Chromatin entanglements undergo specific protein-mediated compaction to fold into mitotic chromosomes.
Abstract
We have analyzed the topological organization of chromatin inside mitotic chromosomes. We show that mitotic chromatin is heavily self-entangled through experiments in which topoisomerase (topo) II is observed to reduce mitotic chromosome elastic stiffness. Single chromosomes were relaxed by 35% by exogenously added topo II in a manner that depends on hydrolysable adenosine triphosphate (ATP), whereas an inactive topo II cleavage mutant did not change chromosome stiffness. Moreover, experiments using type I topos produced much smaller relaxation effects than topo II, indicating that chromosome relaxation by topo II is caused by decatenation and/or unknotting of double-stranded DNA. In further experiments in which chromosomes are first exposed to protease to partially release protein constraints on chromatin, ATP alone relaxes mitotic chromosomes. The topo II–specific inhibitor ICRF-187 blocks this effect, indicating that it is caused by endogenous topo II bound to the chromosome. Our experiments show that DNA entanglements act in concert with protein-mediated compaction to fold chromatin into mitotic chromosomes.
Introduction
Chromosomal DNAs of eukaryote cells are hierarchically folded, beginning with wrapping of DNA into nucleosomes and chromatin fiber. Chromatin organization at larger scales is thought to involve its tethering, e.g., attachments to the nuclear envelope (Marshall et al., 1996), chromatin–chromatin interactions that form loops, or cross-links (Adolph et al., 1977; Paulson and Laemmli, 1977; Marsden and Laemmli, 1979; Sachs et al., 1995; Poirier and Marko, 2002). Although the molecular mechanisms behind tethering of eukaryote chromosomes remain incompletely understood, there is a general prejudice that proteins are responsible.
During cell division, chromosomes undergo condensation from an open, interphase conformation to a compact, cylindrical mitotic form. In mitotic chromosomes, structural maintenance of chromosome (SMC) proteins are thought to play an important chromatin-tethering role, largely on the basis of experiments indicating that condensin SMCs are required for establishment and maintenance of mitotic chromosome structure (Hirano and Mitchison, 1994; Ono et al., 2003, 2004; Hirota et al., 2004; Hirano, 2005). However, in experiments in which condensin subunits were suppressed by conditional knockout or siRNA, persistent mitotic chromosome formation has been observed, suggesting that other factors may contribute to constraint and organization of mitotic chromatin (Hudson et al., 2003; Hirota et al., 2004).
We have developed micromanipulation methods to probe mitotic chromosome structure (Fig. 1 A; Poirier and Marko, 2002) in which structural effects of reagents directly sprayed onto newt (Notophthalmus viridescens) chromosomes after their removal from cells were determined via measurement of chromosome mechanics. We exploit the well-defined and stable elasticity of isolated native chromosomes (Poirier et al., 2000; Pope et al., 2006; Marko, 2008). For the newt cells used in our experiments (Reese et al., 1976), a force of ∼500 pN extends a mitotic chromosome to double its native length (∼20 µm). Such experiments have revealed that mitotic chromosomes are completely dissolved by restriction nucleases (Poirier and Marko, 2002; Pope et al., 2006), indicating that they must have a network organization, with ∼15-kb stretches of chromatin strung between isolated cross-links.
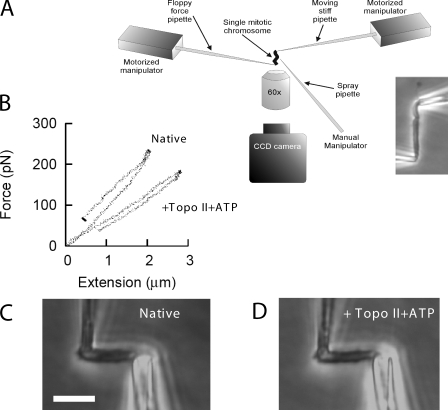
Figure 1.
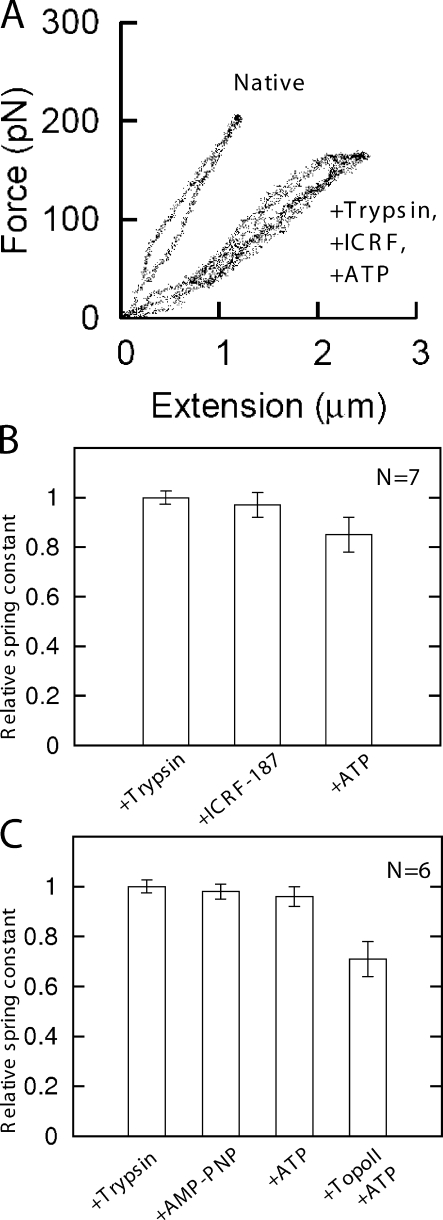
Topo II + ATP reduces the spring constant of mitotic chromosomes. (A) Single-chromosome micromanipulation setup. A mitotic chromosome is suspended between two pipettes and observed in a microscope. One of the pipettes has a long floppy taper used to measure forces in the 10–1,000-pN range via observation of its bending, whereas a second pipette is relatively stiff and does not bend. A third pipette is used to spray reagents. CCD, charge-coupled device. (B) Force versus extension (change in length) of a single mitotic chromosome in its native state shows it to have a spring constant (slope of curve) of 100 pN/µm. The same measurement after spray of topo II + ATP in AB (see Results) for 30 min shows a 50% reduction in spring constant to ∼50 pN/µm. (C) Native chromosome of B. Bar, 10 µm. (D) Chromosome of C after exposure to topo II + ATP reveals little change in morphology despite the large change in spring constant.
Intriguingly, proteases only slowly and partially decondense mitotic chromosomes (Maniotis et al., 1997; Bojanowski et al., 1998; Almagro et al., 2004; Pope et al., 2006). In micromanipulation experiments with reconstituted Xenopus laevis chromatids (Almagro et al., 2004) and for newt chromosomes (Pope et al., 2006), protease treatment reduces chromosome stiffness (spring constant), but chromosomes remain elastic. Although it is possible that in these experiments some well-protected proteins are inaccessible to the proteases, this partial unfolding of chromosomes is difficult to attribute to any inability of enzymes to penetrate to the interior of the chromosome given the dramatic results with nucleases. An alternative hypothesis compatible with these observations is that nonprotein-based mechanisms play a role in stabilizing mitotic chromosomes. A major role for RNA has been eliminated by experiments with RNAase in which no relaxation of mitotic chromosomes occurred (Almagro et al., 2004).
Given the large amount of DNA contour length in a whole mitotic chromosome and also that chromatin is thought to be at least partially constrained by protein interconnections, we hypothesized that DNA in mitotic chromatin might be heavily self-entangled and that entanglements might stabilize mitotic chromosome structure. Previous experiments have shown that topoisomerase (topo) II is required for normal mitotic chromosome condensation (Uemura et al., 1987; Wood and Earnshaw, 1990; Adachi et al., 1991; Hirano and Mitchison, 1991). Topo II, in the presence of ATP, mediates passage of one double-stranded DNA through a second, transiently cleaved DNA segment, thus allowing DNA topology to change. Therefore, previous experiments suggest that establishment of normal mitotic chromosome structure requires changes in DNA topology.
To directly examine the role of DNA entanglements in stabilizing mitotic chromosome structure, we performed chromosome microspraying experiments with recombinant human topo IIα using chromosome elasticity measurements to read out structural changes caused by enzyme activity. This approach allows time sequencing of reactions because the microspraying technique introduces only small volumes of reagents, which quickly diffuse away after the spraying is stopped (Poirier and Marko, 2002; Pope et al., 2006). We used the α-isoform of topo II because it is essential for cell division and bound to chromosomes throughout mitosis, a distribution and functionality not found for the β-isoform (Christensen et al., 2002; Carpenter and Porter, 2004; Linka et al., 2007).
Results
Topo IIα plus hydrolysable ATP relaxes mitotic chromosomes
Fig. 1 B shows force–extension curves for a native chromosome and the same chromosome after 30-min exposure to 80 ng/µl purified topo IIα + 2 mM ATP in topo II activity buffer (AB). Although no visible change in chromosome morphology occurred (Fig. 1, compare C and D), topo II + ATP reduced the chromosome spring constant (Fig. 1 B, see slope of data curves, i.e., change in force divided by change in length). A series of stretch–relax cycles were performed in each case both before and after enzyme exposure to verify that changes in chromosome elasticity occurred only during the enzyme spray period and not during the stretching–relaxation cycles (unpublished data).
In 12 topo II + ATP experiments of this type, a broad range of chromosome relaxations occurred (Fig. 2 A). In three cases, a relatively weak relaxation was achieved (Fig. 2 A, right), but importantly, an increase in chromosome spring constant was never observed in any topo II + ATP trial, which attests to a specific effect of topo II on isolated chromosomes. Exposure of native chromosomes to topo II + ATP for periods >30 min did not cause further chromosome relaxation.
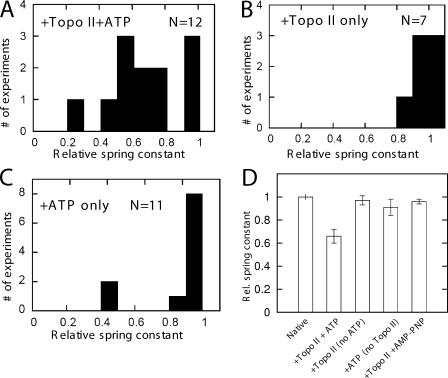
Figure 2.
Relaxation of mitotic chromosomes by topo II requires hydrolysable ATP. (A) Results of 12 experiments of the type shown in Fig. 1, showing the ratio of the spring constant after topo II + ATP treatment to native spring constant. More than 20% relaxation occurred in nine of the trials. (B) Results of seven experiments where topo II in AB (no ATP) was sprayed for 30 min. No large relaxations occurred, and in three runs, a slight increase in stiffness was observed. (C) Histogram for 11 experiments where ATP in AB was used for 30 min; in 8/11 experiments, little or no relaxation occurred, but in 3/11 experiments, significant relaxation occurred. (D) Summary of experiments with different enzyme–nucleotide combinations. Height of bars shows relative spring constants normalized to the native (untreated) chromosome spring constant; error bars indicate standard error for each set of experiments. The 35% relaxation effect observed for topo II + ATP (second bar) does not occur for topo II with no ATP (third bar). A small relaxation effect occurs for ATP with no topo II (fourth bar), suggesting that ATPases on the chromosome are active. Experiments with topo II + AMP-PNP (fifth bar) generated nearly no relaxation.
In the 12 topo II + ATP trials, the mean relaxation in spring constant was 35% (Fig. 2 D, second bar), calculated relative to the initial native chromosome spring constant. Given the relatively small error (Fig. 2 D), the difference between the first two bars of Fig. 2 D is statistically significant (P = 3 × 10−7).
No significant relaxation occurred in any experiments where chromosomes were sprayed with 80 ng/µl topo II in AB with no ATP (seven trials; Fig. 2 B). In this case, we obtained a very narrow distribution of results showing either a small increase or decrease of spring constant, demonstrating that ATP is required for topo II–induced chromosome relaxation. These experiments show that at the concentrations we have used, topo II by itself has little or no effect when sprayed on native mitotic chromosomes.
We next used topo II and AMP-PNP, a nonhydrolysable ATP analogue, which stalls topo II in a closed conformation on DNA after performing at most one strand passage (Roca et al., 1996). When mitotic chromosomes were exposed to topo II + 2 mM AMP-PNP, no experiment showed strong relaxation (six trials; Fig. 2 D). Finally, no relaxation was observed in separate experiments in which topo II AB alone was sprayed onto chromosomes (unpublished data). Thus, significant relaxation of native chromosomes requires hydrolysable ATP and is not the result of ATP-independent binding of topo II to DNA.
ATP alone can relax native chromosomes
We also performed experiments with 2 mM ATP without any topo II with the result that a large relaxation (>40% change) of native mitotic chromosome spring constant was observed in 2/11 trials (Fig. 2 C). The relaxation by ATP was not as robust as in the experiments with topo II + ATP, as the relaxation by ATP alone averaged over all trials was small as a result of the majority of runs showing essentially zero relaxation. However, the distribution of relaxations was much broader than that observed for topo II alone, topo II + AMP-PNP, or buffer alone (unpublished data). We suspected that this relaxation by ATP alone was the result of endogenous topo II on the chromosome (see Relaxation of chromosomes by ATP….). Variability in relaxation driven by ATP alone (Fig. 2 C) relative to the relaxation driven by topo II + ATP (Fig. 2 A) might either reflect that endogenous topo II may have been lost from the periphery of chromosomes during their isolation (Christensen et al., 2002) or that it had already relaxed entanglements near its binding sites to varying degrees in the isolated chromosomes. We will show that if mitotic chromosomes are mildly trypsinized, subsequent ATP treatment leads to a much more reliable relaxation effect.
Topo IIα catalytic activity is required for relaxation of mitotic chromosomes
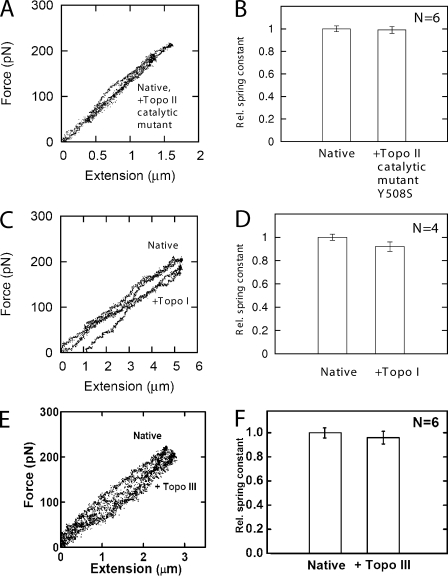
To examine the question of what aspect of topo II activity was responsible for the chromosome relaxation effect (Fig. 2 A), we performed experiments with a catalytic mutant of human topo IIα (Y805S). This mutant cannot cleave DNA, and therefore cannot carry out strand passage, but is able to hydrolyze ATP and close its N-terminal clamp (Oestergaard et al., 2004). Chromosome spray experiments were performed exactly as for wild-type topo IIα (80 ng/µl enzyme with 2 mM ATP in AB). No relaxation was seen in any of six experiments (Fig. 3, A and B), indicating that DNA-cleaving activity is an absolute requirement for the chromosome relaxation observed for wild-type topo IIα.
Figure 3.
Relaxation requires topo II DNA cleavage activity and is not caused by relaxation of DNA torsional stress. (A) Native chromosome spring constant is not changed by exposure to human topo IIα catalytic mutant Y805S + ATP in AB. (B) In a series of six experiments as in A, no change in spring constant was observed after treatment with Y805S + ATP in AB. (C) Native chromosome force response is nearly unchanged by exposure to the human type IB topo I. (D) In a series of four experiments as in C, only a very small relative reduction in chromosome spring constant was observed. (E) Native chromosome force response is unchanged by exposure to the type IA topo III. (F) In a series of six experiments as in E, no change in chromosome spring constant was observed.
Topo I relaxes mitotic chromosomes only very weakly
To determine whether the topo IIα–mediated chromosome relaxation (Fig. 2 A) was caused by topo II’s ability to either remove DNA entanglements or, alternately, was caused by relaxation of DNA supercoils, we performed experiments with purified human topo I, a type IB topo. Type IB topos cleave only one strand of double-helix DNA, and therefore cannot release DNA entanglements, but efficiently relax supercoils in an ATP-independent manner by releasing multiple links per cleavage event (Koster et al., 2005).
Experiments with topo I (40 ng/µl topo I with no ATP; four trials) resulted in only a very weak (∼15%) relaxation effect (Fig. 3, C and D). This effect is at the edge of our ability to reliably detect, although a stiffening effect was not observed in any trial. Importantly, at the enzyme concentrations used during chromosome spraying, only topo II decatenated plasmid DNA, whereas the supercoiling relaxation activity per unit mass of topo I is at least 20-fold higher than that of topo II (Fig. S1). Topo I may very weakly relax mitotic chromosomes but to a degree well below that accomplished by topo IIα. The chromosome relaxation observed in topo IIα experiments (Fig. 2 A) can therefore be ascribed to that enzyme’s ability to remove DNA entanglements, whereas its ability to remove supercoils plays a minor role.
Topo III does not relax mitotic chromosomes
Topo III is a type IA topo that carries out ATP-independent strand exchange of single-stranded DNAs (Mondragón and DiGate, 1999), and its function in vivo is thought to be mainly decatenation of single-stranded DNAs. Topo III can relax supercoiled DNA if single-stranded regions are present, and it will partially relax negatively supercoiled plasmids under physiological conditions in vitro (Fig. S3). Because topo III is needed to maintain genomic integrity (Win et al., 2004) possibly by playing a role in recombinational repair during S phase (Kwan et al., 2003) and is involved in sister chromatid cohesion (Lai et al., 2007), we decided to test whether mitotic chromosomes might be partially stabilized by catenated single-stranded DNA regions. Spray experiments with topo III (80 ng/ul topo III in T3AB with no ATP; six trials) resulted in no statistically significant relaxation (Fig. 3, E and F), which is consistent with our topo I results, indicating that extensive catenated single-stranded DNA regions do not help stabilize mitotic chromosomes by themselves.
Trypsinization of chromosomes stimulates their subsequent relaxation by ATP alone
We now return to the observation that treatment of native chromosomes with ATP alone showed a highly variable relaxation effect (Fig. 2 C). We reasoned that if we reduced the density of protein cross-links, we might be able to observe more robust relaxation by either topoIIα + ATP or perhaps even by ATP alone. The latter would permit us to more convincingly address the question of whether endogenous topo II was responsible for relaxation of chromosomes by ATP alone. We chose to use trypsin to release protein constraints in mitotic chromosomes, which causes chromosome expansion (Maniotis et al., 1997; Bojanowski et al., 1998; Almagro et al., 2004; Pope et al., 2006) driven by internal chromatin osmotic stress (Marko and Siggia, 1997).
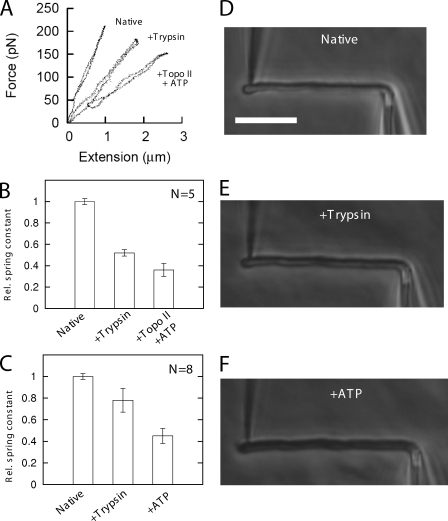
Thus, we performed two reaction–step experiments in which an isolated mitotic chromosome was first treated with 100 nM trypsin to effect ∼50% relaxation (Fig. 4 A, compare native with trypsin curves). When chromosomes were subsequently exposed to topo II + ATP, appreciable relaxation occurred in four out of five trials (Fig. 4, A and B), indicating that significant DNA entanglement remained after trypsinization.
Figure 4.
Trypsinized mitotic chromosomes are relaxed by ATP alone as well as by topo II + ATP. (A) Force–extension experiment showing spring constant of native chromosome, the same chromosome after exposure to trypsin, and after subsequent exposure to topo II + ATP for 30 min. As for native chromosomes (Fig. 1), topo II + ATP causes relaxation. (B) Results of a series of five experiments as in A. Bar heights indicate averaged ratios of spring constants to native spring constants. (C) Results of a series of eight experiments similar to those of A and B but where only ATP was sprayed for 30 min. On average, 40% relaxation occurred, which is a much stronger effect than for ATP on native chromosomes (compare with Fig. 2 D, fourth bar). (D–F) Phase-contrast images show little or no difference between native (D), trypsinized (E), and ATP-treated (F) chromosomes; neither trypsinization nor subsequent relaxation by ATP leads to clearly visible decondensation. Bar, 10 µm.
In separate experiments, we again trypsinized chromosomes and exposed them to 2 mM ATP with the result that on average, a 40% relaxation of the trypsinized chromosomes occurred (Fig. 4 C, difference between second and third bars). More than 20% relaxation of trypsinized chromosomes occurred in six of the eight trials with a larger and more consistent effect than in experiments on native chromosomes (compare with Fig. 2, C and D [+ATP bar]). Therefore, mild trypsinization of mitotic chromosomes stimulates their further relaxation by ATP, suggesting release of protein-shielded entanglements and/or activation of axially located topo II (Earnshaw and Heck, 1985). Despite the rather large chromosome relaxation, there was little or no visually observable change in the level of condensation of chromosomes after either trypsin or ATP reactions (Fig. 4, D–F). In experiments in which chromosomes were first trypsinized and exposed to 80 ng/µl topo II alone (no ATP), no change in spring constant was observed, e.g., as a result of topo II–DNA interactions (unpublished data).
Relaxation of chromosomes by ATP alone is caused by endogenous topo II
To test whether the ATP-driven relaxation of trypsinized chromosomes was caused by topo II rather than some other ATP-dependent enzyme, we performed three reaction–step experiments in which after trypsinization, chromosomes were exposed to 170 µM specific topo II inhibitor ICRF-187 (30-min reaction; sufficient to obtain 90% suppression of strand passage activity as shown previously by Jensen et al., 2006) and finally to 2 mM ATP. Results of a representative experiment are shown in Fig. 5 A, which shows that after ICRF-187 treatment, the chromosome force constant is essentially unchanged by ATP exposure.
Figure 5.
Topo II on the chromosomes combined with externally supplied hydrolyzable ATP is responsible for ATP-driven relaxation of trypsinized chromosomes. (A) Force–extension experiment showing initial native elastic response, elasticity after trypsinization, elasticity after subsequent exposure to the topo II inhibitor ICRF-187, and finally exposure to ATP. Data for measurements after each of these chemical exposures are essentially indistinguishable, indicating first that ICRF-187 does not appreciably stiffen or relax chromosomes, and second, that it blocks any relaxation by subsequent exposure to ATP. (B) Results of seven experiments as in A. ATP causes at most a weak relaxation of a trypsinized chromosome after its exposure to ICRF-187 (compare with large effect of ATP in Fig. 4 C). Bar heights show mean spring constants relative to those of the trypsinized chromosomes. (C) Results of a series of six experiments in which chromosomes were trypsinized, exposed to AMP-PNP, exposed to ATP, and finally exposed to topo II + ATP. Results are normalized to trypsinized chromosomes. AMP-PNP does not change chromosome stiffness (second bar) and blocks relaxation by subsequent ATP exposure (third bar). However, topo II + ATP does relax chromosomes after AMP-PNP exposure (fourth bar).
Results of a series of seven experiments are shown in Fig. 5 B, where spring constants are shown relative to those of the trypsinized chromosome. After trypsinization, exposure to ICRF-187 causes little or no change in elastic response (Fig. 5 B, second bar). Treatment with ATP after ICRF-187 caused only a small change in spring constant (7% standard error; P value of change = 0.04; Fig. 5 B, 12% reduction from second to third bar). This effect was much smaller than that observed without ICRF-187 treatment (compare Fig. 5 [A and B] with Fig. 4 C). We concluded that inhibition of endogenous topo II reduces the ATP-driven relaxation of trypsinized mitotic chromosomes and therefore, that the endogenous topo II is principally responsible for the relaxation.
Hydrolysable ATP is required for relaxation of trypsinized chromosomes by endogenous topo II
The reliable relaxation of trypsinized chromosomes by ATP alone (Fig. 4 C) allowed us to more directly examine the role of ATP hydrolysis in the reaction involving endogenous topo II. We performed a series of four reaction–step experiments in which a native chromosome was first trypsinized and exposed to 2 mM AMP-PNP. In six such experiments, no relaxation was driven by AMP-PNP (results normalized to trypsinized chromosome spring constant; Fig. 5 C, second bar). The third reaction step was treatment of chromosomes by 2 mM ATP, and again no relaxation occurred (Fig. 5 C, third bar), which is consistent with blocking of endogenous topo II turnover by AMP-PNP. In in vitro decatenation assays, we confirmed that binding of AMP-PNP to topo IIα before addition of ATP blocks plasmid decatenation (Fig. S2).
After these steps, subsequent exposure to exogenous topo II + ATP caused a 25% relaxation (P value of change = 2 × 10−4; Fig. 5 C, fourth bar). Thus, even after blockage of endogenous topo II by AMP-PNP, exogenous topo II + ATP is able to relax chromosomes.
Discussion
Topo IIα relaxes whole mitotic chromosomes by resolving double-stranded DNA entanglements
We have shown that topo IIα + ATP reduces native mitotic chromosome spring constants by a mean value of 35% (Fig. 2 A). This effect requires hydrolysable ATP (Fig. 2 D) and DNA cleavage by topo II (Fig. 3, A and B). Experiments with topo I indicate that this relaxation effect is not mainly caused by relaxation of DNA torsional stress or unconstrained supercoiling (Fig. 3, C and D). Furthermore, experiments with topo III (Fig. 3, E and F) indicate that single-stranded DNA catenations do not play a major role in stabilizing mitotic chromatin. We conclude that the relaxation of whole chromosomes by topo IIα + ATP is mainly the result of double-strand passages mediated by that enzyme. Notably, the relaxation of mitotic chromosomes by topo IIα + ATP, although a large fraction of the mechanical stiffness of the chromosome, is not accompanied by easily observed morphological changes (Fig. 1, C and D) and is essentially undetectable by optical microscopy observation alone.
We note that we do not observe large changes in structure or mechanical properties of mitotic chromosomes during extraction of them from the cell (Poirier et al., 2002; Marko, 2008). Nevertheless, we do note that it is possible that some amount of either swelling or condensation of mitotic chromosomes might occur during their extraction and that this might change the degree to which topo II removes DNA entanglements in our experiments relative to what would have been the case inside the cell. However, this does not change our primary conclusion that DNA entanglements play an appreciable role in constraining mitotic chromatin.
Comparison of topo II concentrations used to those in the cell
Our relaxation experiments used a spray of 80 ng/µl topo IIα, corresponding to a monomer concentration of 470 nM (active topo IIα consists of two 170-kD subunits). This is comparable with the total topo IIα concentration in the cell; it has been estimated that there are 105 topo IIα molecules in a human nucleus (Meyer et al., 1997). Given a nuclear volume of roughly 10−13 liters, one obtains an estimate for in vivo concentration of monomer of ∼1 µM. Therefore, our experiments are performed with enzyme concentrations comparable with those found in vivo; it is unlikely that our results are an artifact of use of excessively high enzyme concentration.
Endogenous topo II on the chromosome can relax mitotic chromosomes after proteolysis
ATP alone was observed to generate significant chromosome relaxation in some experiments (Fig. 2 C). We subsequently found that this effect is more reproducible after mild trypsinization of whole mitotic chromosomes, which reduces their stiffness or, equivalently, their elastic modulus (the elastic modulus is the force per unit area that would be required to double the length of an elastic object if its initial linear elastic response continued to that length and provides a shape-independent way of characterizing elastic stiffness). After their trypsinization, exposure of chromosomes to ATP alone causes a reliable relaxation (Fig. 4 C). This relaxation by ATP alone is entirely blocked by a topo II–specific drug (Fig. 5, A and B), and subsequent exposure to topo II + ATP leads to relaxation. This series of experiments indicates that endogenous topo II on the chromosome is able to relax mitotic chromosomes after their proteolysis. A possible explanation for this effect is that the mild trypsinization enables access of endogenous topo II to entanglements and also provides a driving force, i.e., internal osmotic pressure of the chromatin fibers, for entanglement resolution.
DNA entanglements are present at high density in mitotic chromosomes
Our experiments show that DNA topological constraints contribute to the folding of chromatin into mitotic chromosomes; because removal of DNA interlocks relaxes chromosomes, the presence of DNA interlocks in the native chromosome contributes to mitotic chromosome mechanical stability.
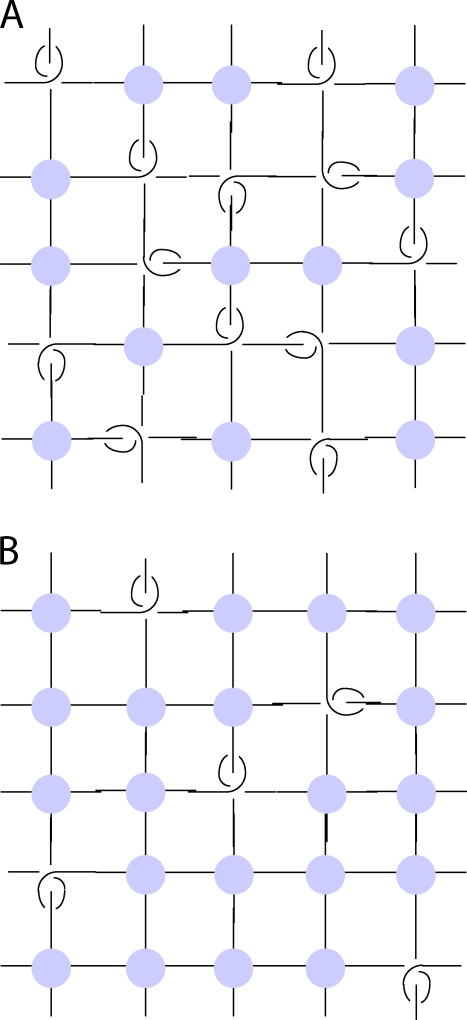
The interior of a mitotic chromatid is a well-solvated network of chromatin held together by (noncovalent) cross-linking elements (i.e., a polymer gel; Poirier and Marko, 2002). Topological constraints on chromatin between cross-link points are expected to be present (Fig. 6 A) as in any polymer network. The elastic modulus of a polymer network (proportional to chromosome spring constant) has been shown to be a weighted sum of the densities of cross-links and entanglements (Everaers and Kremer, 1996). Therefore, removing either protein cross-linkers (trypsinization) or DNA entanglements (topo II + ATP) will reduce the elastic modulus (stiffness) of a polymer network.
Figure 6.
Cross-link topological entanglement network model of interior of mitotic chromatid. (A) Cross-links (gray) and topological constraints (interlinks) both constrain chromatin segments (black lines) and are present at comparable densities in the native chromosome; removal of either the cross-links or entanglements will reduce the elastic response. To simplify the sketch, only local entanglements are shown; the path of the chromatin fiber can also be self-entangled at scales larger than the characteristic network mesh scale. (B) Cross-links at higher density than entanglements eliminate the possibility for entanglement removal to significantly impact network elasticity.
Our finding of an ∼35% relaxation of native chromosomes by topo II + ATP indicates that densities of cross-linkers and entanglements are comparable: if the density of cross-linking was much larger than that of entanglements (Fig. 6 B), strand passages would not significantly impact elasticity. However, if the cross-link density was much less than that of entanglements, addition of topo II in excess should be able to almost entirely eliminate elasticity of native chromosomes.
Theoretical results for the relative contributions of chain stretching and topological entanglements to the net elasticity of random polymer networks (Everaers and Kremer, 1996) allow a rough estimate to be made of the distance between protein cross-links. We have observed a 35% relaxation of native chromosome spring constant by topo II + ATP. Taking this as the contribution of topological constraints to chromosome elasticity (most likely this is an underestimate), previous data (see Table I in Everaers and Kremer, 1996) indicate that there are ∼20 persistence lengths of chromatin or ∼200 nucleosomes between successive network cross-links, corresponding to 30 kb DNA (an estimate of 10 nucleosomes per chromatin persistence length is used; note that the bending stiffness of chromatin fibers is a quite unsettled subject). This inter–cross-link spacing is in rough agreement with the cross-link density estimated from DNA cleavage experiments (Poirier and Marko, 2002).
The effect of ATP alone, which we have shown permits endogenous topo II to relax entanglements, and the stimulation of this effect by trypsinization, are both easily understood using this network model. The density and distribution (Christensen et al., 2002) of endogenous topo II on native isolated chromosomes limits relaxation in ATP-only experiments because DNA-bound topo II can relax entanglements only near to its binding position within the chromosome domain delineated by neighboring cross-links and cannot resolve chromatin entanglements, which are spread over regions larger than the inter–cross-link spacing. Endogenous topo II is likely to have already relaxed entanglement density to this limit in our experiments, causing ATP alone to have a relatively weak effect (Fig. 2, C and D). However, trypsinization reduces the density of cross-links, allowing endogenous topo II to remove topological constraints of originally cross-link–constrained chromatin segments and to reduce chromosome elasticity when given ATP (Fig. 4 C).
Functions of topo II in disentanglement and entanglement of mitotic chromatin
Topo II in the cell does not remove all of the self-entanglement in chromosomes before metaphase, and inside condensing chromatids, it may act to introduce entanglements, which become thermodynamically favorable for tightly packed polymers (Krasnow and Cozzarelli, 1982; Everaers and Kremer, 1996; Arsuaga et al., 2002). Active (ATP dependent) cross-linking, e.g., by condensin I, which rigidifies chromosomes to ensure their mechanical stability during mitosis (Gerlich et al., 2006), could drive topo II to add intrachromatid linking at the same time that it is driving the release of interchromatid links (Marko and Siggia, 1997), explaining observations of topo II involvement in metaphase compaction (Chang et al., 2003; Sakaguchi and Kikuchi, 2004).
An additional mechanism by which entanglements may arise is via chiral structures along chromatin that may be introduced actively by condensin complexes. X. laevis condensin I has been observed to generate positive-writhe knotting of nicked circular DNA in vitro using ATP and topo II (Kimura et al., 1999; Stray et al., 2005). The simplest interpretation of those experiments is that condensin I binds DNA so as to promote positive writhe, which in the presence of a type II topo can drive local knot formation. It has been suggested that this mechanism might contribute to higher-order folding of chromatin in vivo (Kimura et al., 1999). If this occurs in vivo, one would expect extracted chromosomes to contain a high degree of local positive-writhe knotting, increasing overall chromosome elastic modulus by essentially shortening and stiffening inter–cross-link connections. Resolution of these knots by exogenous topo II may contribute to the chromosome relaxation we have observed.
Bimodality of topo II’s topology-changing activities, i.e., its ability to entangle or disentangle DNA depending on local driving forces, is likely essential to ensure the segregation of chromatids by resolving interchromatid interlocks, while at the same time, strengthening them so as to withstand anaphase by adding intrachromatid entanglements. During telophase, dissociation of cross-linkers (or knot stabilizers) such as condensin I or II would generate a driving force for topo II to resolve these topological constraints, allowing unfolding of chromosomes into their interphase form. This process is mimicked by our trypsinization of metaphase chromosomes, where removal of protein constraint of chromatin is seen to increase the ability of either endogenous or exogenous topo II to relax chromosomes.
Thus, we hypothesize that the topological state of chromatin in mitotic chromatids varies throughout mitosis, controlled by modulation of chromatin fiber folding and cross-linking (Marko and Siggia, 1997). This suggests an explanation for the variability of chromosome relaxation by topo II + ATP (Fig. 2 A) and by ATP alone (Fig. 2 C), namely that self-entanglement in chromosomes is changing during mitosis. However, we have not yet systematically explored this possibility, and other factors (e.g., cell to cell or chromosome to chromosome variations in entanglement) may be responsible for the variability of chromosome relaxation that we have observed. A related question is that of precisely how elasticity of whole chromosomes is related to density and types of DNA entanglements. The application of results from the aforementioned studies of polymer networks to our experiments is suggestive but crude, and further work will be required to understand how DNA entanglements play a role in chromosome structure and the mitotic condensation process.
Materials and methods
Recombinant enzymes
Plasmid YEpWOB6 for expression of human topo IIα was transformed into the yeast strain Saccharomyces cerevisiae JEL1. Plasmid pHT143 for expression of human topo I was transformed into the yeast strain S. cerevisiae top-null strain RS190. Crude extracts from yeast cultures expressing topo IIα or topo I were prepared, and the proteins were purified as described previously (Habermeyer et al., 2005). Purity of recombinant enzymes were judged to be at least 95% for topo I and topo IIα as determined by SDS gel electrophoresis followed by Coomassie staining. Escherichia coli topo III was provided by A. Mondragon (Northwestern University, Evanston, IL), which was purified as described previously (Hiasa et al., 1994).
Specific activities of purified topo I and III were verified by gel analysis of relaxation of supercoiled plasmid pUC19 (2.7 kb) as described previously (Figs. S1 and S3; Barthelmes et al., 2004). Activity of topo II was analyzed by decatenation of kinetoplast DNA (interlinked 2.5-kb circles; Topogen; Figs. S1 and S2). Specific activity reactions were run in the same buffers and at similar enzyme concentrations to those used for micromanipulation experiments. Mutant human topo IIα (Y805S) was purified as described previously (Oestergaard et al., 2004).
Cell culture
Sample dishes were prepared using #1 microscope glass onto which ∼25-mm-diameter rubber O-rings were affixed using wax. Newt TVI (N. viridescens) cells were grown (TVI cell line; Reese et al., 1976) in open dishes in cell culture medium, in room air, and at room temperature. Culture medium was 50% vol/vol L-15 with L-glutamine (Cellgro Mediatech), 8% vol/vol fetal bovine serum (Cambrex), 1% vol/vol Pen/Strep (Cambrex), and 1 ng/ml fungizone (Cambrex) in sterile water. Medium was replaced daily. Experiments were performed on ∼80% confluent samples.
Enzyme reaction buffers and microspraying experiments
Topo IIα was diluted in AB (10 mM Bis-Tris-propane/HCl, pH 7.9, 120 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, and 30 ng/µl BSA) either with or without ATP (Thermo Fisher Scientific) or AMP-PNP (Sigma-Aldrich). Solutions were loaded into glass pipettes, pulled, and cut so as to have an ∼5-µm-diameter aperture. The pipettes were mounted on a three-axis manual manipulator (Taurus; World Precision) used to position them in the sample dish. Pressure between 10 and 100 Pa was used to drive spraying of the enzyme mix onto chromosomes. Microspray reactions were performed for 30 min, and flow of enzyme was stopped for several minutes to allow the small amounts of reagents introduced into the ∼1.5 ml sample cell to diffuse away from the chromosome. Previous experiments indicated that enzyme reactions do not continue after spraying is finished and that successive stretch–release cycles after enzyme exposures are all the same (Poirier and Marko, 2002; Pope et al., 2006); we observed the same behavior in the experiments of this study.
Experiments with topo IIα mutant Y805S were performed in the same way, by dilution of enzyme into AB with ATP followed by spraying onto chromosomes. Topo I, ATP, AMP-PNP, and ICRF-187 (Pfizer) were diluted into AB and sprayed similarly. Topo III was diluted into a slightly different AB (T3AB) with 120 mM NaCl, 1 mM MgCl2, 40 mM Tris-HCl, pH 7.5, and 1% BSA but was otherwise sprayed onto chromosomes similarly. Trypsin (Sigma-Aldrich) was diluted in 60% PBS (Cambrex) for spraying experiments.
Microscopy and chromosome micromanipulation
Micromanipulation experiments were performed on the stage of an inverted microscope (IX-70; Olympus) using a 60× 1.4 NA oil immersion objective as previously described (Poirier et al., 2000, 2006) at 23°C. All imaging was performed in the newt cell culture medium using a charge-coupled device camera (WV-BP310; Panasonic) with images acquired by a frame grabber (IMAQ PCI-1408; National Instruments) to a PC.
In brief, samples were placed on the microscope stage, prometaphase cells were identified, and a spray pipette filled with 0.05% vol/vol Triton X-100 in 60% PBS (Thermo Fisher Scientific) positioned by a motorized micromanipulator (MP-285; Sutter Instrument Co.) was used to destabilize the cell membrane via microspraying. After the chromosomes flowed out of the cell, one pipette was used to grab one end of one chromosome using aspiration; then, a second pipette was attached to the other chromosome end.
One of the two pipettes was pulled with a rather short taper so that it was very stiff, whereas the other was pulled with a long taper so as to have a softer end deflection spring constant of ∼200 pN/micron. The stiffer pipette was pulled at a rate of ∼2 µm/min, slowly enough to avoid viscoelastic effects (Poirier et al., 2000), and bending of the softer pipette was observed. Each extension–relaxation measurement was repeated two or three times to ensure their reproducibility and to verify that any changes in chromosome stiffness caused by previous enzyme exposures had ceased. Micromechanical data were collected automatically using online image analysis software written in Labview (National Instruments). We estimate experimental errors in individual spring constant measurements to be <10%, mainly from systematic errors in spring constant calibration (Poirier et al., 2000).
Online supplemental material
Fig. S1 shows relative catalytic activity of topo I and topo IIα. Fig. S2 shows specific kinetoplast DNA decatenation activity of topo IIα using buffer conditions mirroring those used during spray experiments. Fig. S3 shows relaxation activity of topo III. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200910085/DC1.
Acknowledgments
We are grateful to Prof. A. Mondragon for generously providing purified topo III.
This work was supported by the National Science Foundation (grants MCB-0240998 and DMR-0715099), the Chicago Biomedical Consortium with support from The Searle Funds at The Chicago Community Trust, the National Institutes of Health (grant U54-CA143869-01; NU-PS-OC), the Deutsche Forschungsgemeinschaft (grants CH 713/1-1, GRK 1033, and SFB 728), and the Danish Natural Science Research Council (grant 272-05-0274).
Footnotes
Abbreviations used in this paper:
- AB
- activity buffer
- SMC
- structural maintenance of chromosome
- topo
- topoisomerase
References
- Adachi Y., Luke M., Laemmli U.K. 1991. Chromosome assembly in vitro: topoisomerase II is required for condensation. Cell. 64:137–148 10.1016/0092-8674(91)90215-K [DOI] [PubMed] [Google Scholar]
- Adolph K.W., Cheng S.M., Laemmli U.K. 1977. Role of nonhistone proteins in metaphase chromosome structure. Cell. 12:805–816 10.1016/0092-8674(77)90279-3 [DOI] [PubMed] [Google Scholar]
- Almagro S., Riveline D., Hirano T., Houchmandzadeh B., Dimitrov S. 2004. The mitotic chromosome is an assembly of rigid elastic axes organized by structural maintenance of chromosomes (SMC) proteins and surrounded by a soft chromatin envelope. J. Biol. Chem. 279:5118–5126 10.1074/jbc.M307221200 [DOI] [PubMed] [Google Scholar]
- Arsuaga J., Vázquez M., Trigueros S., Sumners D., Roca J. 2002. Knotting probability of DNA molecules confined in restricted volumes: DNA knotting in phage capsids. Proc. Natl. Acad. Sci. USA. 99:5373–5377 10.1073/pnas.032095099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelmes H.U., Habermeyer M., Christensen M.O., Mielke C., Interthal H., Pouliot J.J., Boege F., Marko D. 2004. TDP1 overexpression in human cells counteracts DNA damage mediated by topoisomerases I and II. J. Biol. Chem. 279:55618–55625 10.1074/jbc.M405042200 [DOI] [PubMed] [Google Scholar]
- Bojanowski K., Maniotis A.J., Plisov S., Larsen A.K., Ingber D.E. 1998. DNA topoisomerase II can drive changes in higher order chromosome architecture without enzymatically modifying DNA. J. Cell. Biochem. 69:127–142 [DOI] [PubMed] [Google Scholar]
- Carpenter A.J., Porter A.C. 2004. Construction, characterization, and complementation of a conditional-lethal DNA topoisomerase IIalpha mutant human cell line. Mol. Biol. Cell. 15:5700–5711 10.1091/mbc.E04-08-0732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.J., Goulding S., Earnshaw W.C., Carmena M. 2003. RNAi analysis reveals an unexpected role for topoisomerase II in chromosome arm congression to a metaphase plate. J. Cell Sci. 116:4715–4726 10.1242/jcs.00797 [DOI] [PubMed] [Google Scholar]
- Christensen M.O., Larsen M.K., Barthelmes H.U., Hock R., Andersen C.L., Kjeldsen E., Knudsen B.R., Westergaard O., Boege F., Mielke C. 2002. Dynamics of human DNA topoisomerases IIα and IIβ in living cells. J. Cell Biol. 157:31–44 10.1083/jcb.200112023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W.C., Heck M.M. 1985. Localization of topoisomerase II in mitotic chromosomes. J. Cell Biol. 100:1716–1725 10.1083/jcb.100.5.1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everaers R., Kremer K. 1996. Topological interactions in model polymer networks. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 53:R37–R40 [DOI] [PubMed] [Google Scholar]
- Gerlich D., Hirota T., Koch B., Peters J.M., Ellenberg J. 2006. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr. Biol. 16:333–344 10.1016/j.cub.2005.12.040 [DOI] [PubMed] [Google Scholar]
- Habermeyer M., Fritz J., Barthelmes H.U., Christensen M.O., Larsen M.K., Boege F., Marko D. 2005. Anthocyanidins modulate the activity of human DNA topoisomerases I and II and affect cellular DNA integrity. Chem. Res. Toxicol. 18:1395–1404 10.1021/tx050039n [DOI] [PubMed] [Google Scholar]
- Hiasa H., DiGate R.J., Marians K.J. 1994. Decatenating activity of Escherichia coli DNA gyrase and topoisomerases I and III during oriC and pBR322 DNA replication in vitro. J. Biol. Chem. 269:2093–2099 [PubMed] [Google Scholar]
- Hirano T. 2005. Condensins: organizing and segregating the genome. Curr. Biol. 15:R265–R275 10.1016/j.cub.2005.03.037 [DOI] [PubMed] [Google Scholar]
- Hirano T., Mitchison T.J. 1991. Cell cycle control of higher-order chromatin assembly around naked DNA in vitro. J. Cell Biol. 115:1479–1489 10.1083/jcb.115.6.1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Mitchison T.J. 1994. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 79:449–458 10.1016/0092-8674(94)90254-2 [DOI] [PubMed] [Google Scholar]
- Hirota T., Gerlich D., Koch B., Ellenberg J., Peters J.M. 2004. Distinct functions of condensin I and II in mitotic chromosome assembly. J. Cell Sci. 117:6435–6445 10.1242/jcs.01604 [DOI] [PubMed] [Google Scholar]
- Hudson D.F., Vagnarelli P., Gassmann R., Earnshaw W.C. 2003. Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev. Cell. 5:323–336 10.1016/S1534-5807(03)00199-0 [DOI] [PubMed] [Google Scholar]
- Jensen L.H., Liang H., Shoemaker R., Grauslund M., Sehested M., Hasinoff B.B. 2006. A three-dimensional quantitative structure-activity relationship study of the inhibition of the ATPase activity and the strand passing catalytic activity of topoisomerase IIalpha by substituted purine analogs. Mol. Pharmacol. 70:1503–1513 10.1124/mol.106.026856 [DOI] [PubMed] [Google Scholar]
- Kimura K., Rybenkov V.V., Crisona N.J., Hirano T., Cozzarelli N.R. 1999. 13S condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell. 98:239–248 10.1016/S0092-8674(00)81018-1 [DOI] [PubMed] [Google Scholar]
- Koster D.A., Croquette V., Dekker C., Shuman S., Dekker N.H. 2005. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 434:671–674 10.1038/nature03395 [DOI] [PubMed] [Google Scholar]
- Krasnow M.A., Cozzarelli N.R. 1982. Catenation of DNA rings by topoisomerases. Mechanism of control by spermidine. J. Biol. Chem. 257:2687–2693 [PubMed] [Google Scholar]
- Kwan K.Y., Moens P.B., Wang J.C. 2003. Infertility and aneuploidy in mice lacking a type IA DNA topoisomerase III beta. Proc. Natl. Acad. Sci. USA. 100:2526–2531 10.1073/pnas.0437998100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.S., Seki M., Ui A., Enomoto T. 2007. Rmi1, a member of the Sgs1-Top3 complex in budding yeast, contributes to sister chromatid cohesion. EMBO Rep. 8:685–690 10.1038/sj.embor.7401000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linka R.M., Porter A.C., Volkov A., Mielke C., Boege F., Christensen M.O. 2007. C-terminal regions of topoisomerase IIalpha and IIbeta determine isoform-specific functioning of the enzymes in vivo. Nucleic Acids Res. 35:3810–3822 10.1093/nar/gkm102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniotis A.J., Bojanowski K., Ingber D.E. 1997. Mechanical continuity and reversible chromosome disassembly within intact genomes removed from living cells. J. Cell. Biochem. 65:114–130 [DOI] [PubMed] [Google Scholar]
- Marko J.F. 2008. Micromechanical studies of mitotic chromosomes. Chromosome Res. 16:469–497 10.1007/s10577-008-1233-7 [DOI] [PubMed] [Google Scholar]
- Marko J.F., Siggia E.D. 1997. Polymer models of meiotic and mitotic chromosomes. Mol. Biol. Cell. 8:2217–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden M.P., Laemmli U.K. 1979. Metaphase chromosome structure: evidence for a radial loop model. Cell. 17:849–858 10.1016/0092-8674(79)90325-8 [DOI] [PubMed] [Google Scholar]
- Marshall W.F., Dernburg A.F., Harmon B., Agard D.A., Sedat J.W. 1996. Specific interactions of chromatin with the nuclear envelope: positional determination within the nucleus in Drosophila melanogaster. Mol. Biol. Cell. 7:825–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.N., Kjeldsen E., Straub T., Knudsen B.R., Hickson I.D., Kikuchi A., Kreipe H., Boege F. 1997. Cell cycle–coupled relocation of types I and II topoisomerases and modulation of catalytic enzyme activities. J. Cell Biol. 136:775–788 10.1083/jcb.136.4.775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragón A., DiGate R. 1999. The structure of Escherichia coli DNA topoisomerase III. Structure. 7:1373–1383 10.1016/S0969-2126(00)80027-1 [DOI] [PubMed] [Google Scholar]
- Oestergaard V.H., Knudsen B.R., Andersen A.H. 2004. Dissecting the cell-killing mechanism of the topoisomerase II-targeting drug ICRF-193. J. Biol. Chem. 279:28100–28105 10.1074/jbc.M402119200 [DOI] [PubMed] [Google Scholar]
- Ono T., Losada A., Hirano M., Myers M.P., Neuwald A.F., Hirano T. 2003. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 115:109–121 10.1016/S0092-8674(03)00724-4 [DOI] [PubMed] [Google Scholar]
- Ono T., Fang Y., Spector D.L., Hirano T. 2004. Spatial and temporal regulation of condensins I and II in mitotic chromosome assembly in human cells. Mol. Biol. Cell. 15:3296–3308 10.1091/mbc.E04-03-0242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson J.R., Laemmli U.K. 1977. The structure of histone-depleted metaphase chromosomes. Cell. 12:817–828 10.1016/0092-8674(77)90280-X [DOI] [PubMed] [Google Scholar]
- Poirier M.G., Marko J.F. 2002. Mitotic chromosomes are chromatin networks without a mechanically contiguous protein scaffold. Proc. Natl. Acad. Sci. USA. 99:15393–15397 10.1073/pnas.232442599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier M., Eroglu S., Chatenay D., Marko J.F. 2000. Reversible and irreversible unfolding of mitotic newt chromosomes by applied force. Mol. Biol. Cell. 11:269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier M.G., Eroglu S., Marko J.F. 2002. The bending rigidity of mitotic chromosomes. Mol. Biol. Cell. 13:2170–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope L.H., Xiong C., Marko J.F. 2006. Proteolysis of mitotic chromosomes induces gradual and anisotropic decondensation correlated with a reduction of elastic modulus and structural sensitivity to rarely cutting restriction enzymes. Mol. Biol. Cell. 17:104–113 10.1091/mbc.E05-04-0321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese D.H., Yamada T., Moret R. 1976. An established cell line from the newt Notophthalmus viridescens. Differentiation. 6:75–81 10.1111/j.1432-0436.1976.tb01472.x [DOI] [PubMed] [Google Scholar]
- Roca J., Berger J.M., Harrison S.C., Wang J.C. 1996. DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proc. Natl. Acad. Sci. USA. 93:4057–4062 10.1073/pnas.93.9.4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs R.K., van den Engh G., Trask B., Yokota H., Hearst J.E. 1995. A random-walk/giant-loop model for interphase chromosomes. Proc. Natl. Acad. Sci. USA. 92:2710–2714 10.1073/pnas.92.7.2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi A., Kikuchi A. 2004. Functional compatibility between isoform alpha and beta of type II DNA topoisomerase. J. Cell Sci. 117:1047–1054 10.1242/jcs.00977 [DOI] [PubMed] [Google Scholar]
- Stray J.E., Crisona N.J., Belotserkovskii B.P., Lindsley J.E., Cozzarelli N.R. 2005. The Saccharomyces cerevisiae Smc2/4 condensin compacts DNA into (+) chiral structures without net supercoiling. J. Biol. Chem. 280:34723–34734 10.1074/jbc.M506589200 [DOI] [PubMed] [Google Scholar]
- Uemura T., Ohkura H., Adachi Y., Morino K., Shiozaki K., Yanagida M. 1987. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 50:917–925 10.1016/0092-8674(87)90518-6 [DOI] [PubMed] [Google Scholar]
- Win T.Z., Goodwin A., Hickson I.D., Norbury C.J., Wang S.W. 2004. Requirement for Schizosaccharomyces pombe Top3 in the maintenance of chromosome integrity. J. Cell Sci. 117:4769–4778 10.1242/jcs.01351 [DOI] [PubMed] [Google Scholar]
- Wood E.R., Earnshaw W.C. 1990. Mitotic chromatin condensation in vitro using somatic cell extracts and nuclei with variable levels of endogenous topoisomerase II. J. Cell Biol. 111:2839–2850 10.1083/jcb.111.6.2839 [DOI] [PMC free article] [PubMed] [Google Scholar]