916 10.1128/MCB.02396-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Boukhelifa M., Tribble E., Morin-Kensicki E., Uetrecht A., Bear J.E., Bankaitis V.A. 2008. The Sac1 phosphoinositide phosphatase regulates Golgi membrane morphology and mitotic spindle organization in mammals. Mol. Biol. Cell. 19:3080–3096 10.1091/mbc.E07-12-1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Sánchez I., Sanz-García M., Lazo P.A. 2009. Plk3 interacts with and specifically phosphorylates VRK1 in Ser342, a downstream target in a pathway that induces Golgi fragmentation. Mol. Cell. Biol. 29:1189–1201 10.1128/MCB.01341-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Tai G., Hong W. 2004. Autoantigen Golgin-97, an effector of Arl1 GTPase, participates in traffic from the endosome to the trans-golgi network. Mol. Biol. Cell. 15:4426–4443 10.1091/mbc.E03-12-0872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Tsai M.Y., Wang S., Lu B., Chen R., Iii J.R., Zhu X., Zheng Y. 2009. Requirement for Nudel and dynein for assembly of the lamin B spindle matrix. Nat. Cell Biol. 11:247–256 10.1038/ncb1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie M., Dale H.A., Sannerud R., Saraste J. 2009. The function of the intermediate compartment in pre-Golgi trafficking involves its stable connection with the centrosome. Mol. Biol. Cell. 20:4458–4470 10.1091/mbc.E08-12-1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra P., Salvatore L., Mironov A., Jr., Di Campli A., Di Tullio G., Trucco A., Beznoussenko G., Mironov A., De Matteis M.A. 2007. The biogenesis of the Golgi ribbon: the roles of membrane input from the ER and of GM130. Mol. Biol. Cell. 18:1595–1608 10.1091/mbc.E06-10-0886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P.M., Folkmann A.W., Maia A.R., Efimova N., Efimov A., Kaverina I. 2009. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat. Cell Biol. 11:1069–1080 10.1038/ncb1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miserey-Lenkei S., Couëdel-Courteille A., Del Nery E., Bardin S., Piel M., Racine V., Sibarita J.B., Perez F., Bornens M., Goud B. 2006. A role for the Rab6A' GTPase in the inactivation of the Mad2-spindle checkpoint. EMBO J. 25:278–289 10.1038/sj.emboj.7600929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peränen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., Jackson P.K. 2007. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 129:1201–1213 10.1016/j.cell.2007.03.053 [DOI] [PubMed] [Google Scholar]
- Nebenführ A., Staehelin L.A. 2001. Mobile factories: Golgi dynamics in plant cells. Trends Plant Sci. 6:160–167 10.1016/S1360-1385(01)01891-X [DOI] [PubMed] [Google Scholar]
- Ngo M., Ridgway N.D. 2009. Oxysterol binding protein-related Protein 9 (ORP9) is a cholesterol transfer protein that regulates Golgi structure and function. Mol. Biol. Cell. 20:1388–1399 10.1091/mbc.E08-09-0905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouthas F., Girard P., Lecaudey V., Ly T.B., Gilmour D., Boulin C., Pepperkok R., Reynaud E.G. 2008. In migrating cells, the Golgi complex and the position of the centrosome depend on geometrical constraints of the substratum. J. Cell Sci. 121:2406–2414 10.1242/jcs.026849 [DOI] [PubMed] [Google Scholar]
- Preisinger C., Short B., De Corte V., Bruyneel E., Haas A., Kopajtich R., Gettemans J., Barr F.A. 2004. YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14-3-3ζ. J. Cell Biol. 164:1009–1020 10.1083/jcb.200310061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisinger C., Körner R., Wind M., Lehmann W.D., Kopajtich R., Barr F.A. 2005. Plk1 docking to GRASP65 phosphorylated by Cdk1 suggests a mechanism for Golgi checkpoint signalling. EMBO J. 24:753–765 10.1038/sj.emboj.7600569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D., Mulholland J., Franzusoff A., Segev N., Botstein D. 1992. Characterization of the Saccharomyces Golgi complex through the cell cycle by immunoelectron microscopy. Mol. Biol. Cell. 3:789–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M.A., Bachert C., Puri S., Lanni F., Linstedt A.D. 2006. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat. Cell Biol. 8:238–248 10.1038/ncb1366 [DOI] [PubMed] [Google Scholar]
- Rehberg M., Kleylein-Sohn J., Faix J., Ho T.H., Schulz I., Gräf R. 2005. Dictyostelium LIS1 is a centrosomal protein required for microtubule/cell cortex interactions, nucleus/centrosome linkage, and actin dynamics. Mol. Biol. Cell. 16:2759–2771 10.1091/mbc.E05-01-0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilein A.R., Serpinskaya A.S., Karcher R.L., Dujardin D.L., Vallee R.B., Gelfand V.I. 2003. Differential regulation of dynein-driven melanosome movement. Biochem. Biophys. Res. Commun. 309:652–658 10.1016/j.bbrc.2003.08.047 [DOI] [PubMed] [Google Scholar]
- Ríos R.M., Sanchís A., Tassin A.M., Fedriani C., Bornens M. 2004. GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell. 118:323–335 10.1016/j.cell.2004.07.012 [DOI] [PubMed] [Google Scholar]
- Rivero S., Cardenas J., Bornens M., Rios R.M. 2009. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 28:1016–1028 10.1038/emboj.2009.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roghi C., Allan V.J. 1999. Dynamic association of cytoplasmic dynein heavy chain 1a with the Golgi apparatus and intermediate compartment. J. Cell Sci. 112:4673–4685 [DOI] [PubMed] [Google Scholar]
- Royle S.J., Bright N.A., Lagnado L. 2005. Clathrin is required for the function of the mitotic spindle. Nature. 434:1152–1157 10.1038/nature03502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybakin V., Gounko N.V., Späte K., Höning S., Majoul I.V., Duden R., Noegel A.A. 2006. Crn7 interacts with AP-1 and is required for the maintenance of Golgi morphology and protein export from the Golgi. J. Biol. Chem. 281:31070–31078 10.1074/jbc.M604680200 [DOI] [PubMed] [Google Scholar]
- Sahlender D.A., Roberts R.C., Arden S.D., Spudich G., Taylor M.J., Luzio J.P., Kendrick-Jones J., Buss F. 2005. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J. Cell Biol. 169:285–295 10.1083/jcb.200501162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima D.T., Cabrera-Poch N., Pepperkok R., Warren G. 1998. An ordered inheritance strategy for the Golgi apparatus: visualization of mitotic disassembly reveals a role for the mitotic spindle. J. Cell Biol. 141:955–966 10.1083/jcb.141.4.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short B., Preisinger C., Körner R., Kopajtich R., Byron O., Barr F.A. 2001. A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J. Cell Biol. 155:877–883 10.1083/jcb.200108079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohda M., Misumi Y., Yamamoto A., Yano A., Nakamura N., Ikehara Y. 2001. Identification and characterization of a novel Golgi protein, GCP60, that interacts with the integral membrane protein giantin. J. Biol. Chem. 276:45298–45306 10.1074/jbc.M108961200 [DOI] [PubMed] [Google Scholar]
- Sohda M., Misumi Y., Yoshimura S., Nakamura N., Fusano T., Sakisaka S., Ogata S., Fujimoto J., Kiyokawa N., Ikehara Y. 2005. Depletion of vesicle-tethering factor p115 causes mini-stacked Golgi fragments with delayed protein transport. Biochem. Biophys. Res. Commun. 338:1268–1274 10.1016/j.bbrc.2005.10.084 [DOI] [PubMed] [Google Scholar]
- Stanley H., Botas J., Malhotra V. 1997. The mechanism of Golgi segregation during mitosis is cell type-specific. Proc. Natl. Acad. Sci. USA. 94:14467–14470 10.1073/pnas.94.26.14467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga K., Hattori H., Saito A., Akagawa K. 2005. RNA interference-mediated silencing of the syntaxin 5 gene induces Golgi fragmentation but capable of transporting vesicles. FEBS Lett. 579:4226–4234 10.1016/j.febslet.2005.06.053 [DOI] [PubMed] [Google Scholar]
- Sun Y., Shestakova A., Hunt L., Sehgal S., Lupashin V., Storrie B. 2007. Rab6 regulates both ZW10/RINT-1 and conserved oligomeric Golgi complex-dependent Golgi trafficking and homeostasis. Mol. Biol. Cell. 18:4129–4142 10.1091/mbc.E07-01-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K.H., de Pablo Y., Vincent F., Johnson E.O., Chavers A.K., Shah K. 2008. Novel genetic tools reveal Cdk5's major role in Golgi fragmentation in Alzheimer's disease. Mol. Biol. Cell. 19:3052–3069 10.1091/mbc.E07-11-1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sütterlin C., Hsu P., Mallabiabarrena A., Malhotra V. 2002. Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell. 109:359–369 10.1016/S0092-8674(02)00720-1 [DOI] [PubMed] [Google Scholar]
- Sütterlin C., Polishchuk R., Pecot M., Malhotra V. 2005. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol. Biol. Cell. 16:3211–3222 10.1091/mbc.E04-12-1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Shibata H., Shimakawa M., Miyamoto M., Mukai H., Ono Y. 1999. Characterization of a novel giant scaffolding protein, CG-NAP, that anchors multiple signaling enzymes to centrosome and the golgi apparatus. J. Biol. Chem. 274:17267–17274 10.1074/jbc.274.24.17267 [DOI] [PubMed] [Google Scholar]
- Thyberg J., Moskalewski S. 1999. Role of microtubules in the organization of the Golgi complex. Exp. Cell Res. 246:263–279 10.1006/excr.1998.4326 [DOI] [PubMed] [Google Scholar]
- Valderrama F., Babià T., Ayala I., Kok J.W., Renau-Piqueras J., Egea G. 1998. Actin microfilaments are essential for the cytological positioning and morphology of the Golgi complex. Eur. J. Cell Biol. 76:9–17 [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Zareno J., Whitmore L., Choi C.K., Horwitz A.F. 2007. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J. Cell Biol. 176:573–580 10.1083/jcb.200612043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walenta J.H., Didier A.J., Liu X., Krämer H. 2001. The Golgi-associated hook3 protein is a member of a novel family of microtubule-binding proteins. J. Cell Biol. 152:923–934 10.1083/jcb.152.5.923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Seemann J., Pypaert M., Shorter J., Warren G. 2003. A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. EMBO J. 22:3279–3290 10.1093/emboj/cdg317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J.H., Seemann J. 2009. The mitotic spindle mediates inheritance of the Golgi ribbon structure. J. Cell Biol. 184:391–397 10.1083/jcb.200809090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S., Wang Q., Ruan Q., Liu T., Jhanwar-Uniyal M., Guan K., Dai W. 2004. MEK1-induced Golgi dynamics during cell cycle progression is partly mediated by Polo-like kinase-3. Oncogene. 23:3822–3829 10.1038/sj.onc.1207479 [DOI] [PubMed] [Google Scholar]
- Yadav S., Puri S., Linstedt A.D. 2009. A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Mol. Biol. Cell. 20:1728–1736 10.1091/mbc.E08-10-1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S., Yoshioka K., Barr F.A., Lowe M., Nakayama K., Ohkuma S., Nakamura N. 2005. Convergence of cell cycle regulation and growth factor signals on GRASP65. J. Biol. Chem. 280:23048–23056 10.1074/jbc.M502442200 [DOI] [PubMed] [Google Scholar]
- Yoshino A., Setty S.R., Poynton C., Whiteman E.L., Saint-Pol A., Burd C.G., Johannes L., Holzbaur E.L., Koval M., McCaffery J.M., Marks M.S. 2005. tGolgin-1 (p230, golgin-245) modulates Shiga-toxin transport to the Golgi and Golgi motility towards the microtubule-organizing centre. J. Cell Sci. 118:2279–2293 10.1242/jcs.02358 [DOI] [PubMed] [Google Scholar]
- Yvon A.M., Walker J.W., Danowski B., Fagerstrom C., Khodjakov A., Wadsworth P. 2002. Centrosome reorientation in wound-edge cells is cell type specific. Mol. Biol. Cell. 13:1871–1880 10.1091/mbc.01-11-0539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Atkins J.B., Rompani S.B., Bancescu D.L., Petersen P.H., Tang H., Zou K., Stewart S.B., Zhong W. 2007. The mammalian Golgi regulates numb signaling in asymmetric cell division by releasing ACBD3 during mitosis. Cell. 129:163–178 10.1016/j.cell.2007.02.037 [DOI] [PubMed] [Google Scholar]
- Zolov S.N., Lupashin V.V. 2005. Cog3p depletion blocks vesicle-mediated Golgi retrograde trafficking in HeLa cells. J. Cell Biol. 168:747–759 10.1083/jcb.200412003 [DOI] [PMC free article] [PubMed] [Google Scholar]