An automated, quantitative 4D image analysis method is used to track kinetochore dynamics in metaphase cells.
Abstract
During mitosis in most eukaryotic cells, chromosomes align and form a metaphase plate halfway between the spindle poles, about which they exhibit oscillatory movement. These movements are accompanied by changes in the distance between sister kinetochores, commonly referred to as breathing. We developed a live cell imaging assay combined with computational image analysis to quantify the properties and dynamics of sister kinetochores in three dimensions. We show that baseline oscillation and breathing speeds in late prometaphase and metaphase are set by microtubule depolymerases, whereas oscillation and breathing periods depend on the stiffness of the mechanical linkage between sisters. Metaphase plates become thinner as cells progress toward anaphase as a result of reduced oscillation speed at a relatively constant oscillation period. The progressive slowdown of oscillation speed and its coupling to plate thickness depend nonlinearly on the stiffness of the mechanical linkage between sisters. We propose that metaphase plate formation and thinning require tight control of the state of the mechanical linkage between sisters mediated by centromeric chromatin and cohesion.
Introduction
Mitotic chromosome segregation requires the proper attachment of sister kinetochores to microtubules (MTs) emanating from opposing spindle poles and the generation of force at kinetochores to drive the separation of sister chromatids at anaphase. In all eukaryotes, before the initiation of anaphase, chromosomes congress to the metaphase plate, a central position equidistant between the spindle poles. It has been observed for more than 70 years that in most metazoan and plant cells, chromosomes oscillate on the plate for many minutes until anaphase initiates (Lewis, 1939; Hughes and Swann, 1948). Sister oscillations have been proposed to be driven by the coordinated assembly and disassembly of mechanically coupled kinetochore MT bundles and/or by molecular motors that mediate chromosome–MT attachment at the kinetochore (Skibbens et al., 1993). In addition, the interaction of nonkinetochore MTs with plus end–directed kinesin motors at the chromosome arms exerts antipoleward forces on the whole chromosomes (Rieder et al., 1986; Antonio et al., 2000; Funabiki and Murray, 2000). Therefore, chromosome oscillations and migration to the metaphase plate are the result of the summing of stochastic forces at the kinetochore and chromosome arms along with dynamic growth and shrinkage of MTs at the kinetochore. This generates broadly heterogeneous behaviors of sister pairs that, when examined at any given time or in a small sample set, only provide a limited view of the underlying mechanisms.
Pharmacological treatments or laser ablation producing very short chromosome fragments showed that a single kinetochore is sufficient for chromosome congression, indicating that kinetochores are the dominant factor for chromosome movement (Khodjakov et al., 1997; O’Connell et al., 2008). At the molecular level, both MT dynamics and MT motors like centromere protein E (CENP-E), mitotic centromere-associated kinesin (MCAK), and Kif18A have been implicated in the generation of force at kinetochores. CENP-E plays an important role in chromosome alignment during prometaphase via its ability to transport kinetochores to the spindle equator along MT bundles (Kapoor et al., 2006). Less known are its roles after alignment, where most kinetochores move via end-on attachment. Kif18A (kinesin-8) possesses not only plus end–directed motor activity but also depolymerase activity (Mayr et al., 2007; Varga et al., 2009), and MCAK (kinesin-13) acts as a pure MT depolymerase without motor activity (Hunter et al., 2003). MCAK and Kif18A have been implicated in the proper alignment of chromosomes and in the regulation of their oscillations about the metaphase plate (Kline-Smith et al., 2004; Wordeman et al., 2007; Stumpff et al., 2008). Previous studies of kinetochore oscillation speed and frequency after Kif18A depletion have reported conflicting results (Mayr et al., 2007; Stumpff et al., 2008), possibly because these studies relied on different selections of chromosomes in a dynamically heterogeneous population. The roles of other factors in the regulation of chromosome dynamics during mitosis, e.g., the role of the mechanical properties of the molecular linkers between sister kinetochores, are even less understood.
Results
Automated 4D live cell assay for systematic probing of HeLa kinetochore dynamics
To dissect the mechanisms of sister oscillation and breathing and to deduce their functional implications on metaphase plate alignment, the full range of sister kinetochore dynamics must be sampled and the statistical distributions of motion behaviors must be analyzed under a variety of well-defined molecular perturbations. To this end, we developed a HeLa cell line stably expressing the inner kinetochore protein CENP-A fused to EGFP (Fig. S1). We established a standardized 3D live cell imaging protocol by maximizing signal to noise ratio and spatiotemporal sampling while minimizing phototoxicity (Fig. 1). The acquisition of time-lapse images of EGFP–CENP-A fluorescence at a rate of one 3D stack (20 optical sections separated by 0.5 µm in focal position) every 7.5 s for 5 min yielded >90% kinetochore detectability and unaliased time courses of kinetochore movements while preserving normal progression through mitosis to anaphase in unperturbed cells (see Materials and methods; Fig. 1). Under these imaging conditions, approximately two thirds of the kinetochore pairs were visible in the 3D volume (Video 1).
Figure 1.
Live cell assay that maximizes signal-to-noise ratio while minimizing phototoxicity. To define a standardized protocol for our assay, we established a range of imaging conditions that maintained cell viability, i.e., 80% of cells progressed through mitosis with normal timing. The kinetochore detection results under these imaging conditions were compared with the “ground truth,” determined by detecting kinetochores in very high signal-to-noise ratio (SNR) images (in which detection sensitivity was maximized at the expense of cell viability). With this, imaging conditions that allowed the automatic detection of >90% of kinetochores while maintaining cell viability were established. Although not depicted, we also optimized temporal sampling of the time-lapse imaging protocol (see Materials and methods). We found that recording image stacks every 7.5 s was sufficient to capture the dynamics of sister oscillation and breathing while maintaining satisfactory cell viability (Fig. 2, j and k). NEBD, nuclear envelope breakdown. Bar, 5 µm.
Contributions of MT dynamics and components of the kinetochore and sister cohesion were probed pharmacologically and by siRNA-mediated depletion. For each perturbation, we used reagents and protocols known to fully perturb the system or provide quantitative knockdown, and we validated knockdown levels in individual cells (Fig. S2 and Table S1). We compared each perturbation with untreated wild-type (WT) cells or with WT cells synchronized by thymidine/aphidicolin block and release as appropriate.
Each time-lapse dataset was subjected to an automated image analysis pipeline that identified and tracked sister kinetochore pairs to yield their trajectories over time (see Materials and methods; Fig. 2 a and Videos 2–5). Under experimental conditions in which kinetochores formed a clear metaphase plate, we tracked the plate position over the course of a video by fitting a plane to the detected kinetochore positions. This defined a metaphase plate–centered coordinate system for kinetochore positions, allowing each kinetochore to be classified as aligned or unaligned by determining whether it was an inlier or an outlier with respect to the distribution of kinetochore positions along the normal to the metaphase plate (Fig. 2 a, step 2; and Video 3). The metaphase plate–centered coordinate system was also a translation- and rotation-free coordinate system that assisted in the tracking of kinetochores over the course of a video (Jaqaman et al., 2008). If anaphase onset occurred during the time window of a video, the software identified it as the time point at which the average separation between all sisters in the video started to monotonically increase.
Figure 2.
Automated image analysis and quantitative analysis of sister kinetochore dynamics. (a) The four steps of image analysis to measure sister kinetochore dynamics. White lines show boundaries of the 3D dataset, and the black rectangle indicates the magnified region shown in b. Bar, 2 µm. (b) Basic parameters of kinetochore dynamics: sister separation d and center normal position x. Bar, 0.5 µm. (c) Experimental time courses of sister center normal position x and sister separation d. (top) Green and orange indicate movements in the positive and negative directions along the normal. (bottom) Green and orange indicate phases of increase and decrease in sister separation. In both, black indicates unclassified intervals. Error bars indicate SD of the measured coordinates. (d) Illustration of sister (ΔprL and ΔprR) and center (ΔprC) displacement projections onto the vector connecting sister pairs (black). (e) Sister center normal and projection speeds and sister breathing speed in WT and fixed cells. Error bars indicate mean ± SD, representing cell to cell variation. Red double asterisks highlight values significantly smaller than WT (P < 0.001 by one-tailed t test). (f) Distribution of oscillation half-periods τx in WT. (inset) Illustration of the oscillation half-period τx; green and orange as in c (top). (g) Distribution of breathing half-periods τd in WT. (inset) Illustration of breathing half-period τd; green and orange as in c (bottom). (h) Three models with increasing width of the distribution of sister oscillation half-period. (i) Sister displacement cross-correlation (left) and sister center autocorrelation (right) resulting from the models in h and from a series of random sister positions. The blue and cyan curves were generated by the same half-period distribution but with more or less coupling, respectively, between the two sisters. (j) Sister displacement cross-correlation and sister center autocorrelation in WT and fixed cells calculated by combining all aligned sister kinetochores in each condition. (i–k) Line thickness represents the 95% confidence interval of the correlations about their calculated values (see Materials and methods). (k) Sister breathing autocorrelation in WT and fixed cells. Details as in j. Arrows, mean oscillation half-period; arrowheads, mean oscillation full period; carets, cross-correlation at lag ΔT = 0; stars, −0.5 autocorrelation at lag ΔT = 7.5 s (one frame), which is indicative of a series of random positions.
For each condition, we analyzed a minimum of 17 cells (44 cells on average), displaying an average of 32 sister kinetochore pairs per cell (Table S2). Although most conditions could be analyzed using a default set of image analysis parameters, some conditions changed kinetochore behavior so drastically that they required an adjustment of critical parameters (Table S3). In all cases, the goal was to use parameters that yielded image analysis results of the same quality across conditions, as discussed in more detail in Materials and methods (Fig. S3).
In our analysis of chromosome dynamics, we pooled trajectories of all aligned sisters (or all sisters if there was no metaphase plate) from each condition. Consequently, rare or outlier events were masked by the dynamic properties of the majority. Moreover, we have ignored potential systematic differences between sisters at different locations in the metaphase plate (e.g., at the periphery vs. in the middle) as well as differences between aligned and unaligned sisters. Future analyses of this image dataset may focus on the heterogeneity in sister dynamics to extract further spatial information about the process of chromosome alignment and spindle forces.
Aligned sister kinetochores oscillate and breathe with characteristic speeds
We characterized the dynamics of individual sister kinetochores over the time course of a video using the following parameters: (a) sister separation d (Fig. 2, b and c), (b) sister center position x along the normal to the metaphase plate if there was a metaphase plate (Fig. 2, b and c), and (c) the projections of the sister and center 3D displacements onto the vector connecting sister pairs ΔprL, ΔprR, and ΔprC (Fig. 2 d). The center normal position (x) indicated motion relative to the metaphase plate, whereas the displacement projection parameters were independent of the plate, thus allowing the characterization of kinetochore dynamics in conditions without a metaphase plate. Future versions of this assay may be combined with centrosome labeling, allowing us to monitor kinetochore motion relative to the spindle poles as well.
From the time courses of individual sister pair movements, we derived population-level oscillation and breathing speed measurements: (a) we assembled distributions of the frame to frame changes in sister center normal position Δ x and sister separation Δd. These distributions had a 0 mean value because of the symmetry of sister oscillation and breathing; however, their SDs yielded measures for the average sister center normal speed (i.e., center speed along the normal to the metaphase plate) and the average sister breathing speed (both in micrometers/minute). (b) Sister center speed was also derived from the 3D center displacement projection ΔprC (Fig. 2 d). Because of motion symmetry, the distribution of ΔprC also had 0 mean, whereas its SD yielded a measure for the average sister center projection speed (in micrometers/minute). For conditions without a metaphase plate, this was the only measure of sister center speed.
Assembling the data of 6,605 aligned sisters from 212 3D time-lapse videos of late prometaphase and metaphase WT cells, we measured an average sister breathing speed of 1.31 ± 0.18 µm/min and average sister center normal speed and projection speed of 1.46 ± 0.25 µm/min and 1.56 ± 0.26 µm/min, respectively (for all parameters, the uncertainty estimate reflects the SD of the distribution of single cell averages; Fig. 2 e). We also obtained an average separation between individual sisters of 0.98 ± 0.08 µm, which is in agreement with previous values (Gerlich et al., 2006b). Importantly, all speed values were significantly larger than the apparent speed values measured for fixed cells (Fig. 2 e). These apparent movements of fixed kinetochores reflect the uncertainty of our measurements, mostly associated with the uncertainties in estimating kinetochore positions as a result of image noise but possibly also with contributions from mechanical vibrations of the mounted sample and microscope stage. These results demonstrate that our live cell assay has sufficient sensitivity to resolve sister oscillation and breathing movements over a sampling interval as short as 7.5 s.
Aligned sister kinetochores oscillate and breathe with characteristic periods
For a temporal characterization of kinetochore dynamics, we initially measured oscillation half-periods τx and breathing half-periods τd (in seconds), defined as the durations between consecutive turning points in the time courses of sister oscillation and breathing (Fig. 2, f and g, insets; Dorn et al., 2005). We found that HeLa kinetochores exhibited a wide distribution of oscillation and breathing periods (Fig. 2, f and g, WT distributions). To achieve a metric of the mixture of periods that would allow the extraction of the characteristic behavior of sister oscillation and breathing within each condition and the comparison of dynamics between conditions, we used auto- and cross-correlation methods (see Materials and methods). For sister breathing, we calculated the autocorrelation of the time course of the frame to frame change in sister separation Δd (sister breathing autocorrelation), whereas for sister oscillations about the metaphase plate, we calculated the autocorrelation function of the time course of the frame to frame change in center normal position Δx (sister center autocorrelation). In addition, because some conditions did not form a metaphase plate, we characterized sister movement by calculating the cross-correlation between the time courses of the plate-independent sister displacement projections ΔprL and ΔprR (Fig. 2 d; sister displacement cross-correlation).
For a regular oscillator with a narrow distribution of periods (Fig. 2 h, model 1), auto- and cross-correlations display a series of strong sinusoidal-shaped side lobes (Fig. 2 i, model 1). The minimum of the first negative side lobe indicates the oscillation half-period (Fig. 2, h and i, arrows); the maximum of the first positive side lobe denotes the full period (Fig. 2 i, arrowheads). For oscillators with mixed periods over wider distributions, the side lobes are increasingly attenuated at longer time lags (Fig. 2, h and i, models 2 and 3), and the minima and maxima of the side lobes might get slightly shifted. The value of the sister displacement cross-correlation at lag ΔT = 0 reflects the degree of motion coupling between sisters (Fig. 2 i, carets). Its value ranges between −1 and 1, where 1 indicates perfect coupling, −1 indicates perfect anticoupling, and 0 implies that the two sisters move independently. In contrast to a semiregular oscillator, purely random motion such as diffusion or jitter as a result of noise has no characteristic time scale: its autocorrelation exhibits one negative value, −0.5 at the first lag and 0 otherwise, whereas its cross-correlation is 0 everywhere (Fig. 2 i).
Combining the data of all WT cells, the sister displacement cross-correlation and the sister center autocorrelation functions exhibited a negative lobe with a minimum at ∼35 s and a small positive lobe with a maximum at ∼80 s (Fig. 2 j; arrows and arrowheads, respectively). The value of the cross-correlation at lag ΔT = 0 was 0.66 ± 0.07. These results indicate that sisters oscillate about the metaphase plate with fairly tight coupling and an average half-period of 35–40 s. The sister breathing autocorrelation function also showed a negative side lobe and one small positive lobe with a maximum at 40 s, implying that the breathing half-period was ∼20 s on average (Fig. 2 k). The auto- and cross-correlation curves of WT cells were very different from those of fixed cells, which in turn looked similar to the correlation curves resulting from random motion (Fig. 2, i–k, compare brown lines). The result that significant correlation values in WT cells occurred at lags greater than the sampling interval confirmed that the temporal sampling of our videos was sufficient to capture the dynamics of kinetochore movement and that kinetochore oscillation and breathing contain elements of regularity.
Oscillation and breathing require bipolar attachment to dynamic MT ends
To test whether MT plus end dynamics are the major driving force for kinetochore oscillations and breathing observed in HeLa cells, we analyzed sister kinetochore dynamics after (a) 10 nM taxol treatment, which keeps kinetochores attached to MTs but inhibits MT plus end dynamics, (b) 100 ng/ml nocodazole treatment, which depolymerizes MTs and leaves kinetochores detached, and (c) siRNA-mediated depletion of Hec1 or Nuf2R, two components of the Ndc80 complex that are essential for stable end-on attachment of kinetochores to MTs (DeLuca et al., 2005).
As previously described (Jordan et al., 1993; Waters et al., 1998; DeLuca et al., 2005), loss of tension as a result of taxol treatment, loss of attachment as a result of nocodazole treatment, or depletion of Nuf2R of Hec1 reduced the average sister separation (Fig. 3 a). Taxol treatment also reduced both breathing speed and sister center speed (Fig. 3 b), implying that a substantial component of kinetochore oscillations and breathing in HeLa cells is governed by MT plus end dynamics. As a further confirmation that after taxol treatment the observed kinetochore motion was reduced to random jitter on stationary MT plus ends, the auto- and cross-correlation curves after taxol treatment were similar to those of fixed cells (Fig. 3, c and d; compare with Fig. 2, j and k).
Figure 3.
Nonrandom sister oscillation and breathing require bipolar attachment of kinetochores to dynamic MT plus ends. (a) Sister separation. (b) Sister center normal and projection speeds and sister breathing speed. (c–f) Sister displacement cross-correlation (c and e), sister center autocorrelation (c), and sister breathing autocorrelation (d and f) calculated for each condition by combining all aligned sister kinetochores belonging to that condition. Line thickness represents the 95% confidence interval of the correlations about their calculated values (see Materials and methods). S, synchronization. Error bars indicate mean ± SD, representing cell to cell variation. Green and red double asterisks highlight values significantly larger and smaller than WT, respectively, unless indicated otherwise (i.e., P < 0.001 by one-tailed t test).
Under nocodazole treatment or Hec1 or Nuf2R depletion, the auto- and cross-correlation curves were also indicative of random motion (Fig. 3, c–f). However, the sister center speed after Hec1 or Nuf2R depletion was significantly higher than after nocodazole treatment (Fig. 3 b). This implies that although the motion after nocodazole treatment is purely diffusive, kinetochore dynamics are at least partially motor driven along the sides of MTs after Hec1 or Nuf2R depletion, e.g., by CENP-E or dynein (Kapoor et al., 2006; Gassmann et al., 2008; Vorozhko et al., 2008). Collectively, these results indicate that the oscillation and breathing of sister kinetochores observed in HeLa cells requires end-on attachment to dynamic MTs and that oscillation and breathing are driven by MT plus end dynamics.
Kinetochore-associated depolymerases set the oscillation and breathing speeds
Although kinetochore oscillations may be related to the intrinsic dynamics of MT plus ends, MT motor proteins may play an important role in sliding the kinetochore on MT ends and/or in modulating the growth and shrinkage of MTs. Therefore, we analyzed the effects of CENP-E, MCAK, and Kif18A depletions on sister kinetochore oscillation and breathing.
Our assay revealed that although the depletion of CENP-E resulted in alignment failure of some chromosomes (Fig. S2) in agreement with previous studies (McEwen et al., 2001; Putkey et al., 2002), it did not have any significant effect on the oscillation or breathing of aligned sisters in speed (Fig. 4, a and b) or periodicity (Fig. 4, c and d). Thus, our data show that once kinetochores are aligned at the metaphase plate, their oscillation and breathing are independent of CENP-E. To confirm this conclusion, we treated CENP-E–depleted cells with 10 nM taxol. In contrast to cells with only CENP-E depletion or only taxol treatment, these cells did not form metaphase plates, indicating that chromosome alignment into a metaphase plate required at least the presence of CENP-E or dynamic MTs. However, the speeds and correlation curves for this condition were indistinguishable from those produced under taxol treatment of WT cells (Fig. 4, a–d). These results provide strong additional evidence that CENP-E does not contribute to kinetochore sliding on MT plus ends after end-on attachment and alignment on the metaphase plate.
Figure 4.
Depolymerases set oscillation and breathing speeds, whereas sister linkage modulates oscillation and breathing period. (a and h) Sister separation. (b and i) Sister center normal and projection speeds and sister breathing speed. (c–f, j, and k) Sister displacement cross-correlation and sister center autocorrelation (c, e, and k) and sister breathing autocorrelation (d, f, and j) are shown. Line thickness represents the 95% confidence interval of the correlations about their calculated values (see Materials and methods). Note that all experiments in e and f were performed at the University of Dundee (Dundee, Scotland, UK), not at the Marine Biological Laboratory (Woods Hole, MA). These different experimental settings led to small but highly reproducible changes in the oscillation and breathing periods of WT cells. Therefore, cell perturbations were always compared with the control data acquired at the same site. (g) Sister displacement cross-correlation at lag ΔT = 0. ns, not significant. Error bars indicate mean ± SD, representing cell to cell variation. Green and red double asterisks highlight values significantly larger and smaller than WT, respectively, unless indicated otherwise (i.e., P < 0.001 by one-tailed t test).
We next examined the role of two MT depolymerases, MCAK and Kif18A, on kinetochore dynamics (Desai et al., 1999; Varga et al., 2009). As previously shown (Kline-Smith et al., 2004; Stumpff et al., 2008), depletion of MCAK increased chromosome segregation errors during anaphase, and Kif18A depletion elevated the rate of chromosome alignment failure and led to a mitotic arrest (Fig. S2). Depletion of MCAK also reduced the oscillation speed of aligned kinetochores, whereas depletion of Kif18A increased it (Fig. 4 b), which are both consistent with previous observations (Wordeman et al., 2007; Stumpff et al., 2008). Depletion of either motor led to the disappearance of the positive lobe from the oscillation and breathing correlation plots and to a slight shift of the negative lobe in the oscillation correlation plots (Fig. 4, e and f), indicating that both depletions tend to randomize kinetochore oscillation and breathing (a broadening of the half-period distribution shifts the negative peak; Fig. 2, h and i). However, neither depletion significantly changed the average oscillation and breathing periods, which is partially in disagreement with previous results (Stumpff et al., 2008). We surmise that these differences are the result of a manual selection of sisters as opposed to a comprehensive tracking of a large set of sister kinetochore pairs in 4D. Collectively, our data suggest that the MT depolymerases MCAK and Kif18A primarily set the speed of kinetochore oscillations and breathing.
The mechanical linkage between sisters regulates oscillation and breathing periods
Directional switches in kinetochore oscillations require a switch in the state of a kinetochore from leading to following and vice versa. Recent data suggested that these states may partially depend on the varying level of polar ejection forces transmitted to the kinetochore as a function of the sister pair position along the spindle (Ke et al., 2009). However, force levels at the kinetochores may also be altered, as sisters breathe under the forces exerted by the depolymerases on the two sisters. Therefore, we hypothesized that the mechanical linkage between sisters may be a second key factor in establishing directional switches.
Sister linkage is mediated mainly by cohesin that cross-links chromatids (Nasmyth, 2002) as well as by condensin, which compacts centromeric chromatin (Swedlow and Hirano, 2003; Oliveira et al., 2005; Gerlich et al., 2006a; Ribeiro et al., 2009). To investigate how these components might affect sister oscillation, we first depleted the condensin I subunit CAP-D2. The reduced compaction of chromatin upon CAP-D2 depletion caused a large decrease in the sister cross-correlation at lag 0 (Fig. 4 g), implying a reduced coupling between sister kinetochores. It also increased the average sister separation and the sister breathing speed (Fig. 4, h and i), which is in agreement with previous results (Oliveira et al., 2005; Gerlich et al., 2006a; Ribeiro et al., 2009). Interestingly, CAP-D2 depletion increased not only the characteristic half-period of breathing (Fig. 4 j) but also the half-period of oscillation (Fig. 4 k) compared with WT. It also increased the oscillation speed (Fig. 4 i), although to a lesser extent than Kif18A depletion. These observations indicate that the mechanical linkage between sisters indeed plays a role in regulating the oscillation of aligned sister kinetochores, particularly their directional switching frequency.
We next sought to modulate the level of cohesion between sister kinetochores. We considered depleting cohesin, but the resulting premature sister disjunction would make our assay unusable. Therefore, we depleted instead the enzyme separase, which cleaves cohesin (Uhlmann et al., 2000; Hauf et al., 2001). Complementary to the depletion of CAP-D2, in the absence of separase, we observed a reduction in the oscillation period (Fig. 4 k) and a small reduction in oscillation speed (Fig. 4 i). These changes in kinetochore dynamics were observed in aligned chromosomes before anaphase onset, suggesting that separase modulates the extent of cohesion between sisters already during late prometaphase and metaphase. This implies that there is a dynamic balance of assembly and disassembly of cohesin before anaphase, as has been observed in yeast (Ocampo-Hafalla et al., 2007). In summary, our data suggest that centromere stiffness regulates kinetochore sister oscillations primarily through their directional switching frequency. Whether condensin and cohesin directly collaborate to regulate centromere stiffness and balance the dynamics of kinetochores will require further study.
The metaphase plate gets thinner as cells progress toward anaphase because of a reduction in oscillation speed
How do oscillations and breathing affect the overall architecture of the metaphase plate? Visually, our live cell data suggested that the metaphase plate becomes thinner as cells progress toward anaphase. By quantifying metaphase plate thickness as the SD of the distribution of aligned sister center positions along the normal to the metaphase plate (Fig. 2 b, x axis), we found that cells entering anaphase in the course of a video indeed had systematically thinner plates (Fig. 5 a). Cells that spent a long period in late prometaphase with one or a few faraway unaligned kinetochores near the spindle poles tended to have thinner plates as well (Fig. 5 a). This suggests that thinning is the result of a time-dependent ordering of the metaphase plate that occurs independently of whether cells have attached all their kinetochores or not.
Figure 5.
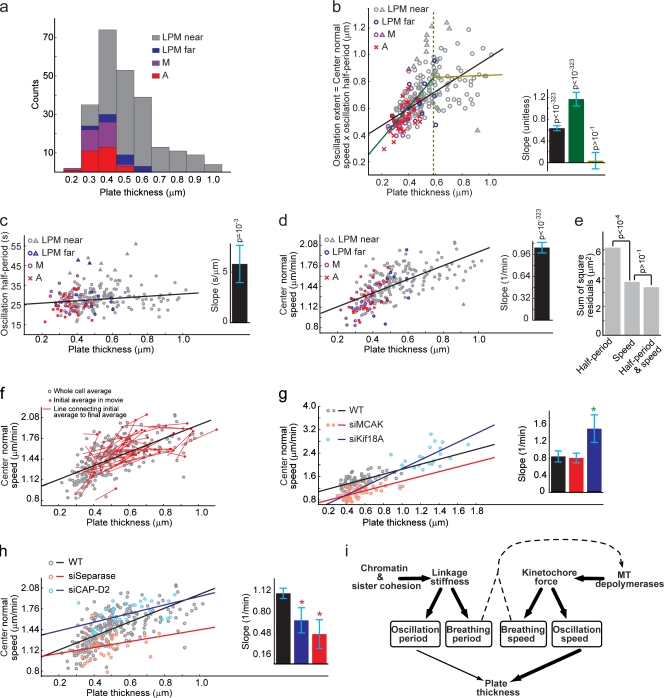
Metaphase plate thinning in preparation for anaphase is induced by a reduction in oscillation speed. (a) Histogram of average plate thicknesses in individual videos. (b) Scatter plot of oscillation extent versus plate thickness. Black line, one-line fit to the scatter plot; dark and light green lines, two-line fit to the scatter plot; dotted green line, transition point between the two scaling regimes. Triangles indicate data points deemed as outliers in the least-median square fit of the straight line. (inset) Line slopes matched by color; error bars indicate mean ± SD as estimated by the least-squares fit; p-values indicate comparison of slopes to 0 (see Materials and methods) to determine their significance. (c and d) Scatter plots of oscillation half-period (c) and center normal speed (d) versus plate thickness. (a–d) M, metaphase cells with no unaligned kinetochores; A, cells that enter anaphase during the course of a video. (e) Sum of square residuals from regressing plate thickness on oscillation half-period, center normal speed, or half-period and center normal speed combined. P-values indicate comparison of model fits using the F-test (Jaqaman and Danuser, 2006). (f) Scatter plot of center normal speed versus plate thickness, showing their variation within individual videos using the 25% of the videos that exhibited the largest decrease in plate thickness over the time course of the video. Gray circles are combined from late prometaphase cells with unaligned kinetochores near the metaphase plate (LPM near), late prometaphase cells with unaligned kinetochores far from the metaphase plate (LPM far) metaphase, and anaphase cells. (g and h) Scatter plots of center normal speed versus plate thickness after depolymerase depletion (g) and sister linkage perturbation (h). Outliers from the least-median square fit are not depicted. Crosses indicate cells that entered anaphase during a video, and circles indicate cells that did not. Green and red asterisks highlight values significantly larger and smaller than WT, respectively (P < 0.05). (i) Proposed model for the regulation of kinetochore breathing and oscillation and of metaphase plate thickness. Sister linkage at the centromere is set by a combination of chromatin stiffness and sister cohesion and determines both oscillation and breathing periods. Activity of MT depolymerases generates force at kinetochores, where net effect of antagonizing depolymerase activities at sister pairs sets baseline oscillation and breathing speeds. Plate thickness is primarily determined by oscillation speed with a minor contribution from oscillation period. Breathing speed and period affect distance between centromere and kinetochore and, thus, potentially the activity of kinetochore and centromere kinases and phosphatases on MT motors. Thick and thin arrows indicate strong and weak dependencies, respectively. The dashed arrow indicates the proposed feedback from breathing characteristics to depolymerase activity.
Plate thickness is expected to be determined primarily by kinetochore oscillations. This means that plate thickness and oscillation extent, defined as the center normal speed multiplied by the oscillation half-period, should be coupled with a proportionality coefficient equal to one. A scatter plot of oscillation extent versus plate thickness averaged over the duration of an entire video indicated that the two variables were indeed proportional to each other (Fig. 5 b). The slope of a straight line fitted to the data using least-median squares was found to be 0.63 ± 0.05 (goodness of fit evaluation; Fig. S4; Jaqaman and Danuser, 2006). This value is significantly different from 0, confirming the coupling between plate thickness and oscillation extent. However, it is also significantly different from 1. We hypothesized that the slope was shifted toward smaller values because of the large data scatter for thicker metaphase plates and possible lack of coupling between the two variables in that regime. Thus, we fitted the scatter plot with two lines with an unknown transition point between them. The fit yielded a transition point at 0.59 ± 0.03 µm, with line slopes of 1.17 ± 0.13 and 0.04 ± 0.15 for the regimes below and above the transition point, respectively (Fig. 5 b). The two-line fit was only marginally better than the one-line fit (an F-test comparing the two fits’ residuals yielded P ≈ 0.22; Jaqaman and Danuser, 2006); nevertheless, the line slope below the transition point was statistically indistinguishable from 1 (P = 0.1). This confirms that plate thickness is indeed tightly coupled to the extent of kinetochore oscillations, especially in the regimen of thinner, more organized plates.
We next asked whether speed, period, or both were altered to induce metaphase plate thinning. We approached this question in three ways. First, we generated scatter plots of half-period versus plate thickness and center normal speed versus plate thickness, fitted each with a straight line, and calculated how different the slope of each line was from zero. With this, we found weak coupling between plate thickness and oscillation half-period (Fig. 5 c) but a strong coupling between plate thickness and center normal speed (Fig. 5 d). In a second approach, we used multivariable regression, where we investigated the dependence of plate thickness on center normal speed alone, oscillation half-period alone, or center normal speed and oscillation half-period together. We compared the residuals from the different regressions using an F-test (Jaqaman and Danuser, 2006). The fit to speeds had much smaller residuals than the fit to periods, and the fit to both variables did not yield any statistically significant improvement beyond fitting to center normal speeds alone (Fig. 5 e). Third, in addition to the analyses based on whole-video averages, we monitored the variation of plate thickness and center normal speed within the 5-min window of each video. We calculated averages of plate thickness and center normal speed in the first 10 frames and the last 10 frames of each video and found that center normal speeds decreased in 78% of the cases where the plate got thinner within a video (Fig. 5 f). These three sets of independent analyses provide strong evidence that plate thinning during metaphase is primarily coupled to a reduction in kinetochore oscillation speed, with only a small change in oscillation period.
Metaphase plate thinning through oscillation speed reduction depends on the mechanical linkage between sisters
Next, we investigated which factors might be responsible for the gradual decrease in oscillation speed throughout mitotic progression toward anaphase. We fitted the scatter plot of oscillation speed versus plate thickness after depletion of MCAK, Kif18A, CAP-D2, or separase with a straight line and compared the resulting slopes with the slope in WT. To our surprise, depletion of MCAK only shifted the vertical position of the regression line without changing its slope (Fig. 5 g), indicating that MCAK sets the baseline speed of oscillation but is not essential for the mechanisms responsible for the progressive speed reduction necessary for plate thinning. Kif18A depletion changed the line’s slope (Fig. 5 g); however, this result needs to be interpreted with caution because the two conditions occupy very different, almost nonoverlapping plate thickness regimes.
The depletion of CAP-D2 or separase markedly reduced the slope of the regression line while maintaining a WT range of plate thicknesses (Fig. 5 h). Thus, the coupling between oscillation speed and plate thickness was reduced after these perturbations. Interestingly, despite the lack of oscillation speed reduction after perturbing sister linkage, plate thinning still occurred (Fig. 5 h). This may be the result of a compensatory mechanism that increases under these conditions the dependence of plate thickness on oscillation period. Indeed, we found that the slope of the line correlating plate thickness with oscillation period increased after CAP-D2 or separase depletion (unpublished data). Collectively, our observations suggest that in WT HeLa cells, the gradual reduction of oscillation speed of aligned kinetochores results from a gradual change in the mechanical linkage between sisters.
Discussion
Our quantitative live cell analyses show that sister oscillation and breathing are complex processes involving the interplay between several finely tuned mechanical and chemical factors (Fig. 5 i). Our data indicate that breathing and oscillation periods mainly depend on the stiffness of the sister linkage (Fig. 4, j and k), whereas the baseline breathing and oscillation speeds are set by MT depolymerases at the two sisters (and possibly other regulators of MT stability and turnover; Fig. 4 b). Once kinetochores are congressed on the metaphase plate, final alignment of kinetochores into a progressively thinning metaphase plate strongly depends on a reduction in oscillation speeds, with a minor contribution from a small reduction in oscillation period (Fig. 5, a–f).
From the standpoint of mechanical control, metaphase plate thinning via decreasing oscillation speeds is more robust than via decreasing oscillation periods, which would require higher directional instability in chromosome movement. Thus, our study provides evidence for an optimal metaphase control circuit that prepares for the onset of anaphase by increasing both global order among chromosomes via alignment into a gradually thinning plate and local order via reduction of net tubulin turnover at the interface between MTs and individual kinetochores.
The MT depolymerases MCAK and Kif18A act antagonistically on sister kinetochores
Our data indicate that the depletion of MCAK reduces oscillation speed and the depletion of Kif18A elevates oscillation speed, whereas neither depletion has a significant effect on oscillation period (Fig. 4, b and e). The lack of effect of either depolymerase depletion on oscillation period (Fig. 4 e) is incompatible with a previous model, which proposed that Kif18A regulates the directional switching of sister kinetochores by accumulating in an MT length–dependent manner on the following kinetochore and causing a directional switch upon reaching a certain threshold concentration (Stumpff et al., 2008). Our observations lead us to propose an alternative model: the opposing effects of MCAK and Kif18A on oscillation speed suggest that these two kinetochore-associated MT depolymerases are mutual antagonists in regulating the speed of sister kinetochore oscillations about the metaphase plate. The speed decrease and increase induced by abrogation of MCAK and Kif18A, respectively, suggest that MCAK preferentially promotes depolymerization of MTs at the leading sister, whereas Kif18A preferentially promotes depolymerization at the following sister, generating resistance to the sister pair movement. Both proteins could affect oscillation speed by controlling the number of MTs in a k-fiber that undergo a catastrophe event and/or the rate of MT depolymerization.
Our model predicts that loss of MCAK, which would reduce the force driving sister kinetochore motion, should decrease the coupling between sisters. However, loss of Kif18A, which would lead to less Kif18A-mediated resistance by the following sister, should increase the coupling between sisters. In agreement with these predictions, the sister displacement cross-correlation at lag ΔT = 0, which reflects the degree of coupling between the motion of sister kinetochores, decreased with MCAK depletion and increased with Kif18A depletion (Fig. 4 g). Although we cannot formally exclude that some of the observed changes in chromosome dynamics originate from off-kinetochore effects of motor depletion, these shifts in oscillation speed and sister coupling together strongly support the model that MCAK promotes depolymerization at the leading sister to drive motion, whereas Kif18A promotes depolymerization at the following sister, functioning as a brake. Consistent with this conclusion, previous localization studies in fixed spindles have shown unequal concentrations of Kif18A (Stumpff et al., 2008) and MCAK phosphorylation (Andrews et al., 2004; Lan et al., 2004) between sister kinetochore pairs. Thus, the state of a kinetochore as a leading or following sister may be controlled by its variable affinity for motor binding and/or phosphorylation.
Gradual stiffening of sister linkage through metaphase mediates the reduction in oscillation speed required for plate thinning
Our regression analyses show that in HeLa cells, the metaphase plate gets thinner throughout prometaphase and metaphase mainly because of a reduction in oscillation speed (Fig. 5, b–f). Depletion of CAP-D2 or separase eliminates this speed reduction (Fig. 5 h), implying that a progressive change in sister linkage throughout late prometaphase and metaphase is responsible for oscillation speed reduction. In support of this model, our data also show that the plate thickness versus oscillation speed relationship after CAP-D2 or separase depletion overlaps with WT at wider and thinner plates, respectively (Fig. 5 h, the CAP-D2 and separase lines intersect the WT line at plate thicknesses of 0.92 µm and 0.1 µm, respectively). Because sister linkage stiffness is expected to be reduced by CAP-D2 depletion and elevated by separase depletion, the distinct regions of overlap between CAP-D2 and separase depletion data with WT data suggest that the mechanical linkage between sisters is weaker in prometaphase and gets stiffer as cells progress through metaphase. These stiffness changes are most probably caused by a progressive change in the activity of proteins controlling sister linkage, as a result of which, the speed of aligned kinetochore oscillations is gradually reduced, leading to a gradual thinning of the metaphase plate.
The mechanism of oscillation speed reduction induced by changes in the mechanical linkage between sisters remains elusive at this point (Fig. 5 i, dashed line). We surmise that variation of sister linkage leads to changes in breathing dynamics, which in turn lead to changes in the activity of MT regulators. One possibility for such mechanochemical transformation of breathing dynamics into the control of oscillation speeds is the reported dependence of the phosphorylation of kinetochore components by aurora B on sister separation (Liu et al., 2009). By analogy to increased kinase activity at the kinetochore with shorter sister separation, smaller breathing periods may also increase the influence of chromatin-associated kinases (and phosphatases) on kinetochore protein phosphorylation. Moreover, phosphorylation-dependent changes in MT dynamics are reinforced by their feedback effect on sister kinetochore breathing. From the observation that the coupling between speed and thickness is dramatically reduced under perturbation of the mechanical linkage between sisters, we conclude that under WT conditions, the oscillation speed–dependent pathway of plate thinning gradually gains importance throughout metaphase over the oscillation period–dependent pathway (Fig. 5 i). This hints at a nonlinear amplification of speed decrease in response to stiffness increase. Further evidence for this amplification through metaphase is the biphasic correlation between plate thickness and oscillation extent observed for WT cells (Fig. 5 b).
Materials and methods
Generation of stable EGFP–CENP-A HeLa cell line
To obtain a stable EGFP–CENP-A cell line, HeLa cells were transfected with pEGFP–CENP-A–IRESpuro vector (provided by D. Gerlich, ETH Zurich, Zurich, Switzerland) using Fugene 6 (Roche). Cells were grown in DME supplemented with 10% FCS, 100 U/ml penicillin, 100 µg/ml streptomycin, and 500 ng/ml puromycin (Invitrogen) at 37°C with 5% CO2 in a humidified incubator. To obtain a homogeneous population of cells with respect to the EGFP–CENP-A kinetochore signal, we performed two rounds of manual single-colony selection and screened the clonal populations by visual inspection (Fig. S1).
Fluorescence time-lapse imaging of EGFP–CENP-A HeLa cells
Cells were seeded in dishes (LabTekII; VWR) and imaged in L-15 medium (Invitrogen) supplemented with 10% FCS (Invitrogen). Time points comprised of 20 z sections 0.5 µm apart were acquired every 7.5 s for 5 min (i.e., 41 time points in a video) with a 100× 1.35 NA objective lens (Olympus) and a camera (CoolSnap HQ or HQ2; Photometrics) on an imaging system (DeltaVision Core, Personal, or Spectris; Applied Precision) fitted with a 37°C environmental chamber. Camera pixels were binned (2 × 2), yielding a pixel size of 129 nm in the lateral direction. The 10-µm z stack covered about two thirds of the metaphase plate (Video 1). The acquisition routines in softWoRx (Applied Precision) were used to control all aspects of image data acquisition.
Imaging conditions were designed to balance three essential imaging requirements (Dorn et al., 2008): (1) exposing cells to as little light as possible to minimize phototoxicity, (2) obtaining images of sufficiently high signal to noise ratio to allow reliable detection of the EGFP–CENP-A signal, and (3) sampling sufficiently fast to measure unaliased kinetochore motion.
(1) Cell viability.
Phototoxic effects on cells, as a result of frequent exposure to too much light, can manifest themselves as delayed anaphase timing, i.e., a delay in the time between nuclear envelope breakdown and anaphase onset, as the result of an activation of the spindle checkpoint (Mikhailov et al., 2002). To assess the effect of various 3D imaging protocols on cell viability, we first determined the normal anaphase timing of WT HeLa cells measured by imaging fields of cells exposed to minimal amounts of light. Specifically, cells were imaged every 5 min over 6 h using a 60× 1.4 NA lens (Olympus) with 25-ms exposure, 32% neutral density filter, and seven z sections 1 µm apart. This yielded a reference anaphase timing of ∼34 min (Fig. 1, top). To assess the effect of fast 3D imaging on cell viability, we varied exposure time, neutral density filter, recovery times between exposures and a number of imaged time points, and compared the resulting anaphase timing distribution with the reference anaphase timing. We found that an imaging protocol of 15-ms exposure time, 32% neutral density filter, and a sampling rate of one z stack (20 z sections 0.5 µm apart) every 7.5 s for 41 time points (resulting in a video length of 5 min), i.e., a total of 820 exposures per video, ensured that ≥80% of the cells exhibited the normal anaphase timing of ∼34 min.
(2) Detection quality.
Automated kinetochore detection (see following paragraph) fails to properly distinguish spot signals from noise if the signal amplitude becomes too similar to the noise amplitude. In such a situation, both the number of false positives, i.e., noise peaks that are mistaken for true kinetochore signal, and the number of false negatives, i.e., kinetochore signals that are missed, become large. Thus, it was essential to determine imaging protocols that minimized both false positives and false negatives while maintaining cell viability. The rate of false positives and negatives was estimated for each imaging protocol by taking multichannel images of EGFP–CENP-A cells, in which in each channel a different imaging protocol was used, and comparing the detection results between the different channels. In one channel, high light exposure that led to flawless spot detection was used to provide the detection ground truth (although such high light exposure did not satisfy the cell viability conditions). In the other two channels, light exposures and neutral density filter combinations that were consistent with cell viability were used. The kinetochore signal was detected in all three channels and compared (Fig. 1, bottom). With this, we found that imaging with 15-ms exposure and 32% neutral density filter, yielding videos with an average signal-to-noise ratio of 3–4, resulted in <10% false positives and negatives (Fig. 1, bottom). This detection quality was acceptable for the subsequent analysis steps (see Automated image analysis).
(3) Unaliased kinetochore movement.
To avoid aliasing and to correctly capture an object’s motion characteristics, the motion must be sampled at least two times faster and preferably three times faster than its characteristic frequency (Dorn et al., 2008). Thus, the characteristic period of the monitored motion must be at least two to three times longer than the interval between consecutive time points used for recording. None of the estimated average sister kinetochore breathing and oscillation periods were <30 s for conditions in which kinetochores were attached to the plus end of dynamic MTs (Figs. 2–4). Thus, acquiring one z stack every 7.5 s yielded unaliased breathing and oscillation time courses that could be used to probe the functions of kinetochore proteins on kinetochore movement. To satisfy cell viability with such fast sampling, we had to limit the video duration to 5 min (i.e., 41 time points). To capture the full range of possible kinetochore motions, we acquired a large number of videos at various points in late prometaphase and metaphase for each condition (Table S2).
siRNA and drug treatment of cells
Cells were transfected using Oligofectamine (Invitrogen) with 60 nM siRNA duplexes for 48 or 72 h using previously published sequences (Table S1) in modified essential media supplemented with 10% FCS, 100 U/ml penicillin, 100 µg/ml streptomycin, and 500 ng/ml puromycin (Invitrogen). Hec1- and Nuf2R-depleted cells were synchronized using a thymidine/aphidicolin block and release (Meraldi et al., 2004). Depletions were validated by immunofluorescence or Western blotting (Fig. S2 and Table S1). An exception was separase depletion, in which a suitable antibody for validation of separase protein levels was not available to us. Thus, we used phenotypic validation; i.e., for the analysis, we only used cells from experiments in which cells did not enter anaphase within 90 min of imaging after treatment with separase siRNA (Waizenegger et al., 2002). Small molecule inhibitors used were 100 ng/ml nocodazole (3 h) and 8 nM taxol (16 h; Jordan et al., 1993).
Automated image analysis
All image and data analysis software was written in MATLAB (The MathWorks, Inc.) with some core functions written in C and was run from the MATLAB command line. Image analysis results were overlaid on the microscopy data and visualized using Imaris and ImarisXT (Bitplane). ImarisXT allowed us to seamlessly integrate the visualization capabilities of Imaris with the MATLAB-based image analysis framework. This visualization made it possible to inspect every video included in the dataset and to qualitatively verify that all image analysis steps were successfully executed. Sister kinetochore dynamics were obtained from the raw 3D time-lapse sequences in the following four basic steps (Fig. 2 a).
Step 1: detection (Video 2)
Detection of local maxima.
We identified local maxima in each frame as voxels whose intensity was maximal in a 3 × 3 × 3–voxel volume after filtering out high frequency noise and subtracting local background from the image. To achieve this, we first approximated the experimental point spread function (PSF) of the imaging setup by fitting a 3D Gaussian to isolated kinetochores. The PSF Gaussian kernel’s SD was estimated to be 118 nm in the lateral direction (x and y) and 402 nm in the axial direction (z). With this, noise removal was achieved by convolving the raw image with the 3D PSF Gaussian kernel. Background was estimated by convolving the image with a 3D Gaussian 15× wider than the 3D PSF Gaussian kernel used for noise removal. The filtered image, in which kinetochore detection was performed, was calculated as the difference between the noise-removed image and the background image. The detection of local maxima yielded its position and amplitude above local background for every maximum. However, even after noise filtering, the detected local maxima consisted of both true kinetochore signals as well as noise.
Identification of significant local maxima representing true kinetochore signal.
The distinction between true kinetochore signal and noise local maxima was achieved by histogram thresholding of the signal amplitude. The histogram of amplitude values from all frames of a video generally had a tall, narrow peak at small amplitude values, representing noise-associated local maxima, and a much smaller and wider peak at high amplitude values, representing true kinetochore signals (Fig. S5). The threshold distinguishing true kinetochore signal from noise was automatically determined using a histogram-cutting algorithm especially suited for thresholding distributions in which the first mode is much larger than the second mode. It was based on the algorithm developed by Rosin (2001), who defined the threshold as the bin i with counts Hi such that the point (i, Hi) is furthest away from the straight line connecting the largest bin of the histogram (i.e., the first large peak) with the first empty bin after the largest amplitude (Fig. S5, a and b). However, the Rosin algorithm generally underestimated the threshold and was not robust. Thus, to improve the threshold estimate, our algorithm looked for one minimum to the left of the Rosin threshold (as long as that minimum was to the right of the large peak) and for two minima to the right of the Rosin threshold (Fig. S5, c). The deepest of the three minima was chosen as the final threshold (Fig. S5, d). Importantly, this thresholding produced a relatively low number of false positives and negatives (∼10%; Fig. 1). False negatives were compensated for by the gap-closing feature of our tracker (Jaqaman et al., 2008), whereas false positives were also eliminated during tracking because they yielded very short tracks (one to three frames long).
Estimation of kinetochore subpixel positions.
We estimated the subpixel positions of the local maxima surviving step by calculating the intensity center of mass from a 5 × 5 × 5–voxel volume around each local maximum. This volume contained 99% of the PSF intensity centered on the local maximum voxel. Using the fixed cell data, the uncertainty in kinetochore positions was estimated to be 0.4 voxels.
Step 2: plane fit (Video 3)
We attempted to fit a plane representing the metaphase plate for five purposes: (1) to determine whether the video displays an arrangement in which the majority of kinetochores is aligned on a metaphase plate, (2) to identify unaligned kinetochores as those far away from the plate, (3) to classify the mitotic stage of the imaged cell in every frame of a video using points 1 and 2, (4) to determine the normal to the metaphase plate in each frame to study kinetochore oscillations about the metaphase plate, and (5) to provide a frame of reference for kinetochore positions that is independent of global spindle translation and rotation.
When kinetochores are aligned on the metaphase plate, their positions are narrowly distributed along the normal to the metaphase plate and widely distributed in the plane of the metaphase plate. Thus, the plane fit can be based on the anisotropy in the scatter of kinetochore positions. Once a plane is fitted, unaligned kinetochores can be sensitively identified by their outlier distance from the metaphase plate. However, when the majority of kinetochores are aligned but a few are unaligned, the unaligned kinetochores will weaken the anisotropy in the scatter of kinetochore positions and will consequently destabilize the plane fit. Therefore, unaligned kinetochores must be identified and excluded from the anisotropy calculation before the plane fit. To resolve this dilemma, we performed the plane fit and unaligned kinetochore identification in the following steps.
Initial identification of inlier and outlier kinetochores based on nearest neighbor distances.
In each frame, we constructed a distribution of the average distances between kinetochores and their 10 nearest neighbors, termed distnn10. In the case in which the majority of kinetochores were aligned but a few (<5 kinetochore pairs) were unaligned, the distribution of distnn10 would reflect the characteristics of the majority aligned kinetochores. With this, kinetochores following the majority distribution were classified as inliers and were used for the plane fit in that frame, whereas kinetochores not following the majority distribution were classified as outliers and were not used in the plane fit. Note that in the case in which all kinetochores in a frame were aligned or no kinetochores had aligned into the metaphase plate yet, the distribution of distnn10 would be homogeneous without any outliers, and no kinetochores would be excluded from the plane fit.
Frame by frame plane fit through inlier kinetochores.
The plane fit was based on eigenvalue decomposition of the variance–covariance matrix of the positions of inlier kinetochores in every frame. The variance–covariance matrix of inlier kinetochore positions represented their scatter in space, and its eigenvalue decomposition can be thought of as fitting a 3D ellipsoid around the kinetochore positions. Eigenvalue decomposition yielded three eigenvectors, representing the orientation of the three ellipsoid axes, and three eigenvalues, reflecting the length of the three axes. For kinetochores aligned on the metaphase plate, the ellipsoid is flat (resembling a disk), with one short axis perpendicular to the metaphase plate and two long axes in the plane of the plate. In this case, eigenvalue decomposition would yield one eigenvalue that is much smaller than the other two, with the corresponding eigenvector representing the normal to the metaphase plate. However, when the majority of kinetochores are unaligned, their scatter is isotropic, and all eigenvalues are of similar magnitude (because the ellipsoid resembles a sphere).
Consequently, to determine whether there is a metaphase plate in a frame, we calculated the ratio of the minimum eigenvalue to the average of the other two eigenvalues. If the ratio was smaller than one third, we concluded that there was a metaphase plate in that frame. The eigenvectors would then define the new metaphase plate–based frame of reference for kinetochore positions in that frame, with the new x axis being the normal to the metaphase plate, and the new y and z axes defining the plane of the metaphase plate. The origin of the metaphase plate–based frame of reference was taken as the center of mass of inlier kinetochores in each frame, thus accounting for global translation.
In practice, identifying the eigenvalue corresponding to the normal to the metaphase plate was complicated by the fact that only two thirds of the plate could be imaged. This compromised the equality of the two in-plane eigenvalues and sometimes reduced their difference from the eigenvalue corresponding to the normal to the metaphase plate. Determining the eigenvector representing the normal to the metaphase plate on a frame by frame basis introduced errors and rotational jumps of the metaphase plate–based kinetochore frame of reference. Therefore, to increase robustness in identifying the metaphase plate, the selection of the eigenvector representing the normal to the plane was made not on a per frame basis but by integrating the eigenvector tripods over the entire video. In brief, (a) we established the correspondence of eigenvectors between frames (only those with sufficient anisotropy) under the assumption that plate rotation is minimal between frames, (b) we calculated the average eigenvalue of each of the three eigenvector sequences, and (c) we took the eigenvector sequence with the smallest average eigenvalue as the sequence of normals to the metaphase plate.
Aligned and unaligned kinetochore classification based on distance from the metaphase plate.
In every frame that had a metaphase plate, we collected the distances between kinetochores and the plate (i.e., their new x coordinates along the normal to the metaphase plate). The distribution of aligned kinetochores could be approximated by a Gaussian with a SD (σ) estimated from the data. Kinetochores whose distance from the metaphase plate was >2.5 σ away from the metaphase were considered outliers to the distribution and were classified as unaligned (Fig. 2 a). All other kinetochores were considered aligned.
Mitotic phase classification.
Based on the aforementioned analysis, we classified video frames as follows: prophase or early prometaphase, no plate present (we did not distinguish further between these two phases); late prometaphase, a plate and unaligned kinetochores present; metaphase, a plate and no unaligned kinetochores present; and anaphase, the frame at which the SD of the distribution of kinetochore distances from the metaphase plate starts to systematically increase. All following frames were considered to be anaphase.
A video with late prometaphase frames, even if followed by metaphase frames, was classified as a late prometaphase video. A video with only metaphase frames was classified as a metaphase video. A video in which the cell entered anaphase was classified as an anaphase video. As a further subclassification of late prometaphase videos, those with all unaligned kinetochores <10 σ away from the metaphase plate were classified as late prometaphase near, and those with at least one kinetochore >10 σ away from the metaphase plate were classified as late prometaphase far.
Step 3: tracking (Video 4)
We constructed kinetochore trajectories using the single particle tracking (SPT) algorithm described previously (Jaqaman et al., 2008). This SPT algorithm robustly followed dense kinetochore configurations and constructed complete kinetochore trajectories by recovering track interruptions caused by temporary kinetochore disappearance resulting from detection failure or kinetochore movement out of the imaging volume. Algorithm performance was improved by tracking kinetochores after subtracting global spindle translation and rotation from their coordinates; i.e., we used kinetochore coordinates in the metaphase plate–based frame of reference. When not available, we only accounted for global translation by subtracting the kinetochore center of mass.
The cost functions described previously (Jaqaman et al., 2008) for weighing competing assignments of kinetochore linking and track segment gap closing were reparameterized to optimally track HeLa kinetochore motion in late prometaphase and metaphase cells under our imaging conditions. Tracking was based on a Brownian motion model; i.e., we did not use the linear motion propagation option of the tracker (Jaqaman et al., 2008), and it did not look for merging and splitting events because kinetochore images tracked in 3D are not expected to merge or split. Two critical parameters had to be adjusted (Table S3): (1) the gap-closing time window, i.e., the maximum time between the disappearance of a kinetochore as a result of detection failure and its reappearance, and (2) the search radius upper limit. The search radius defines the distance beyond which a link between two kinetochores in two consecutive frames is deemed impossible. In our SPT algorithm, it is calculated per kinetochore on the fly given the measured kinetochore motion (Jaqaman et al., 2008). However, to avoid drifts in the self-adaptive estimation, the user must define a search radius upper limit. Because aligned and unaligned kinetochores behave differently, they were given different search radius upper limits. However, note that all kinetochores, aligned and unaligned, were tracked simultaneously, allowing an unaligned kinetochore to become aligned during a video.
Step 4: sister pairing (Video 5)
Sister pairing was based on kinetochore trajectories, making use of both spatial and temporal information to add robustness to the pairing of kinetochores. Kinetochore trajectories were paired using the following three criteria.
Average distance.
On average, sister kinetochores are closer to each other than nonsister kinetochores. Thus, for every pair i and j of kinetochore trajectories, we calculated the average distance dij between them throughout the video. Trajectory pairings with average distance beyond an upper bound (Table S3) were deemed impossible.
Distance variation.
Sister kinetochores generally follow each other more than nonsister kinetochores. This implies that the variance of the distance between sister kinetochores over a video is generally smaller than the variance of the distance between nonsisters. Thus, for every pair i and j of kinetochore trajectories, we calculated the variance vij of the distance between them throughout the video.
Angle with the normal to the metaphase plate.
If there is a metaphase plate, aligned sister kinetochores tend to have the vector connecting sister pairs parallel to the normal to the metaphase plate. Thus, for videos with a metaphase plate, for every pair i and j of kinetochore trajectories, we obtained the vector connecting them over the course of the video, calculated its angle with the normal to the metaphase plate in each frame, and calculated the average angle, αij. Trajectory pairings with average angle beyond an upper bound (Table S3) were deemed impossible.
To account for all three criteria, the cost for pairing any two trajectories i and j was defined as dij × vij if there was no metaphase plate and as dij × vij × αij if there was a metaphase plate. Similar to the tracking algorithm (Jaqaman et al., 2008), all trajectories within the acceptable bounds (Table S3) were allowed to compete with each other, each pair was weighed by its cost, and the combination of trajectory pairings with the minimum global cost was taken as the assignment of sister kinetochore trajectories.
Validation and adjustment of control parameters
Maximum consistency in the image analysis of different conditions was achieved by, on the one hand, using the same parameter settings for as many conditions as possible and, on the other hand, adjusting critical parameters to obtain the same image analysis quality. Image analysis quality was assessed by three criteria: (1) tracking quality was assessed by ensuring that the histogram of kinetochore frame to frame displacements in each condition was not prematurely cut off because of a too-small search radius upper limit and that the fraction of kinetochore trajectories spanning the full video was similar between all conditions (Fig. S3 a); (2) sister identification quality was assessed by ensuring that the histogram of average sister separations in each condition was also not prematurely cut off because of a too-small maximum average distance imposed during trajectory pairing and that it did not contain any tails indicating a too-large maximum average distance (Fig. S3 b); and (3) the overall quality of video analysis was assessed by ensuring that the number of sister pairs identified in each video was similar, between 20 and 40 pairs (Table S2). This measure was comprehensive because the final number of sister pairs in a video is sensitive to all analysis steps.
Statistical methods
Autocorrelation curves.
The autocorrelation function extracts the underlying periodicity and memory in behavior from the time course of a variable, such as the rate of change of sister separation, Δd, or the rate of change of center normal position, Δx. This is achieved by systematically comparing the behavior of the variable at different time points. Mathematically, the autocorrelation function of a time series, x(t), t = 1, 2, …, N, with mean μ and SD σ, is defined as
| (1) |
where indicates the averaging operator. In practice, the autocorrelation function can be calculated by (a) defining a normalized series,
with mean 0 and SD 1, (b) generating for every ΔT the distribution of products
and (c) taking the distribution average, which is equal to γ(ΔT) defined in Eq. 1.
For this study, we imaged multiple cells for each condition, and each cell contained multiple sisters. Thus, we had multiple time series x1(t), x2(t), …, xn(t) representing any process of interest. For example, we had multiple time courses of the rate of change of sister separation Δd representing WT sister breathing or multiple time courses of the rate of change of center normal position Δx representing WT sister center oscillation. This means that we had multiple time series that were different yet equivalent realizations of the same process. These series could be combined to increase both the accuracy and robustness of the autocorrelation calculation. Combining series required a generalization of Eq. 1, which was achieved in the following manner: (a) from each series xi(t), with mean μi and SD σi, we generated a normalized series:
(b) For each time lag ΔT (ΔT = 0, 1, …, ΔTmax), we generated the distribution of products for each series
and we combined the distributions from all n series together to obtain the overall distribution of products:
(c) The average of the overall distribution yielded the autocorrelation function at lag ΔT, γ(ΔT). (d) Because the autocorrelation function was the average of a large number of variables (the products), it follows from the central limit theorem (Papoulis, 1991) that it has a normal distribution with mean = γ(ΔT) and a SD of:
The normality of the autocorrelation function values, predicted by the central limit theorem, holds true even when calculating the autocorrelation for one individual series as short as 40 time points. (e) With this, the 95% confidence interval of the autocorrelation curves (Fig. 3, c and d) was defined as γ(ΔT) ± 2σγ(ΔT), ΔT = 0, 1, …, ΔTmax.
Cross-correlation curves.
The cross-correlation function is similar to the autocorrelation function except that it compares the time courses of two variables to extract the dependency between them. Mathematically, for two time series x(t) and y(t), t = 1, 2, …, N, the time courses of the sister displacement projections, e.g., ΔprL and ΔprR (Fig. 2 d), the cross-correlation at lag ΔT is defined as
| (2) |
where (μx, σx) and (μy, σy) are the mean and SD of x(t) and y(t), respectively. Note that for cross-correlation calculations, the lags ΔT take both positive and negative values; with positive lags, we correlate x(t) with later time points of y(t), and with negative lags, we correlate x(t) with earlier time points of y(t).
As with the autocorrelation function, the cross-correlation can be calculated by (a) defining two normalized series,
and (b) generating at every lag ΔT the distribution of cross products:
The average of this distribution is the cross-correlation at lag ΔT defined in Eq. 2.
To calculate the cross-correlation from multiple equivalent pairs of series (x1(t), y1(t)), (x2(t), y2(t)), …, (xn(t), yn(t)), we followed the aforementioned procedure for the autocorrelation function and generated the overall distribution of cross products:
 |
The cross-correlation κ(ΔT) was calculated as the average of the distribution D(ΔT), and the SD of the cross-correlation, σκ(ΔT), was estimated as the SD of the distribution The cross-correlation SD, like the autocorrelation SD, is again calculated using the central limit theorem (Papoulis, 1991), which holds true even for calculating the cross-correlation between two series each for 40 time points. With this, the 95% confidence interval of the cross-correlation curves (Fig. 3 c) was defined as κ(ΔT) ± 2σκ(ΔT), ΔT = −ΔTmax, −ΔTmax + 1, …, 0, 1, …, ΔTmax.
Hypothesis testing for line fit comparisons.
Line slopes were compared with 0 to determine their significance and with each other to compare kinetochore behavior under different conditions using statistical hypothesis testing. To compare two line slopes m1 ± δm1 and m2 ± δm2 (mean ± SD), we tested the null hypothesis (H0), Δm = m1 − m2 = 0, against the alternative hypothesis (HA), Δm ≠ 0. Under the null hypothesis, the test statistic Δm follows a normal distribution with mean 0 and SD H0 was rejected, and two line slopes representing two conditions were considered significantly different from each other if the test statistic’s P < 0.01.
To test the significance of a line slope m1 ± δm1, it was compared with 0 by performing the same test with the special case of m2 = δm2 = 0. Similarly, to test whether a line slope m1 ± δm1 was significantly different from 1, the same test was performed using m2 = 1 and δm2 = 0.
Online supplemental material
Fig. S1 shows the establishment of a stable EGFP–CENP-A cell line. Fig. S2 shows confirmation of kinetochore protein depletions by siRNA. Fig. S3 shows that critical image analysis parameters were varied to obtain results of similar quality. Fig. S4 shows that the relationship of oscillation extent, oscillation half-period, and center normal speed to plate thickness can be modeled as linear. Fig. S5 shows automatic thresholding of amplitude histogram to distinguish kinetochores signals from noise. Table S1 shows siRNA oligo sequences and primary antibody sources and dilutions. Table S2 shows the number of imaged cells and sister pairs used for analysis. Table S3 shows the adjustable tracking parameters and sister pairing parameters, their default values, and exceptions. Videos 1–5 show the raw 3D time-lapse sequence of an EGFP–CENP-A HeLa cell in late prometaphase (Video 1) and the four steps of image analysis: detection (Video 2), metaphase plate fit (Video 3), tracking (Video 4), and sister pairing (Video 5). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200909005/DC1.
Acknowledgments
This work was carried out in large part over the summers of 2006–2008 at the Marine Biological Laboratory (MBL; Woods Hole, MA). We are grateful to Larry Burdge, Martha Peterson, and Louie Kerr for their assistance in running our laboratories at MBL, Chris Allan, Zion Maffeo, Joshua Kunken, and Robert Loyott for their help with configuring the computing networks, the whole MBL Summer Research Community for their support and thoughtful discussions, and in particular, Lenny Dawidowicz for his enthusiasm and support for our work. We also thank Tomo Tanaka for critical reading of the manuscript and Sam Swift for help with configuring and maintaining microscopes, and we are particularly thankful to Applied Precision for donating the microscopes used for this study.
We thank Nikon (J.R. Swedlow and G. Danuser) and the Laura and Arthur Colwin Endowed Summer Research Fellowship program (J.R. Swedlow, P. Meraldi, and A.D. McAinsh) for financial support. J.R. Swedlow was supported by a Wellcome Trust Senior Research Fellowship (067433). A.C. Amaro, N. Mchedlishvili, and A. Toso are members of the Life Science Zurich Graduate School in Molecular Life Science. P. Meraldi is the recipient of an SNF-Förderungprofessur and a European Young Investigator Award and is supported by a grant from the Swiss National Science Foundation and ETH. Work in the McAinsh laboratory is supported by the Marie Curie Cancer Care. Work in the Danuser laboratory is supported by the National Institutes of Health (R01 GM68956). K. Jaqaman was supported in part by a Paul Sigler/Agouron postdoctoral fellowship from the Helen Hay Whitney Foundation.
J.R. Swedlow, G. Danuser, P. Meraldi, and A.D. McAinsh conceived and initiated this study. A.C. Amaro established the EGFP–CENP-A cell line. K. Jaqaman and J.F. Dorn designed and implemented the image and data analysis software. I.M. Porter, J.F. Dorn, and K. Jaqaman defined the imaging protocol. E.M. King, A.C. Amaro, N. Mchedlishvili, S.E. McClelland, I.M. Porter, M. Posch, A. Toso, and J.R. Winter performed the siRNA treatments and acquired the live cell videos, which were analyzed by K. Jaqaman and H.L. Elliott with help from E.M. King and A.C. Amaro. K. Jaqaman, G. Danuser, J.R. Swedlow, P. Meraldi, and A.D. McAinsh wrote the manuscript with help from E.M. King, A.C. Amaro, J.F. Dorn, and I.M. Porter.
Footnotes
Abbreviations used in this paper:
- CENP-E
- centromere protein E
- MCAK
- mitotic centromere-associated kinesin
- MT
- microtubule
- PSF
- point spread function
- SPT
- single particle tracking
- WT
- wild type
References
- Andrews P.D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., Swedlow J.R. 2004. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 6:253–268 10.1016/S1534-5807(04)00025-5 [DOI] [PubMed] [Google Scholar]
- Antonio C., Ferby I., Wilhelm H., Jones M., Karsenti E., Nebreda A.R., Vernos I. 2000. Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell. 102:425–435 10.1016/S0092-8674(00)00048-9 [DOI] [PubMed] [Google Scholar]
- DeLuca J.G., Dong Y., Hergert P., Strauss J., Hickey J.M., Salmon E.D., McEwen B.F. 2005. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol. Biol. Cell. 16:519–531 10.1091/mbc.E04-09-0852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Verma S., Mitchison T.J., Walczak C.E. 1999. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 96:69–78 10.1016/S0092-8674(00)80960-5 [DOI] [PubMed] [Google Scholar]
- Dorn J.F., Jaqaman K., Rines D.R., Jelson G.S., Sorger P.K., Danuser G. 2005. Yeast kinetochore microtubule dynamics analyzed by high-resolution three-dimensional microscopy. Biophys. J. 89:2835–2854 10.1529/biophysj.104.058461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn J.F., Danuser G., Yang G. 2008. Computational processing and analysis of dynamic fluorescence image data. Methods Cell Biol. 85:497–538 10.1016/S0091-679X(08)85022-4 [DOI] [PubMed] [Google Scholar]
- Funabiki H., Murray A.W. 2000. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 102:411–424 10.1016/S0092-8674(00)00047-7 [DOI] [PubMed] [Google Scholar]
- Gassmann R., Essex A., Hu J.S., Maddox P.S., Motegi F., Sugimoto A., O’Rourke S.M., Bowerman B., McLeod I., Yates J.R., III, et al. 2008. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 22:2385–2399 10.1101/gad.1687508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D., Hirota T., Koch B., Peters J.M., Ellenberg J. 2006a. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr. Biol. 16:333–344 10.1016/j.cub.2005.12.040 [DOI] [PubMed] [Google Scholar]
- Gerlich D., Koch B., Dupeux F., Peters J.-M., Ellenberg J. 2006b. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr. Biol. 16:1571–1578 10.1016/j.cub.2006.06.068 [DOI] [PubMed] [Google Scholar]
- Hauf S., Waizenegger I.C., Peters J.M. 2001. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 293:1320–1323 10.1126/science.1061376 [DOI] [PubMed] [Google Scholar]
- Hughes A.F., Swann M.M. 1948. Anaphase movements in the living cell - a study with phase contrast and polarized light on check tissue cultures. J. Exp. Biol. 25:45 [Google Scholar]
- Hunter A.W., Caplow M., Coy D.L., Hancock W.O., Diez S., Wordeman L., Howard J. 2003. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol. Cell. 11:445–457 10.1016/S1097-2765(03)00049-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman K., Danuser G. 2006. Linking data to models: data regression. Nat. Rev. Mol. Cell Biol. 7:813–819 10.1038/nrm2030 [DOI] [PubMed] [Google Scholar]
- Jaqaman K., Loerke D., Mettlen M., Kuwata H., Grinstein S., Schmid S.L., Danuser G. 2008. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods. 5:695–702 10.1038/nmeth.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan M.A., Toso R.J., Thrower D., Wilson L. 1993. Mechanism of mitotic block and inhibition of cell proliferation by taxol at low concentrations. Proc. Natl. Acad. Sci. USA. 90:9552–9556 10.1073/pnas.90.20.9552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor T.M., Lampson M.A., Hergert P., Cameron L., Cimini D., Salmon E.D., McEwen B.F., Khodjakov A. 2006. Chromosomes can congress to the metaphase plate before biorientation. Science. 311:388–391 10.1126/science.1122142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke K., Cheng J., Hunt A.J. 2009. The distribution of polar ejection forces determines the amplitude of chromosome directional instability. Curr. Biol. 19:807–815 10.1016/j.cub.2009.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Cole R.W., McEwen B.F., Buttle K.F., Rieder C.L. 1997. Chromosome fragments possessing only one kinetochore can congress to the spindle equator. J. Cell Biol. 136:229–240 10.1083/jcb.136.2.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith S.L., Khodjakov A., Hergert P., Walczak C.E. 2004. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol. Cell. 15:1146–1159 10.1091/mbc.E03-08-0581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S.L., Rosasco S.E., Barrett-Wilt G.A., Shabanowitz J., Hunt D.F., Walczak C.E., Stukenberg P.T. 2004. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14:273–286 [DOI] [PubMed] [Google Scholar]
- Lewis W.H. 1939. Changes of viscosity and cell activity. Science. 89:400 [Google Scholar]
- Liu D., Vader G., Vromans M.J.M., Lampson M.A., Lens S.M.A. 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 323:1350–1353 10.1126/science.1167000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr M.I., Hümmer S., Bormann J., Grüner T., Adio S., Woehlke G., Mayer T.U. 2007. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr. Biol. 17:488–498 10.1016/j.cub.2007.02.036 [DOI] [PubMed] [Google Scholar]
- McEwen B.F., Chan G.K., Zubrowski B., Savoian M.S., Sauer M.T., Yen T.J. 2001. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol. Biol. Cell. 12:2776–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Draviam V.M., Sorger P.K. 2004. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 7:45–60 10.1016/j.devcel.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Mikhailov A., Cole R.W., Rieder C.L. 2002. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr. Biol. 12:1797–1806 10.1016/S0960-9822(02)01226-5 [DOI] [PubMed] [Google Scholar]
- Nasmyth K. 2002. Segregating sister genomes: the molecular biology of chromosome separation. Science. 297:559–565 10.1126/science.1074757 [DOI] [PubMed] [Google Scholar]
- O’Connell C.B., Loncarek J., Hergert P., Kourtidis A., Conklin D.S., Khodjakov A. 2008. The spindle assembly checkpoint is satisfied in the absence of interkinetochore tension during mitosis with unreplicated genomes. J. Cell Biol. 183:29–36 10.1083/jcb.200801038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo-Hafalla M.T., Katou Y., Shirahige K., Uhlmann F. 2007. Displacement and re-accumulation of centromeric cohesin during transient pre-anaphase centromere splitting. Chromosoma. 116:531–544 10.1007/s00412-007-0118-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira R.A., Coelho P.A., Sunkel C.E. 2005. The condensin I subunit Barren/CAP-H is essential for the structural integrity of centromeric heterochromatin during mitosis. Mol. Cell. Biol. 25:8971–8984 10.1128/MCB.25.20.8971-8984.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoulis A. 1991. Probability, Random Variables, and Stochastic Processes. Third edition McGraw-Hill, New York: 666 pp. [Google Scholar]
- Putkey F.R., Cramer T., Morphew M.K., Silk A.D., Johnson R.S., McIntosh J.R., Cleveland D.W. 2002. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev. Cell. 3:351–365 10.1016/S1534-5807(02)00255-1 [DOI] [PubMed] [Google Scholar]
- Ribeiro S.A., Gatlin J.C., Dong Y., Joglekar A., Cameron L., Hudson D.F., Farr C.J., McEwen B.F., Salmon E.D., Earnshaw W.C., Vagnarelli P. 2009. Condensin regulates the stiffness of vertebrate centromeres. Mol. Biol. Cell. 20:2371–2380 10.1091/mbc.E08-11-1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Davison E.A., Jensen L.C.W., Cassimeris L., Salmon E.D. 1986. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J. Cell Biol. 103:581–591 10.1083/jcb.103.2.581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin P.L. 2001. Unimodal thresholding. Patt. Recogn. 34:2083–2096 10.1016/S0031-3203(00)00136-9 [DOI] [Google Scholar]
- Skibbens R.V., Skeen V.P., Salmon E.D. 1993. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J. Cell Biol. 122:859–875 10.1083/jcb.122.4.859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J., von Dassow G., Wagenbach M., Asbury C., Wordeman L. 2008. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev. Cell. 14:252–262 10.1016/j.devcel.2007.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedlow J.R., Hirano T. 2003. The making of the mitotic chromosome: modern insights into classical questions. Mol. Cell. 11:557–569 10.1016/S1097-2765(03)00103-5 [DOI] [PubMed] [Google Scholar]
- Uhlmann F., Wernic D., Poupart M.A., Koonin E.V., Nasmyth K. 2000. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 103:375–386 10.1016/S0092-8674(00)00130-6 [DOI] [PubMed] [Google Scholar]
- Varga V., Leduc C., Bormuth V., Diez S., Howard J. 2009. Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell. 138:1174–1183 10.1016/j.cell.2009.07.032 [DOI] [PubMed] [Google Scholar]
- Vorozhko V.V., Emanuele M.J., Kallio M.J., Stukenberg P.T., Gorbsky G.J. 2008. Multiple mechanisms of chromosome movement in vertebrate cells mediated through the Ndc80 complex and dynein/dynactin. Chromosoma. 117:169–179 10.1007/s00412-007-0135-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger I.C., Giménez-Abián J.F., Wernic D., Peters J.M. 2002. Regulation of human separase by securin binding and autocleavage. Curr. Biol. 12:1368–1378 10.1016/S0960-9822(02)01073-4 [DOI] [PubMed] [Google Scholar]
- Waters J.C., Chen R.H., Murray A.W., Salmon E.D. 1998. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 141:1181–1191 10.1083/jcb.141.5.1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L., Wagenbach M., von Dassow G. 2007. MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J. Cell Biol. 179:869–879 10.1083/jcb.200707120 [DOI] [PMC free article] [PubMed] [Google Scholar]